Abstract
Mammalian cells have been shown to internalize oligonucleotide-functionalized gold nanoparticles (DNA-Au NPs or siRNA-Au NPs) without the aid of auxiliary transfection agents and use them to initiate an antisense or RNAi response. Previous studies have shown that the dense monolayer of oligonucleotides on the nanoparticle leads to the adsorption of serum proteins and facilitates cellular uptake. Here, we show that serum proteins generally act to inhibit cellular uptake of DNA-Au NPs. We identify the pathway for DNA-Au NPs entry in HeLa cells. Biochemical analyses indicate that DNA-Au NPs are taken up by a process involving receptor-mediated endocytosis. Evidence shows that DNA-Au NPs entry is primarily mediated by scavenger receptors, a class of pattern-recognition receptors. This uptake mechanism appears to be conserved across species as blocking the same receptors in mouse cells also disrupted DNA-Au NP entry. Polyvalent nanoparticles functionalized with siRNA are shown to enter through the same pathway. Thus, scavenger receptors are required for cellular uptake of polyvalent oligonucleotide functionalized nanoparticles.
INTRODUCTION
Polyvalent DNA-functionalized gold nanoparticles (DNA-Au NP) are structures with a gold nanoparticle core and a high surface density of DNA ligands (1). As a result of this dense functionalization, these hybrid nanostructures have demonstrated applications in programmable materials crystallization (2-3) and non-enzymatic biodiagnostic assays (4). Furthermore, the tailorable recognition properties of DNA and the catalytic and plasmonic properties of Au NPs have been used synergistically in a variety of highly sensitive and selective nucleic acid, small molecule, metal ion, and cancer cell detection strategies (5-11).
Recently, it was reported that DNA-Au NPs surprisingly readily enter a wide variety of cell types (over 50 cell types examined to date) without the aid of transfection agents (12-13). These structures have properties that make them particularly attractive as intracellular gene regulation agents. They resist nuclease degradation (12, 14), exhibit enhanced target binding (15), show minimal interferon β response (16), and enter cells rapidly and efficiently (17). Consequently, they have been used for antisense (12, 18) and siRNA (19-21) based post-transcriptional gene regulation, intracellular detection of biologically relevant molecules (22-23), and intracellular delivery of MRI contrast agents (24) as well as chemotherapeutic agents (25). Given the fundamental ability of these nanomaterials to readily enter cultured cells, the mechanism of cellular uptake is of significant interest. It has shown that subtle changes in surface functionalities can lead to drastic changes in cellular internalization ability of nanoparticles (26-27). We have shown that the densely functionalized shell of polyanionic DNA (19 pmol/cm2 for 13 nm gold particle) is a primary contributor to the cellular uptake of DNA-Au NPs (17), despite the conventional wisdom that negatively charged materials will not enter cells efficiently.
When material surfaces are exposed to biological environments, they are often modified by the adsorption of biomolecules, such as proteins (28). It has been shown that adsorption of proteins to a surface causes conformational changes in the proteins. These changes may expose hidden epitopes that cause unpredictable behavior and result in unusual properties (29). Protein adsorption on a nanoparticle surface has been implicated in facilitating uptake of various nanomaterials, including single-walled nanotubes, nanosized amorphous silica, and citrate capped gold nanoparticles (30-32). Indeed, we have previously shown that serum proteins are adsorbed to the dense monolayer of DNA on the Au NP surface, and that increasing the density of DNA on the Au NP surface increased the number of proteins adsorbed to DNA-Au NPs (17) as well as intracellular concentration of DNA-Au NPs.
In this paper, we explore the role of certain adsorbed serum proteins (BSA and transferrin) in the cellular internalization of the highly negatively charged DNA-Au NPs. We show that the adsorbed serum proteins reduce the cellular uptake of DNA-Au NPs, and that DNA-Au NP uptake is highest in serum-free conditions. Furthermore, using pharmacological methods, we identify scavenger receptors as the membrane proteins primarily responsible for facilitating the cellular uptake of DNA-Au NPs. We show that other oligonucleotide-functionalized nanoparticles, such as siRNA-Au NPs, also follow the same pathway of internalization. For DNA-Au NPs, the density of DNA on nanoparticles determines the extent of their cellular uptake. This entry pathway is conserved in mouse cells and may be a general mechanism of cellular entry of nanomaterials displaying dense monolayers of oligonucleotides.
