Abstract
Intra-molecular and inter-molecular cross-linking of protonated polypeptide ions in the gas phase via ion/ion reactions have been demonstrated using N-hydroxysulfosuccinimide (sulfo-NHS) based reagent anions. The initial step in the ion/ion reaction involves the formation of a long-lived complex between the peptide and reagent, which is a prerequisite for the covalent bioconjugation chemistry. The sulfonate groups on the NHS rings of the homo-bifunctional cross-linking reagents have high affinity for the protonated sites in the peptide and therefore facilitate the long-lived complex formation. In addition to the formation of a long-lived chemical complex, intra-molecular cross-linking also requires two unprotonated primary amine sites within a molecule where the covalent modification takes place. Alternatively, inter-molecular cross-linking demands the availability of one neutral primary amine site in each of the two peptides that are being cross-linked. Nucleophilic displacement of two sulfo-NHS groups by the amine functionalities in the peptide is a signature of the covalent cross-linking chemistry in the gas phase. Upon removal of the two sulfo-NHS groups, two amide bonds are formed between an unprotonated, primary amine group of a lysine side chain in the peptide and the carboxyl group in the reagent.
Keywords: gas phase ion/ion reactions, N-hydroxysulfosuccinimide esters, gas-phase bioconjugation, intra-molecular and inter-molecular cross-linking
INTRODUCTION
Gas phase ion chemistry plays significant roles in biological mass spectrometry in both ionization and structural characterization. The structural characterization and identification of biomolecules using tandem mass spectrometry is highly dependent on the nature of the ion (i.e., the ion-type) that is examined, such as a molecular ion (M+•), protonated molecule (M+H)+, deprotonated molecule (M−H)−, etc. [1] The ion-type subjected to structural interrogation is usually determined by the ionization method. However, means for altering the ion-type after the ionization step have been explored. Ion/ion reactions have been shown to be a particularly flexible overall approach for altering ion-types in the gas-phase.[2,3,4,5] For a given analyte ion-type, a wide range of reactions can be accessed via the proper selection of the oppositely charged reagent.[6] Examples of ion-type manipulation via ion/ion chemistry include, inter alia, charge reduction via proton transfer,[7,8] charge inversion via multiple ion transfers in a single collision,[9,10] the increase of charge via sequential charge inversion reactions,[11,12] metal ion incorporation into peptides either via metal transfer reaction [13,14] or metal cation attachment,[15] and the conversion of multiply charged ions to radical species via electron transfer.[16,17,18] Ion/ion reactions involving these types of ion manipulations have been used in the structural analysis of peptides, proteins,[19,20,21] and oligonucleotides.[22,23]
Covalent bond formation between a reagent ion and an oppositely charged bio-ion via a long-lived reaction complex opens up new possibilities for ion transformation relative to the cation, anion, or electron transfer reaction. Recently, Schiff base chemistry in the gas phase has been reported resulting in covalent bond formation between primary amine groups in multiply protonated peptide ions and an aldehyde-containing reagent anion (i.e. singly deprotonated 4-formyl-1,3-benzenedisulfonic acid).[24] Schiff base formation was noted in conjunction with charge inversion of singly protonated peptide ions reacted with doubly deprotonated 4-formyl-1,3-benzenedisulfonic acid.[25] Subsequent collision-induced dissociation (CID) of the modified peptide anions gave rise to richer informative structural information than did CID of either the singly protonated or singly deprotonated peptide.
Functional groups of various amino acid side-chains in proteins and peptides can be selectively modified in solution to facilitate ionization,[26] quantification,[27,28] or structural characterization [29,30] in mass spectrometry. N-hydroxysuccinimide (NHS) derivatives are popular reagents used for modification of primary amine groups in peptides or proteins in solution. Recently, we demonstrated the utility of NHS or sulfo- N-hydroxysuccinimide (sulfo-NHS) ester derivative ions in the modification of primary amine sites in gaseous polypeptide ions via ion/ion reactions.[31] Loss of the NHS or sulfo-NHS functionalities represents a signature for the covalent modification of the peptide. It was also demonstrated that collisional activation of the covalently modified peptide ions gave rise to sequence informative ions containing the modification.
The ability to introduce covalent modifications to peptide ions in the gas-phase in a selective fashion by ion/ion reactions provides new means for altering ion-types in a mass spectrometer. In this report, we demonstrate that it is possible to introduce a cross-linker into a gaseous peptide ion or between gaseous peptide ions. In recent years, mass spectrometry has been used to elucidate intra-molecular and inter-molecular cross-linking interactions of peptides and proteins. [32] A cross-link between two amino acid side chains imposes a distance constraint on their relative locations thereby providing information regarding the three-dimensional structure of the protein. The NHS and sulfo-NHS functionalities are common building blocks of various amine reactive cross-linkers. [33] For example, chemical cross-linking with bis[sulfosuccinimidyl] suberate (BS3) and bis[sulfosuccinimidyl] glutarate (BS2G) in solution, and Fourier transform ion cyclotron resonance mass spectrometric analysis have been applied in mapping the protein interfaces of the calmodulin/adenylyl cyclase 8 peptide complex. [34] Additionally, solution phase cross-linking methodologies using dithiobis(succinimidylpropionate) (DSP) and an LC MS/MS approach have been used to isolate signal transduction complexes thereby furthering the understanding of signal transduction. [35] More recent reports have detailed the use of amine reactive cross-linkers in solution along with mass spectrometry analysis to elucidate a three-dimensional structure of various proteins.[36,37] The cross-linking techniques are not limited to the study of proteins. For example, the architecture of RNA polymerase II–TFIIF complex has also been revealed by solution phase cross-linking methods and mass spectrometry.[38]
A wide variety of amine reactive cross-linking reagents are commercially available. In this study we demonstrate intra-molecular and inter-molecular cross-linking of gaseous peptide ions with sulfo-NHS based amine-reactive cross-linking reagent anions. Loss of two sulfo-NHS functionalities represents a signature for the covalent modification and cross-linking process. We also demonstrate that collisional activation of the intra-molecularly cross-linked peptides results in fragment ions containing the cross-link between two lysine residues, while collisional activation of inter-molecularly cross-linked peptides results in one peptide cross-linked to fragment ions of the second peptide. While it might be premature to speculate about applications of gas-phase cross-linking given its novelty, intra-molecular peptide cross-linking may provide a new tool for gaseous ion structure characterization while inter-molecular cross-linking may provide a novel means for screening mixtures. The exploration of such applications is the subject of an ongoing study. This paper emphasizes methodologies and criteria for successful cross-linking.
