Abstract
Sequence-specific nucleases represent valuable tools for precision genome engineering. Traditionally, zinc-finger nucleases (ZFNs) and meganucleases have been used to specifically edit complex genomes. Recently, the DNA binding domains of transcription activator-like effectors (TALEs) from the bacterial pathogen Xanthomonas have been harnessed to direct nuclease domains to desired genomic loci. In this study, we tested a panel of truncation variants based on the TALE protein AvrBs4 to identify TALE nucleases (TALENs) with high DNA cleavage activity. The most favorable parameters for efficient DNA cleavage were determined in vitro and in cellular reporter assays. TALENs were designed to disrupt an EGFP marker gene and the human loci CCR5 and IL2RG. Gene editing was achieved in up to 45% of transfected cells. A side-by-side comparison with ZFNs showed similar gene disruption activities by TALENs but significantly reduced nuclease-associated cytotoxicities. Moreover, the CCR5-specific TALEN revealed only minimal off-target activity at the CCR2 locus as compared to the corresponding ZFN, suggesting that the TALEN platform enables the design of nucleases with single-nucleotide specificity. The combination of high nuclease activity with reduced cytotoxicity and the simple design process marks TALENs as a key technology platform for targeted modifications of complex genomes.
INTRODUCTION
Designer nucleases have developed into invaluable tools to modify the genomes of complex organisms. By inserting a DNA double-strand break (DSB) into the target locus such nucleases activate DNA repair, which can be harnessed to knockout genes or to promote gene targeting (1,2). Because the DNA damage response is highly conserved in eukaryotic cells, the concept of DSB-based genome engineering is easily transferrable between highly diverse organisms. Accordingly, designer nuclease-based genome engineering has been successfully established in more than 10 model organisms thus far, including plants (3,4), invertebrates (5), fish (6,7) and mammals (8). Moreover, the genome of multipotent and pluripotent human stem cells has been efficiently modified (9–12), without affecting the differentiation potential of these cells.
Zinc-finger nucleases (ZFNs) comprise the most successful class of engineered nucleases to date. ZFNs consist of two functional domains: a customized zinc-finger array fused to the non-specific endonuclease domain of the well-characterized restriction enzyme FokI. Upon dimerization of the two ZFN subunits in correct spacing and orientation, the nuclease domain cuts the target DNA within the spacer sequence that separates the two target half-sites (13). In a first approximation, each zinc-finger within a tandem array recognizes three bases at the DNA level (14). However, target site overlap and crosstalk between individual fingers in a zinc-finger array considerably complicate the production of sequence-specific ZFNs (15), requiring the use of labor-intense selection procedures to generate zinc-finger arrays with sufficient affinity and specificity (1,16,17). Novel platforms, such as context-dependent assembly (CoDA), have simplified the design process but the quality of such ZFNs may not be sufficient for therapeutic applications (18). Furthermore, although modifications in the FokI cleavage domain have been shown to prevent homodimerization of the individual ZFN subunits (19–22), the genome-wide specificity of ZFNs is still under scrutiny.
The ease of the design process as well as the balance between nuclease activity and associated toxicity are key parameters in the application of any type of designer nuclease (2). Two recent studies identified a novel protein scaffold based on transcription activator-like effector (TALE) proteins isolated from plant pathogens of the Xanthomonas genus to be amenable for engineering of customized DNA binding domains (23,24). TALEs are modular proteins composed of an N-terminal translocation domain, central repeats that collectively mediate sequence-specific DNA binding, and a C-terminal segment that encompasses nuclear localization signals (NLS) and a transcriptional activation domain (25). The central TALE DNA binding domain contains a variable number (characteristically between 12 and 30) of conserved 33–35 residues long repeats arranged in tandem arrays. Polymorphisms between repeats are predominantly found in positions 12 and 13, also referred to as the repeat variable di-residues (RVDs), and RVDs that preferentially recognize one of the four bases in the target site have been defined (23,24,26,27). Hence, this ‘one repeat to one base’ code enables the prediction of the DNA binding sites of natural TALEs or, vice versa, the engineering of customized TALE repeat arrays that recognize a user-defined target sequence. As a result, TALE repeat arrays have attracted great interest as a DNA targeting tool in the context of designer TALE-type transcription factors (dTALEs) (26,28) and TALE nucleases (TALENs) (27,29–34).
Here, we have characterized the cleavage parameters for efficient TALEN-mediated genome editing in human cells. Moreover, we performed a side-by-side comparison between engineered TALENs and well-characterized ZFNs at two endogenous human loci, CCR5 and IL2RG. We show that our designer TALENs can be as effective as ZFNs in terms of genome editing activity but significantly less cytotoxic. Moreover, our results indicate that the TALEN platform enables the design of nucleases with single-nucleotide specificity. Given both the ease with which TALENs can be engineered and their superior toxicity profile, TALENs are likely to have a significant impact on targeted genome engineering in the context of applied as well as basic biology.
MATERIALS AND METHODS
Plasmids
All TALE derivatives were generated using standard cloning procedures. The AvrBs4 and AvrBs3 deletion variants were generated by subcloning a BamHI–BamHI (A4-BB, A3-BB), Eco147I–HincII (A4-EH), NarI–HincII (A4-NH, A3-NH), Eco147I–BclI (A4-EC) and NarI–BclI (A4-NC) fragment of plasmids pENTR-D-avrBs3 and pENTR-D-avrBs4 (26), respectively, into vectors pRK5.AD or pRK5.N (35). Where indicated, TALENs with the obligate heterodimeric KV/EA FokI variants were used (19). All engineered TALEs were subsequently cloned into the A4-NH and A4-NC scaffolds. The sequences of all TALEs are indicated in Supplementary Figure S1. The luciferase-based reporter plasmid (pGLtk.EBEAvrBs4.Luc) is based on plasmid pGLtk (35) and generated by inserting a tandem repeat of EBEAvrBs4. The templates for the in vitro cleavage assays were generated by subcloning an inverted repeat of EBEAvrBs4 separated by variable spacers (from 6–16 bp; Supplementary Table S1) into plasmid pCMV.LacZ∂GFP (16). The dsEGFP reporter constructs used in the episomal gene disruption assay were generated by cloning homodimeric EBEAvrBs4/AvrBs4 or heterodimeric EBEAvrBs4/AvrBs3 elements (Supplementary Table S1), respectively, between the ATG and the 5′-end of a destabilized Enhanced Green Fluorescent Protein (dsEGFP) gene into plasmid pLV.CMV.dsEGFP. Reporter plasmid pLV.CMV.IL2RG-dsEGFP was generated by cloning the IL2RG gene derived from plasmid pRRL.MP.IL2RGpre (kindly provided by Axel Schambach, Hannover Medical School) into pLV.CMV.dsEGFP. All ZFN expression vectors were generated by subcloning a synthesized DNA-binding domain (GeneArt, Regensburg) into the pRK5.N (35) vector backbone, which encodes an N-terminal HA tag followed by a nuclear localization domain, and either of the obligate heterodimeric FokI variants KV/EA (19). The target sites and the recognition α-helices of the EGFP (17), CCR5 (9) and IL2RG-specific (36) ZFNs have been described. The complete sequences and maps of all plasmids can be obtained upon request.
