Abstract
In Crenarchaea, several tRNA genes are predicted to express precursor-tRNAs (pre-tRNAs) with canonical or non-canonical introns at various positions. We initially focused on the tRNAThr species of hyperthermophilic crenarchaeon, Aeropyrum pernix (APE) and found that in the living APE cells three tRNAThr species were transcribed and subsequently matured to functional tRNAs. During maturation, introns in two of them were cleaved from standard and non-standard positions. Biochemical studies revealed that the APE splicing endonuclease (APE-EndA) removed both types of introns, including the non-canonical introns, without any nucleotide modification. To clarify the underlying reasons for broad substrate specificity of APE-EndA, we determined the crystal structure of wild-type APE-EndA and subsequently compared its structure with that of Archaeaoglobus fulgidus (AFU)-EndA, which has narrow substrate specificity. Remarkably, structural comparison revealed that APE-EndA possesses a Crenarchaea specific loop (CSL). Introduction of CSL into AFU-EndA enhanced its intron-cleaving activity irrespective of the position or motif of the intron. Thus, our biochemical and crystallographic analyses of the chimera-EndA demonstrated that the CSL is responsible for the broad substrate specificity of APE-EndA. Furthermore, mutagenesis studies revealed that Lys44 in CSL functions as the RNA recognition site.
INTRODUCTION
RNA splicing, which removes introns and joins exons in a primary transcript, is essential for the maturation of functional RNA. The introns in eukaryotic cytoplasmic and archaeal precursor (pre)-tRNA are removed by an RNA-splicing endonuclease (EndA) (1–3). In eukaryotic pre-tRNA, the introns are included in a typical motif, called the bulge–helix–loop (BHL) motif, and are predominantly located between the nucleotide positions 37 and 38 in the anticodon loop of the tRNA. In contrast, the introns in archaeal pre-tRNAs show several variations according to the classification of archaea (4). The intron in the bulge–helix–bulge (BHB) motif is commonly found in the pre-tRNA of all phyla of archaea and is located not only in the anticodon loop but also in various other positions including the D- and T-loops, variable region and aminoacyl stem (4). Pre-tRNAs from Crenarchaea and Nanoarchaea often have non-canonical introns with BHL or HBh’ motifs as well as canonical introns with BHB motifs (4,5). Therefore, the EndAs from Crenarchaea and Nanoarhcaea should have the ability to remove both canonical and non-canonical introns from various positions of the pre-tRNA. The Crenarchaeal and Nanoarhcaeal EndAs form heterotetramer with α2- and β2-subunits, which is in contrast to the Euryarchaeal EndAs that form homodimer (α2) or homotetramer (α4) and only cleave the canonical BHB intron at standard position (6–12). The α-subunit in the archaeal EndAs is a catalytic subunit and shares homology with the catalytic Sen2- and Sen34-subunits of eukaryotic EndA, implying an evolutionary relationship between the eukaryotic and archaeal EndAs (13,14). Although the catalytic and RNA recognition mechanisms of the α2 type EndA from the hyper-thermophilic Euryarchaeon Archaeaoglobus fulgidus (AFU) have already been described (3,15), the molecular mechanism underlying the broad substrate specificity of the α2β2 EndA remains yet to be elucidated.
Intron-containing tRNA genes found in the Crenarchaea genomes are generally much more than those found in the Euryarchaea genomes (16). The numbers, sizes, motifs and locations of introns varied according to the Crenarchaea species. In the genome of Aeropyrum pernix (APE), an aerobic hyper-thermophilic Crenarchaeota, presence of 14 intron-containing tRNA genes have been predicted from the complete genome sequence determination (17), RT–PCR analysis (18) and bioinformatics studies of the archaeal tRNA-gene database (SPLITSdb) (16). Thus, the introns must be cleaved by APE-EndA for these 14 tRNA species to mature. In the current study, we focused on the removal of introns from pre-tRNA species by APE-EndA and clarified that the Crenarchaea specific loop is responsible for the broad substrate specificity of APE-EndA.
MATERIALS AND METHODS
Strain and growth media
The APE was cultured in the nutrient rich media as previously reported (19). The cells were harvested at the exponential growth phase.
Preparation of total RNA
The APE cells (wet weight, 10 g) were suspended in 25 ml of TE buffer [10 mM Tris–HCl (pH 8.0) and 1 mM EDTA] and then mixed with 25 ml of TE buffer-saturated phenol. The mixture was centrifuged at 15 000g for 15 min at room temperature. The aqueous phase was collected and then mixed with an equal volume of TE buffer-saturated phenol. This manipulation was repeated until the precipitant in the interface between the aqueous and organic phases disappeared. The RNA was recovered from the aqueous phase by ethanol precipitation. The RNA was loaded onto a Q-Sepharose column (GE Healthcare) equilibrated with buffer A [20 mM Tris–HCl (pH 7.6), 10 mM MgCl2] containing 400 mM NaCl2. Small RNA molecules (mainly tRNA) was eluted with buffer A containing 550 mM NaCl2 and recovered by ethanol precipitation.
Northern blot analysis and RT–PCR analysis
To identify the tRNAThr species, we performed northern blot analysis. The small RNA fraction (1.0 A260 unit) was transferred to a Hybond-N+ membrane (GE Healthcare) by electro blotting, and fixed by UV254 nm irradiation. Northern hybridization was performed with the hybridization buffer (GE Healthcare) using a 5′-32P-labeled DNA probe at 48°C over-night. Nucleotide sequences of the DNA probes used in this study are as follows: APE-tRNAThr1 (CGU), 5′-CGA GGC CGG CGC TCT ACC GCT GAG GTA CGG CGG C-3′; APE- tRNAThr2(UGU), 5′-CCT GCT GAG CTA CGG CGG C-3′; APE-tRNAThr3(UGU), 5′-CAG CTT AGC TAC TGC GTG-3′; and APE-tRNAThr4(GGU), 5′-CCA GGG CGG CGC TCT GCC TG-3′. The hybridized bands were monitored with a Fuji Photo Film BAS2000 imaging analyzer. The small RNA fraction was concentrated by agarose gel electrophoresis. For the RT–PCR analysis, total RNA (0.8 A260 unit) was separated by 10% PAGE/7 M urea. The gel between 5.8 S rRNA and tRNA were sliced into four pieces according to the sizes of RNAs and then the RNAs were extracted from the gels. The RNA fractions were treated with DNase I for complete removal of genomic DNA. Each RNA fraction was tested whether the RT–PCR product was amplified or not. After these pilot experiments, the RT–PCR analyses were performed using the RNA fractions containing pre-tRNA or mature tRNA.
Isolation of tRNAThr species from the total RNA
We tried to isolate the A. pernix tRNAThr1(CGU), A. pernix tRNAThr2(UGU), A. pernix tRNAThr3(UGU) and A. pernix tRNAThr4(GGU) from the total RNA using the solid-phase DNA probe column chromatography method (20,21). The following 5′-biotinylated DNA probes were used for this purpose: APE-Thr1, 5′-TCG TCC GTC TCG CGG CCG GAA CA-3′; APE-Thr2, 5′-TCG CCA TCT CGC GGC CGG AGCA-3′; APE-Thr3, 5′-TCG ACT CTG TCG CGG CGG GAA CA-3′; and APE-Thr4, 5′-TCG ACC GTC TCG CGG CGG GAC CA-3′. The isolated tRNAs were analyzed by 10% PAGE/7 M urea and used in the threonine charging activity assay.
Aminoacylation of purified tRNAThr species
The threonyl-tRNA synthetase (Thr-RS) fraction was prepared as follows. Briefly, the S-100 fraction (20 ml) of the wild-type strain was loaded onto a DE52 column (column volume, 10 ml) and Thr-RS was eluted off the column by using a KCl linear gradient (50–350 mM). Fractions containing Thr-RS were identified by monitoring their threonine charging activity. The aminoacylation assay was performed using l-[14C(U)]-threonine (5.55 GBq/mmol, American Radiolabeled Chemicals) using 0.03 A260 units of isolated tRNAThr as described in the reference (22).
