Abstract
The CRISPR/Cas adaptive immune system provides resistance against phages and plasmids in Archaea and Bacteria. CRISPR loci integrate short DNA sequences from invading genetic elements that provide small RNA-mediated interference in subsequent exposure to matching nucleic acids. In Streptococcus thermophilus, it was previously shown that the CRISPR1/Cas system can provide adaptive immunity against phages and plasmids by integrating novel spacers following exposure to these foreign genetic elements that subsequently direct the specific cleavage of invasive homologous DNA sequences. Here, we show that the S. thermophilus CRISPR3/Cas system can be transferred into Escherichia coli and provide heterologous protection against plasmid transformation and phage infection. We show that interference is sequence-specific, and that mutations in the vicinity or within the proto-spacer adjacent motif (PAM) allow plasmids to escape CRISPR-encoded immunity. We also establish that cas9 is the sole cas gene necessary for CRISPR-encoded interference. Furthermore, mutation analysis revealed that interference relies on the Cas9 McrA/HNH- and RuvC/RNaseH-motifs. Altogether, our results show that active CRISPR/Cas systems can be transferred across distant genera and provide heterologous interference against invasive nucleic acids. This can be leveraged to develop strains more robust against phage attack, and safer organisms less likely to uptake and disseminate plasmid-encoded undesirable genetic elements.
INTRODUCTION
Bacteria and Archaea rely on a diversity of defense systems that allow them to survive exposure to foreign genetic elements such as viruses. In their natural habitats, bacteria have evolved a battery of defense mechanisms to prevent phage infection, including prevention of adsorption, blocking of injection, or degradation of foreign nucleic acids (1,2). Recently, an adaptive prokaryotic immune system based on clustered regularly interspaced short palindromic repeats (CRISPR) was identified that provides acquired immunity against viruses and plasmids (3). CRISPR consists of arrays of short conserved repeat sequences interspaced by unique DNA sequences of similar size called spacers, which often originate from phage or plasmid DNA (3–5). CRISPR arrays, together with cas (CRISPR-associated) genes form the CRISPR/Cas adaptive immune system.
The CRISPR/Cas system has the ability to acquire short pieces of DNA (spacers) which provide immunity against subsequent exposures to phages and plasmids that carry matching sequences (3,6,7). The detailed mechanism by which CRISPR/Cas systems provides resistance against foreign DNA is subject to multiple current studies. Although the large majority of bacteria die upon virulent phage infection, a small proportion of the population survives by acquisition of phage-derived spacers (3). CRISPR-encoded immunity is provided by transcription of the repeat-spacer array, followed by transcript processing into small crRNAs (CRISPR RNAs), which are then used in combination with Cas proteins as guides to interfere with invasive DNA (6,7) or RNA (8).
Cas proteins, which often carry functional domains typical of nucleases, helicases, polymerases and nucleotide-binding proteins (9), are involved in multiple stages of CRISPR-based immunity. Notwithstanding their genetic hypervariability and mechanistic idiosyncrasies, CRISPR/Cas systems are grouped into different subtypes (9,10).
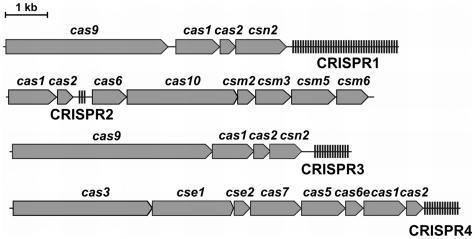
A few model systems have been established in the study of CRISPR/Cas functionality, notably in Escherichia coli (6,11), Staphylococcus aureus (7), Pyrococcus furiosus (8) and Streptococcus thermophilus (3,12). The S. thermophilus DGCC7710 model organism, for which CRISPR/Cas interference has been demonstrated against phages (3,13) and plasmids (12) contains four distinct CRISPR/Cas systems: CRISPR1, CRISPR2, CRISPR3 and CRISPR4 (14) (Figure 1). Direct spacer acquisition activity has been demonstrated for the CRISPR1 and CRISPR3 systems, with the former being more active in this strain (3,12,13). CRISPR1 and CRISPR3, which both belong to Type II CRISPR/Cas systems (15), share a similar architecture, with four cas genes located upstream of the CRISPR spacer array. Both cas1 and cas2 are universal, whereas cas9 (formerly named cas5 and csn1) is the signature gene of the Type II system. It has been shown that cas9 and csn2 are involved in interference and spacer acquisition, respectively (3,12). In silico analysis of phage sequences adjacent to CRISPR1 and CRISPR3 proto-spacers (nucleotide sequences in the target DNA corresponding to the spacers) revealed the presence of conserved PAM (Proto-spacer Adjacent Motif) sequences, NNAGAAW and NGGNG respectively (13,16,17), that are involved in interference. Single point mutations in the proto-spacer or the PAM allow the phages to circumvent CRISPR-mediated immunity (13).