In vitro cleavage assay
In vitro cleavage assays were basically performed as previously described (21). Briefly, TALENs were expressed in vitro using the TnT SP6 Quick Coupled Transcription/Translation System (Promega). The ~1-kb target DNA fragment was generated by Polymerase Chain Reaction (PCR) with Phusion polymerase (Finnzymes), primers #78 and #77 (Supplementary Table S2), and either of the six different plasmids pCMV.LacZ-X-∂GFP (X denoting spacer length) as a template. For in vitro cleavage, 1 µl of each TnT lysate containing one TALEN subunit was mixed with 200 ng of the DNA template and 1 µg of BSA in NEBuffer 4 (New England Biolabs) supplemented with 100 mM NaCl in a total volume of 10 µl. After incubation for 90 min at 37°C the reaction was analyzed on a 1.2% agarose gel.
Transcriptional reporter assay
All cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% Fetal Bovine Serum (FBS) and penicillin/streptomycin (Invitrogen). HEK293T cells were seeded in 24-well plates at a density of 80 000 cells/well. After 24 h, cells were transfected in duplicate using polyethylenimin (PEI) as described before (21). Transfection cocktails included 80 ng of reporter plasmid pGLtk.EBEAvrBs4.Luc, 400 ng of dTALE encoding plasmids and 10 ng of pRL (Promega) coding for Renilla luciferase to normalize for transfection efficiency. The amount of DNA was kept constant by adding pUC118 to 1.2 µg. Cells were harvested 48 h after transfection in 1 × PLB lysis buffer (Promega). Firefly and Renilla Luciferases activities were measured in a luminometer (Berthold Technologies, Bad Wildbach, Germany) using Dual-Luciferase Reporter Assay System (Promega) following the manufacturer’s instructions.
Gene disruption and quantitative cell toxicity assays
For episomal gene disruption, 80 000 HEK293T cells were seeded per well of a 24-well plate. After 24 h, cells were PEI transfected with 50 ng of pLV.CMV.IL2RG-dsGFP reporter plasmid, 400 ng of nuclease (TALEN, ZFN, I-SceI) expression plasmid, 50 ng of a mCherry expression vector (kindly provided by Roger Y. Tsien, UC San Diego) to normalize for transfection efficiency, and pUC118 to 1.2 µg. For chromosomal gene disruption, dsEGFP reporter cells were generated by lentiviral transduction (LV.CMV.EBEAvrBs4xAvrBs3.dsGFP; Supplementary Table S1) with a vector dose that rendered <1% of cells resistant to geneticin-sulfate (0.4 mg/ml), so ensuring that cells contained a single copy target locus (16). Reporter cells were seeded in 24-well plates (80 000 cells/well) and transfected after 24 h with 400 ng (or 1–600 ng) of nuclease expression plasmids, 100 ng of a mCherry expression vector, and pUC118 to 1.2 µg. After 2 and 5 days, the fractions of mCherry-positive and EGFP-negative cells were determined by flow cytometry (FACSCalibur; BD Biosciences). The cell survival rate was calculated as the decrease in the number of mCherry-positive cells from Days 2 to 5, normalized to cells transfected with a non-functional nuclease expression vector (37).
Genotyping
Genomic DNA of transfected cells was extracted using QIAamp DNA mini kit (Qiagen). The genomic region encompassing the nuclease target sites in dsEGFP or the human CCR2, CCR5 and IL2RG loci, respectively, were PCR amplified (Supplementary Table S2), and amplicons cleaned up with QIAquick PCR Purification Kit (Qiagen). The DNA fragments were then subjected to digestion with either XhoI or the mismatch-sensitive T7 endonuclease I (T7E1; New England BioLabs). For T7E1 assay, DNA was denatured at 95°C for 5 min, slowly cooled down to room temperature to allow for formation of heteroduplex DNA, treated with 5 U of T7E1 for 15 min at 37°C, and then analyzed by 2% agarose gel electrophoresis.
Immunoblotting
Western blots were performed as described before (21). TALEN or β-actin were detected with anti-HA tag (1:2000; Novus Biologicals) or anti-β-actin (1:2000; Cell Signaling) antibodies, respectively, and visualized with HRP-conjugated anti-rabbit antibody (Dianova) and West Pico Chemiluminescence substrate (Thermo Scientific).
Statistical analysis
All data sets shown as bar graphs represent the average of at least three independent experiments. Error bars indicate standard error of mean (SEM). Statistical significance was determined using a two-tailed, homoscedastic Student’s t-test.