Protein expression and purification
The APE and AFU genomic DNA (NBRC number, 100138G and 100126G) were supplied by the National Institute of Technology and Evaluation Biological Resource Center (Kisarazu, Japan). The genes encoding the α- and β-subunits of the APE-EndA were individually cloned into the NdeI and BamHI sites of the vector pET-21 a (Novagen). The resultant plasmid harboring the β-subunit gene was digested with the restriction enzymes BglII and XhoI. The DNA fragment, containing the T7 promoter, ribosome-binding site and the β-subunit gene (in order), was cloned between the BamHI and XhoI sites of the pET-21 a harboring the α-subunit gene to obtain a plasmid co-expressing both subunits of APE-EndA. The gene encoding the AFU-EndA was cloned into the NdeI and BamHI sites of the vector pET-30 a (Novagen). These plasmid constructs were used for overexpressing recombinant APE-EndA and AFU-EndA in Escherichia coli Rosetta 2(DE3) strain (Novagen). Escherichia coli cells harboring a plasmid were grown in LB media supplemented with either 100 μg/ml of ampicillin or 50 μg/ml of kanamycin. The cells were suspended in 10 ml buffer containing 50 mM Tris–HCl (pH 7.5), 5 mM MgCl2, 6 mM 2-mercaptoethanol and 50 mM KCl, and then disrupted with an ultrasonic disruptor (model UD-200, Tomy, Japan). Most of the E. coli proteins were denatured by heat treatment at 70°C for 30 min and removed by centrifugation. Recombinant proteins were further purified by two consecutive chromatography including HiTrap Heparin-Sepharose and HiLoad 16/60 Superdex 75 pg columns (GE Healthcare). The mutant genes were generated using the QuickChange site-directed mutagenesis kit (Stratagene), and the mutants were verified by DNA sequencing. The mutant proteins were expressed and purified by following the same procedures used for the wild-type protein.
Intron-cleavage assay by the splicing endonuclease
The transcripts of A. pernix pre-tRNAThr 1(CGU), A. pernix pre-tRNAThr2 (UGU), BHB mini helix of A. pernix pre-tRNAThr2 (UGU) and BHL mini helix of C. symbiosum tRNATyr were prepared using T7 RNA polymerase as described in our previous report (23). In the case of internal labeling with radioisotope, [α-32P]-GTP was added to the transcription mixture. The transcripts were purified on 10% PAGE/7 M urea. Splicing reactions were performed as follows. An amount of 0.2 μg EndA was mixed with 0.5 nmol unlabeled and 32P-labeled (~30 000 dpm) transcript in 100 μl buffer [50 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 6 mM 2-mercaptoethanol, 50 mM KCl] and incubated at 65°C. Aliquots (10 µl) were taken out at 0, 2, 5, 10, 20 and 30 min, and were analyzed by 15% PAGE/7 M urea. Radioactivity on gel was monitored by autoradiography using a Fuji Photo Film BAS2000 imaging analyzer.
To identify the cleavage sites, we further performed northern blot analysis and 5′-end nucleotide analysis. The transcripts were prepared with non-radioisotope labeled nucleotides. After the cleavage reaction, the RNA fragments were separated by 10% PAGE/7 M urea, followed by northern blotting as described above. The sequences of the DNA probes used are as follows: 5′ exon for the A. pernix pre-tRNAThr1 and pre-tRNAThr2, 5′-GCT GAG CTA CGG CGG-3′; 3′ exon for the A. pernix pre-tRNAThr1 and pre-tRNAThr2, 5′-CGC CGG CGG GAT TC-3′; intron for the A. pernix pre-tRNAThr1, 5′-CTC GGA GCT AGC CCG-3′; intron for the A. pernix pre-tRNAThr2, 5′-AGG CCC CGC GCA GG-3′. The hybridized bands were monitored with a Fuji Photo Film BAS2000 imaging analyzer.
In order to determine the 5′-end nucleotide of each fragment, the RNA fragments were excised, labeled with γ-32P-ATP and T4 polynucleotide kinase (New England Biolabs), and then completely digested with nuclease P1 (Wako Pure Chemicals). The resultant nucleotides were separated by the 2D thin layer chromatography using the following solvent system (24): first dimension, isobutylic acid: conc. ammonia: water, 66:1:33, v/v/v; second dimension, isopropyl alcohol: HCl: water, 70:15:15, v/v/v. The 32P-labeled nucleotides were monitored with a Fuji Photo Film BAS2000 imaging analyzer. The standard nucleotides were monitored with UV254 nm irradiation.
Crystallization
Peak enzyme activity containing fractions from the Superdex-75 gel filtration column were pooled and then concentrated to ~10 mg/ml using Amicon Ultra-15 centrifugal filter units. Initial trials for crystallization of APE-EndA and AFU-CSL were performed by hanging-drop vapor diffusion method using the Crystal Screening Kit (Hampton Research). The drop solution was equilibrated against 500 μl of reservoir solution at 22°C. A few crystals were obtained under some test crystallization conditions containing ammonium sulfate as the precipitant. Based on the initial crystallization conditions, the APE-EndA protein solution was mixed with an equal volume of the crystallization solution that contained 0.25 M ammonium sulfate, 0.1 M sodium citrate (pH 5.6), 0.9 M lithium sulfate and 1 mM MgCl2, and the AFU-CSL protein solution was mixed with an equal volume of the crystallization solution that contained 2.2 M ammonium sulfate, 0.2 M potassium sodium tartrate tetrahydrate and 0.1 M sodium citrate (pH 5.6). In the case of APE-EndA, it took only 1 day to obtain a full size cubic-shape (250 × 150 × 100 µm) crystal. In the case of the AFU-CSL, it took a few days to obtain a full size, rectangular-shape (300 × 50 × 50 µm) crystal. Cryo-protection of each crystal was achieved by stepwise transfer to the respective artificial mother liquor containing 20% glycerol. Crystals were then flash-frozen in liquid nitrogen.
Data collection and structure determination
X-ray diffraction data were collected at 100 K on the BL38B1 and BL41XU beamlines at SPring-8 (Hyogo, Japan). Data reduction was performed using the HKL2000 program (25). Structures of APE-EndA and AFU-CSL were determined by molecular replacement using the mutant APE H133A and AFU-EndA (PDB ID code 1RLV) coordinates, respectively, as search models. Molecular replacement was performed using the Phaser program (26). The resulting maps were used for manually building the models using COOT (27). The model was further refined by using CNS (28) and PHENIX (29). The structure of APE-EndA was refined to Rwork/Rfree of 26.0%/31.8% at 2.8 Å resolution (Table 1). The space group of the crystal belonged to P31, where three α2β2 APE-EndA molecules were present in an asymmetric unit and they were structurally almost identical. The final model contained residues 3–170 (chains A, E and I), 7–186 (chains B, F and L), 10–170 (chains C, G and K), 3–186 (chains D, H and L) and 408 water molecules. The structure of AFU-CSL was refined to Rwork/Rfree of 24.7%/28.7% at 2.05 Å resolution (Table 1). The crystal belonged to the space group P212121 with four molecules in the asymmetric unit. The final model contained residues 2–315 (chains A–D) and 302 water molecules. The final models of APE-EndA and AFU-CSL structures were further checked using PROCHECK (30), showing the quality of the refined model. Ramachandran plots (%) of the APE-EndA and AFU-CSL structures were tabulated in Table 1. All structural figures were generated by PyMOL (DeLano Scientific, Palo Alto, CA, USA) and electrostatic potential surface models were calculated by using APBS (31).
Table 1.
Data collection and refinement statistics
| APE-EndA | AFU-CSL | |
|---|---|---|
| Data collection | ||
| Space group | P31 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 135.03, 135.03, 156.24 | 81.77, 104.64, 165,30 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 90 |
| Resolution (Å) | 50 to 2.8 (2.91–2.80) | 50 to 2.05 (2.12–2.05) |
| Rmergea | 6.2 (60.2) | 9.5 (67.3) |
| I/σI | 26.6 (1.8) | 12.8 (3.2) |
| Completeness (%) | 99.8 (99.1) | 99.7 (97.1) |
| Redundancy | 6.9 (5.6) | 14.1 (10.8) |
| Refinement | ||
| Resolution (Å) | 41.2–2.8 | 50–2.05 |
| No. reflections | 75 771 | 84 124 |
| Rworkb/ Rfreec | 26.0/31.8 | 24.7/28.7 |
| No. of atoms | 16 581 | 10 722 |
| Protein | 16 173 | 10 420 |
| Water | 408 | 302 |
| Avg. B-factors (Å2) | 50.5 | 36.4 |
| RMSDs | ||
| Bond lengths (Å) | 0.009 | 0.006 |
| Bond angles (°) | 1.3 | 1.4 |
| Ramachandran plot (%) | ||
| Most favored | 88.0 | 89.0 |
| Additional allowed | 11.5 | 9.2 |
| Generously allowed | 0.5 | 1.7 |
| Disallowed | 0.0 | 0.1 |
The value in the parentheses is for the highest resolution shell.
aRmerge = ΣΣj|<I(h)> – I(h)j|/ΣΣj|<I(h)>|, where <I(h)> is the mean intensity of symmetry-equivalent reflections.
bRwork = Σ (IIFp(obs) – Fp(calc)II)/ΣIFp(obs)I.
cRfree = R factor for a selected subset (10%) of reflections that was not included in earlier refinement calculations.