Figure 1.
CRISPR/Cas systems of S. thermophilus DGCC7710. Cas proteins of the CRISPR1 and CRISPR3 systems belong to Type II, while CRISPR2 and CRISPR4 belong to Type III and Type I, respectively.
The CRISPR2 and CRISPR4 systems present in the S. thermophilus DGCC7710 genome belong to the Type III (Mtube) and Type I (Ecoli), respectively (14,15). Differences between types can be observed in terms of repeat, spacer and cas gene content and sequence. The multiplicity of CRISPR/Cas systems in S. thermophilus is explained by their susceptibility to horizontal gene transfer, and phage selective pressure.
Here we report the first cloning and heterologous expression of a functional CRISPR/Cas system into a different bacterial genus. We demonstrate that the S. thermophilus CRISPR3 system prevents plasmid transformation in E. coli. We show that both the proto-spacer and PAM sequences are necessary for immunity, although mutations distant from the PAM are tolerated. Furthermore, we provide experimental evidence that Cas9 alone is sufficient for plasmid DNA interference, and demonstrate by mutational analysis the importance of the McrA/HNH- and RuvC/RNaseH- nuclease domains.
MATERIALS AND METHODS
Bacterial strains and plasmids
Escherichia coli strain ER2267 [F′ proA+B+ lacIq Δ(lacZ)M15 zzf::mini-Tn10 (KanR)/Δ(argF-lacZ)U169 glnV44 e14−(McrA−) rfbD1? recA1 relA1? endA1 spoT1? thi-1 Δ(mcrC-mrr)114::IS10] (New England Biolabs, Beverly, MA, USA) and E. coli strain RR1 [F- mcrB mrr hsdS20(rB− mB−) leuB6 ara-14 proA2 lacY1 galK2 xyl-5 mtl-1 rpsL20(SmR) glnV44 λ−] (18) were used in the cloning and plasmid transformation experiments, respectively. Escherichia coli cells were grown in LB medium supplemented with ampicillin (50 μg/ml) and/or chloramphenicol (10 μg/ml) when necessary. Plasmid vectors pACYC184 (19) and pUC18 (20) were used for cloning and subcloning procedures. Genomic DNA of Streptococcus thermophilus DGCC7710 strain was kindly provided by Danisco (Dangé-Saint-Romain, France).
Isolation of DNA and recombinant DNA techniques
Plasmids were isolated using GeneJET Plasmid Miniprep Kit (Fermentas, Vilnius, Lithuania). Standard procedures (21) were used for generation of recombinant plasmid. The QIAquick Gel Extraction Kit (Qiagen, Chatsworth, CA, USA) was used for isolation of DNA fragments from agarose gels. DNA sequencing was carried out on an ABI PRISM 377 sequencer by using The BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). All enzymes used for DNA manipulations and corresponding buffers were obtained from Fermentas.
Construction of plasmids
For construction of plasmids used in the transformation assay, the pUC18 vector was cut with EcoRI and dephosphorylated with FastAPTM alkaline phosphatase, oligoduplexes (Supplementary Table S1) containing sticky EcoRI ends were assembled by annealing complementary oligonucleotides (Metabion, Martinsried, Germany), phosphorylated with T4 polynucleotide kinase and ligated using T4 ligase.
Plasmid transformation
Transformations were performed using the CaCl2 heat-shock procedure (21). Escherichia coli RR1 strain, carrying pCRISPR3 plasmid or its derivatives (see sections below), was used as a recipient strain. Cells were grown in LB medium at 37°C until OD600 = 0.4. One milliliter aliquot of bacterial culture was used for each transformation experiment which was performed at 0°C. Cells were recovered by centrifugation, washed using 0.5 ml of ‘Na solution’ [5 mM Tris–HCl (pH 8.0), 100 mM NaCl and 5 mM MgCl2) and resuspended into 0.5 ml ‘Ca solution’ [5 mM Tris–HCl (pH 8.0), 100 mM CaCl2, 5 mM MgCl2] and incubated for 20 min. After incubation, cells were centrifugated, and pellets were resuspended into 50 μl ‘Ca solution’ at 0°C. One nanogram of plasmid DNA was added to competent E. coli cells, incubated for 20 min at 4°C followed by 2 min incubation at 42°C. Then 450 μl of LB medium were added to the transformation mix and incubated at 37°C for 1 h. Finally, transformants were plated on LB agar with appropriate antibiotics. All transformation experiments were repeated at least three times. Bars in the graphs are presented as mean values from three or more independent experiments ± 1 SD.
