Abstract
The interferon-induced enzymes 2′-5′-oligoadenylate synthetase (OAS) and RNase L are key components of innate immunity involved in sensory and effector functions following viral infections. Upon binding target RNA, OAS is activated to produce 2′-5′-linked oligoadenylates (2-5A) that activate RNase L, which then cleaves single-stranded self and non-self RNA. Modified nucleosides that are present in cellular transcripts have been shown to suppress activation of several RNA sensors. Here, we demonstrate that in vitro transcribed, unmodified RNA activates OAS, induces RNase L-mediated ribosomal RNA (rRNA) cleavage and is rapidly cleaved by RNase L. In contrast, RNA containing modified nucleosides activates OAS less efficiently and induces limited rRNA cleavage. Nucleoside modifications also make RNA resistant to cleavage by RNase L. Examining translation in RNase L−/− cells and mice confirmed that RNase L activity reduces translation of unmodified mRNA, which is not observed with modified mRNA. Additionally, mRNA containing the nucleoside modification pseudouridine is translated longer and has an extended half-life. The observation that modified nucleosides in RNA reduce 2-5A pathway activation joins OAS and RNase L to the list of RNA sensors and effectors whose functions are limited when RNA is modified, confirming the role of nucleoside modifications in suppressing immune recognition of RNA.
INTRODUCTION
The antiviral 2-5A system is initiated when double-stranded (ds)RNA is bound by 2′-5′-oligoadenylate synthetases (OAS). There are four OAS genes in humans, OAS1, OAS2, OAS3 and OASL, encoding 8–10 isoforms due to alternative splicing. Activated OAS (other than enzymatically inactive OASL) uses ATP as substrate to produce unique, short 2′-5′-linked oligomers called 2-5A [px5′A(2′p5′A)n; x = 1−3; n ≥ 2] that activate the latent endoribonuclease RNase L. Binding of 2-5A to the N-terminal region of RNase L monomers causes RNase L dimerization and activates the C-terminal nuclease domain. Activated RNase L cleaves single-stranded (ss)RNA preferentially after UU and UA dinucleotide motifs. [For a recent review of the 2-5A system, see ref. (1).]
Although nucleoside modifications are common in RNA, how this influences 2-5A system activity is unknown. RNA contains more than 100 different modified nucleosides. Nucleoside modifications are produced naturally during RNA maturation and are introduced post-transcriptionally in a site-specific manner. Pseudouridine (Ψ) is the most prevalent modified nucleoside found in RNA (2,3). One function of Ψ at specific locations in tRNA and ribosomal RNA (rRNA) is to stabilize crucial secondary structure (4). However, no physiologic role has been identified for the majority of RNA modification sites, and the effect of nucleoside modifications on most RNA-binding proteins has not been established.
In vitro transcribed RNA containing modified nucleosides has been shown to be less stimulatory to several host defense RNA sensors, including protein kinase R (PKR), toll-like receptor (TLR)3, TLR7, TLR8 and retinoic acid-inducible gene I (RIG-I) (5–8). We previously reported the production of in vitro transcribed mRNA in which every uridine is replaced by pseudouridine (Ψ-mRNA) and found that protein expression from Ψ-mRNA is higher than from unmodified in vitro transcribed mRNA (9), and this enhanced translation is due in part to reduced activation of PKR by Ψ-mRNA (6).
Here, we report that the presence of modified nucleosides in RNA has two effects on the 2-5A pathway. Certain unmodified in vitro transcribed mRNAs activate OAS, resulting in rRNA cleavage and reduced translation. Additionally, unmodified RNA is more rapidly cleaved by activated RNase L. In contrast, RNAs containing certain modified nucleosides fail to activate OAS and are resistant to cleavage by RNase L. Modified RNA is therefore identified as a distinguishing pattern for 2-5A system activity.
MATERIALS AND METHODS
Cells and reagents
Human embryonic kidney (HEK) 293T cells were obtained from the American Type Culture Collection and were cultured in DMEM supplemented with 2 mM l-glutamine (Life Technologies), 100 U/ml penicillin, 100 µg/ml streptomycin (Invitrogen) and 10% fetal calf serum (HyClone). Immortalized wild-type (WT) and RNase L−/− mouse embryonic fibroblasts (MEFs) were maintained in RPMI medium supplemented with 2 mM l-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin and 10% fetal calf serum. Oligo RNAs C11U2C7 and C11Ψ2C7 were custom synthesized (Dharmacon) and were 5′-end-labeled using [γ-32P]ATP (PerkinElmer) and T4 polynucleotide kinase (New England Biolabs). Polyinosinic:polycytidylic acid [poly(I:C)] was purchased from Sigma.
mRNA synthesis and purification
RNAs were transcribed as previously described (7), using linearized plasmids encoding firefly luciferase (pT7TS-fLuc) or Renilla luciferase (pT7TS-Ren) and T7 RNA polymerase (Megascript, Ambion). Capped mRNA was generated by performing transcription in the presence of cap analog 3′-O-Me-m7G(5′)ppp(5′)G (New England Biolabs). Metabolically labeled mRNA was generated by performing transcription in the presence of [α-32P]CTP (PerkinElmer). All mRNAs were transcribed to contain 30 nt-long 3′-poly(A) tails. RNA containing modified nucleosides was generated by substituting a modified nucleoside triphosphate [NTP derivatives of Ψ, m6A and s2U (TriLink)] for its cognate unmodified NTP in transcription reactions, as described previously (7,9). Following transcription, the template plasmids were digested with Turbo DNase and RNAs were precipitated with 2.5 M lithium chloride at −20°C for 4 h. RNAs were pelleted by centrifugation, washed with 75% ethanol and then reconstituted in nuclease-free water. The concentration of RNA was determined by measuring the optical density at 260 nm. All RNA samples were analyzed by agarose gel electrophoresis for quality assurance. RNA was high performance liquid chromatography (HPLC) purified on an RNASep cartridge (Transgenomic). Lack of double-stranded contaminants in the purified RNA samples was confirmed by RNA dot–blot performed with dsRNA-specific mAb J2 (English & Scientific Consulting Bt.).