RESULTS
Four putative tRNAThr genes in the APE genome
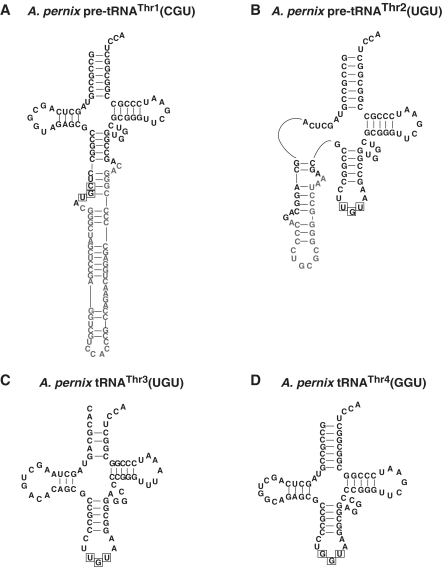
The complete APE genome sequence analysis has revealed that out of the 47 putative tRNA genes, 14 genes are expected to include an intron (17). Of these tRNA (and tRNA-like) genes, we were interested in four tRNAThr genes. Figure 1 shows the predicted pre-tRNAs transcribed from these putative tRNAThr genes. The numbering of the tRNA genes is according to that of the genome sequence analysis (17). The pre-tRNAThr1(CGU) has an intron at the standard position in the anticodon loop, suggesting that this pre-tRNA normally matures via the splicing process. Because the pre-tRNAThr4(GGU) does not have an intron, this tRNA probably matures without the splicing process. Four threonine codons (ACU, ACC, ACG and ACA) can be decoded only by these two tRNA species assuming that there was no modification of anticodons. Consistent with this idea, Haloferax volcanii (classical name, Halobacterium volcanii) has only two tRNAThr species, tRNAThr1(GGU) and tRNAThr2(CGU) (32). Additionally, in the case of APE, two curious tRNAThr genes, tRNAThr2(UGU) and tRNAThr3(UGU) genes, have been reported (17). The pre-tRNAThr2(UGU) has an intron at the non-standard position (D-loop) of the tRNA, whereas the pre-tRNAThr3(UGU) has a disrupted aminoacyl-stem.
Figure 1.
Predicted secondary structures of the tRNAThr species. The predicted secondary structures of (A) pre-tRNAThr1(CGU), (B) pre-tRNAThr2(UGU), (C) tRNAThr3(UGU), and (D) tRNAThr4(GGU). The predicted introns and anticodons are indicated in gray–white and boxed, respectively.
Presence of three tRNAThr species in the living APE cells
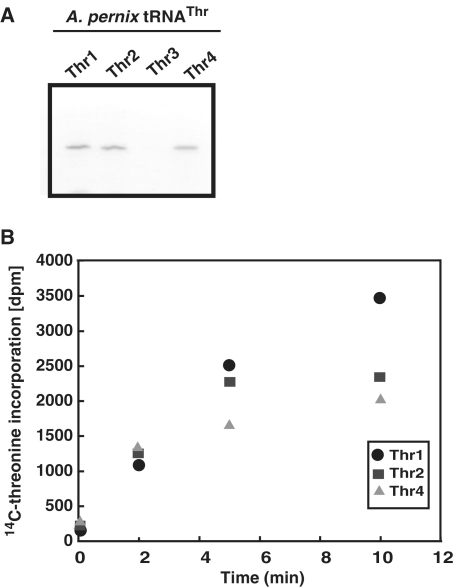
Initially, we examined whether all four of these pre-tRNAThr species were transcribed in the living APE cells. For this purpose, we prepared total RNA from APE cells and subsequently performed northern blot analysis (data not shown). However, because the amount of pre-tRNAs in the total RNA fraction was very small, the pre-tRNA bands were difficult to visualize. Only three mature tRNAThr species [tRNAThr1(CGU), tRNAThr2(UGU) and tRNAThr4(GGU)] were detected (Supplementary Figure S1), suggesting that two pre-tRNAThr species [pre-tRNAThr1(CGU) and pre-tRNAThr2(UGU)] containing an intron are indeed transcribed in the cells. To overcome this problem, we first concentrated the pre-tRNA fractions by agarose gel electrophoresis as described in the ‘Materials and Methods’ section, and then performed the RT–PCR analysis. During the course of this study, Yamazaki et al. (18) reported the existence of three tRNAThr species [tRNAThr1(CGU), tRNAThr2(UGU) and tRNAThr4(GGU)] in APE cells by RT–PCR analysis. Although we independently detected the pre-tRNAThr1(CGU) and pre-tRNAThr2(UGU) species by the RT–PCR analysis, our results coincided with their results. Therefore, we do not present our data in this report. Thus, existences of three tRNAThr species [tRNAThr1(CGU), tRNAThr2(UGU) and tRNAThr4(GGU)] were strongly suggested. In contrast, any band corresponding to the size of tRNAThr3(UGU) was not found on the gel (Supplementary Figure S1). Thus, these results suggested that at least three of the four tRNAThr species [tRNAThr1(CGU), tRNAThr2(UGU) and tRNAThr4(GGU)] are indeed transcribed in the living APE cells. To further confirm the existence of these tRNAThr species in the cells, we attempted to purify all four tRNA species by using the solid-phase DNA probe affinity column chromatography (20,21). As shown in Figure 2A, only three mature tRNA species [tRNAThr1(CGU), tRNAThr2(UGU) and tRNAThr4(GGU)] could be purified from the APE total RNA. However, tRNAThr3(UGU) was not purified. These results are consistent with the results of the northern blot analysis and RT–PCR analysis. Next, we prepared the threonyl-tRNA synthetase fraction from the APE cell extract, and tested the threonine charging activity of the purified tRNAs. As shown in Figure 2B, all three purified tRNAs had threonine charging activity. Based on these experimental results, we concluded that these three tRNA genes [i.e. tRNAThr1(CGU), tRNAThr2(UGU) and tRNAThr4(GGU)] are indeed transcribed in APE cells and their transcripts mature to functional tRNAThr species. Our results also suggested that the tRNAThr3(UGU) gene is a pseudogene. It should be mentioned that our RT–PCR analysis showed existence of the antisense RNA of tRNAThr3(UGU) gene region. This result reinforces the idea: the tRNAThr3(UGU) gene is a pseudogene.
Figure 2.
Isolation of tRNAThr species and threonine charging activity. (A) tRNA species were isolated by solid-phase DNA probe column chromatography as described in Materials and methods section. The purified tRNA species were analyzed by 15% PAGE/7 M urea. The gel was stained with methylene blue. The tRNAThr3(UGU) was not obtained by the solid-phase DNA probe column chromatography. (B) Threonine charging activities of the isolated tRNAs were measured as described in ‘Materials and Methods’ section. Abbreviations used are as follows: Thr1, tRNAThr1(CGU); Thr2, tRNAThr2(UGU); Thr3, tRNAThr3(UGU); and Thr4, tRNAThr4(GGU).