Frameshift mutations/deletions of cas genes
To inactivate cas genes, frameshift mutations or small deletions were created. The pCas9(–) plasmid was constructed by cutting pCRISPR3 with Eco105I–Bpu1102I. The inactivation of cas1, cas2 and csn2 genes was performed in two steps. First, the blunt-ended Eco31I–NheI DNA fragment from pCRISPR3, which encodes all three genes, was subcloned into HindIII-EcoRI pre-cleaved and blunt-ended pUC18 vector. The resulting plasmid pUC-CRISPR3del was used for subsequent inactivation of cas genes. For cas1 inactivation, pUC-CRISPR3del was cleaved with Eco72I–Eco47III and re-ligated with T4 DNA ligase to generate the recombinant pUCΔcas1 plasmid which has a 41-bp deletion. For cas2 gene inactivation, pUC-CRISPR3del was cleaved with XagI, blunt-ended and re-ligated to generate the recombinant plasmid pUCΔcas2 which has a 1-bp insertion leading to a frameshift mutation in cas2. For the csn2 gene inactivation, the pUC-CRISPR3 plasmid was cut with EcoRI, blunt-ended and re-ligated to generate the pUCΔcsn2 plasmid which has a 4-bp insertion in csn2 gene. Mutated genes in all three cases were transferred into the pCRISPR3 plasmid by subcloning BstXI–Eco147I DNA fragments from pUCΔcas1, pUCΔcas2, and pUCΔcsn2 plasmids. Recombinant plasmids were called pCas1(–), pCas2(–) and pCsn2(–), respectively. To obtain pCas9 plasmid lacking cas1, cas2 and csn2 genes, first the Eco47III–BcuI deletion in pUC-CRISPR3 was obtained and then the BstXI-Eco147I DNA fragment from pUCΔ[cas1−csn2] plasmid was subcloned into pCRISPR3 plasmid to yield pCas9 plasmid.
Site-directed mutagenesis
Megaprimer method (22) was used to obtain plasmids carrying a mutated cas9 gene (see ‘Materials and methods’ section in Supplementary Data for the details).
RESULTS
Cloning of the CRISPR3/Cas system in E. coli
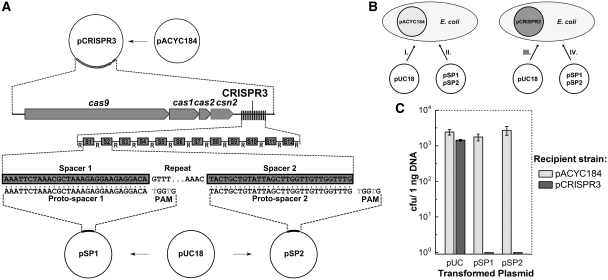
The S. thermophilus DGCC7710 CRISPR3/Cas locus (GenBank HQ712120) spans ~7.6 kb and consists of four cas genes: cas9, cas1, cas2 and csn2, followed by an A+T rich 382-bp leader sequence and an array of 13 repeat-spacer units (Figure 2A). In order to clone the CRISPR3/Cas locus in E. coli, we used three different primer pairs to generate three DNA fragments covering the ~7.6-kb CRISPR3/Cas locus (Supplementary Figure S1). These fragments were cloned separately into the pACYC184 plasmid, and reassembled into a full-length CRISPR3/Cas locus using two subcloning steps. In the final pCRISPR3 plasmid (Figure 2A), the ~7.6-kb fragment containing the natural S. thermophilus promoter and other possible regulatory elements located upstream of Cas9, is inserted 487 bp downstream of the constitutive tet gene promoter (Ptet). It remains to be established whether the components of the recombinant interference system are expressed from Ptet or from the native CRISPR/Cas promoters.
Figure 2.