RESULTS
Minimal DNA binding domain of TALE proteins
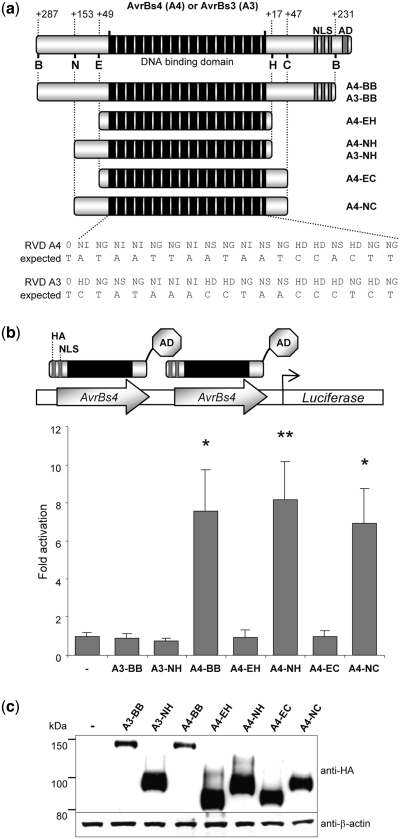
The biological function of many Xanthomonas TALE proteins has been studied extensively and a recent report identified a minimal domain required for efficient DNA binding of the TALE protein Hax3 in human cells (28). In order to assess whether these findings can be transferred to the TALE proteins AvrBs3 and AvrBs4, which were used as the design scaffold here, we created some corresponding N- and C-terminal deletion variants (Figure 1a) and fused them to the transcriptional activation domain of the VP16 protein of the herpes simplex virus (35) (Figure 1b). All variants contained an N-terminal NLS and a hemagglutinin (HA) tag, which allowed us to monitor the expression levels of these dTALEs in HEK293T cells by western blotting (Figure 1c). While the dTALE variants with extended truncations were expressed to high levels, the larger variants A3-BB and A4-BB, respectively, revealed lower steady-state levels. Note, the two-letter code before the hyphen in the nomenclature of the TALE variants refer to specificity: e.g. ‘A4’ in ‘A4-BB’ defines a TALE domain that recognizes the predicted 19-bp binding element of AvrBs4. The two letters following the hyphen denote the TALE truncation variant, e.g. ‘BB’ in ‘A4-BB’ stands for the BamHI–BamHI fragment, as depicted in Figure 1a. The ability of the deletion variants to mediate binding to the predicted 19-bp binding element of AvrBs4 (EBEAvrBs4) was determined in a luciferase-based reporter assay (Figure 1b). While the deletion variants A4-BB, A4-NH and A4-NC activated the luciferase reporter construct containing the EBEAvrBs4 sequence motif, reporter gene activation by A4-EH and A4-EC was indistinguishable from the control. Moreover, the AvrBs3-based variants A3-BB and A3-NH did not activate the reporter with the EBEAvrBs4 motif, indicating that activation of the reporter gene by the TALE-type transcription factors was mediated by a specific interaction of the AvrBs4 DNA binding domain with the matching EBEAvrBs4 sequence motif. Furthermore, these data confirm that a region encompassing more than 100 residues at the N-terminus of the TALE repeat array is essential for efficient binding of dTALE proteins to matching DNA target sequences. In contrast, the residues located C-terminally with respect to the TALE repeat domain seem to be dispensable for dTALE–DNA interaction.
Figure 1.
Minimal DNA binding domain of the AvrBs4 TALE. (a) Schematic of AvrBs4 and AvrBs3 deletion variants. The black rectangles represent the 17.5 central tandem repeat arrays that mediate DNA recognition. The C-terminal nuclear localization signals (NLS) and transcriptional activation domain (AD) are highlighted in gray. Deletion variants are generated using the restriction sites indicated with letters (B: BamHI; N: NarI; E: Eco147I; H: HincII; C: BclI) and reported in the respective names (right). The number of remaining residues at N- and C-termini (relative to the position of the DNA binding domain) and the RVDs with the expected target sequences for AvrBs3 and AvrBs4 are shown. (b) Transcriptional reporter assay. Designer TALE-based transcription factors (dTALEs) consist of the VP16 transcriptional AD fused to the AvrBs4 or AvrBs3 deletion variants. HEK293T cells were transfected with dTALE expression plasmids and the reporter construct containing two upstream binding sites for AvrBs4 (AvrBs4), followed by a minimal promoter element and the luciferase gene. The graph displays luciferase activity normalized for transfection efficiency and relative to transfection with empty vector (–). Significant activation above background is indicated by *P < 0.05 and **P < 0.01. (c) dTALE expression levels. Transfected HEK293T cells were harvested after 48 h and cell lysates probed either with antibodies against HA tag or β-actin. Lysate of non-transfected cells is marked with ‘–’.
Requirements for efficient DNA cleavage by TALENs
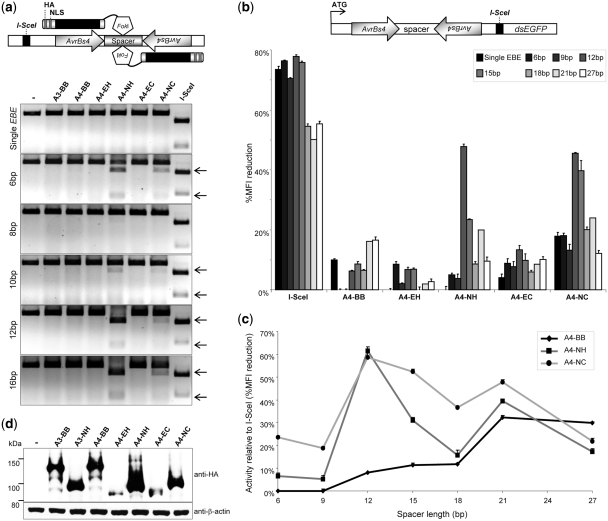
To generate TALE-based nucleases (TALENs), the VP16 transcriptional activation domain was replaced with the catalytic domain of the FokI endonuclease (38). The parameters for TALEN-mediated cleavage were initially determined in an in vitro cleavage assay. A linear DNA fragment containing an inverted repeat of EBEAvrBs4 separated by spacers ranging from 6 to 16 bp was incubated with the in vitro translated TALEN variants (Figure 2a). A recognition site for the meganuclease I-SceI (39) was included in all target DNAs as an internal control. In agreement with the transcriptional reporter assay, TALEN variants A4-NH and A4-NC were able to bind the DNA target and induce cleavage, while A4-EH and A4-EC were not. Highest activity was observed by variant A4-NH at spacers of 6, 12 and 16 bp. Variant A4-NC, which harbors a longer protein linker between the TALE repeat units and the FokI catalytic domain, showed reduced activity. Although variant A4-BB was expected to cleave substrates containing the EBEAvrBs4 motif based on the luciferase reporter assay (Figure 1b), it did not display any cleavage activity. None of the TALEN variants induced a DSB in DNA substrates with a single EBEAvrBs4, confirming that TALEN-mediated in vitro cleavage was sequence specific and mediated by dimerization of two TALEN subunits at the specific target site.
Figure 2.