EXPERIMENTAL SECTION
Materials
Methanol and glacial acetic acid were purchased from Mallinckrodt (Phillipsburg, NJ). Peptide KKKKKKKKKK was custom synthesized by Pepnome Limited (China), peptides KAGK and YGGFLK were synthesized by CPC Scientific (San Jose, CA), peptide KGAILKGAILR was synthesized by SynPep (Dublin, CA), and peptide RKRARKE was purchased from Sigma Aldrich (St. Louis, MO). Reagents BS3 (BS3 = Bis[sulfosuccinimidyl] suberate), and DTSSP (DTSSP = 3,3′-Dithiobis[sulfosuccinimidylpropionate]) were obtained from Thermo Fisher Scientific Inc. (Rockford, IL). All peptide solutions for positive electrospray were prepared in 49.5:49.5:1 (v/v/v) water/methanol/acetic acid mixture at a concentration of ~50 μM. The reagent solutions were prepared at a concentration of approximately 2 mM in equal parts of methanol and water.
Mass Spectrometry
All ion/ion reactions were performed on a prototype version of a triple quadrupole/linear ion trap, QTRAP mass spectrometer [39] (Applied Biosystems/MDS Sciex, Concord, ON, Canada), equipped with a home-built nano-electrospray ionization source.[40] All intra-molecularly cross-linked peptides were generated during a single mutual storage ion/ion reaction.[40,41,42] Briefly, mass resolved peptide and reagent ions of opposite polarity are allowed to react in the Q2 collision cell for various times on the order of hundreds of milliseconds, depending primarily upon the number of reagent anions admitted into Q2. The reaction products are accelerated as they are transferred from Q2 to Q3 in order to remove any residual sulfo-NHS groups. Once in Q3, the intra-molecularly cross-linked peptide is subjected to collisional activation and the fragment ions are analyzed using mass selective axial ejection (MSAE).[43] The inter-molecularly cross-linked peptides were generated in a process involving two ion/ion reactions. Independently resolved peptide and reagent ions are reacted using a transmission mode ion/ion reaction in Q2.[44] The formed peptide-reagent complex is accelerated from Q2 to Q3 in order to remove one sulfo-NHS group. The remaining complex in Q3 is allowed to react with a second peptide in a mutual storage mode ion/ion reaction. The newly formed complex in the second ion/ion reaction is subjected to collisional activation to remove the second sulfo-NHS group. MS3 is then performed on the inter-molecularly cross-linked peptides, and the fragment ions are analyzed by MSAE.
RESULTS AND DISCUSSION
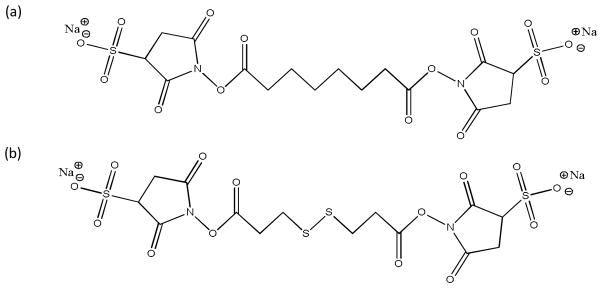
Various homo-bifunctional NHS and sulfo-NHS based cross-linking reagents are commercially available for cross-linking two primary amine sites within a molecule or between two molecules. Generally, these reagents consist of a spacer arm with two identical reactive groups on each end. The sulfo groups on each of the NHS rings are acidic sites that facilitate negative ion formation of the reagent in ESI and they also serve as “sticky” groups in forming a long-lived chemical complex with the protonated polypeptide ions.[45] The reagents used in this work were supplied as sodium salts. The anions derived from the reagents are therefore indicated as [reagent-Na]− for singly charged anions and [reagent-2Na]2− for doubly charged anions, where the reagent is either BS3 or DTSSP. The structures of both reagents are shown in Scheme 1.
Scheme 1.
Structures of anionic reagents examined: (a) BS3 (Bis[sulfosuccinimidyl] suberate) and (b) DTSSP (3,3′-Dithiobis[sulfosuccinimidylpropionate]).
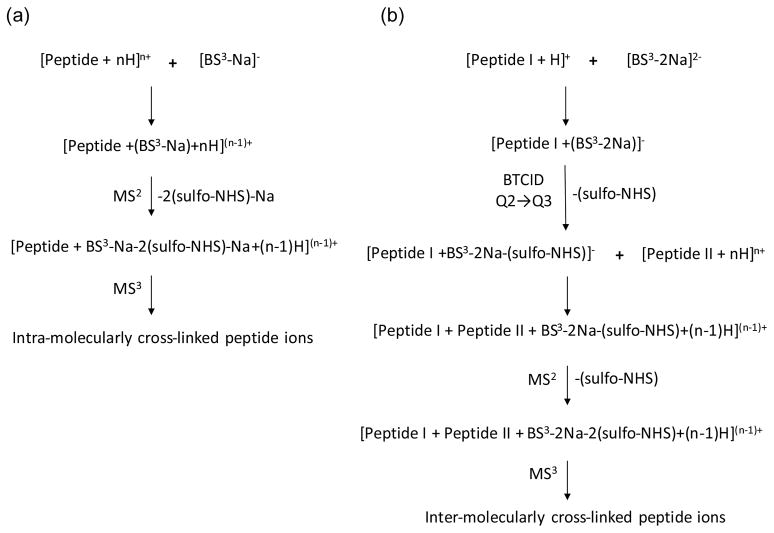
The ion/ion reactions described in this report lead to the formation of either intra-molecularly cross-linked or inter-molecularly cross-linked peptide ions. The overall processes taking place during the cross-linking of two primary amine sites with sulfo-NHS-based reagents are depicted in Scheme 2. The initial step in intra-molecular cross-linking involves the formation of a long-lived complex between the reagent and the peptide ions as shown in Scheme 2(a). Collisional activation of the complex induces the loss of two sulfo-NHS groups from the reagent. The cross-linking occurs via nucleophilic attack by two neutral primary amine sites at the carbonyl carbons of the ester functionalities within the reagent. The sulfo-NHS groups of the cross-linker serve as leaving groups in this process. The loss of two sulfo-NHS groups, therefore, represents a signature for the cross-linking, which is supported by subsequent activation of the cross-linked complex (see below). Loss of the intact cross-linking reagent occurs when no covalent attachment occurs (see below).
