Abstract
Mutations in Pol γ represent a major cause of human mitochondrial diseases, especially those affecting the nervous system in adults and in children. Recessive mutations in Pol γ represent nearly half of those reported to date, and they are nearly uniformly distributed along the length of the POLG1 gene (Human DNA Polymerase gamma Mutation Database); the majority of them are linked to the most severe form of POLG syndrome, Alpers–Huttenlocher syndrome. In this report, we assess the structure–function relationships for recessive disease mutations by reviewing existing biochemical data on site-directed mutagenesis of the human, Drosophila and yeast Pol γs, and their homologs from the family A DNA polymerase group. We do so in the context of a molecular model of Pol γ in complex with primer–template DNA, which we have developed based upon the recently solved crystal structure of the apoenzyme form. We present evidence that recessive mutations cluster within five distinct functional modules in the catalytic core of Pol γ. Our results suggest that cluster prediction can be used as a diagnosis-supporting tool to evaluate the pathogenic role of new Pol γ variants.
INTRODUCTION
Mitochondrial DNA polymerase, Pol γ, is the sole known DNA polymerase in animal mitochondria and is responsible for mitochondrial DNA (mtDNA) replication and repair (1,2). The human enzyme is a heterotrimer consisting of a catalytic subunit, Pol γA, and dimer of an accessory subunit, Pol γB (3). Pol γA is a member of the family A DNA polymerase group to which bacterial DNA Pol I belongs (1). It comprises three domains: an N-terminal domain containing 3′→5′ exonuclease (exo) activity, a spacer domain and a C-terminal domain, containing 5′→3′ DNA polymerase (pol) activity in three subdomains termed the palm, fingers and thumb. Pol γA also bears a 5′-deoxyribose phosphate lyase activity (4), but the location of its active site is unknown. The accessory subunit, Pol γB, serves as a processivity factor, enhancing the DNA-binding affinity and catalytic activities of Pol γA (5,6). The crystal structure of human Pol γ in its apoenzyme form was solved recently (PDB code 3IKM) (7).
Mutations in Pol γ represent a major cause of human mitochondrial diseases, especially those affecting the nervous system in adults and in children. Dominant mutations typically cause adult-onset myopathies and encephalopathies (8,9), whereas recessive mutations result in severe adult or juvenile onset ataxia-epilepsy syndromes (MIRAS, SCA-E, SANDO) (10–12), or devastating early-childhood Alpers syndrome (Alpers–Huttenlocher syndrome) characterized by intractable epilepsy, psychomotor retardation and liver failure that leads to early death (13,14). On the molecular level, POLG syndromes are accompanied by either tissue-specific mtDNA depletion, deletions or combination of both (15,16). To date, more than 145 POLG1 disease mutations have been identified (Human DNA Polymerase gamma Mutation Database, http://tools.niehs.nih.gov/polg/), of which more than half are recessive. The functional consequences of dominant mutations, which affect primarily catalytic residues in the pol domain, have been explained by effective competition for the DNA substrate by the mutant Pol γ with the wild-type enzyme (7,17,18), and by site-specific stalling of mutant DNA polymerase (18). Studies that explain effects of individual recessive mutations on protein function are limited (19–22). The majority of the reported recessive mutations are linked to the most severe form of POLG syndrome, Alpers–Huttenlocher syndrome. Manifestation of Alpers disease typically requires the presence of at least two recessive, compound heterozygous mutations, and is accompanied primarily with mtDNA depletion (16,23). The detailed mechanisms of pathogenesis and mtDNA depletion are unknown.
The assignment of a newly identified amino acid substitution as a pathogenic mutation is generally based on the absence of the variant in the normal population, conservation of the site among species and segregation in families. These assignments are particularly challenging because of the extensive variation in the amino acid sequences of Pol γA in the normal population. To date, data accumulated on recessive disease mutations show that they are almost uniformly distributed among the three structural domains of the catalytic subunit and in most cases, it is unclear which property of the enzyme is affected, and how it contributes to mtDNA deletion or depletion.