In vitro OAS activation and measurement of functional 2-5A
Recombinant hexahistidine-tagged human OAS1 p42 was a gift of Rune Hartmann (University of Aarhus, Denmark) (10). Recombinant human RNase L and p3[2′p5′A]2A (trimer 2-5A) were prepared as described (11,12). Dual-labeled fluorescent probe 6-FAM-UUA UCA AAU UCU UAU UUG CCC CAU UUU UUU GGU UUA-BHQ-1 was custom synthesized by Integrated DNA Technologies. In vitro OAS1 activation was performed as described (13). Briefly, 20 µg/ml of OAS1 was activated with 2.0 µg/ml RNA for the indicated time in buffer consisting of 20 mM HEPES pH 7.5, 20 mM Mg(OAc)2, 20 mM KCl, 1 mM EDTA and 10 mM ATP. Reactions were stopped by heating to 95°C for 3 min. Rate of functionally active 2-5A produced was measured using a fluorescence resonance energy transfer (FRET) based assay as described previously (14,15). Synthesized p3[2′p5′A]2A (trimer 2-5A) (12) was purified using HPLC and used for generating standard curves (13).
rRNA cleavage
One day prior to transfection, WT or RNase L−/− MEF cells were seeded into 96-well plates at a density of 5.0 × 104 cells/well and treated with 1000 U/ml interferon-αA/D (Sigma). Poly(I:C) or mRNAs were complexed with lipofectin as described (7). Cells were exposed to 50 µl DMEM containing lipofectin-complexed RNA (2.5 µg) for 1 h, which was then replaced with complete medium and further cultured. At 3 h post-transfection, total RNA was recovered from cells using Trizol (Invitrogen). RNA was separated by agarose gel electrophoresis, stained with SybrGold reagent (Invitrogen) and detected using UV fluorescence and a GelDoc 2000 imager (Bio-Rad Laboratories).
In vitro RNA cleavage by RNase L
Recombinant human RNase L was prepared as described (14). For oligo RNAs, 12.5 nM RNase L was activated on ice with 100 nM trimer 2-5A for 30 min in RNase L cleavage buffer (25 mM Tris–HCl pH 7.4, 100 mM KCl, 10 mM MgCl2, 50 mM ATP and 7 mM β-mercaptoethanol). Then 100 nM 5′-end-labeled oligo RNA [32P]pC11U2C7 or [32P]pC11Ψ2C7 was added and reactions were incubated at 30°C. At the indicated times, reactions were stopped by the addition of urea-TBE loading buffer (Bio-Rad) and heating to 95°C for 3 min. Aliquots were separated by 15% polyacrylamide gel electrophoresis, gels were dried, and samples were imaged using a phosphor storage screen (Molecular Dynamics) and detected using a Typhoon PhosphorImager (GE Healthcare). Cleavage of mRNA was performed similarly, using 10 nM RNase L, 10 nM trimer 2-5A and 100 nM of metabolically 32P-labeled firefly luciferase mRNA. Reactions were stopped by heating to 95°C for 5 min. The mRNA was recovered by phenol:chloroform extraction and detected by northern blotting.
RNA stability in rabbit reticulocyte lysate
Equal mass (25 ng/µl) or equal molar (40 µM) mRNAs-encoding firefly and Renilla luciferases were incubated in 15 µl rabbit reticulocyte lysate (RRL) (Promega) at 30°C. At the indicated times, a 2 µl aliquot was removed and the RNA was recovered using Trizol for subsequent detection by northern blotting.
RNA stability in cell culture
HEK293T, WT MEF or RNase L−/− MEF cells were nucleofected with 5 µg mRNA using nucleofector program T-020 and nucleofector V kit (Lonza). After 15 min recovery in RPMI, cells were plated in complete media and incubated at 37°C. At the indicated time, RNA was recovered from cells using Trizol for subsequent detection by northern blotting.
Northern blotting
RNA was isolated from RRL or cells using Trizol. Samples were processed and analyzed on northern blots as previously described (16). Probes were derived from plasmids and were specific for the coding region of firefly luciferase or Renilla luciferase.
Half-life calculation
After performing northern blots, images were scanned from film and ImageJ (version 1.44 p) was used to measure the density of the band corresponding to the full-length mRNA. For each data point, the log10 was taken and the values were plotted as a function of time. The slope of best fit line (k) was used to calculate the mRNA half-life using the equation t1/2 = 0.693/k (17).
Detection of reporter proteins in RNA-transfected cells
Cells were seeded into 96-well plates at a density of 2–5 × 104 cells/well one day prior to transfection. Cells were exposed to 50 µl DMEM containing lipofectin-complexed RNA (0.25 µg) for 1 h, which was then replaced with complete medium and further cultured. Cells were lysed in 25 µl Firefly-, Renilla- or Dual-Luciferase-specific lysis reagents (Promega). Aliquots of 2 µl were assayed with the corresponding enzyme substrates (Promega) and a LUMAT LB 950 luminometer (Berthold) at a 10-s measuring time.
Translation of mRNA in mice
All mice were cared for according to institutional guidelines at the University of Pennsylvania under a protocol approved by the Institutional Animal Care and Use Committee. WT C57Bl/6 (NCI) and C57Bl/6 backcrossed RNase L−/− mice at 9–16 weeks of age received tail vein injections of 1 µg RNA complexed with lipofectin in 60 µl DMEM. At the indicated time, mice were sacrificed and spleens were isolated. Each spleen was bisected and spleen fragments were homogenized in 200 µl cell culture lysis reagent (Promega). Luciferase activity was detected in a 40 µl aliquot of lysed spleen using 200 µl luciferase assay substrate (Promega) and a LUMAT LB 950 luminometer at a 10-s measuring time.
Immunoprecipitation
HEK293T cells were seeded into 96-well plates at a density of 5.0 × 104 cells/well 1 day prior to transfection. Cells were exposed to 50 µl DMEM containing lipofectin-complexed RNA (0.25 µg) for 1 h, which was then replaced with complete medium and further cultured. Cells were incubated in methionine/cysteine-free medium (Invitrogen) for 1 h, then pulsed with complete medium supplemented with 35S-methionine/cysteine (140 mCi/ml) (PerkinElmer) for 3 h prior to lysis in 50 µl RIPA buffer supplemented with protease inhibitor cocktail (Sigma). Renilla luciferase was immunoprecipitated from lysates using an anti-Renilla luciferase antibody (PM047, Medical & Biological Laboratories) and protein G-coated Dynabeads (Invitrogen) and separated by 15% polyacrylamide gel electrophoresis. Gels containing the labeled samples were treated with 1 M sodium salicylate, dried and a fluorogram was generated by exposure to BioMax MS film (Kodak).