Scheme 2.
Sequence of events in the ion/ion reaction between polypeptide cation(s) and a sulfo-NHS-based reagent anion leading to: (a) intra-molecularly cross-linked peptide and (b) inter-molecularly cross-linked peptides in the gas phase.
The sequence of events for inter-molecular cross-linking of peptides is demonstrated in Scheme 2(b). Singly protonated peptide ions and doubly charged reagent anions form a chemical complex with a negative overall charge. As the complex is accelerated between the collision cell (Q2) and the mass analyzer (Q3), a sulfo-NHS group is lost resulting in a covalently modified peptide at a single primary amine site. Note that the second sulfo-NHS group is still attached to the cross-linking reagent and it has not undergone the covalent modification chemistry. This covalently modified peptide-reagent complex is further reacted with a multiply protonated second peptide. Collisional activation of the peptide-reagent-peptide complex results in loss of the second sulfo-NHS group, giving rise to inter-molecularly cross-linked peptides. An MS3 experiment that subjects the cross-linked peptides to collisional activation results in one peptide cross-linked to fragment ions from the second peptide. Inter-molecular cross-linking of two peptides is also possible by eliminating the beam type CID step in Scheme 2 (b), where the [Peptide I+(BS3-2Na)]− complex is directly reacted with [Peptide II+nH]n+ ion (see Scheme S1 in Supplementary Information). However, with this procedure we have noted that, in addition to the formation of a [Peptide I+Peptide II+BS3-2Na+(n-1)H](n-1)+ complex, there also appears a reaction channel that leads to the transfer of the entire reagent from Peptide I to Peptide II. As a result, the formation of the peptide I-reagent-peptide II complex following this scheme is not as efficient as following Scheme 2(b).
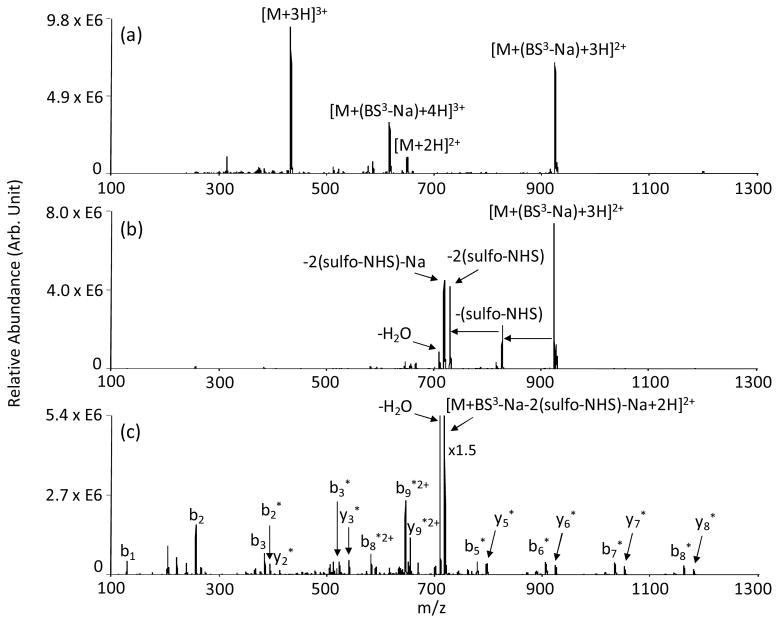
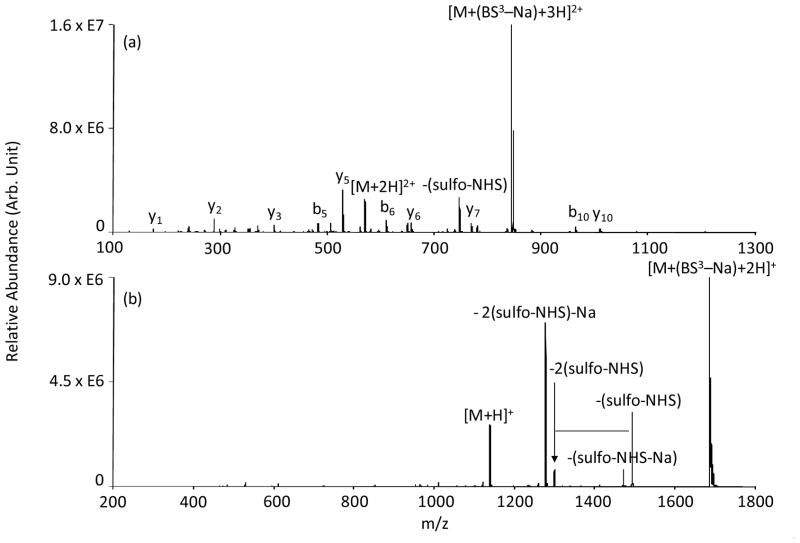
An illustration of the overall ion/ion reaction phenomenology from Scheme 2(a) observed when intra-molecular cross-linking occurs is shown by the spectra in Figure 1, which relate to the reaction between triply protonated KKKKKKKKK cations and singly charged BS3 anions. This reagent, with its hydrocarbon chain spacer arm, is resistant to cleavage and the formed cross-links are irreversible. Mutual storage of [KKKKKKKKKK+3H]3+ and deprotonated BS3 reagent ions results in the [KKKKKKKKKK+(BS3-Na)+3H]2+ complex, as shown in Figure 1(a). Evidence for a minor contribution from proton transfer is also observed, as reflected by the signal for the [M+2H]2+ ion in Figure 1(a), likely resulting from a “proton hopping” channel.[46] (A relatively small population of [KKKKKKKKKK+4H]4+ was also present and gave rise to the [KKKKKKKKKK+(BS3-Na)+4H]3+ complex in Figure 1(a).) As demonstrated previously, [31] ion trap collisional activation of a complex formed between a multiply protonated peptide and a sulfo-NHS-based reagent anion results in a sulfo-NHS neutral molecule loss when covalent modification takes place, whereas loss of the intact neutral reagent is noted when covalent modification does not take place. Given that the BS3 reagent contains a sulfo-NHS group on each end of the hydrocarbon spacer arm, two sulfo-NHS neutral molecule losses would signify cross-linking taking place. Indeed, collisional activation of [KKKKKKKKKK+(BS3-Na)+3H]2+, as shown in Figure 1(b), results in loss of two sulfo-NHS groups. Note that the counter-ion for the sulfate of one of the sulfo-NHS groups can be either a proton or a sodium ion. Hence, two peaks associated with loss of both sulfo-NHS groups appear in Figure 1(b) that correspond to both counter-ions being protons or to one counter-ion being a proton and one being a sodium ion. Subsequent activation of the latter double sulfo-NHS loss peak generates sequence informative fragment ions containing the