APE-EndA has broad substrate specificity
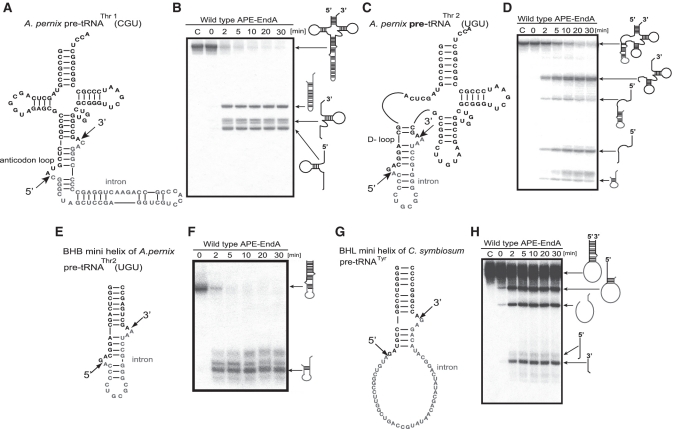
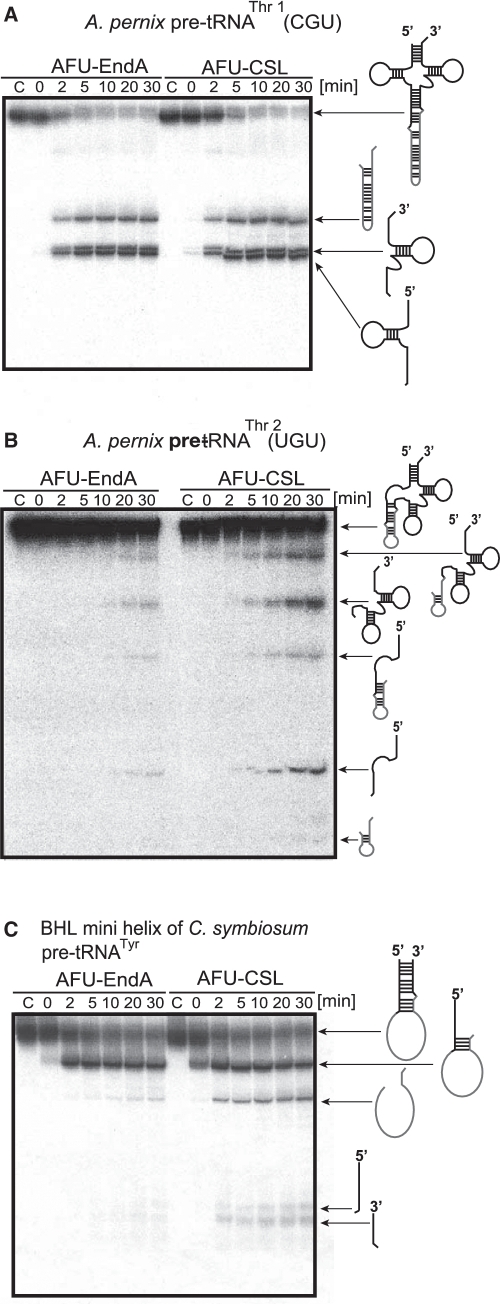
Maturation of pre-tRNAThr1(CGU) and pre-tRNAThr2(UGU) involved removal of introns from different positions (i.e. standard position in the anticodon loop and non-standard position in the D-loop) of the tRNAs. This raised a question whether APE-EndA could cleave these introns efficiently. To test this idea, we constructed an expression system to produce recombinant APE-EndA in E. coli. The recombinant APE-EndA was purified to homogeneity as judged by SDS–polyacrylamide gel electrophoresis (Supplementary Figure S2). The intron-cleavage activity of the APE-EndA was examined using pre-tRNAThr1(CGU) and pre-tRNAThr2(UGU) transcripts (Figure 3). Because the RNA transcripts were internally labeled with α-32P-GTP, their band intensities were different according to the respective RNA sequences. The cleavage sites were identified by northern hybridization and nucleotide analysis of the 5′ termini (data not shown). Briefly, the pre-RNA molecules were prepared with non-radioisotope labeled nucleotides and treated with the APE-EndA. The RNA fragments were identified by northern hybridization. The 5′-end of each fragment, which was purified by polyacrylamide gel electrophoresis, was labeled with γ-32P-ATP and T4 polynucleotide kinase and then digested with nuclease P1. The resultant nucleotides were separated on 2D thin layer chromatography and radioisotope-labeled nucleotides were analyzed by autoradiography. Extra up-shifted bands were observed in some splicing products (for example, the 3′-half fragments in Figure 3B). These extra bands were produced by the so-called N+1 reaction of T7 RNA polymerase, which one or more non-templated nucleotides are added to 3′ terminus of the nascent RNA by run-off transcription (33,34). The observed fragments and cleavage sites are illustrated in Figure 3. Figure 3A shows the pre-tRNAThr1 transcript, which has a canonical intron with the BHB motif at the standard position of the anticodon loop. Removal of this intron was very fast: >90% of the intron (500 pmol) was excised off within 2 min by 5 pmol of APE-EndA under the assay condition (Figure 3B), thus suggesting that our purified recombinant APE-EndA has significant enzyme activity. Next, we used the pre-tRNAThr2 transcript, which has a canonical intron in the D-loop of the pre-tRNA (Figure 3C). As shown in Figure 3D, APE-EndA removed the intron from the non-standard position (D-loop) although the cleavage rate was considerably slower than that was observed for the anticodon loop (Figure 3B): at 2 min, ~70% of the pre-tRNA still remained intact. Probably the steric hindrance of the 3D core of the pre-tRNA was responsible for this reduced cleavage rate, because the same intron, located in a mini-helix, was excised off relatively more rapidly (Figure 3E and F). We have also examined the cleavability of the BHL intron in the mini-helix, which mimicked the anticodon arm of the Cenarchaeum symbiosum pre-tRNATyr (Figure 3G). Because the BHL introns in A. pernix pre-tRNAs are very short (5–7 nt), detection of the cleaved intron was difficult. Therefore, in this experiment, we selected the mini-helix of the C. symbiosum pre-tRNATyr as the substrate. As shown in Figure 3G and H, the BHL intron was removed from the mini-helix, although the cleavage rate in this case was slow compared to the rate when the intron was present in the BHB motif. During the course of our study, the crystal structure of APE-EndA and its enzymatic properties were reported by Yoshinari's group (35). In that study, the 6× His-tagged APE-EndA, however, hardly removed the intron with BHL motif located in the cbf5 pre-mRNA: the junction between the 3′ exon and intron was barely cleaved within 20 min under the test condition (2.5 pmol enzyme and 10 pmol RNA). In contrast, our result showed that the junction between the 3′ exon and intron was clearly cleaved in 2 min under our test condition (5 pmol enzyme and 500 pmol RNA). Because the sequences of BHL substrates used in both studies were largely different, these differences in the cleavage activities of BHL introns might be caused by the substrate sequence itself. Otherwise, this apparent discrepancy in results might be due to the presence of 6× His tag in the recombinant APE-EndA used in the former study, in which the extra His residues have added extra positive charges at the N-terminal region of the β-subunit and thereby could have disturbed the correct binding of the substrate RNA. This idea is consistent with our experimental results described in a later section suggesting that the positive charges on the enzyme surface are very important for the substrate selectivity. Taken together, these results suggest that the APE-EndA has broad substrate specificity, and can remove canonical (BHB) as well as non-canonical (BHL) introns in the various positions of pre-tRNA. Our results also demonstrated that the APE-EndA does not require the entire pre-tRNA structure for exerting its activity (Figure 3F and H). Moreover, it is also clear from the results described above is that the removal of intron from a non-standard position, at least from the D-loop, does not require any additional protein subunit, guide RNA and modified nucleotide in the pre-tRNA.
Figure 3.
Splicing activity and specificity of APE-EndA. (A) Predicted secondary structure of A. pernix pre-tRNAThr1(CGU). (B) Time-dependent cleavage of A. pernix pre-tRNAThr1(CGU). (C) Predicted secondary structure of A. pernix pre-tRNAThr2 (UGU). (D) Time-dependent cleavage of A. pernix pre-tRNAThr2(UGU). (E) Predicted secondary structure of the BHB mini helix of A. pernix pre-tRNA Thr2(UGU). (F) Time-dependent cleavage of BHB mini helix. (G) Predicted secondary structure of the BHL mini helix of C. symbiosum pre-tRNATyr. (H) Time-dependent cleavage of the BHL mini helix of C. symbiosum pre-tRNATyr. Short arrows in (A), (C), (E) and (G) indicate the splicing sites for each pre-tRNA. Reaction mixtures were separated on 15% polyacrylamide/7 M urea gels. In each gel, ‘C’ (first lane) indicates control (no enzyme). The cleavage products are shown using schematic models at the right hand side of the gel.