The present study aims to assess the structure–function relationships for recessive disease mutations, by reviewing existing biochemical data on site-directed mutagenesis of the human, Drosophila and yeast Pol γs, and their homologs from the family A DNA polymerase group; we do so in the context of the recently solved crystal structure of human Pol γ (7), onto which we have modeled primer–template DNA (ptDNA). We present evidence that recessive mutations cluster within five distinct functional modules in the catalytic core of Pol γ that we designate as ‘Alpers Clusters 1–5’. We note that our analysis of the recessive mutations found in compound heterozygous form in Alpers patients reveals that a severe disease manifestation is typically caused by a combination of at least two mutations from different clusters. Our results suggest that cluster prediction can be used as a diagnosis-supporting tool to evaluate the pathogenic role of new Pol γ variants.
APPROACH TO COMPARATIVE STRUCTURAL ANALYSIS
We docked ptDNA into the putative DNA-binding channel of the apoenzyme form of the human Pol γ structure (PDB code 3IKM) (7) by superposition of the closed ternary complex of T7 Pol bound to ptDNA and dNTP (PDB code 1T8E) (24). The coordinates of the resulting Pol γ: DNA model are provided in Supplementary Data. To ensure reliable positioning for subsequent structural analysis of Alpers recessive mutations, we evaluated three different alignments of the Pol γ and T7 palm domains that are presented in Supplementary Figure S1. First, we aligned the palm domain of Pol γA (PDB code 3IKM, chain A, residues 815–910 and 1095–1239) with that of T7 Pol (PDB code 1T8E, chain A, residues 409–487 and 611–704). Additionally, we aligned the two structures by superposition of their 12β-loop-13β motifs from the palm subdomain, encompassing residues G1127-E1144 in Pol γA and F646-E663 in the T7 catalytic core [for secondary structure element assignment see (25)]. This part of the central β-sheet in the palm subdomain is the most structurally conserved element between DNA polymerases in the family A DNA polymerase group (25), and it superposed with an RMSD of 1.906 Å for Pol γA and T7. Alignment showed that the functionally significant secondary structure elements (SSEs) of the pol domain of Pol γA, including helixes αO, αL and αQ, as well as the loop corresponding to the 7β-loop-8β motif in T7 Pol, are tilted away from the active site, as compared to the analogous SSEs from T7 Pol and Klenow. This suggests conformational rearrangements in the Pol γ catalytic core upon DNA binding. To model the ptDNA in the putative DNA-binding channel of Pol γA, and to identify residues that interact with the DNA duplex, we performed a comparative analysis of the T7 Pol (24) and Klentaq (26) DNA complexes in both the open and closed states. We sought to identify SSEs in the pol domain that fulfill the requirements of interaction with DNA and preservation of conserved positions between the enzymes. As a result, the Q-helix and the 7β-loop-8β motif were identified, and a third alignment of Pol γA and T7 Pol was based on superposition of their αQ helices (residues M1093-E1122 in Pol γ and P606-E635 in T7 Pol), and the hairpins formed by the 7β-loop-8β (residues T846-V855 in Pol γ and P422-T431 in T7 Pol). The RMSD for the superposed hairpins was 1.397 Å.
CLUSTERING OF ALPERS DISEASE MUTATIONS WITHIN THE CATALYTIC CORE OF POL γ
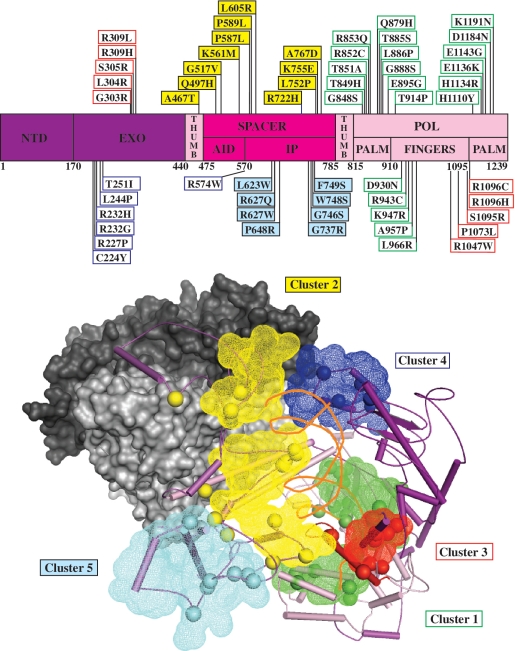
Recessive mutations are distributed along the length of the POLG1 gene sequence (http://tools.niehs.nih.gov/polg/), but they cluster within the tertiary structure of Pol γA to distinct regions within the catalytic core (Figure 1). Five functional modules, termed clusters 1–5, were assigned within Pol γA as follows: Cluster 1 (in green) locates within the pol domain and comprises largely residues affecting DNA polymerase activity; Cluster 2 (in yellow) represents residues lining the upstream DNA-binding channel; Cluster 3 (in red) is associated with a novel structural motif in the fingers subdomain, which we propose confers partitioning of the DNA substrate between the pol and exo sites; Cluster 4 (in blue) lies on the periphery of the exo domain and mediates interactions with the distal Pol γB, stabilizing the Pol γ:DNA complex; and Cluster 5 (in cyan) locates within the spacer domain and represents a region that we propose is involved in replisome interactions.