BS3 hydrocarbon spacer arm (see Figure 1(c)). Given the large number of lysine side chains in excess of the total charge of the peptide, which gives rise to the possibility for many reactive neutral amine sites, a variety of intra-molecularly cross-linked structures are possible. As demonstrated in Figure 1(c), a series of cross-linked b- and y-type ions exist, starting with b2 and y2 as the smallest fragment ions containing the hydrocarbon spacer arm. In the case of b2 ion, the side chain of Lys-2 is cross-linked either to the side chain of Lys-1 or the N-terminus of the peptide. Alternatively, the modified y2 ion containing the C-terminus of the peptide results from the cross-linking of Lys-9 and Lys-10 residues. A similar fragmentation pattern was observed in the ion trap CID spectrum of the [KKKKKKKKKK+DTSSP-Na-2(sulfo-NHS)-Na+2H]2+ complex formed after an ion/ion reaction between [KKKKKKKKKK+3H]3+ and the singly charged DTSSP reagent anion. (See Figure S1 in Supplementary Information).
Figure 1.
Product ion spectra derived from (a) ion/ion reaction between [KKKKKKKKKK+3H]3+ and [BS3-Na]−, (b) ion trap CID of [KKKKKKKKKK+(BS3-Na)+3H]2+ and (c) MS3 of [KKKKKKKKKK+BS3-Na-2(sulfo-NHS)-Na+2H]2+. * denotes ions intra-molecularly cross-linked with BS3.
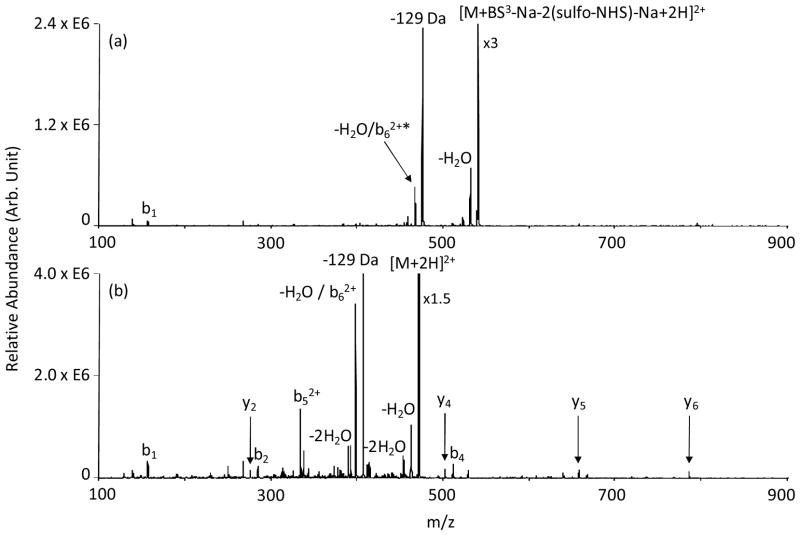
The peptide KKKKKKKKKK was chosen for initial study to maximize the possibility for cross-linking. However, the presence of multiple primary amine sites is expected to give rise to a mixture of products, thereby precluding the use of CID for demonstrating specificity in cross-linking. When the number of possible protonation sites in a peptide is limited, fewer cross-linked structures are possible. For example, in the peptide RKRARKE, two excess protons are expected to be sequestered largely at the arginine residues, leaving the two lysine residues unprotonated and available for cross-linking. The ion/ion reaction between triply protonated RKRARKE and [BS3-Na]− reagent anions results in the formation of a peptide-reagent complex. Ion trap CID of the complex resulted in two sulfo-NHS neutral molecule losses (Figure S2 in Supplementary Information). Isolation and activation of the species formed from the losses of two sulfo-NHS groups (one with a proton counter-ion and one with a sodium counter-ion), as shown in Figure 2(a), gives rise to essentially exclusive loss of the C-terminal glutamic acid residue, indicated as a loss of 129 Da, with the cross-linker present in the product ion. A smaller signal that can arise from formation of the b6-ion containing the cross-linker and, perhaps, from water loss from the species formed via glutamic acid loss is also noted. Ion trap CID of the doubly protonated unmodified RKRARKE peptide (Figure 2(b)) also shows major loss of the elements of C-terminal glutamic acid, as supported by experiments involving methyl esterification of the peptide, but also shows various b- and y-type ions from cleavages along the peptide backbone. The formation of the latter ions is suppressed in the intra-molecularly cross-linked peptide. The loss of the elements of C-terminal glutamic acid has been noted both for protonated [47] and deprotonated peptides [48]. The presence of arginine [49] or fixed charge sites [50] in peptide cations, in particular, enhances the probability of this loss when E is at the C-terminus. The process is believed to involve a rearrangement that gives rise to loss of a pyroglutamic acid molecule [48]. Similar fragmentation behavior is observed in the ion trap CID spectrum of [RKRARKE+DTSSP-Na-2(sulfo-NHS)-Na+2H]2+ derived from the ion/ion reaction between [RKRARKE+3H]3+ and [DTSSP-Na]− (Figure S3 in Supplementary Information).
Figure 2.
Product ion spectra derived from (a) ion trap CID of [RKRARKE+BS3-Na-2(sulfo-NHS)-Na+2H]2+ and (b) ion trap CID of [RKRARKE+2H]2+. * denotes ions intra-molecularly cross-linked with BS3.