Statistical analysis
All data are reported as mean ± standard error of the mean (SEM). Statistical differences between treatment groups were calculated by the Student's t-test using Microsoft Excel. For all statistical testing, a P < 0.05 was considered significant.
RESULTS
RNA containing nucleoside modifications activates OAS less than unmodified RNA
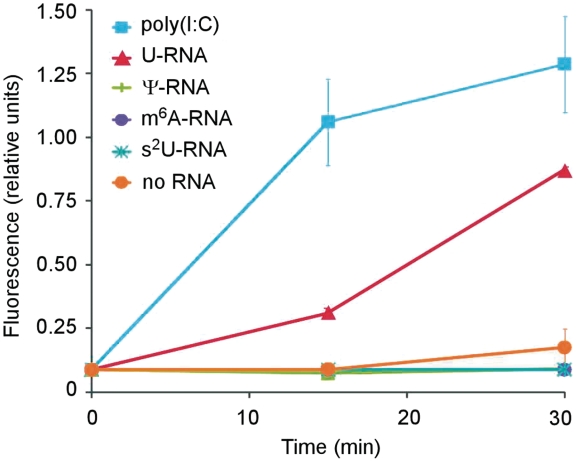
We first compared the activation of purified human OAS1 by unmodified RNA or RNA with identical sequence containing the modified nucleosides Ψ, N6-methyladenosine (m6A) or 2-thiouridine (s2U). The amount of functionally active 2-5A (trimer or higher) produced was quantified using a FRET-based RNase L activation assay (12,13). The unmodified RNA efficiently activated OAS1. In contrast, RNA containing Ψ, m6A or s2U were poor activators of OAS and did not induce production of functionally active 2-5A (Figure 1). Because Ψ is the most prevalent modified nucleoside (2,3) and has also been shown to reduce the RNA activation of other RNA sensors (5–8), subsequent experiments focused on the comparison of unmodified RNA to Ψ-containing RNA with identical sequence.
Figure 1.
OAS activation by RNA containing modified nucleosides. Purified human OAS1 p42 was activated with RNAs containing either unmodified (U) or the indicated nucleoside modifications and the functionally active 2-5A produced was quantified using a FRET-based assay as described in ‘Materials and Methods’ section. Data shown are mean of three replicates.
Pseudouridine-containing RNA induces less rRNA cleavage than unmodified RNA
OAS activation by unmodified RNA leads to activation of RNase L, which mediates the effector function of the 2-5A system by cleaving ssRNA. RNase L-mediated cleavage at exposed loops of rRNAs in intact ribosomes results in well-defined cleavage patterns in rRNA (18). Therefore, the integrity of rRNA following RNA transfection was examined. Lipofectin-complexed RNA was transfected to WT and RNase L−/− MEF cells, and total RNA was subsequently recovered and examined by agarose gel electrophoresis and UV imaging. Cells mock transfected with no RNA were included as a negative control. In WT cells, delivery of unmodified in vitro transcribed RNA induced cleavage of rRNA, but significantly less rRNA was cleaved when the transfected RNA contained Ψ. Transfection of the same set of RNAs into RNase L−/− MEF cells did not generate the specific rRNA degradation profile (Figure 2).
Figure 2.
Induction of rRNA cleavage by in vitro transcribed RNA. Unmodified (U) or Ψ-containing RNA encoding firefly luciferase was complexed to lipofectin and delivered to WT or RNase L−/− MEF cells, as was a no RNA (−) control. At 3 h following transfection, total RNA was recovered from cells. RNA aliquots were separated in an agarose gel and visualized by UV fluorescence. Arrowheads indicate RNase L-specific rRNA cleavage products. Bar graph above bands shows densitometric measurement ± SD of the ratio of cleavage products to uncleaved 18S rRNA, normalized to cells not treated with RNA. Representative data from one of three independent experiments is shown.
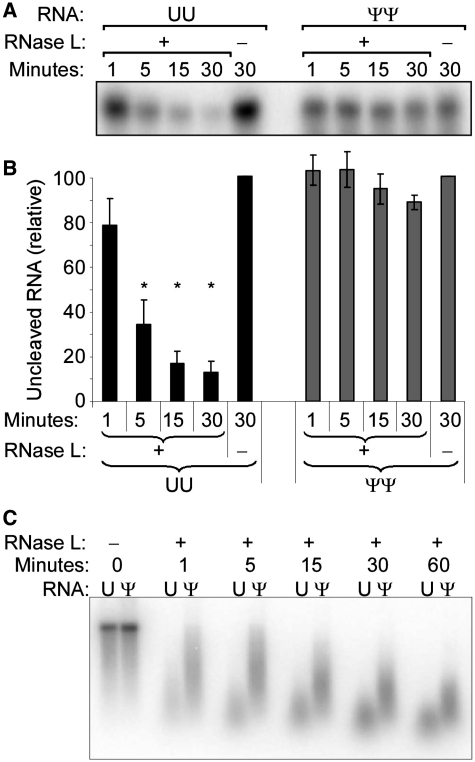
RNase L cleaves uridine-containing RNA more readily than Ψ-containing RNA
Activated RNase L cleaves preferentially after UpNp in ssRNA. Therefore, to compare the ability of RNase L to cleave Ψ-containing RNA, purified recombinant human RNase L was activated with trimer 2-5A and mixed with 5′-[32P] end-labeled oligo RNA containing a single RNase L cleavage site (C11U2C7 or C11Ψ2C7). The oligo RNA containing unmodified uridine was rapidly cleaved, while there was no significant cleavage of the oligo RNA containing Ψ (Figure 3A and B). Full-length firefly luciferase mRNA metabolically labeled with 32P was then analyzed for cleavage by RNase L. Both unmodified and Ψ-RNA could be cleaved by RNase L. However, consistent with the results obtained with oligo RNAs, Ψ-containing RNA was cleaved less efficiently by RNase L than unmodified RNA (Figure 3C).
Figure 3.
Cleavage of Ψ-containing RNA by RNase L. Purified RNase L was activated on ice by trimer 2-5A prior to mixing with RNA substrates. (A) Cleavage of oligo RNAs [32P]pC11U2C7 (UU) or [32P]pC11Ψ2C7 (ΨΨ) by RNase L. Reactions were stopped at the indicated time by addition of loading buffer, and reactions were separated by PAGE and visualized by phosphor-storage radiography. Representative data from one of three independent experiments is shown. (B) Quantification of band intensities. Values were normalized to the values obtained in the 30 min reaction not containing RNase L. Data represents average ± SEM of n = 3 experiments. Asterisks indicate P < 0.05. (C) Cleavage of metabolically [32P]-labeled unmodified (U) or Ψ-containing firefly luciferase mRNA by RNase L. Reactions were stopped at indicated times. Aliquots of isolated RNA from each reaction were separated by PAGE and visualized by phosphor-storage radiography. Representative data from one of three independent experiments is shown.