EXPERIMENTAL PROCEDURES
Synthesis of gold nanoparticles, DNA and RNA
Citrate stabilized gold nanoparticles (13 ± 1 nm diameter) were prepared as described previously (33). DNA oligonucleotides were synthesized on an Expedite 8909 Nucleotide Synthesis System (ABI) using standard solid-phase phosphoramidite synthesis techniques. RNA oligonucleotides were synthesized using TOM-RNA reagents (Glen Research) on a MerMade 6 system (Bioautomation) using manufacturer recommended cleavage and deprotection stategy. All oligonucleotides were purified using reverse-phase high performance liquid chromatography (RP-HPLC) on a Varian Microsorb C18 column (10 μm, 300 × 10 mm) with 0.03 M triethylammonium acetate (TEAA), pH 7 and a 1%/min gradient of 95% CH3CN/5% 0.03 M TEAA at a flow rate of 3 mL/min, while monitoring the UV signal of nucleic acids at 254 nm. After purification, the oligonucleotides were lyophilized and stored at −80° C until use. The following oligonucleotide sequences were synthesized: RNA sequences: 5′-ACC CUG AAG UUC AUC UGC ACC ACC G – (hexaethyleneglycol)2- propylthiol- 3′, 5′-CGG UGG UGC AGA UGA ACU UCA GGG UCA-3′. DNA sequences: 5′-AGT AGA GGC AGG GAT GAT G AAA AAA AAA A- propylthiol- 3′, 5′-Fluorescein - AGT AGA GGC AGG GAT GAT G AAA AAA AAA A- propylthiol- 3′, 5′-TTT TGG GGT TTT GGG GTT TTG GGG TTT TGG GG –propylthiol- 3′, 5′-TTT TGG GGT TTT GGG GTT TTG GGG TTT TGG GG - 3′, 5′ (DTPA) AAA AAA AAA AGT AGT AGG GAC GGA GAT GA 3′ and 5′ (Tosyl-T)-AAA AAA AAA A (S-S) TCA TCT CCG TCC CTA CTA CG 3′. Tosyl-modified thymidine phosphoramidite was synthesized as described previously (34) and added to the oligonucleotide manually as described previously (35). DNA or RNA functionalized gold nanoparticles were prepared following published methods (12, 19). Mixed monolayer nanoparticles with variable DNA density were prepared following a published protocol (17) but with following modified oligonucleotide as diluent strand: 5′ (Propyl Spacer)19- AAA AAA AAA A- propylthiol- 3′. DNA loading onto mixed monolayer nanoparticles was determined using a fluorescein modified DNA strand.
Preparation of protein-DNA conjugates
All proteins used in this study were purchased from Sigma Aldrich and/or Fisher Scientific. The protein was mixed with the tosyl modified oligonucleotide at a 3 fold molar excess in 0.1M borate buffer, pH 8.5. The mixture was shaken at room temperature overnight. Unreacted oligonucleotides were separated from protein-DNA conjugates using 30000 MWCO spin columns (Chemicon).
Preparation of gold nanoparticle conjugates with variable protein loading
Oligonucleotide modified gold nanoparticle conjugates were made as described previously (1). DNA-Au NPs were loaded with various numbers of proteins by mixing with variable volumes of protein-DNA conjugates in phosphate buffered saline (PBS) (Hyclone), heating the mixture to 65°C, and cooling it to room temperature. The conjugates were centrifuged at 4°C and re-dispersed in 1x PBS three times to remove excess protein-DNA conjugates. The average number of protein molecules per gold nanoparticle was determined using previously published methods (17). Briefly, the protein loaded Au NPs were dissolved with potassium cyanide (KCN, 2 mM final concentration). The KCN digested samples were treated with a Quant-iT fluorescent reagent (Invitrogen) as described by the manufacturer. The resulting fluorescence was measured at room temperature using a fluorescence microplate reader (Fluo Dia T70, Photal) with excitation at 486 nm and recording emission at 570 nm. Estimation of the number of proteins in solution was calculated by comparing the fluorescence values of these unknowns to those obtained from a standard curve of known protein concentrations under the same assay conditions. Finally, the average number of proteins per nanoparticle was calculated by dividing this number by the initial nanoparticle concentration prior to KCN digestion.
Cell Culture
The uptake of gold nanoparticles (DNA-Au NPs or siRNA-Au NPs) was studied using a model cell line. HeLa (human cervical carcinoma) cells were obtained from American Type Culture Collection (ATCC) and maintained in minimum essential medium (EMEM) supplemented with 10% heat-inactivated FBS and 1% penicillin/streptomycin at 5% CO2 and 37°C. In the serum free conditions, FBS was omitted from the EMEM formulation. HeLa cells were acclimated to serum free culture by slowly reducing serum concentration from 10% to 0%. Mouse C166 cells (ATCC) were cultured in DMEM with 10% FBS at 5% CO2 and 37 °C.
Serum Pre-incubation of DNA-Au NPs
DNA-Au NPs were prepared as described above. The nanoparticle conjugates were added to MEM buffer containing 10% FBS and incubated at 37 °C for at least 12 hours. Following the incubation, excess serum was removed by three centrifugation, supernatant removal and re-dispersion steps. The nanoparticle conjugates were added to HeLa cells in serum containing and serum-free MEM to measure cellular uptake.
Cellular Uptake Studies and Nanoparticle Calculations
Hela cells were added to 96-well plates (25000 cells/well) 24 hours prior to nanoparticle addition. Filter sterilized nanoparticle conjugates were added directly to the cell culture media of adherent cells in 5 and 10 nM concentrations. Forty-eight hours after nanoparticle addition, the cells were washed twice with PBS, collected, counted and their viability measured using the Guava Viacount reagent on an EasyCyte flow cytometer (Millipore). The gold content of the cell digest was determined by inductively coupled plasma mass spectrometry (ICP-MS) (Thermo-Fisher). To prepare samples for ICP-MS, the cells were dissolved with nitric acid at 60 °C overnight, diluted in a matrix consisting of 2% HNO3, 2% HCl and 1 ppb indium (internal standard). The number of nanoparticles in each sample was calculated based on the concentration of Au found in the sample.