Selection of the target site for the protein engineering
We determined the crystal structure of the wild-type APE-EndA at 2.8 Å resolution to obtain structural information (Supplementary Figure S3A and Table 1; PDB code: 3P1Z). As described above, during the course of our study, the structure of 6× His-tagged mutant APE-EndA (His133Ala) (PDB code: 3AJV) was reported (35). Our crystal structure is almost identical to this previously published mutant APE-EndA structure except for the regions around the mutation site (His133 residue in the α-subunit) and the His-tagged N-terminal end of the β-subunit. Because of the structural similarities between our wild-type and previously published mutant APE-EndAs, we have presented here only minimum information on the structure of the wild-type APE-EndA. As shown in Supplementary Figure S3A, the APE-EndA is composed of two α (pink and light gray) and two β- (cyan and green) subunits, and forms a heterotetrameric α2β2-subunit complex. The overall shape of the α2β2 APE-EndA is like a rectangular parallelepiped, and its structure is similar to the previously reported EndA structures (12,36–39). However, the main chain of the β7 strand and the connection loop between the β7 and β8 strands (Val124-Phe136) of our structure is slightly deviated from the structure reported for the mutant (35). The Cα atom of His133 in the wild-type APE-EndA is ~3.4 Å away from the Cα atom of Ala133 of mutant APE-EndA in the superimposed structures (data not shown). This may be due to the more hydrophilic nature of the His residue because the imidazole ring of His133 protrudes into the solvent.
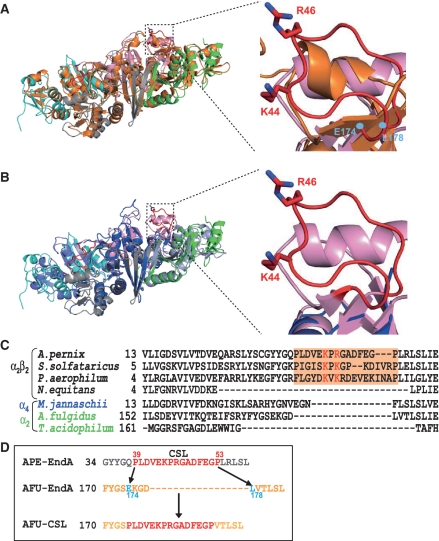
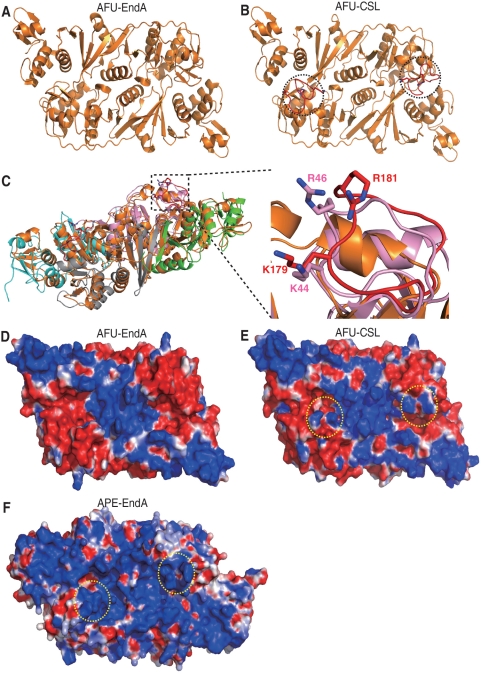
In order to understand the mechanism of broad substrate specificity of APE-EndA, we compared the structure and amino acid sequence of APE-EndA with those of the α2 Euryarchaeal-EndA, which has relatively narrow substrate specificity (8,9,11). It was previously shown that three catalytic residues (tyrosine, histidine and lysine) and two substrate recognition residues (two arginines) are involved in the reaction mechanism of the α2 Euryarchaeal-EndA (AFU-EndA) (15). In the AFU-EndA and RNA complex, a tyrosine residue, a histidine residue and a lysine residue form the catalytic triad and two arginine residues sandwich the adenine base located in the first bulge of the BHB motif by cation–π interactions (3). Our current structure-based amino acid sequence alignment strongly suggests that in APE-EndA the residues Y125, H133 and K164 form the catalytic triad and the residues R157 and W183 are possibly involved in substrate recognition (Supplementary Figures S3B and S4). Thus, one of the arginine residues involved in substrate recognition by AFU-EndA is substituted with a tryptophan residue (W183) in APE-EndA. Notably, this tryptophan residue is conserved in the Crenarchaeal, Nanoarchaeal and Eukaryotic EndAs (40). A tryptophan residue might be an alternative for the arginine residue because its indole ring could interact with the nucleotide by hydrophobic interaction instead of the cation–π interaction. As shown in Supplementary Figure S3B, these five residues (Y125, H133, R157, K164 and W183; shown in black) are located in the predicted catalytic pocket of APE-EndA. Furthermore the importance of the H133 residue for the enzyme activity was recently confirmed by the Yoshinari's group (35). Because these catalytic and substrate recognition residues are conserved in all EndA families, they are probably not involved in determining the broad substrate specificity of APE-EndA. Upon close examination of the Crenarchaeal EndA structure, it was found that the connecting region between the β3- and β4-strands in the α-subunit is quite different from the other members of the three EndA families (Figure 4 and Supplementary Figure S4). Our structural results showed that this connecting region forms a loop structure close to the catalytic pocket (Supplementary Figure S3B, highlighted in red). This loop is structurally conserved only in Crenarchaea, and known as the Crenarchaea specific loop [CSL; classical name, Crenarchaea extra specific loop (12)] (35). In Figure 4A and B, the CSL (red) region in the APE-EndA (pink) is superimposed onto the corresponding region of AFU (orange) and Nanoarchaeum equitans (NEQ, blue)-EndAs. As shown, the CSL is replaced by a β-turn in the AFU-EndA structure and a short disordered loop region in the NEQ-EndA. In addition, two positive amino acid residues (K44 and R46 in the APE-EndA) located in the CSL are conserved only in the Crenarchaeal EndA (Figure 4C).
Figure 4.
Structural comparison of the CSL region of APE-EndA with the corresponding regions of other EndAs. (A) Left: Superimposed structures of archaeal EndAs. The α-subunits of APE-EndA are indicated in pink and light grey. The β-subunits of APE-EndA are indicated in green and cyan. The α-subunits of AFU-EndA are in orange. Right: Close-up view of the structure of CSL region (red) of APE-EndA (pink) superimposed on the structure of the corresponding regions of AFU-EndA (orange). The AFU-EndA residues (E174 and L178), where the CSL peptide was inserted to create the AFU-CSL chimera, are indicated in cyan. The RMSD (root mean square deviation) is 2.5 Å for 538 Cα atoms of AFU-EndA. (B) Left: Superimposed structures of archaeal EndAs. The α-subunits of APE-EndA are indicated in pink and light grey. The β-subunits of APE-EndA are indicated in green and cyan. The α-subunits of NEQ-EndA are indicated in blue. The β-subunits of NEQ-EndA are indicated in light blue. Right: Close-up view of the structure of CSL region (red) of APE-EndA (pink) superimposed on the structure of the corresponding region of NEQ-EndA [α-subunits (blue)]. The RMSD is 2.2 Å for 488 Cα atoms of NEQ-EndA. (C) Amino-acid sequence alignment of α-subunits around the CSL region (highlighted in orange). Full names of the archaea species are as follows; Aeropyrum pernix, Sulfolobus solfataricus, Pyrobaculum aerophilum, Nanoarchaeum equitans, Methanocaldococcus jannaschii, Archaeoglobus fulgidus and Thermoplasma acidophilum. Two conserved positively charged residues are shown in red. (D) Schematic diagram illustrating creation of the AFU-CSL chimera: the peptide between the amino acids E174 to L178 (cyan) of AFU-EndA was replaced with the CSL peptide (red) of APE-EndA.
Insertion of CSL in AFU-EndA confers APE-EndA like broad substrate specificity
We next examined the possibility that the CSL might play a key role in determining the substrate specificity. As shown in Figure 4A and D, we created an AFU-EndA mutant protein (AFU-CSL) in which the EKGDL sequence, from amino acid positions 174–178, of AFU-EndA were replaced by the CSL sequence of APE-EndA. We then analyzed the substrate specificity of AFU-CSL mutant. As shown in Figure 5A, the AFU-CSL cleaved the BHB intron from the anticodon loop in the similar manner as the wild-type AFU-EndA. The wild-type AFU-EndA, however, hardly cleaved the BHB intron from the D-loop of the pre-tRNAThr2 (Figure 5B, left). In contrast, the AFU-CSL effectively cleaved the BHB intron from the D-loop (Figure 5B, right). Additionally, whereas the wild-type AFU-EndA did not remove the intron from the BHL motif in the mini-helix as reported (11) (Figure 5C, left), the AFU-CSL cleaved the BHL intron from the mini-helix just as the APE-EndA did (Figure 5C, right). Thus, these experimental results clearly demonstrated that the insertion of CSL conferred APE-EndA-like broad substrate-specificity to AFU-EndA, which has narrow substrate specificity.
Figure 5.