Nucleoside-modified RNA has an increased half-life
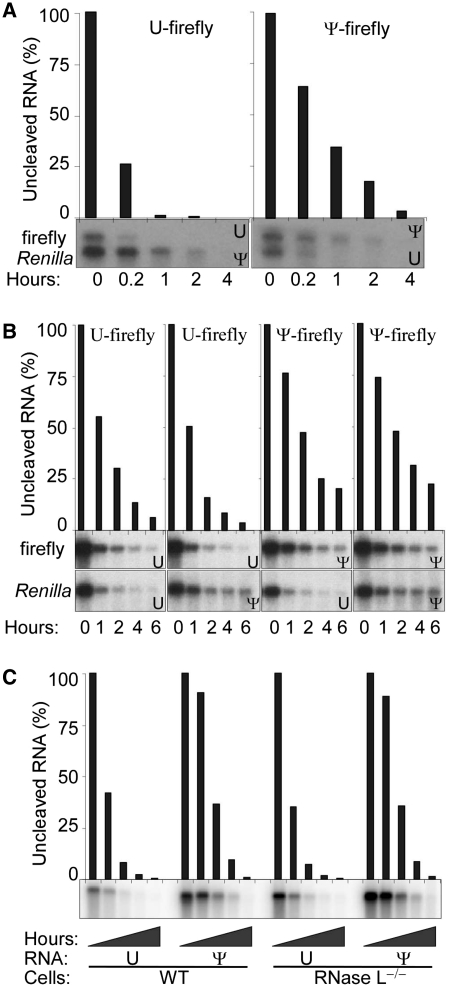
We next examined the stability of unmodified and Ψ-containing RNA by northern blot analysis. Both unmodified and Ψ-RNA were equally stable at room temperature through experimental time courses and indefinitely in storage at −20°C. Unmodified and Ψ-RNAs were added to RRL or transfected to HEK293T cells. When transfecting cells using cationic lipids, a portion of RNA complexed with transfection reagents persist as an extracellular, nuclease-protected fraction. Therefore, for these experiments, we used nucleofection to deliver naked RNA and confirmed the rapid degradation of extracellular RNA by serum nucleases in the culture media. Total RNA was subsequently re-isolated and aliquots were examined by northern blot to compare degradation rates of the reporter RNAs. Two reporter RNAs, firefly and Renilla luciferase, were studied simultaneously to ensure that stability differences were not a result of differences in delivery conditions. Ψ-modified RNAs had longer half-lives than unmodified RNAs in RRL (Figure 4A). Similarly, in HEK293T cells the half-life of Ψ-modified firefly luciferase RNA increased ~2-fold to 6.1 h compared to 3.2 h for unmodified RNA (Figure 4B).
Figure 4.
Stability of Ψ-containing RNA. Unmodified (U) or Ψ-containing RNAs encoding firefly or Renilla luciferases were mixed 1:1 and either added to RRL (A) or nucleofected to HEK293T cells (B). At the indicated time points, RNA was recovered and detected by northern blotting. Radiolabeled DNA probes corresponding to the coding sequence of firefly and Renilla luciferases were mixed prior to northern hybridization (A) or used separately to probe duplicate northern blots from aliquots of the recovered RNA (B). Bar graphs above the images show densitometric measurement of the uncleaved firefly luciferase mRNA. Data shown is representative of at least five independent experiments. (C) Unmodified (U) or Ψ-containing in vitro transcribed RNA encoding firefly luciferase was delivered to WT or RNase L−/− MEF cells by nucleofection. Cells were lysed at 0.2, 1, 3, 6 or 24 h following transfection, total RNA was recovered, and luciferase RNA was assessed by northern blotting. Radiolabeled DNA corresponding to the firefly luciferase coding sequence was used as probe. Bar graph above bands shows densitometric measurement of the uncleaved RNA. Representative data is shown from one of three independent experiments.
Subsequently, the influence of RNase L on the stability of unmodified and Ψ-containing RNA was also compared using RNase L−/− MEF cells. As in HEK293T cells, the RNA was delivered by nucleofection. Total RNA was recovered from cell culture and firefly and Renilla luciferase RNA were assessed by northern blot. In both WT and RNase L−/− MEF cells, Ψ-modified RNA half-life was increased by 50% to 3.8 h compared to 2.5 h for unmodified RNA (Figure 4C).
Translational advantage of Ψ-RNA is reduced in the absence of RNase L
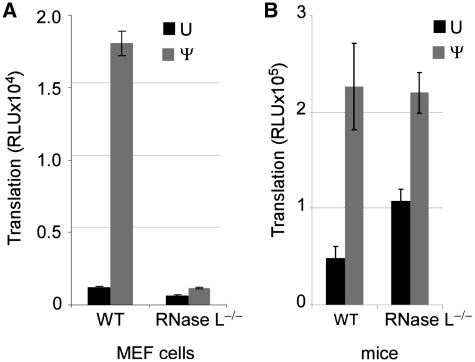
Considering that Ψ-modification of RNA reduced activation of OAS1 and RNA induced rRNA degradation, and that RNA containing Ψ was cleaved by RNase L less efficiently, we asked how the absence of RNase L influences translation of unmodified and Ψ-containing mRNA. Thus, mRNAs encoding luciferase were transfected into WT and RNase L−/− MEF cell lines and translation was assessed by measuring luciferase activity. In WT cells, more protein was translated from the Ψ-containing mRNA than from the unmodified mRNA. In the RNase L−/− cell line, there was lower level translation of both mRNAs and the translational advantage of Ψ-mRNA over unmodified mRNA was dramatically reduced (Figure 5A).
Figure 5.
Translation of unmodified and Ψ-containing mRNA in WT and RNase L−/− cells and mice. Unmodified (U) or Ψ-containing in vitro transcribed mRNA encoding firefly luciferase was complexed to lipofectin and delivered to WT and RNase L−/− MEF cells or mice. Luciferase activity was measured in aliquots of cell or spleen lysate. (A) MEF cells lysed 5 h following transfection. Values presented are luciferase relative light units (RLU) in 2 µl of the total 20 µl cell lysate. Error bars indicate SEM of quadruplicate wells from one representative of at least six independent experiments. (B) Lipofectin-complexed mRNA was delivered by tail vein injection into mice. Mice were sacrificed at 4 h post-transfection and their spleens were homogenized in lysis buffer. Values presented are luciferase RLU in 1/5 of the total 200 µl spleen lysate. Error bars represent SEM of n = 3 mice.