Treatments with Chemical Inhibitors
All chemicals were purchased from Sigma-Aldrich. HeLa cells were treated with different concentrations of methyl-β-cyclodextrin (MβCD), cytochalasin D (CytoD) or bafilomycin A (Baf A) for 60 minutes. DNA-Au NPs were then added in the presence of these chemicals. Controls include incubations of DNA-Au NPs in the presence of the solvent dimethylsulfoxide (DMSO) and in the absence of any chemicals. To probe the role of scavenger receptors, cells were similarly incubated in the presence of known ligands of scavenger receptors, poly-inosinic acid (Poly I) and Fucoidan. Compounds that are not known to inhibit scavenger receptors, poly-adenosinic acid (Poly A), galactose, lipopolysacchride (LPS), and untreated cells were used as controls. Twenty four hours after the addition of nanoparticles, cells were washed with phosphate buffered saline, trypsinized, counted using Viacount (Millipore) reagent and prepared for ICP-MS.
Knockdown of Scavenger Receptors
HeLa cells were plated in 48-well plate at 50000 cells/well. The cells were transfected with commercially bought siRNA (100 nM, 50 nM and 25 nM) against scavenger receptors A and scavenger receptors B1 (Santa Cruz) with Dharmafect (Dharmacon) following the manufacturer’s protocol. After 48 hours, the media was replaced with DNA-Au NP containing media (10 nM) for 24 hours. An equal volume of lipofectamine was used as delivery agent control. The cells were prepared for ICP-MS to determine the cellular gold content as described above.
RESULTS AND DISCUSSION
Serum proteins reduce cellular uptake of DNA-Au NPs
Previous work has shown that the cellular uptake of DNA-Au NPs correlates positively with the density of oligonucleotides on the nanoparticles, and that when DNA-Au NPs are added to serum containing media, they are coated with serum proteins (17). We hypothesized that if adsorption of serum protein promotes cellular uptake, then covalent attachment of serum proteins may increase uptake even further. To test this hypothesis, we attached bovine serum albumin (BSA) and transferrin, two highly abundant serum proteins, in a covalent manner on DNA-Au NPs. BSA is also the most abundant protein in serum and is known to coat polymer nanoparticles in complex media (36). Protein-DNA conjugates were made by covalently linking a tosyl-terminated oligonucleotide to amine groups on BSA or transferrin. These conjugates were then mixed at different ratios with DNA-Au NPs functionalized with the complementary oligonucleotide sequence to load variable numbers of proteins around the nanoparticles (Scheme 1). These nanoconjugates loaded with the same number of oligonucleotides but different numbers of proteins were then added to HeLa cells and their cellular internalization was measured by ICP-MS (see experimental procedures).
Scheme 1.
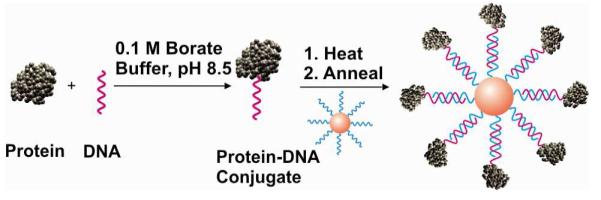
Stable loading of proteins on DNA functionalized gold nanoparticles. The proteins are covalently attached to tosyl-modified oligonucleotides, which are then hybridized to nanoparticles functionalized with a complementary sequence. The variable loading of proteins is achieved by using different ratios of protein-DNA conjugate to DNA-Au NPs.
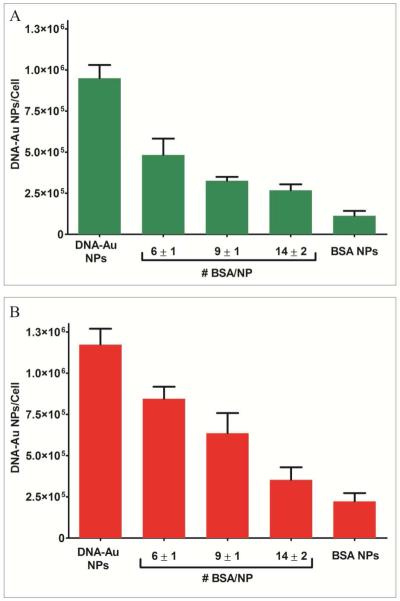
The results show that as BSA loading onto DNA-Au NPs increases, the number of nanoparticles taken up by cells decreases (Figure 1 and Supporting Information). For example, as the number of BSA molecules per DNA-Au NP increased from 0 to 14 ± 2, the number of DNA-Au NP internalized by cells in serum containing conditions decreased by up to 80%, from 1.0 × 106 ± 0.1 × 106 to 2.5 × 105 ± 0.7 × 105 Au NPs per cell. A 70% decrease was observed when transferrin was used to coat DNA-Au NPs instead of BSA. (Supporting Information, Figure 1). These results suggest that the most abundant components of serum lower uptake when they are bound strongly to DNA-Au NPs. These results are surprising since previously we have observed that the high density of DNA on nanoparticles resulted in increased adsorption of proteins as well as cellular uptake and when DNA density was reduced with oligo (ethylene glycol) (OEG) passivation, protein adsorption and cellular uptake were reduced (17). One explanation of this apparent discrepancy is that the dense monolayer of DNA on nanoparticles may generally promote both cellular uptake and protein. However, adsorbed serum proteins do not necessarily promote cellular uptake but may be displaced by some cellular proteins that affect nanoparticle uptake. When nanoparticles are functionalized with lower density of DNA, their binding to all proteins is reduced as is their cellular uptake. In this experiment, since serum proteins are strongly bound to nanoparticles, they cannot be easily displaced by cellular proteins that affect nanoparticle uptake. Furthermore, pre-incubation of DNA-Au NPs in serum and subsequent addition to cell culture also reduced cellular uptake by similar amount (Supporting Information, Figure 2a). Thus it is unlikely that the binding of other less abundant serum proteins promotes cellular uptake of nanoparticles. Overall, these results suggest that serum protein generally reduce cellular uptake of DNA-Au NPs and point to other factors as positive regulators of cellular entry.