Crenarchaea specific loop (CSL) is involved in broad substrate specificity. Time-dependent cleavage activity of AFU-CSL: (A) A. pernix pre-tRNAThr1(CGU), (B) A. pernix pre-tRNAThr2(UGU) and (C) BHL mini helix of C. symbiosum pre-tRNATyr. In each gel, ‘C’ (first lane) indicates the control (no enzyme) sample. In each case, the bands (on the gel) and their corresponding products (on the right hand side of the gel) are indicated.
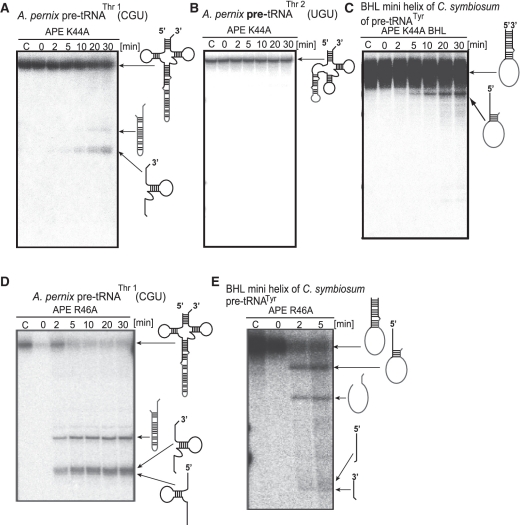
We postulated that the residues K44 and R46 located in the CSL might be involved in substrate recognition. To confirm this idea, we prepared two APE-EndA mutant proteins (APE K44A and R46A) by individually substituting the K44 and R46 residues with alanine. As expected, substitution of K44 with alanine caused severe loss of enzyme activity: the APE K44A mutant could hardly excise the BHB intron at the anticodon loop (Figure 6A), and was unable to excise the BHB intron at the D-loop (Figure 6B). Furthermore, the BHL intron was not completely removed: a faint band corresponding to the 5′ exon and intron joint product was observed, suggesting slight cleavage of the 3′ exon and no cleavage of the 5′ exon (Figure 6C). It should be mentioned that the exposure time of the autoradiograph shown in Figure 6C was prolonged to visualize the faint band corresponding to the 5′ exon and intron joint product. These results clearly showed that the K44 residue is very important for the enzymatic activity of APE-EndA. It is noteworthy that the importance of the K44 residue for enzymatic activity has been reported during the course of our study (35). In contrast, substitution of R46 residue with alanine did not affect its substrate selectivity significantly (Figures 6D and E), suggesting that the R46 residue is not important for the substrate specificity of APE-EndA.
Figure 6.
Splicing activity and specificity of the APE K44A and R46A mutants. Time-dependent cleavage activity of APE K44A: (A) A. pernix pre-tRNAThr1(CGU), (B) A. pernix pre-tRNAThr2(UGU), (C) BHL mini helix of C. symbiosum pre-tRNATyr. Time-dependent cleavage activity of APE R46A: (D) A. pernix pre-tRNAThr1(CGU) and (E) BHL mini helix of C. symbiosum pre-tRNATyr. In each gel, ‘C’ (fist lane) indicates the control (no enzyme) sample. Products corresponding to each band on the gel are shown on the right hand side of the gels.
Structures of AFU-CSL and model of its complex with RNA
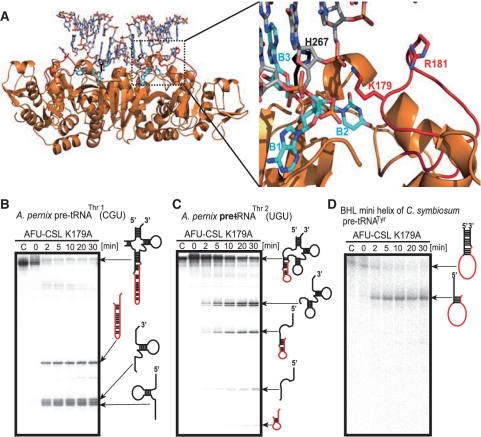
Although the K44 residue in the CSL is very important for the intron cleavage activity, its precise role remained unclear. Therefore, we analyzed the crystal structure of AFU-CSL, a chimeric protein. The purified AFU-CSL protein was crystallized and its structure was determined at 2.05 Å resolution (Table 1). Figure 7A shows the structure of the wild-type AFU-EndA (PDB ID code, 1RLV) (37). The overall structure of AFU-CSL was identical to that of the wild-type AFU-EndA except the inserted CSL regions, indicated by dotted circles (Figure 7B). The conformation of the CSL region in AFU-CSL was almost identical to that in the APE-EndA (Figure 7C). To predict the RNA binding site, electrostatic potential surface models of the wild-type AFU-EndA (Figure 7D), AFU-CSL (Figure 7E) and APE-EndA (Figure 7F) were generated. As shown, the catalytic pockets are located in the center of positive charges (Figure 7D–F). Interestingly, residues K179 and R181 of AFU-EndA, corresponding to the K44 and R46 residues of APE-EndA (Figure 7C), add new positive charges to AFU-EndA (area shown with dotted circle) symmetrically like the positive charges of APE-EndA (Figure 7D and E). We next constructed the docking model of the AFU-CSL and substrate RNA based on the reported AFU-EndA and RNA complex structure (Figure 8A). The RNA in this complex contains the BHB motif (3). In our docking model, the K179 residue is situated near the 3′ phosphate group adjacent to the bulge structure of the RNA. In contrast, the R181 residue is placed away from the RNA in our model, in accordance with the result of the R46 to Ala substitution of APE-EndA. This model suggests that the K179 residue captures the 3′ phosphate group adjacent to the bulge structure (or 3′ phosphate of the third nucleotide in the loop structure), fixes the substrate, and thereby results in determining the substrate specificity.
Figure 7.
Structure of AFU-CSL. (A) Ribbon diagram of the overall structure of AFU-EndA (orange, PDB ID code 1RLV). (B) The crystal structure of AFU-CSL (orange). Two dotted circles indicate the inserted CSL (red). (C) Left: Superimposed structures of archaeal EndAs. The α-subunits of APE-EndA are indicated in pink and light grey. The β-subunits of APE-EndA are indicated in green and cyan. The α-subunits of AFU-CSL are in orange. Right: Close-up view of the structure of CSL region (red) of AFU-CSL (orange) superimposed on the structure of the corresponding region of APE-EndA (pink). The RMSD is 2.6 Å for 628 Cα atoms of AFU-CSL. (D) Electrostatic potential surface models of AFU-EndA: red and blue colors indicate negative and positive charges, respectively. (E) Electrostatic potential surface models of AFU-CSL. Two dotted circles (yellow) show the K179 and R181 residues of AFU-CSL. (F) Electrostatic potential surface models of APE-EndA. Two dotted circles (yellow) show the K44 and R46 residues of APE-EndA.
Figure 8.
The conserved Lys residue of CSL is responsible for broad substrate specificity. (A) Model of the complex formed between the AFU-CSL and an RNA substrate (stick model, light gray) that contains a BHB motif (left). The dotted square shows the active site. Close-up view of the active site of the enzyme-RNA complex (right). Stick models in cyan and black colors show the bulge structure (B1–B2–B3) of the BHB motif and the catalytic residue H267, respectively. The K179 and R181 residues located in the inserted CSL peptide are shown in red. (B–D) Time-dependent cleavage activity of AFU-CSL K179A mutant: (B) A. pernix pre-tRNAThr1(CGU), (C) A. pernix pre-tRNAThr2(UGU) and (D) BHL mini helix of C. symbiosum pre-tRNATyr. ‘C’ indicates the control (no enzyme) sample.
To confirm the importance of the K179 residue in AFU-CSL function, we substituted the K179 residue with alanine. This mutant protein (AFU-CSL K179A) removed the BHB intron from the anticodon loop (Figure 8B) and the D-loop (Figure 8C). However, AFU-CSL K179A could not remove the BHL intron correctly (Figure 8D): the cleavage did not occur at the loop region of the BHL motif, and only generated a product that corresponded to 5′ exon remaining joined to the intron. Thus, these results suggest that the K179 residue plays a key role in the broad substrate specificity.