The initial studies of gas-phase modification of peptide ions via ion/ion reactions using sulfo-NHS-based reagents indicated that the presence of an unprotonated primary amine site is a necessary condition. For intra-molecular cross-linking to occur, it follows that two unprotonated primary amines must be available and they must come within sufficient proximity for the cross-linker to reach them both. The previous examples were expected to meet these criteria. Studies with the peptide KGAILKGAILR were performed to further examine intra-molecular cross-linking and specifically to examine the issue of available unprotonated primary amine sites. When doubly and triply protonated KGAILKGAILR cations were reacted with [BS3-Na]− reagent anions, a peptide-reagent complex was formed (Figure S4 in Supplementary Information). Collisional activation of the [KGAILKGAILR+(BS3-Na)+3H]2+ complex resulted in loss of one sulfo-NHS group and a loss of the intact reagent giving rise to the [KGAILKGAILR+2H]2+ ion (see Figure 3(a)). Since three out of the four possible basic sites in the peptide (i.e., the arginine, two lysines, and N-terminus) are protonated, nominally only one site remains available for modification. Loss of one sulfo-NHS group is observed but no evidence for loss of two sulfo-NHS groups is noted. Loss of the intact neutral reagent from the complex suggests that no covalent reactions took place in at least some of the complexes. The appearance of a number of unmodified y-type and b-type ions and the relative absence of any modified fragments may also reflect fragmentation from unmodified forms of the peptide. The fact that some of the peptide ions were unmodified may reflect a degree of proton sharing between basic sites, which is likely for the N-terminus and Lys-1, as any lone-pair electron association with an excess proton renders an amine site less nucleophilic. When doubly protonated KGAILKGAILR was reacted, the ion trap CID spectrum in Figure 3(b) was observed, which shows major signals for loss of two sulfo-NHS groups, loss of one sulfo-NHS group, and the loss of the neutral reagent. While the signal due to loss of a single sulfo-NHS group may arise from some ions that would undergo another sulfo-NHS group loss upon further activation, the appearance of the product arising from loss of the intact reagent suggests that at least some of the signal due to single sulfo-NHS loss arises from species that underwent only one covalent reaction. Hence, the data of Figure 3(b) probably reflects a mixture of outcomes comprised of cross-linking, which appears to be the dominant process in this case, single covalent modification, and purely non-covalent interaction.
Figure 3.
Product ion spectra derived from (a) ion trap CID of [KGAILKGAILR+(BS3-Na)+3H]2+ and (b) ion trap CID of [KGAILKGAILR+(BS3-Na)+2H]+.
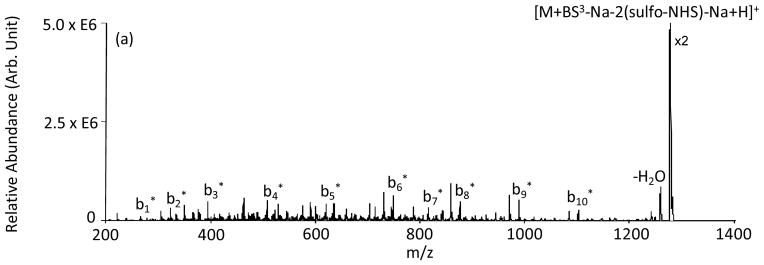
The MS3 spectrum of the [KGAILKGAILR+BS3-Na-2(sulfo-NHS)-Na+2H]2+ species that resulted from the loss of two sulfo-NHS groups gave a complete series of cross-linked b-type ions (Figure 4). The appearance of the b1-b5 ions suggests that, at least for some of the peptide ions, the side chain of Lys-1 residue was cross-linked to the N-terminus of the peptide.
Figure 4.
Product ion spectrum derived from MS3 of [KGAILKGAILR+BS3-Na-2(sulfo-NHS)-Na+H]+. * denotes ions intra-molecularly cross-linked with BS3.
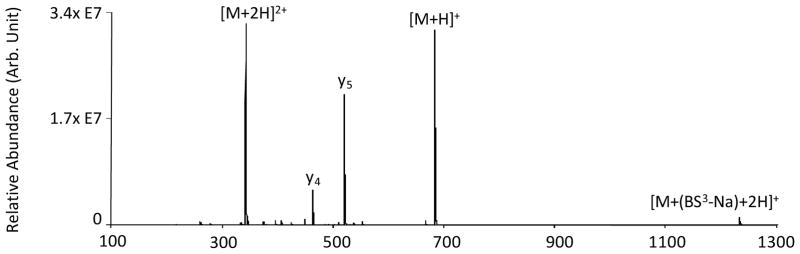
In contrast to the previous examples where complex formation resulted in covalent cross-linking modification, the ion/ion reaction between doubly protonated YGGFLK and [BS3-Na]− reagent anions resulted primarily in proton transfer with minimal complex formation, as shown in Figure 5. With the excess protons nominally associated with the N-terminus and the lysine side-chain, neither of the primary amine sites is expected to be particularly nucleophilic, which renders the ion relatively unreactive to covalent modification. (Note that the y-type ions are likely to have been generated by CID upon transfer from Q2 to Q3.)
Figure 5.
Product ion spectrum derived from the ion/ion reaction between [YGGFLK+2H]2+ and [BS3-Na]−.