Figure 1.
Cellular internalization of DNA-Au NPs when coated with increasing amounts of BSA. The method for loading a variable number of proteins on DNA-Au NPs is shown in Scheme 1. Cells were grown in media containing 10% FBS with 5 nM (A) and 10 nM (B) of DNA-Au NPs.
DNA-Au NP uptake is highest in serum-free culture
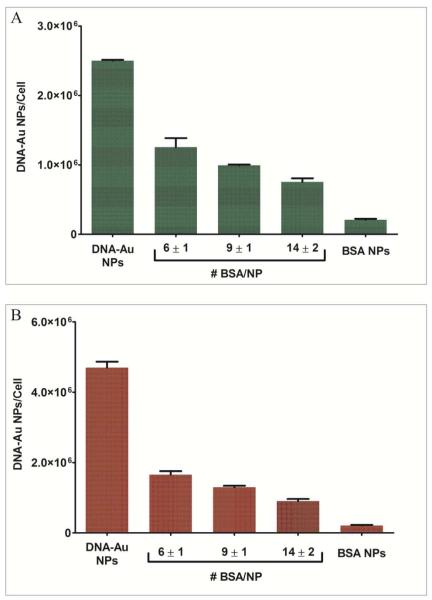
To confirm that serum proteins generally reduce nanoparticle uptake, we performed previously described experiments in serum-free media and compared the results to the results from normal serum-containing cultures. In all cases, the number of DNA-Au NPs internalized by cells in serum-free conditions was significantly higher than in serum-containing conditions. For example, at a 5 nM DNA-Au NPs solution concentration, 2.5 × 106 ± 0.02 × 106 DNA-Au NPs/cell were internalized in serum-free conditions while 1.0 × 106 ± 0.1 × 106 DNA-Au NPs/cell were internalized when the media contained 10% serum, a 150% increase (Supporting Information, Figure 2b). Furthermore, this increase was observed for DNA-Au NPs that were pre-incubated with serum, or DNA-Au NPs loaded with covalently-immobilized BSA (Figure 2) or transferrin (Supporting Information, Figure 3).
Figure 2.
(A) Cellular uptake of DNA-Au NPs loaded with varying number of BSA molecules at 5nM (A) and 10nM (B) solution concentration of DNA-Au NPs in serum-free conditions. Serum-free conditions yield significantly higher number of nanoparticles per cell (150% increase) compared with normal 10% serum cultures (shown in Figure 1).
While the number of DNA-Au NPs internalized is still high in serum containing media, a 150% or higher uptake is observed in serum-free cultures for all of the particles tested, with the exception of fully BSA- or transferrin coated nanoparticles that contained no oligonucleotides, which did not exhibit such an increase. These results are consistent with the conclusion that serum proteins are responsible for decreasing the uptake of DNA-Au NPs. Since it was shown previously that serum proteins adsorb to DNA-Au NPs (17), it is likely that the adsorbed proteins compete with the binding interactions required for nanoparticles to be internalized by cells. These binding interactions between oligonucleotides on the DNA-Au NPs and cellular proteins promote internalization through receptor-mediated endocytosis.
Pharmacological inhibitors of DNA-Au NP uptake machinery
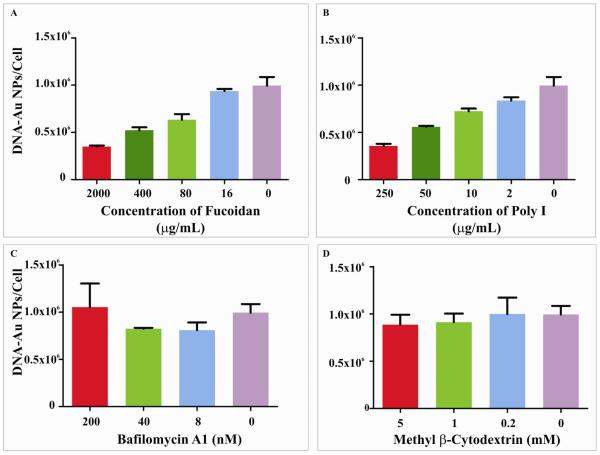
DNA-Au NP entry was explored using a pharmacological approach that tested the effect of inhibitors of cellular uptake mechanisms on nanoparticle uptake. Bafilomycin A (Baf A) can inhibit the acidification of endosomes, thus preventing their maturation and fusion into lysosomes (33). Baf A treatment did not inhibit nanoparticle uptake (Figure 3C). Therefore, the endosomal–lysosomal acidification process is not critical for nanoparticle entry in HeLa cells. Similarly, methyl-β-cyclodextrin (MβCD) and cytochalasin-D, inhibitors of caveolae mediated endocytosis and phagocytosis, respectively, largely did not affect nanoparticle entry into cells (Figure 3D, Supporting Information, Figure 4A). Cytochalasins cause the depolymerization of the actin cytoskeleton and inhibit uptake through the caveolae and macropinocytosis without affecting clathrin-mediated endocytosis whereas MβCD inhibits phagocytosis by the disruption of lipid rafts (32). These experiments suggest that the broader classes of endocytotic process such as caveolae mediated endocytosis, macropinocytosis or phagocytosis are not the primary modes of entry of DNA-Au NPs into HeLa cells.