DISCUSSION
In Crenarchaeal genomes, curious tRNA genes have been reported (16). Several reported tRNA (or tRNA-like) genes are predicted to produce unusual transcripts and/or pre-tRNAs with a canonical or non-canonical intron at various positions. In the current study, we confirmed that three tRNAThr species [tRNAThr1(CGU), tRNAThr2(UGU) and tRNAThr4(GGU)] are indeed transcribed in the living APE cells and mature to functional tRNAs. Our biochemical studies demonstrated that the purified APE-EndA cleaved the intron at non-standard position as well as the intron at standard position. Thus, the removal of intron from non-standard position (at least, in D-loop) does not require any other protein, RNA and/or modified nucleotides in pre-tRNA. The rate of intron cleavage from the D-loop is considerably slower than that from the standard position (anticodon loop) due to the steric hindrance of the 3D core of the tRNA. Thus, the maturation of tRNAThr2(UGU) seems to be slower than the maturation of tRNAThr1(CGU) or tRNAThr4(GGU). In fact, we found that the intensity of the hybridized band of tRNAThr2(UGU) is weak (Supplementary Figure S1). As described above, the threonine codons (ACU, ACC, ACG and ACA) could be decoded only by tRNAThr1(CGU) and tRNAThr4(GGU) without assuming modification of anticodons. However, the G–A base pair is weak at high temperatures, the environment in which APE lives. Therefore, the existence of tRNAThr2(UGU) probably reinforces decoding of the ACA threonine codon in the APE cells.
The APE-EndA removed the intron with BHB or BHL motif irrespective of its position in the pre-tRNA. The APE-EndA also removed the intron with non-canonical BHL motif. To understand the structural basis of its function, we solved the crystal structure of wild-type APE-EndA. The α-subunit of APE-EndA showed structural similarities with the catalytic subunits of the other EndAs (α2, α4 and α2β2 types). The structure based amino-acid sequence alignment suggested that the CSL region is responsible for the observed broad substrate specificity of the Crenarchaeal EndA. Indeed, insertion of CSL conferred broad substrate specificity to AFU-EndA, suggesting that the CSL plays a key role in determining the substrate specificity. We also showed that a conserved lysine residue is very important for the intron cleavage activity. Our docking model based on the AFU-CSL crystal structure suggested that the conserved lysine residue captures the 3′ phosphate group adjacent to the bulge structure. In fact, the electrostatic potential surface models showed that positive charges were added around the catalytic pocket of the AFU-EndA. The importance of the lysine residue in substrate recognition was further confirmed by mutational analysis.
For the last 5 years, the broad substrate specificity of the Crenarchaeal EndA has remained an enigma in the field (6,8,9,35). Results described in this study showed that the interaction between the CSL region of EndA and the RNA determines the substrate specificity. Because the bulge structures of the BHB motif are stabilized by the stem structures, the α2 and α4 EndAs may not be required for holding the 3′ phosphate group adjacent to the bulge structure. In contrast, the loop structure of the BHL motif is probably more flexible compared to the bulge structure of the BHB motif, because both BHL and BHB introns are cleaved by α2β2 EndA. Therefore, the α2β2 EndA requires additional RNA recognition site, namely, the lysine residue in the CSL region. Structure of the CSL has a flexible feature because it showed >60% (particularly >70% at positively charged residues) high temperature flexibility factors. Because of this flexibility, the Crenarhcaeal EndA probably could cleave any type of non-canonical introns. In fact, a search of the SPLITSdb database (16) revealed the diversity of non-canonical introns found in the Crenarchaea pre-tRNAs (especially in the archaeal order, Thermoproteales). Acquisition of the CSL region into EndA consequently might have given rise to the intron diversity.
Our results might possibly explain how introns could have distributed to various pre-tRNA positions in Crenarchaea. We showed that, in contrast to AFU-EndA, the AFU-CSL effectively cleaved the BHB intron at the D-loop (Figure 5B). Therefore, inclusion of CSL not only adds diversity to intron types but also distributes tRNA introns to various positions. This idea also exemplifies that the architecture of EndA and variation of introns in tRNA coevolved in arachaea (9). It has been proposed that the archaeal EndA is subfunctionalized (11), which occurred as a result of gene duplication (41). In this hypothesis, the α-subunit gene of α4 EndA was first duplicated, one of which was then subfunctionalized to encode the β-subunit. The CSL region of Crenarhcaeal EndA may have been acquired during the subfunctionalization process.
About 68.7% pre-tRNAs of Thermoproteales, the thermoacidophilic Crenarchaeal order, contain introns at various positions (16). They also include multiple intron containing pre-tRNAs. Although the double introns of Thermofilum pendens pre-tRNAProш were properly removed by α2β2 EndA (40), it however remains unknown whether the tRNAProш functions in vivo. The CSL region of T. pendens EndA probably plays an important role in the cleavage of multiple introns.
Inclusion of CSL in EndA may be an advantage for the survival of Crenarchaea, because it has been suggested that the role of tRNA intron is to provide protection against integration of mobile genetic elements, such as conjugative plasmids and viruses (42). In fact, many Crenarchaea infecting viruses have been found from the hot acidic environment (43). Additionally, genomic sequences of two archaeal proviruses were shown to be integrated into the 5′- and 3′-distal regions of the tRNA genes of the Euryarchaeal species (44), suggesting the possibility that the tRNA intron might have originated from the viruses’ genome. Therefore, it is possible that by incorporating the CSL region into the Crenarchaeal EndA helps Crenarchaea to defend against such archaeal viral infections.
Finally, our findings presented here may provide a glimpse into the molecular recognition and co-evolution of proteins and RNAs at the early stages of life on Earth.
ACCESSION NUMBERS
PDB 3P1Z, PDB 3P1Y.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: The Sumitomo Foundation (No. 090333 to A.H.); a Grant-in-aid for Science Research on Priority Areas (No. 20034041 to H.H.); Ministry of Education, Science, Sports, and Culture of Japan, Grant-in-aid for Science Research (No. 23350081 to H.H.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the staff members of the beam-line facility at SPring-8 (Hyogo, Japan) for their technical support during data collection. The synchrotron radiation experiments were performed at the BL38B1 and BL41XU in the SPring-8 with the approval of the Japan Synchrotron Radiation Research Institute (JASRI) (Proposal Nos 2008B2177, 2009B1125 and 2010A1263). The authors are also grateful to Dr T. Kanai and Dr T. Endoh (Kyoto University) for helping in the culture of APE cells. Finally, the authors thank Prof. Y. Endo (Ehime University) for encouraging this study.