A similar pattern of translation occurred in the spleens of mice following injection of mRNA. Either WT C57Bl/6 or RNase L−/− mice were given lipofectin-complexed luciferase mRNA by tail vein injection. Luciferase activity was assessed in spleen lysate 4 h later. In WT mice, Ψ-containing mRNA was translated at higher levels than unmodified mRNA. In RNase L−/− mice, translation of Ψ-containing mRNA reached the same level as observed in WT mice, but translation of unmodified mRNA was increased relative to WT (P < 0.05), (Figure 5B).
Translation of pseudouridine-containing mRNA continues for a longer time than unmodified mRNA
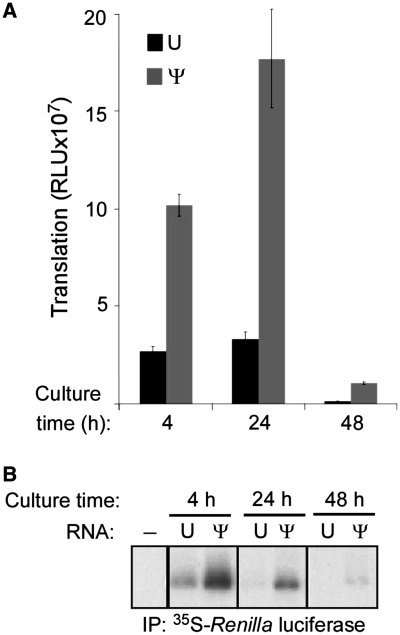
Having seen that the presence of modified nucleosides in mRNA increases its half-life and translation efficiency, we compared the translation over time, to determine how modified nucleosides influence the duration of translation. RNA was complexed to lipofectin and delivered to cells that were subsequently pulsed with 35S-methionine/cysteine for 3 h at 1, 21 and 45 h post-transfection. Translation of the mRNA was assessed by immunoprecipitating the encoded Renilla luciferase protein and measuring 35S incorporation. There was a higher level of total translation of Ψ-containing mRNA at each time point (Figure 6A), and ongoing translation of Ψ-containing mRNA continued after at least 48 h when detectable translation of unmodified mRNA ceased (Figure 6B).
Figure 6.
Translation of Ψ-containing mRNA in cell culture. Unmodified (U) and Ψ-containing mRNA were complexed to lipofectin and delivered to HEK293T cells. (A) Renilla luciferase activity was assessed in aliquots of cell lysate. Data displayed is mean ± SEM from four replicates, each performed in duplicate. (B) Cells were pulsed for the last 3 h with 35S-methionine/cysteine prior to lysis at the indicated time points. Renilla luciferase protein was immunoprecipitated from cell lysates, separated by PAGE and then visualized by fluorography. Data shown is one of four replicates and is representative of three independent experiments.
DISCUSSION
We investigated how the enzymes of the 2-5A system interact with RNA and the role that modified nucleosides play in altering activation and effector function. Our data show that in vitro transcribed unmodified RNA activates OAS1, but this activation is reduced when the RNA contains modified nucleosides. OAS activation by unmodified RNA leads to RNase L-mediated rRNA cleavage, which is reduced by Ψ-RNA. Furthermore, RNase L cleaves unmodified RNA more efficiently than Ψ-containing RNA. Experiments using RNase L−/− MEF cells and RNase L−/− mice demonstrate that translation of unmodified mRNA is decreased in the presence of the intact 2-5A system, but that the translation level of Ψ-mRNA is largely independent of the 2-5A system. In addition, the presence of Ψ increases the half-life of in vitro transcribed RNA in cells and lysates. Finally, we demonstrate that Ψ-containing mRNA is translated for a longer duration than unmodified mRNA.
RNA sensing in the 2-5A pathway is performed by the OAS family of proteins. Activation of OAS was originally characterized as requiring >8 bp of uninterrupted helix in >30-bp long dsRNA (19), but subsequently other structures with significant single-strandedness have been proven to be potent OAS activators, including aptamers (20), viral RNAs (21–24) and some cellular RNAs (13,25). However, dsRNA generated from homopolymers containing 2′-O-methylated nucleosides (26,27) or 5-methyluridines (28) did not activate OAS. Here, we report that unmodified in vitro transcribed RNA activated OAS1 to generate 2-5A, but this was substantially reduced when RNA contained Ψ, m6A or s2U. Recently, the consensus sequence nnWWnnnnnnnnnWGn (W = U or A) was demonstrated to be important for OAS1 activation by dsRNA, and this interaction was dependent on the minor groove and free OH groups on the critical base pairs (29). The requirement that three out of the four critical base pairs in this sequence must be U:A highlights the importance of uridine for OAS1 activation. However, pseudouridine forms hydrogen bounds with adenosine in the same manner that uridine does (Figure 7), and the imino group of Ψ is oriented toward the major groove (30), so how Ψ disrupts OAS1 activation remains unclear. The presence of Ψ stabilizes secondary structure and adds rigidity to both ss and dsRNA [reviewed in (4)]. In this capacity, Ψ could affect OAS activation by altering the equilibrium structure of the RNA, rather than directly affecting OAS binding.
Figure 7.
Structures and base pairing of uridine and pseudouridine. In pseudouridine, uracil is linked to ribose via C5 instead of the N1 linkage found in uridine (C5 and N1 are indicated in bold type). Hydrogen bonds between adenosine and uridine or pseudouridine are indicated by dotted lines. Additional hydrogen bonding potential of pseudouridine is indicated by dashed arrow.