Figure 3.
Endocytotic uptake of DNA-Au NPs in HeLa cells is mediated by scavenger receptors. Treatment with agonists of scavenger receptors, fucoidan (A) and polyinosinic acid (poly I, B), prior to nanoparticle addition results in up to 60% inhibition of the cellular uptake of DNA-Au NPs. Pharmacological agents that inhibit other modes of cellular entry, such as bafilomycin A1 and methyl β-cytodextrin, did not inhibit nanoparticle uptake (C, D, Supporting Information, Figure 4).
DNA-Au NP Uptake is blocked by agonists of pattern-recognition receptors
The role of pattern-recognition receptors in DNA-Au NP uptake was evaluated using a chemical approach. Scavenger receptors are a group of structurally unrelated molecules known to mediate the endocytosis of certain polyanionic ligands, including nucleic acids (37) and are implicated in uptake of phosphorothioate-modified oligonucleotides in animals (38). We examined whether molecules known to interact with scavenger receptors competed for binding of the DNA-Au NPs, and thus inhibited cellular uptake. Poly I and fucoidan, well known ligands of the scavenger-receptor family, inhibited nanoparticle uptake by >60% as measured by ICP-MS (Figure 3 A-B). In contrast, chemically related molecules that interact with other receptors, but do not inhibit scavenger receptors (such as lipopolysacchrides (LPS), polyadenylic acid (poly A), ssDNA or the monosaccharide galactose), did not affect DNA-Au NP uptake (Supporting Information, Figure 4). There was a slight decrease in viability of cells when treated with the highest concentrations of poly I and fucoidan. However, this decrease in viability was not correlated with decrease in cellular uptake as treatments with related chemicals that cause similar drop in viability, poly A and lipopolysaccharides, did not lead to decrease in uptake (Supporting Information, Table 1). Thus the decrease in cellular uptake of nanoparticles is most likely caused by binding of poly I and fucoidan to scavenger receptors which prevents DNA-Au NPs from binding to the same receptors. We also measured the stability of DNA-Au NPs in poly I and fucoidan solutions to rule out any possibility of drop in stability of nanoparticles causing the decrease in uptake (Supporting Information, Figure 9).
While RNAi downregulation of class A (SR-A) or class B (SR-B1) scavenger receptors (Supporting Information, Figure 10) did not result in inhibition of nanoparticle uptake (Supporting Information, Figure 5), the inhibition observed by the pharmacological treatments suggests that multiple types of scavenger receptors with overlapping functions participate in DNA-Au NP uptake. To explore this possibility, mixed siRNAs targeting SR-A and SR-B1 were transfected into HeLa cells. This mixture did not result in a significant reduction in nanoparticle uptake. Recent reports have shown that long dsRNA is internalized by scavenger receptor-mediated endocytosis in S2 cells (39-40). However, these reports were also unable to show that down regulation of individual scavenger receptors significantly reduced dsRNA uptake or resultant RNAi. It is possible that the simultaneous targeting the two classes of receptors by RNAi is not effective enough to impair the DNA-Au NP binding to the remaining receptors. It is also possible that unidentified members of this family of transporter proteins are responsible for this uptake function.
DNA density on nanoparticles determines the extent of inhibition
We next sought to understand the molecular basis of binding of DNA-Au NPs to scavenger receptors. It has been shown that poly I forms complex quadraplex-like structures at physiologic temperatures and that the binding of polynucleotides to scavenger receptors depends on the formation of stable four-stranded structures (41-42). Thus it is likely that other polynucleotide sequences predicted to fold into a structure that can bind scavenger receptors would lower cellular uptake of nanoparticles. Indeed, treatment with telomere-like DNA sequences (which are predicted to fold into quadraplex-like structures) prior to addition of DNA-Au NPs reduced cellular uptake of nanoparticles (Supporting Information, Figure 6). Higher concentrations of this synthetic ligand were necessary to inhibit nanoparticle entry, which is likely due to the weaker binding of the synthetic DNA strand to scavenger receptors compared to poly I or fucoidan.