CRISPR3/Cas system of S. thermophilus provides immunity against plasmid transformation in E. coli cells. (A) Schematic representation of CRISPR3/Cas system cloning and construction of the plasmids for interference assay. Streptococcus thermophilus CRISPR3/Cas system was cloned into E. coli plasmid pACYC184. Plasmids for interference assays were obtained by inserting a proto-spacer and PAM into pUC18 plasmid. (B) Schematic representation of the plasmid transformation interference assay. Escherichia coli RR1 recipient strains carrying plasmids pCRISPR3 and pACYC184 with and without the S. thermophilus CRISPR3/Cas system, respectively, were transformed with plasmids pSP1 and pSP2 carrying proto-spacers and PAMs or pUC18. (C) Interference of plasmid transformation by S. thermophilus CRISPR3/Cas system in E. coli cells. Transformation efficiency is expressed as cfu per nanogram of plasmid DNA (mean ± SD).
The CRISPR3/Cas system prevents plasmid transformation in E. coli
To test the functional activity of the CRISPR3 system in the heterologous E. coli host, we used a plasmid DNA transformation assay (see ‘Materials and methods’ section). Using pUC18, which is compatible with pACYC184 in E. coli, we engineered pSP1 and pSP2 (Figure 2), which contained proto-spacer sequences identical to spacers SP1 and SP2 in the CRISPR3 array, together with the corresponding PAM 5′-TGGTG-3′ downstream of the proto-spacer sequence (Supplementary Table S1), and tested the plasmid transformation efficiency in recipient E. coli cells carrying either pCRISPR3 or pACYC184 (Figure 2B). Typically, 1 ng of plasmid DNA per milliliter of E. coli bacterial culture in LB medium (OD600 = 0.4) was used for transformation. When the recipient strain carrying pACYC184 plasmid was transformed using pSP1, pSP2 or pUC18, the number of transformants exceeded 103 colony forming units (cfu) (Figure 2C). However, when the recipient strain carrying the pCRISPR3 plasmid was transformed with the same set of plasmids, no colony was obtained for the pSP1 and pSP2 plasmids (Figure 2C), but the control plasmid (pUC18), which lacks a proto-spacer, yielded 103 cfu. This is consistent with the presence of CRISPR-encoded specific immunity against spacers SP1 and SP2 in the pCRISPR3 plasmid. Interestingly, pUC18 contains 95 additional 5′-GGTG-3′ sequences corresponding to the PAM of the CRISPR3 system but no corresponding proto-spacer sequences. This is consistent with the necessity to have both a proto-spacer and an adjoining PAM in order for CRISPR-encoded immunity to occur. Altogether these results indicate that the heterologous plasmid pCRISPR3 interferes with transformation of pSP1 and pSP2 plasmids, both bearing corresponding proto-spacer and PAMs, but not with the control pUC18 plasmid. When a recipient strain carrying plasmid pACYC184 is used, no interference with plasmid transformation is observed.
To estimate the efficiency of plasmid transformation, we performed similar experiments using 100 ng of plasmids. Again, in the recipient strain carrying pCRISPR3, no colony was obtained but >105 cfu of transformants were obtained with the control vector (pUC18). The same number of transformants was obtained when the recipient strain containing pACYC184 was transformed with 100 ng of pSP1. This means that CRISPR3/Cas system in E. coli reduces the efficiency of plasmid transformation with more than five orders of magnitude. This is quantitatively comparable to the level of phage interference previously established in S. thermophilus, where CRISPR-based immunity reduces phage-sensitivity and efficiency of plaquing by four to five orders of magnitude per spacer (3,13). These results indicate that CRISPR/Cas systems can be transferred between distant bacterial genera and used heterologously to provide immunity against plasmids.
The CRISPR3/Cas system provides resistance against phage lambda in E. coli
In order to investigate the interference scope of the heterologous S. thermophilus CRISPR3/Cas system in E. coli, we assessed whether it could provide resistance against phages in addition to interference against plasmids. We inserted in vitro a spacer (anti-λvir) targeting a lambda phage sequence adjacent to the corresponding PAM (16) into the heterologous CRISPR3 locus on the pCRISPR3 plasmid (Supplementary Figure S4) and subsequently tested whether the engineered CRISPR system could confer resistance against phage lambda (see ‘Materials and methods’ section in Supplementary Data). Results showed that E. coli cells containing the CRISPR3/Cas system with the anti-λvir spacer were less sensitive to phage lambda by approximately three orders of magnitude, in comparison to the control cells that carried a wild-type CRISPR3 locus without the anti-λvir spacer (Supplementary Figure S5). These preliminary results indicate that the heterologous CRISPR3/Cas system can also provide immunity against phages in E. coli, and sets the stage for further studies that will assess the interference potential of heterologous CRISPR/Cas systems.