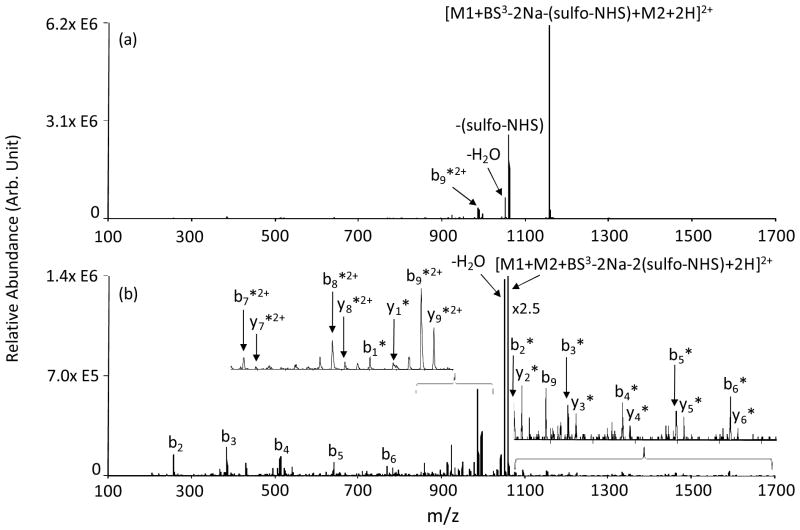
The evidence for intra-molecular cross-linking suggested an attempt at intermolecular cross-linking of peptide ions using the BS3 reagent and the process shown in Scheme 2(b). This process involves two consecutive ion/ion reactions each followed by a CID step in order to facilitate loss of two sulfo-NHS groups. The first peptide is covalently modified at a single Lys primary amine site following collisional activation of the peptide-reagent complex from an ion/ion reaction with a BS3-related anion. The resulting peptide ion, modified at a single site, is further reacted with the second peptide, which also has an available neutral primary amine site. The resulting [peptide I-BS3-(sulfo-NHS)-peptide II] complex is exposed to collisional activation to eliminate the second sulfo-NHS group, thereby cross-linking the two peptides. Figure 6(a) shows the results of the collisional activation of the [YGGFLK+KKKKKKKKKK+BS3-2Na-(sulfo-NHS)+2H]2+ complex, which primarily leads to loss of a second sulfo-NHS group. This complex was initially formed from the ion/ion reaction between [YGGFLK+BS3-2Na-(sulfo-NHS)]− and [KKKKKKKKKK+3H]3+. In the inter-molecular cross-linking scenario, singly-protonated YGGFLK is charge inverted to [YGGFLK+(BS3-2Na)]− in the reaction with [BS3-2Na]2−. The [YGGFLK+(BS3-2Na)]− complex is collisionally activated upon acceleration from the collision cell and into the Q3 analyzer quadrupole, which results in loss of a sulfo-NHS group and modification of YGGFLK at an available primary amine site giving rise to [YGGFLK+BS3-2Na-(sulfo-NHS)]−. MS3 of the second sulfo-NHS loss peak as shown in Figure 6(b) results in a series of b- and y-type ions from KKKKKKKKKK cross-linked with the entire YGGFLK peptide. A similar fragmentation pattern was observed in the ion trap CID of [YGGFLK+RKRARKE+BS3-2Na-2(sulfo-NHS)+3H]2+ formed from the ion/ion reaction between [YGGFLK+BS3-2Na-(sulfo-NHS)]− and [RKRARKE+3H]3+ (Figure S5 in Supplementary Information). When more than one primary amine site is available for modification in the first peptide, as for example in KAGK, collisional activation of the [KAGK+KKKKKKKKKK+BS3-2Na-2(sulfo-NHS)+3H]2+ complex results in intra- and intermolecular cross-linking, the former evident by the appearance of [KAGK+BS3-2Na-2(sulfo-NHS)]+ and the latter by the loss of two sulfo-NHS groups from the complex (see Figure S6 in Supplementary Information).
Figure 6.
Product ion spectra derived from (a) ion trap CID of [YGGFLK+KKKKKKKKKK+BS3-2Na-(sulfo-NHS)+2H]2+ and (b) MS3 of [YGGFLK+KKKKKKKKKK+BS3-2Na-2(sulfo-NHS)+2H]2+. * denotes ions inter-molecularly cross-linked with BS3.
CONCLUSIONS
Intra-molecular and inter-molecular cross-linking of multiply protonated polypeptide ions can be achieved in the gas phase using homobifunctional N-hydroxysulfosuccinimide based cross-linking reagents. These reagents are readily available commercially due to their frequent use in condensed phase chemistry. One of the main criteria for effecting this covalent modification chemistry in the gas phase involves the formation of a long-lived complex between the reagent and the peptide. Sulfo-NHS esters are suitable reagents for this purpose because of the strong interaction of the sulfonate group in the reagent and the protonated sites in the peptide. The “sticky” nature of the sulfo groups attached to each of the NHS rings in the reagent enables the formation of a long-lived complex with the peptide, satisfying the criterion just mentioned. In addition to long-lived complex formation, two unprotonated primary amine sites within a peptide are required in order for intra-molecular cross-linking to take place. Alternatively, intermolecular cross-linking demands the presence of only one neutral primary amine site in each of the two peptides that are being cross-linked. The neutral primary amine sites serve as nucleophiles and are involved in the removal of the sulfo-NHS groups. The elimination of two sulfo-NHS groups from a peptide-reagent complex (i.e. intra-molecular cross-linking) or from a peptide-reagent-peptide complex (i.e. inter-molecular cross-linking) represent a signature of the covalent cross-linking chemistry in the gas phase. Further extension of this type of gas-phase modification to peptide and protein complexes (i.e., formation of a non-covalently-bound complex ion followed by reaction with the cross-linking reagent in the gas-phase) promises to provide new insights into structural differences/similarities between condensed-phase and gas-phase complexes via comparisons with complexes subjected to cross-linking in the condensed-phase and followed by ionization.
Supplementary Material
Acknowledgments
This research was supported by the Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy, under Award DE-FG02-00ER15105 and the National Institutes of Health under Grant GM 45372.