Figure 1.
Alpers disease mutations cluster within functional modules in the catalytic subunit of Pol γ. Upper panel: schematic diagram of the POLG1 gene showing the distribution of recessive Alpers disease mutations (Human DNA Polymerase gamma Database, http://tools.niehs.nih.gov/polg/). AID and IP in the spacer domain refer to the accessory (subunit) interacting and intrinsic processivity subdomains, respectively, that are discussed in the text; NTD refers to the N-terminal domain. Lower panel: tertiary structural representation of the apoenzyme form of Pol γ [PDB code 3IKM, (7)] with modeled DNA, identifying the positions of five functional modules (shown in mesh) that are defined by clusters of amino acid residues (shown as spheres) affected by Alpers disease mutations as follows: Cluster 1, green; Cluster 2, yellow; Cluster 3, red; Cluster 4, blue; Cluster 5, cyan. The domains of Pol γA are shown as surface representations, and in part as secondary structural elements (SSEs) that are colored as depicted in A. The proximal and distal accessory subunits are shown as surface representations in light and dark gray, respectively. Primer–template DNA was docked as described in the text and is displayed as orange ribbons.
ALPERS CLUSTER 1: RESIDUES AFFECTING 5′-3′ DNA POLYMERASE ACTIVITY
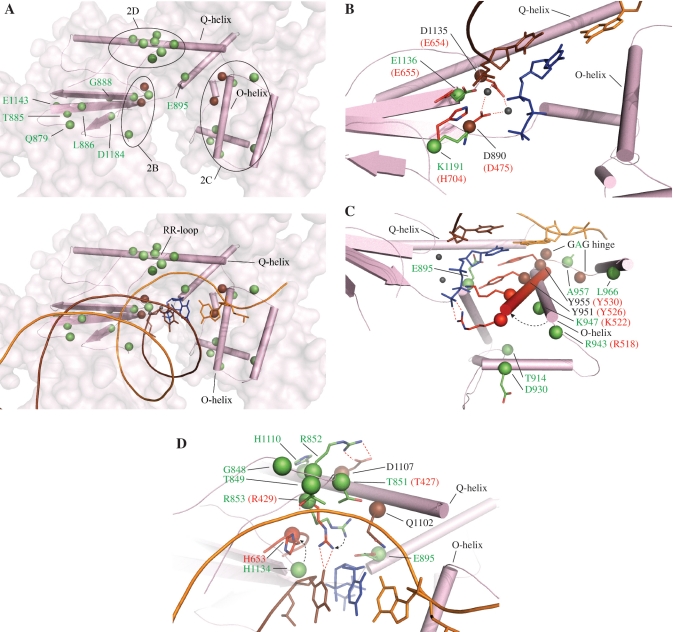
Catalysis of template-directed nucleotide polymerization in DNA polymerases proceeds via a phosphoryl transfer reaction assisted by two Mg2+ ions (27,28). Incorporation of the correct dNMP is mediated by a nucleotide-induced conformational shift of the fingers subdomain between an open and closed state; the closed state results in phosphoryl transfer when correct Watson–Crick base pairs between the incoming nucleotide and a templating base can fit stably in the active site (29,30). Similarities in the overall mechanism of DNA polymerases suggest a conserved structure of their active sites and provide a basis for comparative structural analysis and extrapolation of site-directed mutagenesis data from one DNA polymerase to its close homologs. The amino acid residues that we attribute to Alpers Cluster 1 contribute to nucleotide polymerization per se, to shaping the overall architecture of the pol active site, or to positioning the primer template within the active site (Figure 2A).