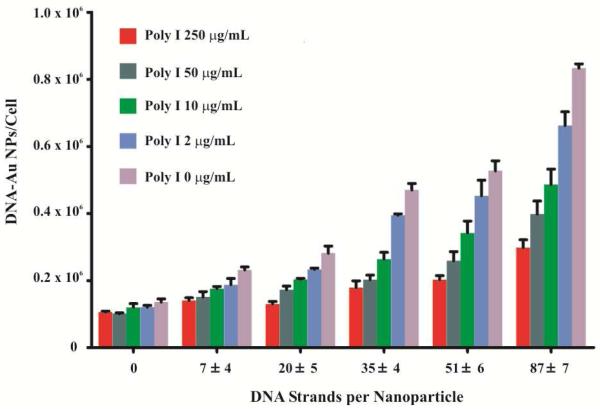
Thus, we hypothesized that the dense monolayer of DNA on the nanoparticle surface mimics the complex structure that Poly I forms in order to bind to scavenger receptors. Indeed, reducing the density of DNA on nanoparticles decreased the Poly I dependent inhibition of nanoparticle uptake (Figure 4). However, since we also reduced the ability of nanoparticles to bind scavenger receptors, the total nanoparticles internalized by cell decreased as well. Interestingly, the extent of inhibition was greater for nanoparticles with higher surface density of DNA. For nanoparticles with 87 ± 7 DNA per nanoparticle, the cellular uptake decreased 64% (from 8.33 × 105 to 2.98 × 105 DNA-Au NPs per cell) as poly I concentration increased from 0 to 250 μg/mL. By contrast, the nanoparticles with 7 ± 4 DNA per nanoparticle, the cellular uptake decreased 38% (from 2.32 × 105 to 1.42 × 105 DNA-Au NPs per cell) with the same poly I treatment. This difference suggests that the nanoparticles with higher density of oligonucleotides are more dependent on scavenger receptors and thus show more inhibition over the range of concentrations of poly I that competes for binding to the same receptors.
Figure 4.
Polyinosinic acid dependent inhibition of DNA-Au NPs is dependent on the density of DNA on nanoparticles. When the cellular uptake of DNA-Au NPs is lowered with decreasing DNA density, the ability of poly I to inhibit cellular internalization of DNA-Au NPs is lowered as well.
These experiments strongly suggest that the dense monolayer of DNA on nanoparticles mimics the complex structures of other polynucleotides, such as poly I or long dsRNA, and serves as a ligand for scavenger receptors. This hypothesis is further strengthened by experiments showing that the cellular uptake of siRNA-Au NPs is inhibited by fucoidan and poly I but not by the other pharmacological treatments (Supporting Information, Figure 7). For siRNA-Au NPs, the inhibition was >85% versus the untreated samples at highest concentrations of the agonists suggesting that these nanomaterials depend on scavenger receptor binding for cellular entry. Identical experiments in a mouse cell line also yielded the same results suggesting that the entry pathway in mouse cells is conserved (Supporting Information, Figure 8). Recently, our group has shown that iron oxide nanoparticles also enter cells with high efficiency when functionalized with dense monolayer of oligonucleotides (43). Therefore, it is likely that nanomaterials displaying dense monolayer of oligonucleotides can engage these receptors and be internalized by mammalian cells using the scavenger receptor pathway.
CONCLUSIONS
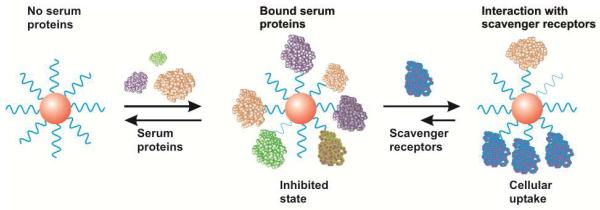
We have shown that serum proteins act to reduce the cellular uptake of oligonucleotide-functionalized nanoparticles and that highest nanoparticle uptake is observed in serum-free cultures. Using a pharmacological approach, we demonstrated that scavenger receptors mediate the cellular entry of oligonucleotide-functionalized gold nanoparticles. We have shown that the interaction of DNA-Au NPs and scavenger receptors is dependent on the density of oligonucleotides on nanoparticles. This interaction can be partially blocked by synthetic polynucleotides that mimic the structures of other ligands of scavenger receptors. Thus, our results show that a determining factor for cellular uptake of DNA-Au NPs is the polyvalent interactions of DNA-Au NPs with scavenger receptors on cell surface (Scheme 2) suggesting a general mechanism by which nanostructures with dense monolayers of oligonucleotides can be internalized by cells.
Scheme 2.
The proposed mechanism of cellular uptake of DNA-Au NPs. In serum containing media, DNA-Au NPs are coated with proteins which reduce nanoparticle interactions with scavenger receptors. Cellular uptake is mediated by the displacement of serum proteins by these receptors. In the absence of serum proteins, these interactions are more easily facilitated, resulting in greater uptake as a result of high DNA density. If the binding to scavenger receptors is inhibited by addition of competitive ligands, then the cellular uptake of nanoparticles is also reduced.
Supplementary Material
ACKNOWLEDGEMENTS
C.A.M. acknowledges a Cancer Center for Nanotechnology Excellence (NCI-CCNE) award for support of this research. P.C.P. was supported by a Ryan Fellowship and Malkin Foundation Fellowship. A.E.P. was supported by a Ryan Fellowship. W.L.D. is grateful for support from the Chicago Biomedical Consortium.