References
- 1.McLuckey SA, Mentinova M. Ion/Neutral, Ion/Electron, Ion/Photon, and Ion/Ion Interactions in Tandem Mass Spectrometry: Do We Need Them All? Are They Enough? J Am Soc Mass Spectrom. 2011 doi: 10.1007/s13361-010-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogorzalek Loo RR, Udseth HR, Smith RD. A New Approach for the study of Gas-Phase Ion-Ion Reactions Using Electrospray Ionization. J Am Soc Mass Spectrom. 1992;3:695–705. doi: 10.1016/1044-0305(92)87082-A. [DOI] [PubMed] [Google Scholar]
- 3.Pitteri SJ, McLuckey SA. Recent Developments in the Ion/Ion Chemistry of High-Mass Multiply Charged Ions. Mass Spectrom Rev. 2005;24:931–958. doi: 10.1002/mas.20048. [DOI] [PubMed] [Google Scholar]
- 4.McLuckey SA, Stephenson JL., Jr Ion/ion Chemistry of High Mass Multiply Charged Ions. Mass Spectrom Rev. 1998;17:369–407. doi: 10.1002/(SICI)1098-2787(1998)17:6<369::AID-MAS1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 5.Scalf M, Westphall MS, Krause J, Kaufman SL, Smith LM. Controlling Charge States of Large Ions. Science. 1999;283:194–197. doi: 10.1126/science.283.5399.194. [DOI] [PubMed] [Google Scholar]
- 6.McLuckey SA. The Emerging Role of Ion/Ion Reactions in Biological Mass Spectrometry: Considerations for Reagent Ion Selection. Eur J Mass Spectrom. 2010;16:429–439. doi: 10.1255/ejms.1031. [DOI] [PubMed] [Google Scholar]
- 7.Stephenson JL, Jr, McLuckey SA. Ion/Ion Reactions in the Gas-Phase: Proton Transfer Reactions Involving Multiply-Charged Proteins. J Am Chem Soc. 1996;118:7390–7397. [Google Scholar]
- 8.Scalf M, Westphall MS, Smith LM. Charge Reduction Electrospray Mass Spectrometry. Anal Chem. 2000;72:52–60. doi: 10.1021/ac990878c. [DOI] [PubMed] [Google Scholar]
- 9.Ogorzalek Loo RR, Udseth HR, Smith RD. Evidence of Charge Inversion in the Reaction of Singly Charged Anions with Multiply Charged Macroions. J Phys Chem. 1991;95:6412–6415. [Google Scholar]
- 10.Emory JF, McLuckey SA. Charge Inversion of Polypeptide Anions Using Protein and Dendrimer Cations as Charge Inversion Reagents. Int J Mass Spectom. 2008;276:102–109. [Google Scholar]
- 11.He M, McLuckey SA. Two Ion/ion Charge Inversion Steps to Form a Doubly-Protonated Peptide from a Singly-Protonated Peptide in the Gas Phase. J Am Chem Soc. 2003;125:7756–7757. doi: 10.1021/ja0354521. [DOI] [PubMed] [Google Scholar]
- 12.He M, McLuckey SA. Increasing the Negative Charge of a Macroanion in the Gas Phase via Sequential Charge Inversion Reactions. Anal Chem. 2004;76:4189–4192. doi: 10.1021/ac496087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newton KA, McLuckey SA. Gas-Phase Peptide/Protein Cationizing Agent Switching via Ion/Ion Reactions. J Am Chem Soc. 2003;125:12404–12405. doi: 10.1021/ja036924e. [DOI] [PubMed] [Google Scholar]
- 14.Gunawardena HP, O’Hair RA, McLuckey SA. Selective Disulfide Bond Cleavage in Gold(I) Cationized Polypeptide Ions Formed via Gas-Phase Ion/Ion Cation Switching. J Proteome Res. 2006;5:2087–2092. doi: 10.1021/pr0602794. [DOI] [PubMed] [Google Scholar]
- 15.Payne AH, Glish GL. Gas-Phase Ion/Ion Interactions Between Peptides or Proteins and Iron Ions in a Quadrupole Ion Trap. Int J Mass Spectrom. 2001;204:47–54. [Google Scholar]
- 16.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and Protein Sequence Analysis by Electron Transfer Dissociation Mass Spectrometry. Proc Natl Acad Sci US A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herron WJ, Goeringer DE, McLuckey SA. Gas-phase Electron Transfer Reactions from Multiply-Charged Anions to Rare Gas Cations. J Am Chem Soc. 1995;117:11555–11562. [Google Scholar]
- 18.Coon JJ, Shabanowitz J, Hunt DF, Syka JEP. Electron Transfer Dissociation of Peptide Anions. J Am Soc Mass Spectrom. 2005;16:880–882. doi: 10.1016/j.jasms.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 19.Reid GE, Shang H, Hogan JM, Lee GU, McLuckey SA. Gas-Phase Concentration, Purification, and Identification of Whole Proteins from Complex Mixtures. J Am Chem Soc. 2002;124:7353–7362. doi: 10.1021/ja025966k. [DOI] [PubMed] [Google Scholar]
- 20.Coon JJ, Ueberheide B, Syka JEP, Dryhurst DD, Ausio J, Shabanowitz J, Hunt DF. Protein Identification Using Sequential Ion/Ion Reactions and Tandem Mass Spectrometry. Proc Natl Acad Sci US A. 2005;102:9463–9468. doi: 10.1073/pnas.0503189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Huang TY, McLuckey SA. Top-Down Protein Identification/Characterization of a priori Unknown Proteins via Ion Trap Collision-Induced Dissociation and Ion/Ion Reactions in a Quadrupole/Time-of-Flight Tandem Mass Spectrometer. Anal Chem. 2009;81:1433–1441. doi: 10.1021/ac802204j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang TY, Liu J, Liang X, Hodges BDM, McLuckey SA. Top-down Analysis of siRNA via Ion Trap Collision-Induced Dissociation. Anal Chem. 2008;80:8501–8508. doi: 10.1021/ac801331h. [DOI] [PubMed] [Google Scholar]
- 23.Huang TY, Liu J, McLuckey SA. Top-down Tandem Mass Spectrometry of a tRNA via Ion Trap Collision-Induced Dissociation. J Am Soc Mass Spectrom. 2010;21:890–898. doi: 10.1016/j.jasms.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 24.Han H, McLuckey SA. Selective Covalent Bond Formation in Polypeptide Ions via Gas-Phase Ion/Ion Reaction Chemistry. J Am Chem Soc. 2009;131:12884–12885. doi: 10.1021/ja904812d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hassell KM, Stutzman JR, McLuckey SA. Gas Phase Bio-conjugation of Peptides via Ion/Ion Charge Inversion: Schiff Base Formation on the Conversion of Cations to Anions. Anal Chem. 2010;82:1594–1597. doi: 10.1021/ac902732v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mirzaei H, Regnier FE. Enhancing Electrospray Ionization Efficiency of Peptides by Derivatization. Anal Chem. 2006;78:4175–4183. doi: 10.1021/ac0602266. [DOI] [PubMed] [Google Scholar]