Figure 2.
Alpers Cluster 1 mutations affect the 5′–3′ DNA polymerase activity of Pol γ. Amino acid residues affected by Alpers Cluster 1 mutations in POLG1 are shown as green spheres. Other Pol γ residues that are discussed in the text are shown in brown, and T7 residues are shown in red. Pol domain SSEs are shown in pink according to the schematic shown in Figure 1; ptDNA is indicated by orange (template) and brown (primer) strands. The incoming dNTP is shown in blue. Mg2+ ions are shown as small gray spheres. A, upper panel, overview of the positions of Alpers Cluster 1 mutations with dashed black lines indicating the regions described in the text and depicted in B–D; A, lower panel, overview of the positions of Alpers Cluster 1 mutations relative to ptDNA and incoming dNTP; B–D, positioning of Pol γ residues in the apoenzyme form [PDB code 3IKM (7)] and T7 Pol residues in its closed ternary complex [PDB code 1T8E (25)] relative to ptDNA and incoming dNTP, with the dashed arrow showing the expected movements of the Pol γ residues upon formation of a closed complex: B, Alpers mutations surrounding the Mg2+-binding residues; C, Alpers mutations affecting O-helix movement and dNTP binding, with the T7 Pol O-helix in its closed conformation superimposed in red; D, Alpers mutations affecting the RR loop and the surrounding residues.
Catalytic residues
The largest group of recessive Alpers mutations map to the pol domain in POLG1 (Figure 1), and a number of the affected amino acids are likely involved in catalysis by Pol γ. Residues E1136 and K1191 surround the highly conserved residues D890 and D1135 (Figure 2B), which locate at the base of the pol active site and have been shown to coordinate the two magnesium ions required for catalysis (7). In vitro mutagenesis of residue E883 in Klenow (E1136 in Pol γ) resulted in decreased pol rate, kcat and affinity for dNTPs and DNA (31,32), which fits with the recessive nature of this mutation and its effect on mtDNA copy number in vivo (16,23). Comparative structural analysis of Pol γ and its close homologs revealed that E895 likely confers discrimination against ribonucleotides, and it is involved in aligning the incoming dNTP within the catalytic site, together with the catalytic residues Y955 and Y951 [Figure 2C (33)]. Interestingly, E895 also occupies part of the dNTP-binding pocket, and whereas E895G is a recessive mutation, Y955C is dominant. In vitro mutagenesis of Pol γ showed that the Y955C variant exhibited dramatically reduced catalytic activity, but had the same affinity for DNA as the wild-type enzyme (33). In a disease situation, these effects cannot be compensated for by the wild-type allele and as a result, Y955C causes a dominantly inherited adult-onset mitochondrial myopathy (autosomal dominant progressive external ophthalmoplegia) associated with mtDNA deletions (34). E1136 is part of one of three conserved sequence motifs in the palm subdomain of family A DNA polymerases (the PolC motif, Figure 2B). Its counterparts E655 in the T7 Pol (25) (Figure 2B) and E786 in the Klentaq (26) structures are not in contact with primer or incoming dNTP, but mutagenesis of Klenow showed that this residue is critical for catalysis (31). The ternary complex of T7 Pol reveals an electrostatic interaction between E655 and H704, and suggests that the role of E655 is to position H704 in close proximity to the phosphate moiety of the 3′ nt in the primer strand, and it was shown to be essential for catalysis (25). K1191 of Pol γ occupies the equivalent position of H704 in T7 Pol, suggesting that it would interact with E1136 in a similar manner upon binding of the primer template to facilitate catalysis.