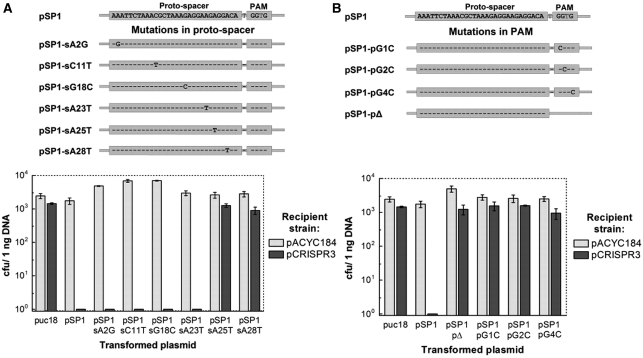
Position-dependent tolerance for mismatches in CRISPR-based plasmid interference
Phage challenge assays in S. thermophilus DGCC7710 have revealed that to ensure phage interference, 100% identity between the spacer sequence from the CRISPR array and the corresponding proto-spacer sequence in the phage DNA is required (3,13). To assess whether sequence identity between a spacer and the target proto-spacer sequence is important for the efficiency of plasmid transformation, we engineered plasmids pSP1-sA2G, pSP1-sC11T, pSP1-sG18C, pSP1-sA23T, pSP1-sA25T and pSP1-sA28T (Supplementary Table S1) which carried single mutations at distinct positions across the proto-spacer sequence (A2G, C11T, G18C, A23T, A25T and A28T, respectively), and analyzed transformation efficiency of the recipient strain containing pCRISPR3 (Figure 3A). Consistent with the phage challenge assay, single point mutations A25T and A28T in the proto-spacer abolished recipient strain ability to prevent plasmid transformation. Unexpectedly, single mismatches at the proto-spacer positions 2, 11, 18, 23 had no effect on the recipient strain ability to interfere with plasmid transformation (Figure 3A). Taken together our data suggest that CRISPR3/Cas system tolerates single nucleotide mismatches between spacer and proto-spacer at certain positions. In light of recent results indicating that cleavage of invading plasmid and phage DNA occurs 3-bp upstream of the PAM sequence in CRISPR1 (12), and given the relatedness of CRISPR1 and CRISPR3 systems, it is likely that the mutations at positions 25 and 28 impact cleavage, whereas distal mutations would not.
Figure 3.
Impact of proto-spacer and PAM mutations on CRISPR-encoded plasmid immunity. (A) Effect of mutations in the proto-spacer region on the plasmid transformation efficiency. Mutations are shown schematically above the figure. (B) Effect of mutations in the PAM region on the plasmid transformation efficiency.
The PAM sequence is important for plasmid interference
PAM sequences are of crucial importance for the phage interference by the CRISPR1 system of S. thermophilus (13,16). To test whether the PAM sequence is important in prevention of plasmid DNA transformation by the CRISPR3/Cas system, we engineered pSP1-pΔ plasmid so as to carry proto-spacer SP1 without its adjacent PAM sequence. Transformation efficiency of E. coli recipient cells with or without CRISPR3 system by the pSP1-pΔ plasmid was similar (Figure 3C). This indicates that the PAM sequence is required for CRISPR3 interference of plasmid transformation, and that the sole presence of a proto-spacer is not sufficient. To test whether all three conserved residues of the predicted PAM sequence 5′-NGGNG-3′ are equally important, we constructed plasmid variants pSP1-pG1C, pSP1-pG2C and pSP1-pG4C (Supplementary Table S1) were three guanine residues in the PAM sequence 5′-GGTG-3′ were replaced by cytosine residues. Transformation experiments revealed that E. coli cells containing pCRISPR3 or pACYCY184 plasmids were efficiently transformed (103 cfu) by the plasmids containing single mutations in the PAM region (Figure 3C). These data clearly demonstrate that while CRISPR3/Cas system tolerates single mutations at certain positions in the proto-spacer region, a conserved PAM sequence is required for the plasmid DNA interference in E. coli.