Optimal spacer length for DNA cleavage by TALEN. (a) In vitro cleavage activity. TALENs consist of the TALE repeat units preceded by an HA tag and an NLS. The C-terminal catalytic domain of the FokI endonuclease (FokI) mediates cleavage after dimerization of two TALEN subunits. The linear DNA substrate containing an inverted AvrBs4 repeat was incubated with TALENs in vitro and the extent of cleavage analyzed by agarose gel electrophoresis. The respective spacer lengths (left) and the positions of the cleavage products (arrows) are indicated. (b) Cleavage activity in cellula. HEK293T cells were co-transfected with the respective TALEN expression plasmids and reporter plasmids that harbors an inverted repeat of AvrBs4 integrated into the 5′-end of a gene encoding a destabilized EGFP (dsEGFP). The position of a binding site for I-SceI used for internal reference is shown. The graph displays reduction of EGFP mean fluorescent intensity (MFI) relative to a non-functional nuclease, as determined by flow cytometry. The respective spacer lengths separating the AvrBs4 sites are indicated. (c) Relative TALEN activity in relation to spacer length. The graph displays TALEN activity (from [b]) relative to I-SceI at the various spacers. (d) TALEN expression levels. Transfected HEK293T cells were harvested after 48 h and cell lysates probed either with antibodies against HA tag or β-actin. Lysate of non-transfected cells is marked with ‘–’.
To verify these results in cells, a fast quantitative reporter assay was developed (Figure 2b). An inverted EBEAvrBs4 repeat separated by spacers ranging from 6 to 27 bp was cloned in between the translational start codon ATG and the open reading frame (ORF) encoding a destabilized EGFP (dsEGFP). A recognition site for I-SceI was included as an internal control. Nuclease mediated cleavage will either lead to rapid degradation of the episomal target plasmid or, alternatively, to error-prone DNA repair by the non-homologous end-joining (NHEJ) pathway. The resulting reduction in dsEGFP fluorescence intensity is hence a measure for nuclease activity. In good agreement with the in vitro data, expression of TALEN variants A4-NH and A4-NC led to a considerable reduction of the mean fluorescence intensity (MFI) in HEK293T cells transfected with reporters that contain target sites separated by 12 and 15 bp, respectively, but not at other spacer lengths. Note that some TALEN variants reduced EGFP expression from the reporter containing a single AvrBs4 recognition site (single EBE; black bars), suggesting that efficient binding of these TALENs to the recognition sequence was sufficient to reduce the MFI between 0 and 20% by interfering with transcription. A similar observation was made when expressing a mutated I-SceI (data not shown). Interestingly, while the A4-NH variant had a restricted activity profile with a pronounced peak at a 12-bp spacer, A4-NC was less selective in terms of spacer length requirements and showed approximately equal activity on spacers from 12 to 21 bp (Figure 2c). Variant A4-BB displayed notable cleavage activity over background only on targets with 21 and 27-bp spacers, while expression of I-SceI reduced dsEGFP expression from all reporter plasmids. These results support the assumption that variants with longer linkers need longer spacers to accommodate the additional amount of protein. Assessment of TALEN expression (Figure 2d) consistently revealed reduced steady-state levels of the less active TALENs A4-EH and A4-EC, which may partially—but not exclusively—explain the lack of activity of these variants.
Together with the transcriptional reporter data, these results suggest that the structural requirements for TALEN-mediated DNA cleavage are divergent from dTALE-mediated gene regulation, i.e. while the dTALE variant based on A4-BB activated luciferase expression, the corresponding TALEN did not efficiently cut the DNA target. Furthermore, variants with a short C-terminal linker peptide that connects the TALE repeats with the FokI cleavage domain, such as the 17- and 47-residue linkers in A4-NH and A4-NC, respectively, provide a better scaffold to generate designer TALENs.
Profiling TALEN activity versus cytotoxicity
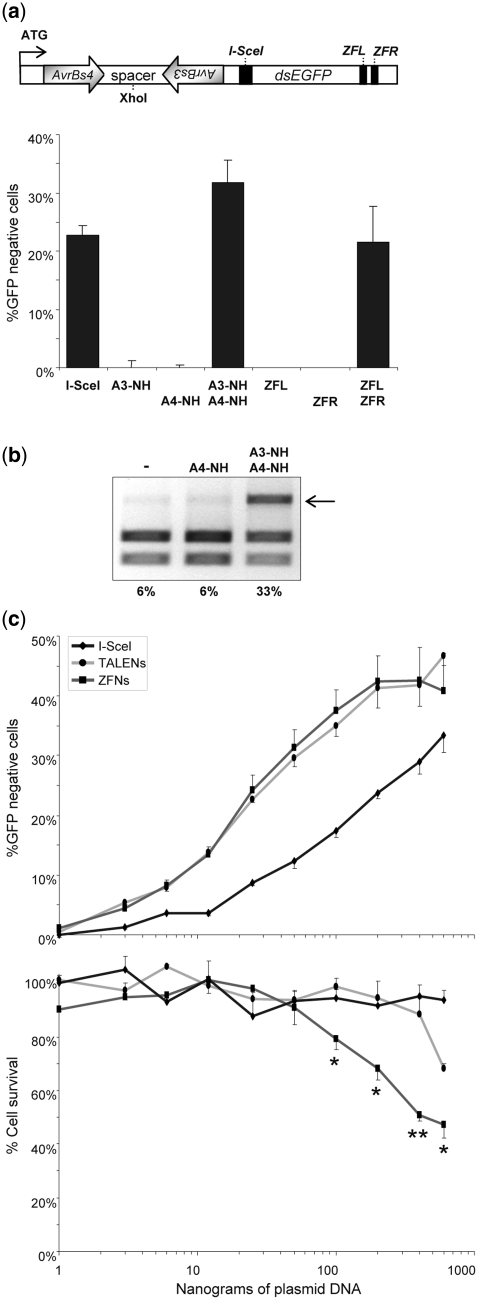
To characterize the target site requirements for efficient cleavage by a heterodimeric TALEN in a chromosomal context, a HEK293-based reporter cell line with an integrated dsEGFP cassette was generated. The dsEGFP ORF contained distinct recognition sites for the TAL effectors AvrBs3 (EBEAvrBs3) and AvrBs4 (EBEAvrBs4) in opposite orientation (Figure 3a). In order to maintain the ORF and keep an optimal distance between the EBEs, a 13-bp spacer was chosen. Again, a recognition site for I-SceI was included as an internal control. Transfection of these cell lines with expression plasmids coding for I-SceI or an EGFP-specific ZFN pair (17) induced gene disruption in ~22% of reporter cells, as determined by flow cytometry. Gene disruption by co-expression of TALEN subunit A3-NH and A4-NH was induced in >30% of the transfected cells. Of note, TALENs that contain an obligate heterodimeric FokI domain (19), as compared to the wild-type FokI version used in this experiment, were slightly less active (Supplementary Figure S2). For both TALENs and ZFNs gene disruption activity was strictly dependent on the presence of both nuclease subunits.
