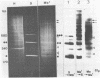
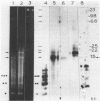
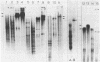
Abstract
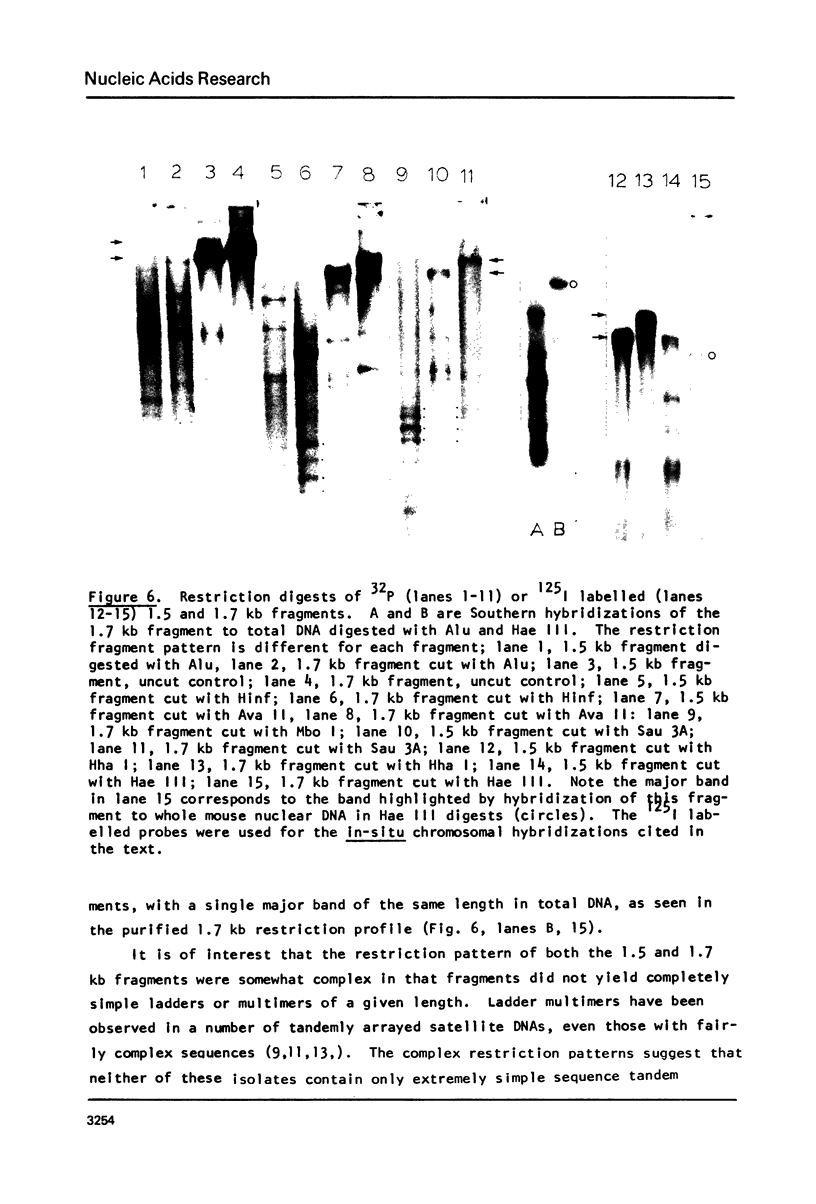
Mouse DNA cleaved with Eco R11 (bst NI) displays two prominent restriction bands of 1.5 and 1.7 kb in agarose gels stained with ethidium bromide. These constitute novel subsets of repeated DNA in the mouse. Sequential Hoechst 33258-CsCl gradient fractionation of mouse DNA, yielding more GC rich main band DNA, and AT rich satellite DNA, revealed that both these fragments copurified with GC rich main band DNA. They were not detected in purified satellite preparations. Together these restriction bands constituted larger than or equal to 0.2% of main band DNAs. Hybridization of 32p labelled satellite DNA to blots of Eco R11 restricted mouse DNA showed positive hybridization only to smaller satellite restriction fragments, indicating satellite DNA had little or no homology with either the 1.5 or 1.7 kb fragments. The 1.5 and 1.7 kb fragments were isolated from gels and labelled with 32p by nick translation. Using a series of restriction endonucleases each of these two fragments showed different cleavage patterns. Filter hybridization confirmed that these two fragments were distinct subsets as they did not cross hybridize with each other. They also did not hybridize to other more minor repeated non-satellite DNA bands noted in ethidium bromide stained gels. Neither of them could be assigned to ribosomal genes as they did not hybridize to 32p kinase labelled 18S and 28S RNA. Isolation of DNA from male and female mice showed comparable amounts of both the 1.5 and 1.7 kb fragments. Thus neither was Y chromosome specific. From restriction patterns, and preliminary chromosome hybridization studies, these fragments are thought to represent "interspersed" repeated sequences rather than very long tandem (satellite like) centromeric arrays. The relationship between these repeated sequence subsets, their evolution and detailed organization, and their representation in different mouse species, remain to be determined.