- 27.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 28.Ross PL, Huang YLN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Quantitative Analysis of Complex Protein Mixtures Using Isotope-Coded Affinity Tags. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Beardsley RL, Sharon LA, Reilly JP. Peptide de novo Sequencing Facilitated by a Dual-Labeling Strategy. Anal Chem. 2005;77:6300–6309. doi: 10.1021/ac050540k. [DOI] [PubMed] [Google Scholar]
- 30.Madsen JA, Brodbelt JS. Simplifying Fragmentation Patterns of Multiply Charged Peptides by N-Terminal Derivatization and Electron Transfer Collision Activated Dissociation. Anal Chem. 2009;81:3645–3653. doi: 10.1021/ac9000942. [DOI] [PubMed] [Google Scholar]
- 31.Mentinova M, McLuckey SA. Covalent Modification of Gaseous Peptide Ions with N-Hydroxysuccinimide Ester Reagent Ions. J Am Chem Soc. 2010;132:18248–18257. doi: 10.1021/ja107286p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sinz A. Chemical Cross-Linking and Mass Spectrometry to Map Three-Dimensional Protein Structures and Protein-Protein Interactions. Mass Spectrom Rev. 2006;25:663–682. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- 33.Chakravarti B, Lewis SJ, Chakravarti DN, Raval A. Three Dimensional Structures of Proteins and Protein Complexes from Chemical Cross-Linking and Mass Spectrometry: A Biochemical and Computational Overview. Curr Proteomics. 2006;3:1–21. [Google Scholar]
- 34.Schmidt A, Kalkhof S, Ihling C, Cooper DMF, Sinz A. Mapping Protein Interfaces by Chemical Cross-Linking and Fourier Transform Ion Cyclotron Resonance Mass Spectrometry: Application to a Calmodulin/Adenylyl Cyclase 8 Peptide Complex. Eur J Mass Spectrom. 2005;11:525–534. doi: 10.1255/ejms.748. [DOI] [PubMed] [Google Scholar]
- 35.Freed JK, Smith JR, Li P, Greene AS. Isolation of Signal Transduction Complexes Using Biotin and Crosslinking Methodologies. Proteomics. 2007;7:2371–2374. doi: 10.1002/pmic.200700219. [DOI] [PubMed] [Google Scholar]
- 36.Tubb MR, Silva RAGD, Fang J, Tso P, Davidson WS. A Three-Dimensional Homology Model of Lipid-Free Apolipoprotein A-IV Using Cross-Linking and Mass Spectrometry. J Biol Chem. 2008;283:17314–17323. doi: 10.1074/jbc.M800036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh P, Panchaud A, Goodlett DR. Chemical Cross-Linking and Mass Spectrometry as a Low-Resolution Protein Structure Determination Technique. Anal Chem. 2010;82:2636–2642. doi: 10.1021/ac1000724. [DOI] [PubMed] [Google Scholar]
- 38.Chen Z, Jawhari A, Fischer L, Buchen C, Tahir S, Kamenski T, Rasmussen M, Lariviere L, Bukowski-Wills JC, Nilges M, Cramer P, Rappsilber J. Architecture of the RNA Polymerase II-TFIIF Complex Revealed by Cross-Linking Mass Spectrometry. EMBO J. 2010;29:717–726. doi: 10.1038/emboj.2009.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hager JW. A New Linear Ion Trap Mass Spectrometer. Rapid Commun Mass Spectrom. 2002;16:512–526. doi: 10.1002/rcm.1020. [DOI] [PubMed] [Google Scholar]
- 40.Liang X, Xia Y, McLuckey SA. Alternately Pulsed Nano-Electrospray Ionization/Atmospheric Pressure Chemical Ionization for Ion/Ion Reactions in an Electrodynamic Ion Trap. Anal Chem. 2006;78:3208–3212. doi: 10.1021/ac052288m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia Y, Wu J, Londry FA, Hager JW, McLuckey SA. Mutual Storage Mode Ion/Ion Reactions in Hybrid Linear Ion Trap. J Am Soc Mass Spectrom. 2005;16:71–81. doi: 10.1016/j.jasms.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 42.Xia Y, Liang X, McLuckey SA. Pulsed Dual Electrospray Ionization for Ion/Ion Reactions. J Am Soc Mass Spectrom. 2005;16:1750–1756. doi: 10.1016/j.jasms.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 43.Londry FA, Hager JW. Mass Selective Axial Ion Ejection from a Linear Quadrupole Ion Trap. J Am Soc Mass Spectrom. 2003;14:1130–1147. doi: 10.1016/S1044-0305(03)00446-X. [DOI] [PubMed] [Google Scholar]
- 44.Liang X, McLuckey SA. Transmission Mode Ion/Ion Proton Transfer Reactions in a Linear Ion Trap. J Am Soc Mass Spectrom. 2007;18:882–890. doi: 10.1016/j.jasms.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He M, Emory JF, McLuckey SA. Reagent Anions for Charge Inversion of Polypeptide/Protein Cations in the Gas Phase. Anal Chem. 2005;77:3173–3182. doi: 10.1021/ac0482312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wells JM, Chrisman PA, McLuckey SA. Formation and Characterization of Protein-Protein Complexes in Vacuo. J Am Chem Soc. 2003;125:7238–7249. doi: 10.1021/ja035051l. [DOI] [PubMed] [Google Scholar]
- 47.Thorne GC, Ballard KD, Gaskell SJ. Metastable Decomposition of Peptide [M+H]+ Ions via Rearrangement Involving Loss of the C-terminal Amino Acid Residue. J Am Soc Mass Spectrom. 1990;1:249–257. [Google Scholar]
- 48.Li Zhong, Yalcin T, Cassady CJ. C-terminal Amino Acid Residue Loss for Deprotonated Peptide Ion Containing Glutamic Acid Aspartic Acid or Serine Residues at the C-terminus. J Mass Spectrom. 2006;41:939–949. doi: 10.1002/jms.1053. [DOI] [PubMed] [Google Scholar]
- 49.Farrugia JM, O’Hair RAJ. Involvement of Salt Bridges in a Novel Gas Phase Rearrangement of Protonated Arginine-Containing Dipeptides which Precedes Fragmentation. Int J Mass Spectrom. 2003;222:229–242. [Google Scholar]
- 50.Sadogopan N, Watson JT. Mass Spectrometric Evidence for Mechanisms of Fragmentation of Charge-Derivatized Peptides. J Am Soc Mass Spectrom. 2001;12:399–409. doi: 10.1016/S1044-0305(01)00211-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.