Figure 3.
Comparison of activity versus toxicity profiles of TALEN and ZFN. (a) Designer nuclease-mediated gene disruption. The HEK293-based reporter cells harbor an integrated dsEGFP gene that contains an inverted heterodimeric AvrBs4/AvrBs3 target sequence separated by 13-bp spacer in the 5′-end of the open reading frame. The position of the diagnostic XhoI site is indicated. For internal reference, a binding site for I-SceI was placed downstream of the TALEN target site. The graph shows the percentage of EGFP-negative cells 5 days after transfection with the nuclease expression vectors (TALEN, ZFN or I-SceI). (b) Molecular characterization. Genomic DNA was extracted 5 days after transfection and PCR amplicons encompassing the target sites were used as templates for digestion with XhoI. An arrow indicates the position of the XhoI-resistant DNA fragment. The numbers below designate the percentages of XhoI-resistant PCR fragments (note background level of ~6%). (c) Activity versus toxicity. The EGFP reporter cells were transfected with increasing amounts (1–600 ng) of nuclease expression vectors (TALEN, ZFN or I-SceI) and a mCherry-encoding plasmid. The percentage of EGFP and mCherry-positive cells was determined by flow cytometry after 2 and 5 days. The graphs display gene disruptions activities (EGFP-negative cells at Day 2; top) and nuclease-associated cytotoxicities (fraction of mCherry-positive cells at Day 5 as compared to Day 2 after transfection; bottom), relative to cells transfected with a mock plasmid. Statistically significant differences in toxicities between TALEN and ZFN are indicated by *P < 0.05 or **P < 0.01, respectively.
The extent of TALEN-mediated target site cleavage in these reporter cells was subsequently confirmed by genotyping (Figure 3b). A genomic fragment encompassing the TALE target sites EBEAvrBs3 and EBEAvrBs4 was amplified by PCR and subjected to digestion with XhoI. The XhoI restriction site is located in the spacer sequence separating the two target half-sites, which is expected to be cleaved by the TALEN. Given that NHEJ-mediated DNA repair after a TALEN-induced DSB will frequently lead to disruption of the target site, the corresponding PCR amplicons will lack the XhoI recognition site and therefore become resistant to XhoI cleavage. The fraction of XhoI-resistant PCR products reflecting the cleavage activity was in good agreement with the percentage of EGFP-negative cells (Figure 3a), confirming high activity of the A3-NH/A4-NH TALEN pair.
To compare directly the activities and nuclease-associated toxicities of TALENs and ZFNs, the same reporter cell line was transfected with increasing amounts of expression vectors ranging from 1 to 600 ng of each subunit (Figure 3c). Note that while the TALEN pair was equipped with a wild-type FokI domain, the ZFNs contain an obligate heterodimeric cleavage domain that was shown to reduce nuclease-associated toxicity (19,21). The activity profiles of both classes of nucleases were comparable for any of the DNA amounts transfected and reached a maximal gene disruption activity at about 45% EGFP-negative cells. However, assessment of cell survival 5 days after transfection revealed a significant increase in cytotoxicity for ZFN transfected cells at high vector doses.
Together with the previous data, these results suggest that the optimal spacer length for TALENs based on the A4-NH scaffold is 12 or 13 bp, while the A4-NC architecture, which contains a longer linker between the DNA binding domain and the C-terminal FokI domain, was more flexible with respect to spacer length. In summary, our data demonstrate that TALEN-mediated gene disruption at chromosomal loci can be as efficient as knockouts created by ZFNs but with reduced nuclease-associated toxicity.
Efficient disruption of endogenous genes
Based on the above findings, we aimed at generating TALEN pairs that target endogenous genes in the human genome. To this end, we designed TALENs to target sites in the CCR5 and IL2RG loci (Figures 4 and 5) that overlap with previously published ZFN recognition sites (20,40). The corresponding TALE repeat domains were cloned into both the A4-NC (47-residue linker) and A4-NH (17-residue linker) scaffolds and termed GC-NC or GC-NH (targeted to the IL2RG locus) and C5-NC or C5-NH (targeted to the CCR5 locus). Note that while the ZFN half-sites are separated by 5 bp, the TALEN target sites contain 15-bp spacers.
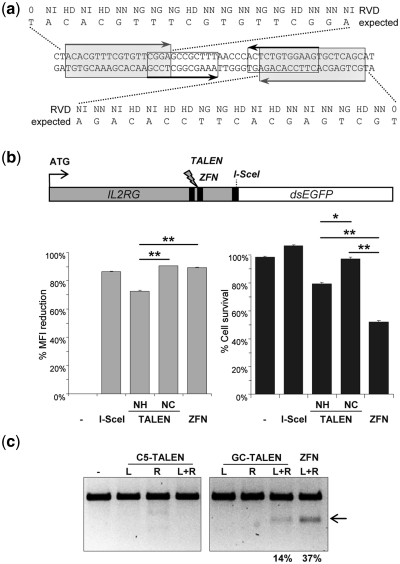
Figure 4.
Modifications at the human IL2RG locus. (a) Target sites in human IL2RG gene. Target sites are highlighted by gray shaded (TALEN) or black (ZFN) boxes, respectively. The RVDs of the engineered TALEs as well as the expected target sequences are indicated. (b) Comparison of the activities and toxicities of IL2RG-specific designer nucleases. HEK293T cells were co-transfected with expression plasmids-encoding IL2RG-specific TALEN and ZFN, respectively, and a reporter plasmid harboring an IL2RG-dsEGFP fusion gene. The graphs display reduction of the mean fluorescent intensity (MFI) of IL2RG-dsEGFP (left) and relative cell survival as compared to cells expressing a nonfunctional nuclease indicated with ‘–’ (right). NH and NC designate the TALEN scaffold used. Statistically significant differences in activity and toxicity between the TALEN and ZFN tested are indicated by *(P < 0.05) or **(P < 0.01), respectively. (c) Disruption of endogenous human IL2RG locus. After transfection with TALEN (NC scaffold) and ZFN expression vectors, genomic DNA was extracted and used as a template for PCR amplification. Amplicons encompassing the target sites were digested with the mismatch-sensitive T7 endonuclease 1 (T7E1). Arrows indicate the expected positions of the T7E1 digestion products. Numbers at the bottom designate the average percentage of modified alleles (n = 2). TALENs targeting the CCR5 locus (C5-TALEN) were used as negative controls. L and R refer to the nuclease subunits, binding to the left or right target half-sites, respectively.