In contrast to the rigid pocket formed by the palm active site residues, the opposite half of the active site is highly flexible to allow a fast transition between open and closed conformations of the fingers subdomain. This conformational asymmetry provides the structural basis for the high affinity of DNA polymerases for the primer–template junction: the template must be bound firmly and consistently each catalytic cycle to assure the fingers sufficient freedom to examine each incoming nucleotide. Several Alpers mutations have been reported in this region of the fingers, scattered mainly along the O-helix (Figure 2C). Biochemical studies on the A957S mutant showed mild defects in kcat and dNTP binding, but a modest increase in DNA-binding affinity (33). A957 locates in the loop between helices O and O1, within one of the three hinge regions, the ‘GAG’ hinge (residues G956/A957/G958), which in related DNA polymerases is known to confer the flexibility necessary for closing and opening of helixes O and O1 (27,35). Glycine to alanine substitution in the GAG hinge in T7 Pol resulted in complete loss of catalytic activity, which can be attributed to complete or diminished ability to shift between the open and closed states (36). Mutations affecting the hinge region likely introduce steric clashes that would limit the extent of conformational shifts. Interestingly, different amino acid replacements for A957 in Pol γ do not have similar consequences: A957S causes dominantly inherited adult-onset myopathy with multiple mtDNA deletions, whereas an A957P substitution has been reported only as a recessive, compound heterozygous mutation in patients with Alpers syndrome and mtDNA depletion. A957S showed an increased affinity for DNA (33), a consequence that is consistent with a slower open–close rate, and could potentially cause pol stalling or dissociation. In contrast, the severe defect caused by A957P suggests that the presence of proline in the hinge abolishes the flexibility between helixes O and O1, a defect that we predict will result in decreased affinity for both dNTP and DNA and may prohibit the formation of a productive ternary complex. The loop between helixes O and O1 has been shown to form the pre-insertion site in Geobacillus stearothermophilus Klenow to accommodate the ssDNA template and prevent its premature entry into the active site (35). However, the available library of DNA polymerase structures reveals variability in the pre-insertion site, making it difficult to assign a critical role. Residue K947, which is associated with Alpers syndrome, is located on the O-helix adjacent to R943. The corresponding residues in T7 Pol (K522 and R518, respectively) contact the triphosphate moiety of the incoming nucleotide in the closed ternary complex (PDB code 1T8E). These electrostatic interactions provide most of the driving force for O-helix movement, and mutations in these positions will weaken interactions.
Architectural residues
Optimal catalysis requires not only the residues interacting directly with dNTP and primer template, but also those that maintain the architecture of the pol active site. We suggest that the pathogenic role of the Alpers mutations Q879H, T885S, L886P, G888S, E1143 and D1184N can be attributed to modifications in the local β-sheet architecture surrounding the catalytic residues in the palm (Figure 2A, upper panel). Defects are predicted to reduce pol rate, and it appears that the closer the mutated architectural residue is to the catalytic site, the more pronounced its effect. In support of this, in vitro mutagenesis of Q879 and T885, located a distance of ~15 Å from the catalytic residues, caused only a 2-fold decrease in pol rate (20). In contrast, mutagenesis of E883 located at the beginning of the strand β13 within the catalytic site in Klenow (E1136 in Pol γA) reduced kcat 26-fold (31).
The Alpers mutation D930N (Figure 2C) was studied in vivo in yeast, and led to complete loss of mtDNA (37). The corresponding residue in T7 Pol (D504) interacts with triphosphate-binding residue R518 (R943 in Pol γ) in the closed conformation (PDB code 1T8E), and therefore contributes indirectly to correct dNTP binding. Residues T914 and L966 located in the fingers subdomain (Figure 2C) are conserved only among Pol γs, and are linked to Alpers syndrome. Our structural analysis predicts that mutations in these residues will likely compromise the stability and conformation, and/ or the transition rate between the open and closed states of the fingers. In sum, we propose that the primary effect of mutations surrounding the O-helix residues is altered affinity for dNTPs, which decreases pol rate.