Footnotes
Supporting Information Available: Experimental procedures, nanoparticle characterization data and results from additional experiments. This information is available free of charge via the Internet at http://pubs.acs.org/
Literature Cited
- (1).Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- (2).Park SY, Lytton-Jean AK, Lee B, Weigand S, Schatz GC, Mirkin CA. DNA-programmable nanoparticle crystallization. Nature. 2008;451:553–556. doi: 10.1038/nature06508. [DOI] [PubMed] [Google Scholar]
- (3).Nykypanchuk D, Maye MM, van der Lelie D, Gang O. DNA-guided crystallization of colloidal nanoparticles. Nature. 2008;451:549–552. doi: 10.1038/nature06560. [DOI] [PubMed] [Google Scholar]
- (4).Nam J-M, Thaxton CS, Mirkin CA. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- (5).Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science. 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- (6).Park SJ, Taton TA, Mirkin CA. Array-based electrical detection of DNA with nanoparticle probes. Science. 2002;295:1503–1506. doi: 10.1126/science.1067003. [DOI] [PubMed] [Google Scholar]
- (7).Taton TA, Mirkin CA, Letsinger RL. Scanometric DNA array detection with nanoparticle probes. Science. 2000;289:1757–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- (8).Liu J, Lu Y. Fast colorimetric sensing of adenosine and cocaine based on a general sensor design involving aptamers and nanoparticles. Angew. Chem., Int. Ed. 2005;45:90–94. doi: 10.1002/anie.200502589. [DOI] [PubMed] [Google Scholar]
- (9).Liu J, Lu Y. A colorimetric lead biosensor using DNAzyme-directed assembly of gold nanoparticles. J. Am. Chem. Soc. 2003;125:6642–6643. doi: 10.1021/ja034775u. [DOI] [PubMed] [Google Scholar]
- (10).Liu J, Lu Y. Colorimetric Cu2+ detection with a ligation DNAzyme and nanopairticles. Chem. Commun. 2007:4872–4874. doi: 10.1039/b712421j. [DOI] [PubMed] [Google Scholar]
- (11).Medley CD, Smith JE, Tang Z, Wu Y, Bamrungsap S, Tan W. Gold Nanoparticle-Based Colorimetric Assay for the Direct Detection of Cancerous Cells. Anal. Chem. 2008;80:1067–1072. doi: 10.1021/ac702037y. [DOI] [PubMed] [Google Scholar]
- (12).Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AKR, Han MS, Mirkin CA. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- (13).Giljohann David A., Seferos Dwight S., Daniel Weston L., Massich Matthew D., Patel Pinal C., Mirkin Chad A. Gold Nanoparticles for Biology and Medicine. Angew. Chem., Int. Ed. 2010;49:3280–3294. doi: 10.1002/anie.200904359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Seferos DS, Prigodich AE, Giljohann DA, Patel PC, Mirkin CA. Polyvalent DNA Nanoparticle Conjugates Stabilize Nucleic Acids. Nano Lett. 2009;9:308–311. doi: 10.1021/nl802958f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Lytton-Jean AK, Mirkin CA. A thermodynamic investigation into the binding properties of DNA functionalized gold nanoparticle probes and molecular fluorophore probes. J. Am. Chem. Soc. 2005;127:12754–12755. doi: 10.1021/ja052255o. [DOI] [PubMed] [Google Scholar]
- (16).Massich MD, Giljohann DA, Seferos DS, Ludlow LE, Horvath CM, Mirkin CA. Regulating Immune Response Using Polyvalent Nucleic Acid-Gold Nanoparticle Conjugates. Mol. Pharmaceutics. 2009;6:1934–1940. doi: 10.1021/mp900172m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Giljohann DA, Seferos DS, Patel PC, Millstone JE, Rosi NL, Mirkin CA. Oligonucleotide loading determines cellular uptake of DNA-modified gold nanoparticles. Nano Lett. 2007;7:3818–3821. doi: 10.1021/nl072471q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Patel PC, Giljohann DA, Seferos DS, Mirkin CA. Peptide antisense nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17222–17226. doi: 10.1073/pnas.0801609105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Giljohann DA, Seferos DS, Prigodich AE, Patel PC, Mirkin CA. Gene Regulation with Polyvalent siRNA-Nanoparticle Conjugates. J. Am. Chem. Soc. 2009;131:2072–2073. doi: 10.1021/ja808719p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Lee JS, Green JJ, Love KT, Sunshine J, Langer R, Anderson DG. Gold, Poly(beta-amino ester) Nanoparticles for Small Interfering RNA Delivery. Nano Lett. 2009;9:2402–2406. doi: 10.1021/nl9009793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Lee SH, Bae KH, Kim SH, Lee KR, Park TG. Amine-functionalized gold nanoparticles as non-cytotoxic and efficient intracellular siRNA delivery carriers. Int J Pharm. 2008;364:94–101. doi: 10.1016/j.ijpharm.2008.07.027. [DOI] [PubMed] [Google Scholar]