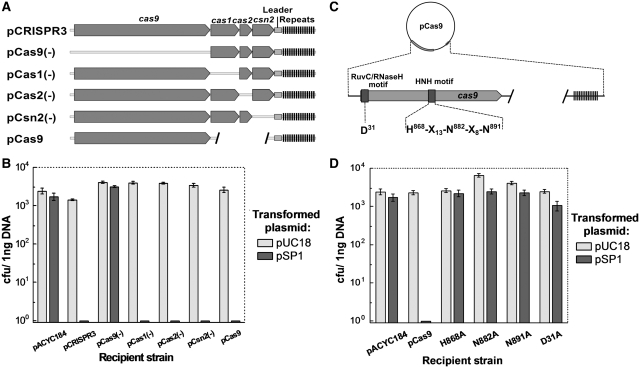
Only Cas9 is required for plasmid interference in E. coli
It was previously shown that inactivation of cas9 from the CRISPR1 system in S. thermophilus resulted in the loss of phage resistance, while inactivation of csn2 (cas7) did not alter phage resistance but impaired the ability to incorporate new spacers (3). To determine the role of individual Cas proteins in the interference step, we engineered frameshift mutants of individual cas genes, generating plasmids pCas9(–), pCas1(–), pCas2(–), pCsn2(–) (Figure 4A) and monitored the efficiency of the pSP1 plasmid transformation into corresponding recombinant E. coli recipient cells. Mutants lacking cas1, cas2 or csn2 retained the ability to interfere with plasmid transformation (Figure 4B). However, E. coli cells carrying pCas9(–) plasmid with mutated cas9, were efficiently transformed both by pSP1 and pUC18 plasmids. This is consistent with previous results indicating that cas9 and csn2 are involved in interference and spacer acquisition, respectively (3).
Figure 4.
Mutational analysis of cas genes. (A) Schematic representation of plasmids carrying mutant variants of cas genes. Individual genes were disrupted by frameshift mutations or small deletions. Three cas genes (cas1, cas2, csn2) were removed by deletion. (B) cas9 gene alone prevents plasmid DNA transformation. (C) Mutagenesis of Cas9 protein. Conserved domains and mutated amino acids are indicated. (D) Mutations in the conserved RuvC/RNaseH− and McrA/HNH domains inactivate cas9.
To further confirm that Cas9 alone can provide interference with plasmid transformation, we deleted the three other cas genes (cas1, cas2, csn2) in pCRISPR3 to generate a pCas9 plasmid which contains only cas9, the leader sequence, and a repeat-spacer region (Figure 4A). The recipient strain containing pCas9 plasmid retained the ability to interfere with pSP1 plasmid transformation (Figure 4B), indicating that Cas9 is the only Cas protein required to provide resistance against foreign DNA. This suggests that Cas1, Cas2 and Csn2 might rather be involved in novel spacer acquisition.
Mutational analysis of Cas9
Cas9 proteins are predicted to contain RuvC/RNaseH and McrA/HNH signature motifs (9). The HNH motif is characteristic of many nucleases that act on double-stranded DNA including colicins (23,24), restriction enzymes (25) and homing endonucleases (26). RuvC/RNaseH fold includes proteins that show wide spectra of nucleolytic functions, acting both on RNA and DNA (RNaseH, RuvC, DNA transposases and retroviral integrases, and PIWI domain of Argonaut proteins) (27). To test whether the conserved amino-acid residues D31 (RuvC/RNaseH motif) (Supplementary Figure S2), H865, N882 and N891 (McrA/HNH motif) (Supplementary Figure S3) are important for Cas9 function, alanine replacement mutants were constructed in the pCas9 plasmid by site-directed mutagenesis to generate recombinant plasmids pD31A, pH868A, pN882A and pN891A, respectively. Plasmid transformation assays revealed that all three mutations in the HNH motif and a single mutation in RuvC/RNaseH motif abolish the Cas9-dependent plasmid interference (Figure 4D). Accordingly, the amino-acid residues of the HNH motif and the conserved aspartate in the N-terminal part of protein play an essential role in the plasmid DNA interference by Cas9 in vivo.
DISCUSSION
The role of Cas proteins in the defense mechanism
CRISPR/Cas loci are highly diverse and fall within different categories depending on the type, number of cas genes and architecture of the cas operon (9,10). The diversity of CRISPR/Cas systems implies mechanistic differences which remain to be established. Meanwhile, two different pathways which target invading genetic elements via the crRNAs and act on DNA (6,7) or RNA (8) targets are emerging. It was shown recently that the S. thermophilus CRISPR1/Cas system specifically cleaves plasmid and bacteriophage double-stranded DNA within the proto-spacer in vivo, at specific sites (12). This endonuclease activity seems to require Cas9 but it remains to be established whether other Cas proteins of the CRISPR1/Cas system contribute to the cleavage. The CRISPR3/Cas system of S. thermophilus DGC7710 belongs to the same Type II (Nmeni) group, and contains a similar set of cas genes (Figure 1), suggesting that both systems may be mechanistically similar. We show here that CRISPR3/Cas module cloned into E. coli is functionally active and provides host cell with interference against plasmid and phage.