Residues conferring a high affinity of the pol site for the primer–template junction
Docking of DNA onto the Pol γ structure shows that the DNA-binding channel of the enzyme encompasses ~20 bp of DNA duplex, whereas those in T7 Pol and Klentaq are relatively short and interact with ~8 bp of DNA duplex (24–26). In Pol γ, similar to other family A DNA polymerases, the DNA-binding channel starts with residues located in the β7-loop-β8 motif in the palm, which form extensive contacts with the first four base pairs in the ptDNA. In Pol γA, this fragment encompasses residues 842–856 and appears as a loop in the crystal structure with the current resolution of 3.24 Å (7). Because the β7-loop-β8 motif has two arginines in its tip, R852 and R853, the former of which is conserved only among Pol γs, we designated it as the ‘RR loop’ (Figure 2D). Several recessive mutations affect the RR loop: G848S, T849H, T851A, R852C and R853Q, as well as the surrounding residues H1134R and H1110Y. In vitro mutagenesis of G848, T851, R852 and R853 in human Pol γ (20), and R668 in Klenow ((31,32), corresponding to R853 in Pol γ), caused a dramatic decrease in catalytic activity and DNA-binding affinity. Polesky et al. (32) highlighted the complex effect of these mutations, noting that a specific amino acid can bind a critical part of the substrate, contributing to the overall affinity of the enzyme for ptDNA, and to catalysis by positioning the substrate in the active site. Comparative structural analysis of Pol γ homologs solved in ternary complexes with bound DNA prompts us to propose that the primary function of the RR loop is in binding and alignment of ptDNA within the DNA-binding channel. H1134 locates adjacent to the RR loop in the tertiary structure (Figure 2D). The corresponding histidines in Klenow (31,32), T7 Pol (25) and Klentaq (26) were shown to be essential for DNA binding, and for coordination of the first nucleotide in the primer strand. Analysis of Pol γ suggests that Q1102 from the Q-helix (Q615 in T7 Pol) and R853 (R429 in T7 Pol) from the RR loop coordinate the first base pair in the ptDNA, and therefore may be crucial in sensing a mispair. T851 (T427 in T7 Pol) likely participates indirectly by positioning Q1102. T849 is predicted to coordinate the DNA duplex by interaction with the phosphate moiety of the third nucleotide from the 3′-end of the primer. In the Pol γ apoenzyme structure, the side chains of the residues from the RR loop are oriented differently than the orthologous residues in the T7 Pol ternary complex, which suggests that DNA and dNTP binding affect the positions of the residues that coordinate them. For example, R853 in the Pol γ apoenzyme interacts with the metal-binding residue D1135 as was noted by Stumpf and Copeland (17) and with E895, but in complex with DNA, R853 would shift to interact with the base of the 3′ nt of the primer strand, as in T7 Pol (24–26). A distinguishing feature of the RR loop, in comparison with the orthologous β7-loop-β8 motif in other family A DNA polymerase group members, is the electrostatic interaction between R852 and D1107, which may confer additional stability to the RR loop in Pol γ (Figure 2D). Disease mutations in this domain affect primarily DNA binding and positioning in the active site channel, manifesting defects in both DNA-binding affinity and in catalytic efficiency.