Activation of OAS leads to production of 2-5A, which binds to the latent endoribonuclease RNase L, the effector enzyme of the 2-5A pathway. Activated RNase L cleaves various ssRNA including specific sites of rRNAs accessible in the intact ribosome, resulting in RNase L-specific cleavage products visible by gel electrophoresis (18). In WT MEF, unmodified RNA induced rRNA cleavage, which was reduced if RNA contained Ψ. However, none of the RNAs caused rRNA cleavage in RNase L−/− cells, confirming that the 2-5A system is required for RNA-induced rRNA cleavage. High levels of 2-5A result in global rRNA cleavage by RNase L (31), and when sustained ultimately lead to apoptosis (32,33). In comparison, the level of rRNA cleavage induced here by transfection of in vitro transcribed RNA is relatively small, and may not be expected to induce high levels of apoptosis. On the other hand, this level of rRNA cleavage is sufficient to have a profound impact on translation of the reporter mRNA. We propose that unmodified RNA induces local OAS and RNase L activation, as demonstrated with viral RNAs and ssRNA covalently linked to dsRNA (34,35). Accordingly, locally activated RNase L cleavage likely reduces translation of unmodified mRNA through local cleavage of rRNA without inducing global rRNA cleavage and apoptosis.
The presence of Ψ has been shown to enhance the stability of RNA secondary structures, but has not previously been demonstrated to cause resistance to nucleases. RNA containing Ψ was cleaved efficiently by RNase A, RNase H (36), RNase T1, RNase T2, nuclease P1 and snake venom phosphodiesterase, although there is some indication that pancreatic diesterase and snake venom phosphodiesterase may cleave Ψ-RNA with reduced efficiency (37). A previous report based on cleavage of a C11N2C7 oligo RNA showed that RNA containing 2′-deoxy-2′-α-fluorouridine was bound by RNase L but cleaved slowly, whereas RNA containing 2′-O-methyluridine was not bound by RNase L (38). Here, we used a similar approach and demonstrated that purified RNase L readily cleaved the oligo ssRNA C11U2C7 but not when the cleavage site contained Ψ. We also extended those findings to the examination of long in vitro transcribed RNA and showed that unmodified RNA was cleaved by purified RNase L, but cleavage of Ψ-RNA proceeded more slowly. The cleavage of Ψ-RNA despite inactivity toward C11Ψ2C7 is not surprising considering the substrate specificity of RNase L. RNase L cleaves preferentially after UpNp, with highest activity following UU, UA and AU, but it is also capable of cleaving RNA following dinucleotide motives that avoid U (e.g. AA, AC and CA) (39–41).
We also examined the effect of Ψ-modification on the stability of in vitro transcribed RNA. In RRL and in cell culture, Ψ-RNA was degraded more slowly than unmodified RNA. Previous experiments also suggested that Ψ-RNA is retained longer following injection in mice (9). Despite the rapid cleavage of unmodified RNA by RNase L in vitro, the half-life of unmodified RNA did not increase to the level of Ψ-RNA in RNase L−/− cells. This suggests that in addition to RNase L, other intracellular nucleases also cleave unmodified RNA more efficiently than Ψ-containing RNA.
As seen in previous reports (6,9), in WT cells, there was significantly higher translation of Ψ-mRNA than unmodified mRNA. In contrast, in RNase L−/− MEF cells the translational advantage of Ψ-mRNA over the unmodified mRNA was limited. Similarly, the translational advantage of Ψ-mRNA was reduced in RNase L−/− mice relative to WT mice. Notably, however, the absolute translation level of Ψ-mRNA remained equal in WT and RNase L−/− mice, while the translation of unmodified mRNA increased in RNase L−/− mice. This indicates that neither the presence of RNase L nor Ψ-mRNA alone significantly affects translation of Ψ-mRNA, but rather that unmodified RNA causes translational inhibition through RNase L activation. Moreover, these results are consistent with the in vitro activation of OAS1 by unmodified RNA that we observed. Furthermore, Ψ-mRNA continued to be actively translated for a longer duration than unmodified mRNA. In RNase L−/− cells and mice, the translational advantage of Ψ-mRNA is reduced, despite the observation that the absence of RNase L in cells does not significantly alter the stability of either U-RNA or Ψ-mRNA. Therefore, we propose that in cells the mechanism by which the 2-5A system enhances translation of Ψ-mRNA is not primarily through reduced degradation of the Ψ-mRNA itself, but instead through decreased rRNA cleavage resulting from diminished OAS activation. Thus, OAS activation by unmodified mRNA results in RNase L activation, which reduces translation due to rRNA cleavage rather than through cleavage of the activating transfected mRNA.
In addition to viral RNA, select cellular mRNAs from prostate cancer cells have been shown to activate OAS (13). Additionally, cleavage of cellular and viral RNAs by RNase L produces short RNAs, which can activate the cytoplasmic RNA sensor RIG-I, leading to interferon production (42–44). The presence and effects of modified nucleosides in these RNase L-generated short RNAs has not been investigated. Because RNA containing modified nucleosides activates OAS less and is less efficiently cleaved by RNase L, if viral infection or cancer development were to alter the level of nucleoside modification, it could lead to modified RIG-I activation and ultimately change immune responsiveness and disease progression. Consistent with this possibility, some viral mRNAs are hyper-methylated compared to mammalian mRNA (45,46). Viral-encoded 2′-O-methyltransferases extensively modify the 5′-ends of their capped mRNA. This modification is critical for the virus to avoid interferon induction and evade detection by the immune system. (47,48). Additionally, a recent study of non-small-cell lung cancer identified the upregulation of small nucleolar RNAs (snoRNAs), which function in directing pseudouridylation and 2′-O-methylation of RNA (49).
Nucleases play a central role in host defense through destruction of pathogenic nucleic acids. The 2-5A system functions to detect and degrade danger-associated intracellular RNAs. Activation of RNase L also leads to reduced translation due to rRNA cleavage and when sustained, results in apoptosis, further limiting replication of the pathogens. Here, we identify that some unmodified RNAs serve as a molecular pattern recognized by OAS and RNase L. The 2-5A system activity is decreased when RNA contains nucleoside modifications, which reduce both OAS activation and cleavage by RNase L. Other RNA sensors, including PKR, TLR3, TLR7, TLR8 and RIG-I (5–8) also exhibit reduced activation by RNA containing modified nucleosides. Therefore, this work supports the proposal made by us and others that RNA sensors recognize certain RNAs that contain unmodified nucleosides as a danger-associated molecular pattern, as part of the extensive system of innate host defenses against pathogenic RNA, but that nucleoside modification suppresses RNA immunogenicity.
FUNDING
National Institutes of Health (R01AI50484 and R21DE019059 to D.W.; T32GM07229, T32DK07748 and T32RR007063 to B.R.A.; R01NS029331 and R42HL87688 to K.K.; R01CA044059 to R.H.S). Funding for open access charge: National Institutes of Health (grant R42HL87688 to K.K.).