Full text
PDF


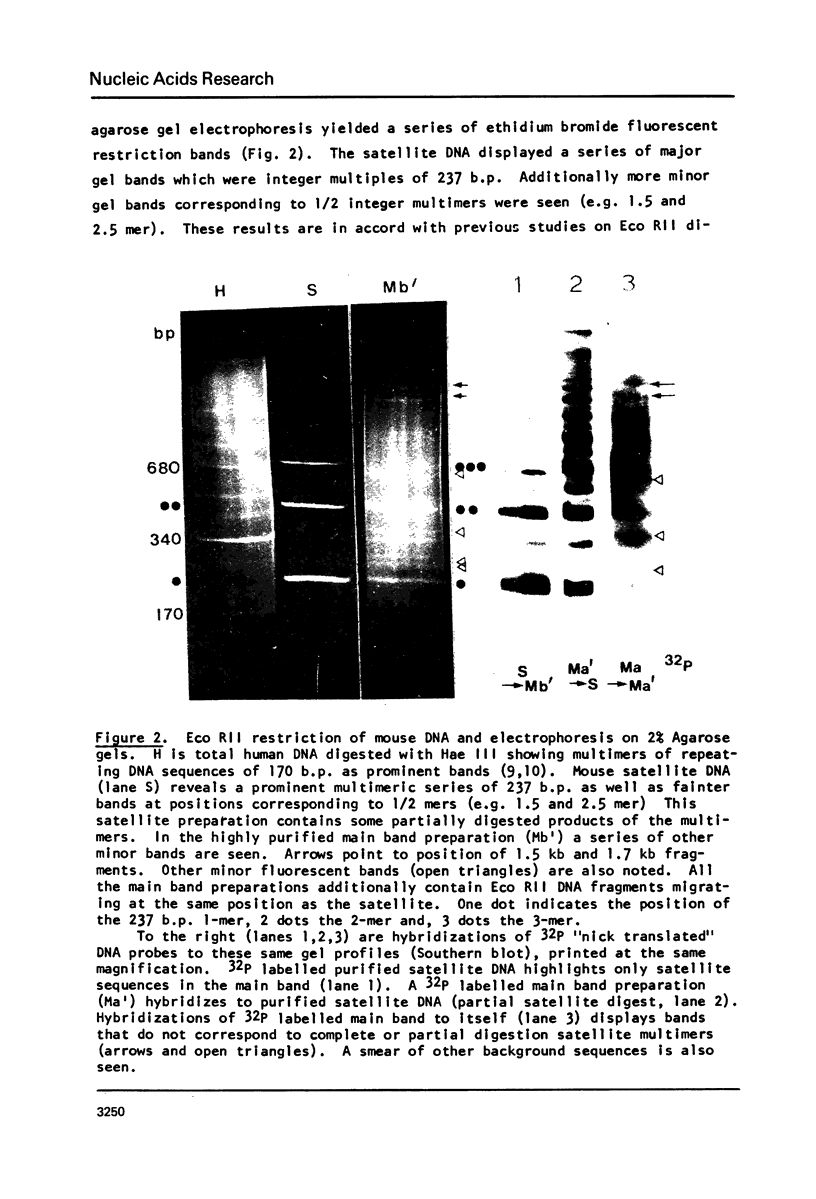







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biro P. A., Carr-Brown A., Southern E. M., Walker P. M. Partial sequence analysis of mouse satellite DNA evidence for short range periodicities. J Mol Biol. 1975 May 5;94(1):71–86. doi: 10.1016/0022-2836(75)90405-2. [DOI] [PubMed] [Google Scholar]
- Brown S. D., Dover G. A. Conservation of segmental variants of satellite DNA of Mus musculus in a related species: Mus spretus. Nature. 1980 May 1;285(5759):47–49. doi: 10.1038/285047a0. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Hearst J. E. Organization of highly repeated sequences in mouse main-band DNA. J Mol Biol. 1976 Jan 25;100(3):227–256. doi: 10.1016/s0022-2836(76)80061-7. [DOI] [PubMed] [Google Scholar]
- Comings D. E., Avelino E. DNA loss during Robertsonian fusion in studies of the tobacco mouse. Nat New Biol. 1972 Jun 14;237(76):199–199. doi: 10.1038/newbio237199a0. [DOI] [PubMed] [Google Scholar]
- Cooke H. Repeated sequence specific to human males. Nature. 1976 Jul 15;262(5565):182–186. doi: 10.1038/262182a0. [DOI] [PubMed] [Google Scholar]
- KIT S. Equilibrium sedimentation in density gradients of DNA preparations from animal tissues. J Mol Biol. 1961 Dec;3:711–716. doi: 10.1016/s0022-2836(61)80075-2. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. A simplified method for preparation of mouse satellite DNA. Anal Biochem. 1977 Apr;78(2):561–568. doi: 10.1016/0003-2697(77)90118-x. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. Complex and simple sequences in human repeated DNAs. Chromosoma. 1978 Mar 22;66(1):1–21. doi: 10.1007/BF00285812. [DOI] [PubMed] [Google Scholar]