- (22).Seferos DS, Giljohann DA, Hill HD, Prigodich AE, Mirkin CA. Nano-flares: probes for transfection and mRNA detection in living cells. J. Am. Chem. Soc. 2007;129:15477–15479. doi: 10.1021/ja0776529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Zheng D, Seferos DS, Giljohann DA, Patel PC, Mirkin CA. Aptamer Nano-flares for Molecular Detection in Living Cells. Nano Lett. 2009;9:3258–3261. doi: 10.1021/nl901517b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Song Y, Xu X, MacRenaris Keith W., Zhang X-Q, Mirkin Chad A., Meade Thomas J. Multimodal Gadolinium-Enriched DNA-Gold Nanoparticle Conjugates for Cellular Imaging. Angew. Chem., Int. Ed. 2009;48:9143–9147. doi: 10.1002/anie.200904666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Dhar S, Daniel WL, Giljohann DA, Mirkin CA, Lippard SJ. Polyvalent oligonucleotide gold nanoparticle conjugates as delivery vehicles for platinum(IV) warheads. J. Am. Chem. Soc. 2009;131:14652–14653. doi: 10.1021/ja9071282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Zhu ZJ, Ghosh PS, Miranda OR, Vachet RW, Rotello VM. Multiplexed Screening of Cellular Uptake of Gold Nanoparticles Using Laser Desorption/Ionization Mass Spectrometry. J. Am. Chem. Soc. 2008;130:14139–14143. doi: 10.1021/ja805392f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Verma A, Uzun O, Hu YH, Hu Y, Han HS, Watson N, Chen SL, Irvine DJ, Stellacci F. Surface-structure-regulated cell-membrane penetration by monolayer-protected nanoparticles. Nat. Mater. 2008;7:588–595. doi: 10.1038/nmat2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Dobrovolskaia MA, Patri AK, Zheng J, Clogston JD, Ayub N, Aggarwal P, Neun BW, Hall JB, McNeil SE. Interaction of colloidal gold nanoparticles with human blood: effects on particle size and analysis of plasma protein binding profiles. Nanomedicine: Nanotechnology. 2009;5:106–117. doi: 10.1016/j.nano.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Linse S, Cabaleiro-Lago C, Xue WF, Lynch I, Lindman S, Thulin E, Radford SE, Dawson KA. Nucleation of protein fibrillation by nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 2007;104:8691–8696. doi: 10.1073/pnas.0701250104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Chithrani BD, Ghazani AA, Chan WCW. Determining the Size and Shape Dependence of Gold Nanoparticle Uptake into Mammalian Cells. Nano Lett. 2006;6:662–668. doi: 10.1021/nl052396o. [DOI] [PubMed] [Google Scholar]
- (31).Dutta D, Sundaram SK, Teeguarden JG, Riley BJ, Fifield LS, Jacobs JM, Addleman SR, Kaysen GA, Moudgil BM, Weber TJ. Adsorbed proteins influence the biological activity and molecular targeting of nanomaterials. Toxicol. Sci. 2007;100:303–315. doi: 10.1093/toxsci/kfm217. [DOI] [PubMed] [Google Scholar]
- (32).Bajaj A, Samanta B, Yan HH, Jerry DJ, Rotello VM. Stability, toxicity and differential cellular uptake of protein passivated-Fe3O4 nanoparticles. J. Mater. Chem. 2009;19:6328–6331. [Google Scholar]
- (33).Frens G. Controlled nucleation for the regulation of the particle size in monodisperse gold solutions. Nature Phys. Sci. 1973;241:20–22. [Google Scholar]
- (34).Herrlein MK, Nelson JS, Letsinger RL. A Covalent Lock for Self-Assembled Oligonucleotide Conjugates. J. Am. Chem. Soc. 1995;117:10151–10152. [Google Scholar]
- (35).Storhoff JJ, Elghanian R, Mucic RC, Mirkin CA, Letsinger RL. One-Pot Colorimetric Differentiation of Polynucleotides with Single Base Imperfections Using Gold Nanoparticle Probes. J. Am. Chem. Soc. 1998;120:1959–1964. [Google Scholar]
- (36).Cedervall T, Lynch I, Foy M, Berggad T, Donnelly SC, Cagney G, Linse S, Dawson KA. Detailed identification of plasma proteins adsorbed on copolymer nanoparticles. Angew. Chem., Int. Ed. 2007;46:5754–5756. doi: 10.1002/anie.200700465. [DOI] [PubMed] [Google Scholar]
- (37).Greaves DR, Gordon S. Recent insights into the biology of macrophage scavenger receptors. J. Lipid Res. 2005;46:11–20. doi: 10.1194/jlr.R400011-JLR200. [DOI] [PubMed] [Google Scholar]
- (38).Bijsterbosch MK, Manoharan M, Rump ET, DeVrueh RLA, vanVeghel R, Tivel KL, Biessen EAL, Bennett CF, Cook PD, vanBerkel TJC. In vivo fate of phosphorothioate antisense oligodeoxynucleotides: Predominant uptake by scavenger receptors on endothelial liver cells. Nucleic Acids Res. 1997;25:3290–3296. doi: 10.1093/nar/25.16.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Saleh MC, van Rij RP, Hekele A, Gillis A, Foley E, O’Farrell PH, Andino R. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat. Cell Biol. 2006;8:793–802. doi: 10.1038/ncbl439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Ulvila J, Parikka M, Kleino A, Sormunen R, Ezekowitz RA, Kocks C, Ramet M. Double-stranded RNA is internalized by scavenger receptor-mediated endocytosis in Drosophila S2 cells. J. Biol. Chem. 2006;281:14370–14375. doi: 10.1074/jbc.M513868200. [DOI] [PubMed] [Google Scholar]
- (41).Arnott S, Chandras R, Marttila CM. Structures for Polyinosinic Acid and Polyguanylic Acid. Biochem. J. 1974;141:537–543. doi: 10.1042/bj1410537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Pearson AM, Rich A, Krieger M. Polynucleotide Binding to Macrophage Scavenger Receptors Depends on the Formation of Base-Quartet-Stabilized 4-Stranded Helices. J. Biol. Chem. 1993;268:3546–3554. [PubMed] [Google Scholar]
- (43).Cutler JI, Zheng D, Xu XY, Giljohann DA, Mirkin CA. Polyvalent Oligonucleotide Iron Oxide Nanoparticle “Click” Conjugates. Nano Lett. 2010;10:1477–1480. doi: 10.1021/nl100477m. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.