Figure 5.
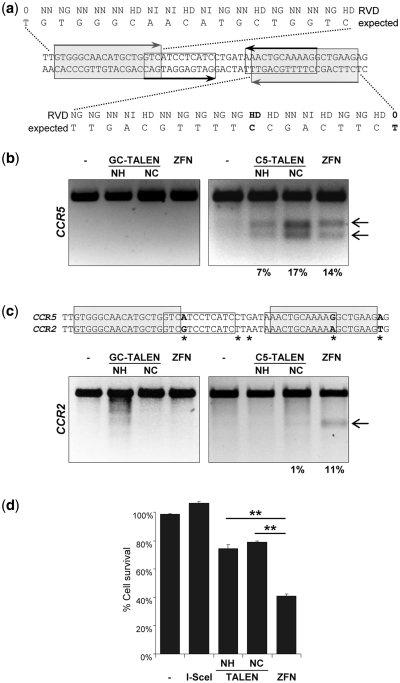
Modifications at the human CCR5 locus. (a) Target sites in human CCR5 gene. Target sites are highlighted by gray shaded (TALEN) or black (ZFN) boxes, respectively. The RVDs of the engineered TALEs as well as the expected target sequences are shown. Bold letters designate the nucleotides not conserved in the CCR2 locus. (b) Disruption of endogenous human CCR5 locus. After transfection with TALEN and ZFN expression vectors, genomic DNA was extracted and PCR amplicons encompassing the CCR5 target sites digested with T7E1. Arrows point out the expected positions of the T7E1 digestion products. Numbers at the bottom indicate the average percentage of modified alleles (n = 3). TALENs targeting the IL2RG locus (GC-TALEN) were used as a negative control. NH and NC designate the TALEN scaffold. (c) Off-target activity at CCR2 locus. Asterisks in the alignment of CCR5 and CCR2 designate the mismatches between the two target sequences, bold letters the nucleotides affecting binding of the designer nucleases. PCR amplicons encompassing the CCR2 target locus were digested with T7E1. Arrows point out the expected positions of the T7E1 digestion products. Numbers at the bottom designate the average percentage of modified alleles (n = 2). (d) Cytotoxicity of CCR5-specific designer nucleases. The graph displays nuclease-associated cytotoxicities relative to cells expressing a nonfunctional nuclease (–). Statistically significant differences in toxicities between TALENs and ZFN are indicated by **P < 0.01.
For quantitative comparison of the activities and toxicities of the IL2RG-specific TALEN (designated ‘GC’ for gamma chain) and ZFN, an IL2RG-dsEGFP reporter construct was generated (Figure 4b). A recognition site for I-SceI was placed in between the two ORFs as a reference. Co-transfection of the reporter with I-SceI, the IL2RG-specific TALENs or ZFN expression vectors, respectively, reduced the MFI of IL2RG-dsEGFP between 69% and 90%. This demonstrated that all of the tested nucleases were highly active, although the TALEN variant GC-NC with the longer linker was somewhat more active than GC-NH. Concomitant assessment of cytotoxicity, on the other hand, revealed an almost 2-fold increase in cell survival when comparing the GC-NC TALEN versus the ZFN pair, respectively. To assess activity of GC-NC at the genome level, HEK293T were transfected with the respective TALEN or ZFN expression vectors (Figure 4c). The extent of NHEJ induced insertions/deletions after target site cleavage was quantified using the mismatch-sensitive T7E1 endonuclease (41). A direct comparison indicated that the engineered IL2RG-specific TALEN pair was about half as active as the well-established ZFN, with targeted allelic modification frequencies of 14% for the TALEN and 37% for the ZFN. A CCR5-specific TALEN served as a negative control at the IL2RG locus.
Then the TALENs designed to target the CCR5 locus, C5-NC and C5-NH (Figure 5a), were transfected into HEK293T. Genomic DNA was isolated and subjected to T7E1-based genotyping. A side-by-side comparison indicated that the engineered C5-NC TALEN was as active as the well-established ZFN, with allelic modifications at 17 and 14%, respectively (Figure 5b). As seen before for the IL2RG-specific TALENs, the C5-NH TALEN with the shorter linker was not as active as C5-NC.
Specificity of designer nucleases is an important parameter. A major off-target locus of the CCR5-specific ZFN has been identified in CCR2 (40,42), which shares a high degree of sequence identity with the CCR5 locus (Figure 5c). The ZFN target site in CCR5 differs from the corresponding site in CCR2 in two positions, one in each target half site. In contrast, the entire 19-bp target sequence of the left TALEN subunit is also found in CCR2 while the right target half-site varies at only one position bound by a TALE repeat unit and an additional one recognized by the 0th repeat (Figure 5a). T7E1-based genotyping at the CCR2 locus revealed that expression of the CCR5-specific ZFN-induced mutations at 11% of CCR2 alleles, while only 1% of CCR2 alleles were mutated after TALEN expression. Parallel assessment of cytotoxicity revealed a 2-fold increase in cell survival when comparing the CCR5-specific TALEN with the ZFN pair (Figure 5d).
DISCUSSION
Designer nucleases have evolved into invaluable tools for targeted genome engineering. In particular, ZFNs have been successfully employed in a broad variety of research fields, ranging from fundamental to applied science. A major drawback of ZFNs, however, is the elaborate and time-consuming experimental selection process (1,17). Although simplified methods, such as modular assembly and CoDA have been reported (18,43,44), the quality of ZFNs generated by such platforms is controversial (45–47) or not determined yet. In this study, we have characterized the cleavage parameters for 17.5 repeats containing TALENs based on the AvrBs4 scaffold. This TALEN scaffold mediates binding to a total of 19 bp, including the invariant thymine (in position–1) that precedes the RVD-defined nucleotides in the TALE target box (24). By employing different in vitro and cellular reporter assays, we have defined a TALEN architecture that allows for efficient DNA cleavage. Depending on the length of the linker that connects the TALE DNA binding domain with the FokI cleavage domain, we found that spacers of 12–15 bp between the two target half-sites are optimal for high TALEN cleavage activity.