- Manuelidis L., Manuelidis E. E. Amount of satellite DNA in four experimentally induced tumors of the central nervous system. Quantitative changes in a glioblastoma producing C-type particles. J Natl Cancer Inst. 1976 Jan;56(1):43–50. doi: 10.1093/jnci/56.1.43. [DOI] [PubMed] [Google Scholar]
- Manuelidis L., Manuelidis E. E. Conservation of repeated DNA sequences in aneuploid human tumor cells. Chromosoma. 1979 May 10;72(3):257–269. doi: 10.1007/BF00331088. [DOI] [PubMed] [Google Scholar]
- Manuelidis L., Manuelidis E. E. Localization of mouse satellite DNA on chromosomes of experimentally induced glioblastomas; non-centromeric lable in one glioblastoma producing C-type particles. Int J Cancer. 1976 May 15;17(5):659–669. doi: 10.1002/ijc.2910170516. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. Repeating restriction fragments of human DNA. Nucleic Acids Res. 1976 Nov;3(11):3063–3076. doi: 10.1093/nar/3.11.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff W. F., Jr, White E. L., Benjamin R., Huang R. C. Low molecular weight RNA species from chromatin. Biochemistry. 1975 Aug 12;14(16):3715–3724. doi: 10.1021/bi00687a031. [DOI] [PubMed] [Google Scholar]
- Orgel L. E., Crick F. H. Selfish DNA: the ultimate parasite. Nature. 1980 Apr 17;284(5757):604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- Pardue M. L., Gall J. G. Chromosomal localization of mouse satellite DNA. Science. 1970 Jun 12;168(3937):1356–1358. doi: 10.1126/science.168.3937.1356. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Long range periodicities in mouse satellite DNA. J Mol Biol. 1975 May 5;94(1):51–69. doi: 10.1016/0022-2836(75)90404-0. [DOI] [PubMed] [Google Scholar]
- Waring M., Britten R. J. Nucleotide sequence repetition: a rapidly reassociating fraction of mouse DNA. Science. 1966 Nov 11;154(3750):791–794. doi: 10.1126/science.154.3750.791. [DOI] [PubMed] [Google Scholar]