Conflict of interest statement. K.K. and D.W. have formed a small biotech company RNARx that receives funding from the National Institutes of Health to explore the use of nucleoside-modified mRNA for gene therapy.
REFERENCES
- 1.Chakrabarti A, Jha BK, Silverman RH. New insights into the role of RNase L in innate immunity. J. Interferon. Cytokine. Res. 2011;31:49–57. doi: 10.1089/jir.2010.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lane BG. Historical perspective on RNA nucleoside modifications. In: Grosjean H, Benne R, editors. Modification and Editing of RNA. Washington DC: ASM Press; 1998. pp. 1–20. [Google Scholar]
- 3.Rozenski J, Crain P, McCloskey J. The RNA Modification Database: 1999 update. Nucleic Acids Res. 1999;27:196–197. doi: 10.1093/nar/27.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49:341–351. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- 5.Nallagatla SR, Bevilacqua PC. Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. RNA. 2008;14:1201–1213. doi: 10.1261/rna.1007408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson BR, Muramatsu H, Nallagatla SR, Bevilacqua PC, Sansing LH, Weissman D, Karikó K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38:5884–5892. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karikó K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 8.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann K-K, Schlee M, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 9.Karikó K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartmann R, Justesen J, Sarkar SN, Sen GC, Yee VC. Crystal structure of the 2′-specific and double-stranded RNA-activated interferon-induced antiviral protein 2′-5′-oligoadenylate synthetase. Mol. Cell. 2003;12:1173–1185. doi: 10.1016/s1097-2765(03)00433-7. [DOI] [PubMed] [Google Scholar]
- 11.Dong B, Xu L, Zhou A, Hassel BA, Lee X, Torrence PF, Silverman RH. Intrinsic molecular activities of the interferon-induced 2-5A-dependent RNase. J. Biol. Chem. 1994;269:14153–14158. [PubMed] [Google Scholar]
- 12.Thakur CS, Jha BK, Dong B, Das Gupta J, Silverman KM, Mao H, Sawai H, Nakamura AO, Banerjee AK, Gudkov A, et al. Small-molecule activators of RNase L with broad-spectrum antiviral activity. Proc. Natl Acad. Sci. USA. 2007;104:9585–9590. doi: 10.1073/pnas.0700590104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molinaro RJ, Jha BK, Malathi K, Varambally S, Chinnaiyan AM, Silverman RH. Selection and cloning of poly(rC)-binding protein 2 and Raf kinase inhibitor protein RNA activators of 2′,5′-oligoadenylate synthetase from prostate cancer cells. Nucleic Acids Res. 2006;34:6684–6695. doi: 10.1093/nar/gkl968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thakur CS, Xu Z, Wang Z, Novince Z, Silverman RH. A convenient and sensitive fluorescence resonance energy transfer assay for RNase L and 2′,5′ oligoadenylates. Methods Mol. Med. 2005;116:103–113. doi: 10.1385/1-59259-939-7:103. [DOI] [PubMed] [Google Scholar]
- 15.Elbahesh H, Jha BK, Silverman RH, Scherbik SV, Brinton MA. The Flv(r)-encoded murine oligoadenylate synthetase 1b (Oas1b) suppresses 2-5A synthesis in intact cells. Virology. 2011;409:262–270. doi: 10.1016/j.virol.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karikó K, Ni H, Capodici J, Lamphier M, Weissman D. mRNA is an endogenous ligand for Toll-like receptor 3. J. Biol. Chem. 2004;279:12542–12550. doi: 10.1074/jbc.M310175200. [DOI] [PubMed] [Google Scholar]
- 17.Gallie DR, Tanguay RL, Leathers V. The tobacco etch viral 5′ leader and poly(A) tail are functionally synergistic regulators of translation. Gene. 1995;165:233–238. doi: 10.1016/0378-1119(95)00521-7. [DOI] [PubMed] [Google Scholar]
- 18.Wreschner DH, James TC, Silverman RH, Kerr IM. Ribosomal RNA cleavage, nuclease activation and 2-5A(ppp(A2′p)nA) in interferon-treated cells. Nucleic Acids Res. 1981;9:1571–1581. doi: 10.1093/nar/9.7.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minks MA, West DK, Benvin S, Baglioni C. Structural requirements of double-stranded RNA for the activation of 2′,5′-oligo(A) polymerase and protein kinase of interferon-treated HeLa cells. J. Biol. Chem. 1979;254:10180–10183. [PubMed] [Google Scholar]
- 20.Hartmann R, Norby PL, Martensen PM, Jorgensen P, James MC, Jacobsen C, Moestrup SK, Clemens MJ, Justesen J. Activation of 2′-5′ oligoadenylate synthetase by single-stranded and double-stranded RNA aptamers. J. Biol. Chem. 1998;273:3236–3246. doi: 10.1074/jbc.273.6.3236. [DOI] [PubMed] [Google Scholar]
- 21.Torrence PF, Johnston MI, Epstein DA, Jacobsen H, Friedman RM. Activation of human and mouse 2-5A synthetases and mouse protein P1 kinase by nucleic acids. Structure-activity relationships and correlations with inhibition of protein synthesis and interferon induction. FEBS Lett. 1981;130:291–296. doi: 10.1016/0014-5793(81)81142-8. [DOI] [PubMed] [Google Scholar]
- 22.Desai SY, Patel RC, Sen GC, Malhotra P, Ghadge GD, Thimmapaya B. Activation of interferon-inducible 2′-5′ oligoadenylate synthetase by adenoviral VAI RNA. J. Biol. Chem. 1995;270:3454–3461. doi: 10.1074/jbc.270.7.3454. [DOI] [PubMed] [Google Scholar]
- 23.Mordechai E, Kon N, Henderson EE, Suhadolnik RJ. Activation of the interferon-inducible enzymes, 2′,5′-oligoadenylate synthetase and PKR by human T-cell leukemia virus type I Rex-response element. Virology. 1995;206:913–922. doi: 10.1006/viro.1995.1014. [DOI] [PubMed] [Google Scholar]
- 24.Sharp TV, Raine DA, Gewert DR, Joshi B, Jagus R, Clemens MJ. Activation of the interferon-inducible (2′-5′) oligoadenylate synthetase by the Epstein-Barr virus RNA, EBER-1. Virology. 1999;257:303–313. doi: 10.1006/viro.1999.9689. [DOI] [PubMed] [Google Scholar]