Similar to our approach, Miller et al. (27) produced a set of TALENs with truncated TALE domains. Their variants with 28 or 63 residue long C-terminal linkers proved to be the most active configurations and are similar to our A4-NH and A4-NC scaffolds with 17- and 47-residue linkers, respectively. In the context of TALE transcription factors, Zhang et al. (28) showed that 147 residues N-terminally of the TALE repeat units are essential for efficient DNA binding. This is in good agreement with our A4-NH and A4-NC scaffolds (+153 amino acids) as well as the architecture used by Miller et al. (27). Although we barely detected activity of our longer TALEN variant A4-BB, TALENs similar to this scaffold have been used in other studies (32,33). Given that the respective TALEN activities in these reports were determined using different methods, in different organisms, and at target sites with longer spacers that may accommodate the longer linkers, it is difficult to compare the results directly. Nonetheless, our data imply that a very long stretch of residues between the TALE repeat units and the FokI nuclease domain may place the catalytic center in an unfavorable position that impairs activity. Alternatively, lower nuclease activity of the longer TALEN variants is simply a result of decreased protein stability, as shown in our immunoblots.
Some of the aforementioned studies also explored in detail the spacer length requirements between the two target half-sites. In accordance with studies performed on ZFNs (48,49), the protein linker that connects the DNA binding domain with the nuclease domain influences selectivity with regard to DNA spacer length and hence ultimately specificity. While TALENs with long linkers (>200 amino acids) seem to work on a very broad range of spacers spanning 14–40 bp (29,30), the shorter linkers (17–63 residues) used in our study and in Miller et al. (27) restrict activity to spacers of 12–22 bp. Nonetheless, these results are in sharp contrast to ZFNs where activity can be restricted to 5/6-bp spacers using a short 4-residue linker (49), so more effort in optimizing the architecture of TALEN has to be invested to further restrict the activity range of these nucleases in the future.
The design process to generate TALENs is based on a simple modular assembly strategy. As opposed to ZFNs, in which an individual zinc-finger module contacts in a first approximation a 3-bp target subsite (14), each TALE repeat unit recognizes a single nucleotide (23,24). We have used the four RVDs NN to target guanine, NI for adenine, NG for thymine and HD for cytosine. Some recent reports showed a stronger preference of the NK repeat to bind guanine than NN (26,27), so it will be interesting to see whether exchanging the respective RVDs will increase activity and further reduce toxicity of designer TALENs.
Overall, the success rate for generating TALENs by simple modular assembly seems very high. We have used the natural TALEs AvrBs4 and AvrBs3 in the context of a TALEN pair to disrupt an EGFP marker and designed two additional TALEN pairs that recognize target sites in two endogenous sites in the human genome. These nucleases induced modifications in 14 or 17% of the targeted IL2RG or CCR5 alleles, respectively, or 45% of the EGFP reporter. These numbers are comparable to data reported by Miller et al. (27) who generated TALENs based on a different TALE backbone and different numbers of TALE repeats. The authors targeted the human CCR5 and NTF3 genes with efficiencies of up to 27% and report that all of their TALEN pairs designed to recognize sites with a 12–21 bp spacer yielded at least 5% gene editing (27). Furthermore, Cermak et al. (32) described design guidelines based on natural TALEs that may further improve the success frequency.
To enable a side-by-side analysis with ZFNs, the endogenous target sites in CCR5 and IL2RG were chosen to overlap with binding sites of previously described ZFNs (20,40). On the whole, our designed TALENs showed similar gene disruption activities as some extensively optimized ZFNs, such as the CCR5-specific ZFN pair that has been employed in two clinical trials (NCT00842634, NCT01044654). On the other hand, the TALENs used here were generally less cytotoxic than their ZFN counterparts, suggesting better specificity. In fact, in a side-by-side analysis of the CCR5-specific designer nucleases, the TALEN showed significantly reduced off-target activity at the CCR2 locus as compared to the ZFN. Remarkably, the CCR5-specific TALEN induced mutations at 17% of CCR5 alleles and at only 1% of the highly homologous CCR2 locus. In contrast, activity of the CCR5-specific ZFN was almost comparable at the two loci, with mutation frequencies of 14% at CCR5 and 11% at CCR2. For both ZFN and TALEN the respective target sites differ at two positions each: 2 out of 24 nt for the ZFN and 2 out of 38 nt for the TALEN pair. One of the two mismatches between CCR5 and CCR2 coincides with the 5′ terminal T of the right TALE binding box (Figure 5a). Previous studies have shown that this T in position −1 of the recognition site pairs with the postulated 0th TALE repeat and is critical for the TALE–DNA interaction (24,50). Although further studies are needed, it seems that the conserved 5′ T nucleotide of the EBE sites are particularly well-suited to discriminate two nearly identical target sites.
As mentioned above, all TALEN pairs used in this study recognize a 38-bp target site while the binding sites for the corresponding ZFNs were 18 or 24 bp long. While much more work will be required to come to definitive conclusions, it is tempting to speculate that the generally longer recognition sites of TALENs as compared to ZFNs go along with higher specificities and therefore less toxicity.
Depending on the specific needs, other functional domains can be fused to the TALE repeat units to create artificial proteins able to modify not only the genome, but also the epigenome or transcriptome in a targeted fashion. The high success rates of the TALE modular assembly strategy to produce functional designer nucleases or artificial transcription factors (26–28,32,33,51) suggest that context-dependent effects between individual TALE repeat units are negligible. The high number of repeat units with their high degree of homology does complicate the assembly of such DNA binding domains when using standard cloning approaches. However, recently introduced Golden Gate based strategies overcome these limitations (28,32,33,51). Given the high interest in sequence-specific genome surgery, it is conceivable that off-the shelf TALENs for each human gene will be available soon.
In conclusion, the TALEN scaffold presented here enables genome editing with high efficiency and precision. A side-by-side comparison between our TALENs and well-characterized ZFNs showed that the TALE platform enables the design of artificial nucleases that are as effective as the ZFNs in terms of activity but likely more specific and less cytotoxic. Although further characterization with regard to specificity will be necessary for clinical applications of TALENs, the simple 1:1 code, i.e. one TALE repeat unit recognizing 1 nt, will greatly facilitate the design of customized DNA binding domains for basic and applied sciences in the future.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Commission’s 7th Framework Programme (PERSIST–222878 to T.C.); 2Blades foundation (to T.L.). Funding for open access charge: European Commission’s 7th Framework Programme (PERSIST–222878).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jessica Wenzl and Juri Lafera for technical assistance, Heimo Riedel for critical discussions, and Axel Schambach and Roger Y. Tsien for plasmids.