The S. thermophilus CRISPR3/Cas Cas9 is a large protein comprised of 1388 amino acid residues. In silico analysis identified a McrA/HNH-nuclease motif and a RuvC/RNaseH-like nuclease signature in the Cas9 protein sequence (9,10). We provide experimental evidence that the alanine replacement of the conserved D31 (RNaseH/RuvC-motif), and H865, N882 and N891 (HNH-motif) residues abolishes Cas9-mediated plasmid interference in E. coli. Since HNH-domains are often found in nucleases that act on double-stranded DNA, we suggest that the HNH-domain of Cas9 is involved in the DNA degradation step. In the CRISPR1/Cas system, cleavage of target DNA occurs at both DNA strands within the proto-spacer sequence (12). If Cas9 of the CRISPR3/Cas system is a monomer in solution and contains a single HNH-motif, it has to dimerise or employ a second active site to cleave both DNA strands. It is possible that HNH- and RuvC/RNaseH-like catalytic sites may act on different DNA strands to achieve a double strand break; a similar strategy is exploited by some restriction endonucleases (28). The RuvC-like nuclease domain is identified in proteins that act on both DNA and RNA, including RNaseH, RuvC, and PIWI domain of Argonaut proteins (27). Therefore, we cannot exclude that the N-terminal RNaseH/RuvC domain in Cas9 might be involved in crRNA maturation. Studies of Cas9 nuclease and ribonuclease functions both in vivo and in vitro are currently on-going.
Our studies of the S. thermophilus CRISPR3/Cas system demonstrate that different CRISPR/Cas systems follow different strategies to achieve cleavage of invading DNA. Indeed, in contrast to a single S. thermophilus Cas9 protein which is potentially involved in both crRNA maturation and DNA cleavage steps, in E. coli a large nucleic acid–protein complex which include crRNA, Cascade and Cas3 is thought to be involved in the degradation of foreign DNA (6).
‘Vaccination’ of E. coli against plasmids and phages by the heterologous CRISPR3/Cas system
There is strong evidence suggesting that CRISPR/Cas systems can move between distinct species in Bacteria and Archaea via horizontal gene transfer (29–31). First, CRISPR/Cas content of closely related species or strains might differ, while on the other hand, evolutionary distant species (even across Bacteria and Archaea) might harbor similar CRISPR systems (10,29,32). For example, genome sequencing of the three closely related S. thermophilus strains revealed that CNRZ1066 and LMG18311 possess two CRISPR/Cas systems (CRISPR1/Cas and CRISPR2/Cas) (16), whereas the LMD-9 strain has an additional Type II system, CRISPR3/Cas (16). Furthermore, S. thermophilus DGCC7710 contains a fourth CRISPR/Cas system (Figure 1) which belongs to Type I (14). It is likely that in S. thermophilus the CRISPR4/Cas system has been acquired recently by horizontal gene transfer, especially since this species is naturally competent for transformation (33). Secondly, there is often a codon bias and/or a marked GC-content difference between CRISPR/Cas loci and the rest of the chromosome (30). In addition, mobile genetic elements such as insertion sequences are usually located in the vicinity of CRISPR/Cas systems, while some CRISPR/Cas systems are located on large plasmids (>40 kb) (29) and even prophages (34).
Here, we provide the first experimental evidence showing that the CRISPR3/Cas system of the Gram-positive S. thermophilus species can be cloned into a plasmid and transferred to a Gram-negative E. coli host. Furthermore, we show that the heterologous system provides resistance against incoming plasmids and phages that carry matching proto-spacer sequences and PAMs. This finding illustrates that CRISPR/Cas systems may function as mobile gene cassettes that overcome barriers between distant species such as incompatibility of promoters and other regulation signals. This successful transfer of a functional CRISPR/Cas system into a phylogenetically distant host opens novel possibilities for practical applications, notably the transfer of active CRISPR/Cas systems between species in order to ‘vaccinate’ bacteria against viruses or plasmids. In light of recent results indicating that CRISPR-encoded immunity can target antibiotic resistance genes (12), there is great interest in transferring active CRISPR/Cas systems in order to strengthen the immunity of select species or strains against the uptake and dissemination of antibiotic resistance genes.
ACCESSION NUMBER
Accession Number: GenBank HQ712120.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
FUNDING
Programme Gilibert between the Lithuanian Ministry of Education and Science and by French Ministry of Foreign and European affairs (MAEE); French Ministry of Higher education and Research (MESR); grant #R100 (3rd priority Strengthening of Capacities of Researchers and Scientists of the 2007–2013 Operational Programme for Human Resources Development implemented by the Research Council of Lithuania). Funding for open access charge: Danisco.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to acknowledge Dr Mindaugas Zaremba and Dr Giedrius Sasnauskas for helpful discussions and comments on the article.