REFERENCES
- 1.Abelson J, Trotta C, Li H. tRNA splicing. J. Biol. Chem. 1998;273:12685–12688. doi: 10.1074/jbc.273.21.12685. [DOI] [PubMed] [Google Scholar]
- 2.Fabbri S, Fruscoloni P, Bufardeci E, Di Nicola Negri E, Baldi M, Attardi D, Mattoccia E, Tocchini-Valentini G. Conservation of substrate recognition mechanisms by tRNA splicing endonucleases. Science. 1998;280:284–286. doi: 10.1126/science.280.5361.284. [DOI] [PubMed] [Google Scholar]
- 3.Xue S, Calvin K, Li H. RNA recognition and cleavage by a splicing endonuclease. Science. 2006;312:906–910. doi: 10.1126/science.1126629. [DOI] [PubMed] [Google Scholar]
- 4.Marck C, Grosjean H. Identification of BHB splicing motifs in intron-containing tRNAs from 18 archaea: evolutionary implications. RNA. 2003;9:1516–1531. doi: 10.1261/rna.5132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Randau L, Münch R, Hohn M, Jahn D, Söll D. Nanoarchaeum equitans creates functional tRNAs from separate genes for their 5′- and 3′-halves. Nature. 2005;433:537–541. doi: 10.1038/nature03233. [DOI] [PubMed] [Google Scholar]
- 6.Calvin K, Hall M, Xu F, Xue S, Li H. Structural characterization of the catalytic subunit of a novel RNA splicing endonuclease. J. Mol. Biol. 2005;353:952–960. doi: 10.1016/j.jmb.2005.09.035. [DOI] [PubMed] [Google Scholar]
- 7.Randau L, Pearson M, Söll D. The complete set of tRNA species in Nanoarchaeum equitans. FEBS Lett. 2005;579:2945–2947. doi: 10.1016/j.febslet.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 8.Randau L, Calvin K, Hall M, Yuan J, Podar M, Li H, Söll D. The heteromeric Nanoarchaeum equitans splicing endonuclease cleaves noncanonical bulge-helix-bulge motifs of joined tRNA halves. Proc. Natl Acad. Sci. USA. 2005;102:17934–17939. doi: 10.1073/pnas.0509197102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tocchini-Valentini G, Fruscoloni P, Tocchini-Valentini G. Coevolution of tRNA intron motifs and tRNA endonuclease architecture in Archaea. Proc. Natl Acad. Sci. USA. 2005;102:15418–15422. doi: 10.1073/pnas.0506750102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshinari S, Itoh T, Hallam S, DeLong E, Yokobori S, Yamagishi A, Oshima T, Kita K, Watanabe Y. Archaeal pre-mRNA splicing: a connection to hetero-oligomeric splicing endonuclease. Biochem. Biophys. Res. Commun. 2006;346:1024–1032. doi: 10.1016/j.bbrc.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 11.Tocchini-Valentini G, Fruscoloni P, Tocchini-Valentini G. The dawn of dominance by the mature domain in tRNA splicing. Proc. Natl Acad. Sci. USA. 2007;104:12300–12305. doi: 10.1073/pnas.0705537104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshinari S, Shiba T, Inaoka D, Itoh T, Kurisu G, Harada S, Kita K, Watanabe Y. Functional importance of crenarchaea-specific extra-loop revealed by an X-ray structure of a heterotetrameric crenarchaeal splicing endonuclease. Nucleic Acids Res. 2009;37:4787–4798. doi: 10.1093/nar/gkp506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trotta C, Miao F, Arn E, Stevens S, Ho C, Rauhut R, Abelson J. The yeast tRNA splicing endonuclease: a tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell. 1997;89:849–858. doi: 10.1016/s0092-8674(00)80270-6. [DOI] [PubMed] [Google Scholar]
- 14.Kleman-Leyer K, Armbruster D, Daniels C. Properties of H. volcanii tRNA intron endonuclease reveal a relationship between the archaeal and eucaryal tRNA intron processing systems. Cell. 1997;89:839–847. doi: 10.1016/s0092-8674(00)80269-x. [DOI] [PubMed] [Google Scholar]
- 15.Calvin K, Li H. RNA-splicing endonuclease structure and function. Cell Mol. Life Sci. 2008;65:1176–1185. doi: 10.1007/s00018-008-7393-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugahara J, Kikuta K, Fujishima K, Yachie N, Tomita M, Kanai A. Comprehensive analysis of archaeal tRNA genes reveals rapid increase of tRNA introns in the order thermoproteales. Mol. Biol. Evol. 2008;25:2709–2716. doi: 10.1093/molbev/msn216. [DOI] [PubMed] [Google Scholar]
- 17.Kawarabayasi Y, Hino Y, Horikawa H, Yamazaki S, Haikawa Y, Jin-no K, Takahashi M, Sekine M, Baba S, Ankai A, et al. Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 1999;6:83–101. doi: 10.1093/dnares/6.2.83. 145–152. [DOI] [PubMed] [Google Scholar]
- 18.Yamazaki S, Kikuchi H, Kawarabayasi Y. Characterization of a whole set of tRNA molecules in an aerobic hyper-thermophilic Crenarchaeon, Aeropyrum pernix K1. DNA Res. 2005;12:403–416. doi: 10.1093/dnares/dsi023. [DOI] [PubMed] [Google Scholar]
- 19.Sako Y, Nomura N, Uchida A, Ishida Y, Morii H, Koga Y, Hoaki T, Maruyama T. Aeropyrum pernix gen. nov., sp. nov., a novel aerobic hyperthermophilic archaeon growing at temperatures up to 100 degrees C. Int. J. Syst. Bacteriol. 1996;46:1070–1077. doi: 10.1099/00207713-46-4-1070. [DOI] [PubMed] [Google Scholar]
- 20.Awai T, Kimura S, Tomikawa C, Ochi A, Ihsanawati, Bessho Y, Yokoyama S, Ohno S, Nishikawa K, Yokogawa T, et al. Aquifex aeolicus tRNA (N2,N2-guanine)-dimethyltransferase (Trm1) catalyzes transfer of methyl groups not only to guanine 26 but also to guanine 27 in tRNA. J. Biol. Chem. 2009;284:20467–20478. doi: 10.1074/jbc.M109.020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yokogawa T, Kitamura Y, Nakamura D, Ohno S, Nishikawa K. Optimization of the hybridization-based method for purification of thermostable tRNAs in the presence of tetraalkylammonium salts. Nucleic Acids Res. 2010;38:e89. doi: 10.1093/nar/gkp1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pachmann U, Zachau HG. Yeast seryl tRNA synthetase: two sets of substrate sites involved in aminoacylation. Nucleic Acids Res. 1978;5:961–973. doi: 10.1093/nar/5.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hori H, Yamazaki N, Matsumoto T, Watanabe Y, Ueda T, Nishikawa K, Kumagai I, Watanabe K. Substrate recognition of tRNA (Guanosine-2′-)-methyltransferase from Thermus thermophilus HB27. J. Biol. Chem. 1998;273:25721–25727. doi: 10.1074/jbc.273.40.25721. [DOI] [PubMed] [Google Scholar]
- 24.Keith G. Mobilities of modified ribonucleotides on two-dimensional cellulose thin-layer chromatography. Biochimie. 1995;77:142–144. doi: 10.1016/0300-9084(96)88118-1. [DOI] [PubMed] [Google Scholar]
- 25.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 26.McCoy A, Grosse-Kunstleve R, Adams P, Winn M, Storoni L, Read R. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.Brunger A. Version 1.2 of the crystallography and NMR system. Nat. Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 29.Adams P, Grosse-Kunstleve R, Hung L, Ioerger T, McCoy A, Moriarty N, Read R, Sacchettini J, Sauter N, Terwilliger T. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 30.Laskowski R, MacArthur M, Moss D, Thoronton J. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1992;26:283–291. [Google Scholar]
- 31.Baker N, Sept D, Joseph S, Holst M, McCammon J. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta R. Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids, and the sequences of 33 class I tRNAs. J. Biol. Chem. 1984;259:9461–9471. [PubMed] [Google Scholar]
- 33.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krupp G. RNA synthesis: strategies for the use of bacteriophage RNA polymerases. Gene. 1988;72:75–89. doi: 10.1016/0378-1119(88)90129-1. [DOI] [PubMed] [Google Scholar]
- 35.Okuda M, Shiba T, Inaoka DK, Kita K, Kurisu G, Mineki S, Harada S, Watanabe Y, Yoshinari S. A conserved lysine residue in the Crenarchaea-specific loop is important for the crenarchaeal splicing endonuclease activity. J. Mol. Biol. 2011;405:92–104. doi: 10.1016/j.jmb.2010.10.050. [DOI] [PubMed] [Google Scholar]
- 36.Li H, Trotta C, Abelson J. Crystal structure and evolution of a transfer RNA splicing enzyme. Science. 1998;280:279–284. doi: 10.1126/science.280.5361.279. [DOI] [PubMed] [Google Scholar]
- 37.Li H, Abelson J. Crystal structure of a dimeric archaeal splicing endonuclease. J. Mol. Biol. 2000;302:639–648. doi: 10.1006/jmbi.2000.3941. [DOI] [PubMed] [Google Scholar]
- 38.Kim Y, Mizutani K, Rhee K, Nam K, Lee W, Lee E, Kim E, Park S, Hwang K. Structural and mutational analysis of tRNA intron-splicing endonuclease from Thermoplasma acidophilum DSM 1728: catalytic mechanism of tRNA intron-splicing endonucleases. J. Bacteriol. 2007;189:8339–8346. doi: 10.1128/JB.00713-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitchell M, Xue S, Erdman R, Randau L, Söll D, Li H. Crystal structure and assembly of the functional Nanoarchaeum equitans tRNA splicing endonuclease. Nucleic Acids Res. 2009;37:5793–5802. doi: 10.1093/nar/gkp537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trotta C, Paushkin S, Patel M, Li H, Peltz S. Cleavage of pre-tRNAs by the splicing endonuclease requires a composite active site. Nature. 2006;441:375–377. doi: 10.1038/nature04741. [DOI] [PubMed] [Google Scholar]
- 41.Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Randau L, Söll D. Transfer RNA genes in pieces. EMBO Rep. 2008;9:623–628. doi: 10.1038/embor.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ortmann A, Wiedenheft B, Douglas T, Young M. Hot crenarchaeal viruses reveal deep evolutionary connections. Nat. Rev. Microbiol. 2006;4:520–528. doi: 10.1038/nrmicro1444. [DOI] [PubMed] [Google Scholar]
- 44.Krupovic M, Bamford D. Archaeal proviruses TKV4 and MVV extend the PRD1-adenovirus lineage to the phylum Euryarchaeota. Virology. 2008;375:292–300. doi: 10.1016/j.virol.2008.01.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.