REFERENCES
- 1.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 2.Händel EM, Cathomen T. Zinc-finger nuclease based genome surgery: it's all about specificity. Curr. Gene. Ther. 2011;11:28–37. doi: 10.2174/156652311794520120. [DOI] [PubMed] [Google Scholar]
- 3.Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, Voytas DF. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature. 2009;459:442–445. doi: 10.1038/nature07845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, Mitchell JC, Arnold NL, Gopalan S, Meng X, et al. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature. 2009;459:437–441. doi: 10.1038/nature07992. [DOI] [PubMed] [Google Scholar]
- 5.Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- 6.Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat. Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, et al. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 10.Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ, et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, Crooks GM, Kohn DB, Gregory PD, Holmes MC, et al. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat. Biotechnol. 2010;28:839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith J, Bibikova M, Whitby FG, Reddy AR, Chandrasegaran S, Carroll D. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 2000;28:3361–3369. doi: 10.1093/nar/28.17.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 15.Cathomen T, Joung KJ. Zinc-finger nucleases: the next generation emerges. Mol. Ther. 2008;16:1200–1207. doi: 10.1038/mt.2008.114. [DOI] [PubMed] [Google Scholar]
- 16.Cornu TI, Thibodeau-Beganny S, Guhl E, Alwin S, Eichtinger M, Joung JK, Cathomen T. DNA-binding specificity is a major determinant of the activity and toxicity of zinc-finger nucleases. Mol. Ther. 2008;16:352–358. doi: 10.1038/sj.mt.6300357. [DOI] [PubMed] [Google Scholar]
- 17.Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat. Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat. Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 20.Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 21.Söllü C, Pars K, Cornu TI, Thibodeau-Beganny S, Maeder ML, Joung JK, Heilbronn R, Cathomen T. Autonomous zinc-finger nuclease pairs for targeted chromosomal deletion. Nucleic Acids Res. 2010;38:8269–8276. doi: 10.1093/nar/gkq720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, Miller JC, Urnov FD, Gregory PD, Holmes MC. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat. Methods. 2011;8:74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- 23.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 24.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 25.Bogdanove AJ, Schornack S, Lahaye T. TAL effectors: finding plant genes for disease and defense. Curr. Opin. Plant Biol. 2010;13:394–401. doi: 10.1016/j.pbi.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 26.Morbitzer R, Römer P, Boch J, Lahaye T. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc. Natl Acad. Sci. USA. 2010;107:21617–21622. doi: 10.1073/pnas.1013133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 28.Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat. Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. TAL effector nucleases create targeted DNA double-strand breaks. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li T, Huang S, Jiang WZ, Wright D, Spalding MH, Weeks DP, Yang B. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2011;39:359–372. doi: 10.1093/nar/gkq704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahfouz MM, Li L, Shamimuzzaman M, Wibowo A, Fang X, Zhu JK. De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc. Natl Acad. Sci. USA. 2011;108:2623–2628. doi: 10.1073/pnas.1019533108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr218. (Epub ahead of print 14 April 2011; doi: 10.1093/nar/gkr218) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li T, Huang S, Zhao X, Wright DA, Carpenter S, Spalding MH, Weeks DP, Yang B. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr188. (Epub ahead of print 31 March 2011; doi: 10.1093/nar/gkr188) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alwin S, Gere MB, Guhl E, Effertz K, Barbas CF, 3rd, Segal DJ, Weitzman MD, Cathomen T. Custom zinc-finger nucleases for use in human cells. Mol. Ther. 2005;12:610–617. doi: 10.1016/j.ymthe.2005.06.094. [DOI] [PubMed] [Google Scholar]
- 36.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 37.Cornu TI, Cathomen T. Quantification of zinc finger nuclease-associated toxicity. Methods Mol. Biol. 2010;649:237–245. doi: 10.1007/978-1-60761-753-2_14. [DOI] [PubMed] [Google Scholar]
- 38.Wah DA, Hirsch JA, Dorner LF, Schildkraut I, Aggarwal AK. Structure of the multimodular endonuclease FokI bound to DNA. Nature. 1997;388:97–100. doi: 10.1038/40446. [DOI] [PubMed] [Google Scholar]
- 39.Rouet P, Smih F, Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc. Natl Acad. Sci. USA. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19:1279–1288. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee HJ, Kim E, Kim JS. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res. 2010;20:81–89. doi: 10.1101/gr.099747.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mandell JG, Barbas CF., 3rd Zinc Finger tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006;34:W516–W523. doi: 10.1093/nar/gkl209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim S, Lee MJ, Kim H, Kang M, Kim JS. Preassembled zinc-finger arrays for rapid construction of ZFNs. Nat. Methods. 2011;8:7. doi: 10.1038/nmeth0111-7a. [DOI] [PubMed] [Google Scholar]
- 45.Ramirez CL, Foley JE, Wright DA, Muller-Lerch F, Rahman SH, Cornu TI, Winfrey RJ, Sander JD, Fu F, Townsend JA, et al. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat. Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim J-S, Lee HJ, Carroll D. Genome editing with modularly assembled zinc-finger nucleases. Nat. Methods. 2010;7:91. doi: 10.1038/nmeth0210-91b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joung JK, Voytas DF, Cathomen T. Reply to “Genome editing with modularly assembled zinc-finger nucleases”. Nat. Methods. 2010;7:91–92. doi: 10.1038/nmeth0210-91b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG, Chandrasegaran S. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol. Cell Biol. 2001;21:289–297. doi: 10.1128/MCB.21.1.289-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Händel EM, Alwin S, Cathomen T. Expanding or restricting the target site repertoire of zinc-finger nucleases: the inter-domain linker as a major determinant of target site selectivity. Mol. Ther. 2009;17:104–111. doi: 10.1038/mt.2008.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Römer P, Strauss T, Hahn S, Scholze H, Morbitzer R, Grau J, Bonas U, Lahaye T. Recognition of AvrBs3-like proteins is mediated by specific binding to promoters of matching pepper Bs3 alleles. Plant Physiol. 2009;150:1697–1712. doi: 10.1104/pp.109.139931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morbitzer R, Elsaesser J, Hausner J, Lahaye T. Assembly of custom TALE-type DNA binding domains by modular cloning. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr151. (Epub ahead of print 18 March 2011; doi: 10.1093/nar/gkr151) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.