REFERENCES
- 1.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 2.Sturino JM, Klaenhammer TR. Engineered bacteriophage-defence systems in bioprocessing. Nat. Rev. Microbiol. 2006;4:395–404. doi: 10.1038/nrmicro1393. [DOI] [PubMed] [Google Scholar]
- 3.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 4.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 5.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 6.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pul U, Wurm R, Arslan Z, Geissen R, Hofmann N, Wagner R. Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol. Microbiol. 2010;75:1495–1512. doi: 10.1111/j.1365-2958.2010.07073.x. [DOI] [PubMed] [Google Scholar]
- 12.Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadàn AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 13.Deveau H, Barrangou R, Garneau JE, Labonté J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 15.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horvath P, Romero DA, Coûté-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 18.Bolivar F, Rodriguez RL, Greene PJ, Betlach MC, Heyneker HL, Boyer HW, Crosa JH, Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 19.Chang AC, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch EF, Maniatis TM. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
- 22.Barik S. Site-directed mutagenesis in vitro by megaprimer PCR. Methods Mol. Biol. 1996;57:203–215. doi: 10.1385/0-89603-332-5:203. [DOI] [PubMed] [Google Scholar]
- 23.Kleanthous C, Kuhlmann UC, Pommer AJ, Ferguson N, Radford SE, Moore GR, James R, Hemmings AM. Structural and mechanistic basis of immunity toward endonuclease colicins. Nat. Struct. Biol. 1999;6:243–252. doi: 10.1038/6683. [DOI] [PubMed] [Google Scholar]
- 24.Ko TP, Liao CC, Ku WY, Chak KF, Yuan HS. The crystal structure of the DNase domain of colicin E7 in complex with its inhibitor Im7 protein. Structure. 1999;7:91–102. doi: 10.1016/s0969-2126(99)80012-4. [DOI] [PubMed] [Google Scholar]
- 25.Saravanan M, Bujnicki JM, Cymerman IA, Rao DN, Nagaraja V. Type II restriction endonuclease R.KpnI is a member of the HNH nuclease superfamily. Nucleic Acids Res. 2004;32:6129–6135. doi: 10.1093/nar/gkh951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen BW, Landthaler M, Shub DA, Stoddard BL. DNA binding and cleavage by the HNH homing endonuclease I-HmuI. J. Mol. Biol. 2004;342:43–56. doi: 10.1016/j.jmb.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 27.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 28.Armalyte E, Bujnicki JM, Giedriene J, Gasiunas G, Kosinski J, Lubys A. Mva1269I: a monomeric type IIS restriction endonuclease from Micrococcus varians with two EcoRI- and FokI-like catalytic domains. J. Biol, Chem. 2005;280:41584–41594. doi: 10.1074/jbc.M506775200. [DOI] [PubMed] [Google Scholar]
- 29.Godde JS, Bickerton A. The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J. Mol. Evol. 2006;62:718–729. doi: 10.1007/s00239-005-0223-z. [DOI] [PubMed] [Google Scholar]
- 30.Horvath P, Coûté-Monvoisin AC, Romero DA, Boyaval P, Fremaux C, Barrangou R. Comparative analysis of CRISPR loci in lactic acid bacteria genomes. Int. J. Food Microbiol. 2009;131:62–70. doi: 10.1016/j.ijfoodmicro.2008.05.030. [DOI] [PubMed] [Google Scholar]
- 31.Portillo MC, Gonzalez JM. CRISPR elements in the Thermococcales: evidence for associated horizontal gene transfer in Pyrococcus furiosus. J. Appl. Genet. 2009;50:421–430. doi: 10.1007/BF03195703. [DOI] [PubMed] [Google Scholar]
- 32.Makarova KS, Aravind L, Grishin NV, Rogozin IB, Koonin EV. A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002;30:482–496. doi: 10.1093/nar/30.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fontaine L, Dandoy D, Boutry C, Delplace B, de Frahan MH, Fremaux C, Horvath P, Boyaval P, Hols P. Development of a versatile procedure based on natural transformation for marker-free targeted genetic modification in Streptococcus thermophilus. Appl. Environ. Microbiol. 2010;76:7870–7877. doi: 10.1128/AEM.01671-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeno-Tarraga AM, Wang H, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 2006;38:779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.