- 25.Nilsen TW, Maroney PA, Robertson HD, Baglioni C. Heterogeneous nuclear RNA promotes synthesis of (2′,5′)oligoadenylate and is cleaved by the (2′,5′)oligoadenylate-activated endoribonuclease. Mol. Cell Biol. 1982;2:154–160. doi: 10.1128/mcb.2.2.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minks M, West D, Benvin S, Greene J, Ts'o P, Baglioni C. Activation of 2′,5′-oligo(A) polymerase and protein kinase of interferon-treated HeLa cells by 2′-O-methylated poly (inosinic acid):poly(cytidylic acid) J. Biol. Chem. 1980;255:6403–6407. [PubMed] [Google Scholar]
- 27.Sarkar SN, Bandyopadhyay S, Ghosh A, Sen GC. Enzymatic characteristics of recombinant medium isozyme of 2′-5′ oligoadenylate synthetase. J. Biol. Chem. 1999;274:1848–1855. doi: 10.1074/jbc.274.3.1848. [DOI] [PubMed] [Google Scholar]
- 28.Baglioni C, Minks MA, De Clercq E. Structural requirements of polynucleotides for the activation of (2′ - 5′) An polymerase and protein kinase. Nucleic Acids Res. 1981;9:4939–4950. doi: 10.1093/nar/9.19.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kodym R, Kodym E, Story MD. 2′-5′-Oligoadenylate synthetase is activated by a specific RNA sequence motif. Biochem. Biophys. Res. Commun. 2009;388:317–322. doi: 10.1016/j.bbrc.2009.07.167. [DOI] [PubMed] [Google Scholar]
- 30.Hall KB, McLaughlin LW. Properties of pseudouridine N1 imino protons located in the major groove of an A-form RNA duplex. Nucleic Acids Res. 1992;20:1883–1889. doi: 10.1093/nar/20.8.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li XL, Blackford JA, Hassel BA. RNase L mediates the antiviral effect of interferon through a selective reduction in viral RNA during encephalomyocarditis virus infection. J. Virol. 1998;72:2752–2759. doi: 10.1128/jvi.72.4.2752-2759.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castelli JC, Hassel BA, Wood KA, Li XL, Amemiya K, Dalakas MC, Torrence PF, Youle RJ. A study of the interferon antiviral mechanism: apoptosis activation by the 2-5A system. J. Exp. Med. 1997;186:967–972. doi: 10.1084/jem.186.6.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou A, Paranjape J, Brown TL, Nie H, Naik S, Dong B, Chang A, Trapp B, Fairchild R, Colmenares C, et al. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J. 1997;16:6355–6363. doi: 10.1093/emboj/16.21.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nilsen TW, Baglioni C. Mechanism for discrimination between viral and host mRNA in interferon-treated cells. Proc. Natl Acad. Sci. USA. 1979;76:2600–2604. doi: 10.1073/pnas.76.6.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baglioni C, De Benedetti A, Williams GJ. Cleavage of nascent reovirus mRNA by localized activation of the 2′-5′-oligoadenylate-dependent endoribonuclease. J. Virol. 1984;52:865–871. doi: 10.1128/jvi.52.3.865-871.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao X, Yu YT. Detection and quantitation of RNA base modifications. RNA. 2004;10:996–1002. doi: 10.1261/rna.7110804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naylor NR, Ho WY, Gilham PT. Selective chemical modifications of uridine and pseudouridine in polynucleotides and their effect on the specificities of ribonuclease and phosphodiesterases. J. Am. Chem. Soc. 1965;87:4209–4210. doi: 10.1021/ja01096a050. [DOI] [PubMed] [Google Scholar]
- 38.Ueno Y, Yamada Y, Nakanishi M, Kitade Y. A specific substrate-inhibitor, a 2′-deoxy-2'-fluorouridine-containing oligoribonucleotide, against human RNase L. Bioorg. Med. Chem. 2003;11:5069–5073. doi: 10.1016/j.bmc.2003.08.025. [DOI] [PubMed] [Google Scholar]
- 39.Wreschner DH, McCauley JW, Skehel JJ, Kerr IM. Interferon action - sequence specificity of the ppp(A2′p)nA-dependent ribonuclease. Nature. 1981;289:414–417. doi: 10.1038/289414a0. [DOI] [PubMed] [Google Scholar]
- 40.Floyd-Smith G, Slattery E, Lengyel P. Interferon action: RNA cleavage pattern of a (2′-5′)oligoadenylate–dependent endonuclease. Science. 1981;212:1030–1032. doi: 10.1126/science.6165080. [DOI] [PubMed] [Google Scholar]
- 41.Carroll SS, Chen E, Viscount T, Geib J, Sardana MK, Gehman J, Kuo LC. Cleavage of oligoribonucleotides by the 2′,5′-oligoadenylate- dependent ribonuclease L. J. Biol. Chem. 1996;271:4988–4992. doi: 10.1074/jbc.271.9.4988. [DOI] [PubMed] [Google Scholar]
- 42.Malathi K, Paranjape JM, Bulanova E, Shim M, Guenther-Johnson JM, Faber PW, Eling TE, Williams BRG, Silverman RH. A transcriptional signaling pathway in the IFN system mediated by 2′-5′-oligoadenylate activation of RNase L. Proc. Natl Acad. Sci. USA. 2005;102:14533–14538. doi: 10.1073/pnas.0507551102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malathi K, Dong B, Gale M, Jr, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malathi K, Saito T, Crochet N, Barton DJ, Gale M, Jr, Silverman RH. RNase L releases a small RNA from HCV RNA that refolds into a potent PAMP. RNA. 2010;16:2108–2119. doi: 10.1261/rna.2244210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bokar JA. The biosynthesis and functional roles of methylated nucleosides in eukaryotic mRNA. In: Grosjean H, editor. Fine-Tuning of RNA Functions by Modification and Editing. Vol. 12. Berlin Heidelbergpp: Springer; 2005. pp. 141–177. [Google Scholar]
- 46.Narayan P, Rottman FM. Methylation of mRNA. Adv. Enzymol. Relat. Areas Mol. Biol. 1992;65:255–285. doi: 10.1002/9780470123119.ch7. [DOI] [PubMed] [Google Scholar]
- 47.Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong H, et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liao J, Yu L, Mei Y, Guarnera M, Shen J, Li R, Liu Z, Jiang F. Small nucleolar RNA signatures as biomarkers for non-small-cell lung cancer. Mol. Cancer. 2010;9:198. doi: 10.1186/1476-4598-9-198. [DOI] [PMC free article] [PubMed] [Google Scholar]