CLUSTER 2: RECESSIVE MUTATIONS AFFECTING THE UPSTREAM DNA-BINDING CHANNEL
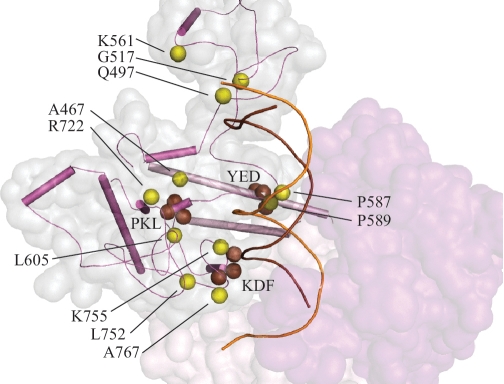
Processivity of a DNA polymerase depends both on DNA-binding affinity and on the rate of DNA polymerization. Structural analysis shows that the mutations affecting interaction of Pol γ with the upstream DNA duplex are located within the spacer domain (Figure 1). This would argue that the AID [accessory (subunit)-interacting domain] and IP (intrinsic processivity) subdomains adopt a different conformation in Pol γ: DNA complexes. To evaluate the potential impact of recessive spacer region mutations on DNA binding, we considered results of in vitro mutagenesis of spacer domain in Drosophila Pol γ (38) and in Saccharomyces cerevisiae Mip1 (22) (Figure 3). Analysis of the Pol γ structure with modeled ptDNA reveals that residues K755 from the IP subdomain and Q497 in the K-tract of the AID subdomain interact with upstream DNA. In the human Pol γ:DNA model, the L752P and A767D substitutions would affect the structure in the tip of IP subdomain, which contacts the minor groove of the DNA duplex in a similar way as reported for T7 Pol [residues T354-V363 (25)]. In the Pol γ:DNA complex, L752 and A767 neighbor the K768/D769/F770 triplet from fly Pol γ; triple alanine substitution of KDF in fly Pol γ resulted in a 1.4-fold decrease in DNA-binding affinity (38). We predict that the Alpers A767D mutation likely causes similar consequences for DNA binding. Mutations of P587 and P589 in the spacer domain are also linked to Alpers disease. These residues constrain the β–hairpin loop between the IP and AID subdomains and are positioned close to the Y452/E453/E454 triplet that lies within the thumb [corresponding to the YED triplet in Drosophila Pol γ (38)]. We postulate that the P587L and P589L mutations may suffer the same consequences as for the triple alanine substitution of YED in the fly enzyme, which caused reduced processivity, most likely as a result of misalignment of the ptDNA, with respect to the pol catalytic site (38). Furthermore, a triple alanine mutant of P556A/K557A/L558A in the fly was shown to be completely deficient in DNA binding and pol activity (38); this defect is likely due to disruption of the hydrophobic structure of the spacer domain, which shapes the DNA-binding channel wall. Notably, the residue affected in the common A467T human disease allele is also part of this PKL hydrophobic center, and its biochemical properties most likely derive from similar structural alterations that interrupt hydrophobicity with the introduction of a hydroxyl group. Whereas we and others have found that the A467T catalytic core alone exhibits highly reduced DNA-binding affinity and pol activity (19,21), we showed that these defects are mitigated partially by its association with the accessory subunit (21). This likely results from partial stabilization of the overall conformation of mutant catalytic core within the reconstituted Pol γ holoenzyme.
Figure 3.
Alpers Cluster 2 mutations affect the upstream DNA-binding channel of Pol γ. Amino acid residues affected by Alpers Cluster 2 mutations in POLG1 are shown as yellow spheres. Other Pol γ residues that are discussed in the text are shown in brown. Spacer domain SSEs are shown in magenta and pol domain SSEs are in pink according to the schematic shown in Figure 1; ptDNA is indicated by orange (template) and brown (primer) strands. The spacer domain is also shown as a transparent surface representation in pale gray and the exo domain is shown in purple.
CLUSTER 3: MUTATIONS ASSOCIATED WITH A NOVEL, POL γ-SPECIFIC FUNCTIONAL MODULE, CONFERRING PARTITIONING OF DNA SUBSTRATE BETWEEN THE POL AND EXO ACTIVE SITES
Comparative analysis of the pol domains from human Pol γ, T7 Pol and T7 RNA Pol reveals a remarkable structural similarity in their overall folds. In fact, the only significant difference between the analogous pol domains lies in the region that connects the P-helix of the fingers subdomain and Q-helix of the palm (Supplementary Figure S2), which contains a novel module (Figure 4) whose amino acid sequence is highly conserved in Pol γs from yeast to man. This, and the finding that several recessive Alpers mutations cluster in this region (Figure 1), argue that it serves an important role. Indeed, in our Pol γ:DNA model, this module extends into the DNA-binding channel in a position very near the pol active site (<10 Å from the Mg2+-binding catalytic residues). In bacteriophage N4 RNA polymerase (PDB code 2PO4) (39) and in T7 RNA polymerase [PDB code 1ARO (40); Supplementary Figure S2], the fragment connecting the fingers with the helix corresponding to the Q-helix in Pol γ participates in specific recognition of the transcriptional promoter; it is aptly termed a specificity loop (39–41). In Pol γ, we have adopted the term ‘partitioning loop’ based upon the functional role we propose it serves. We discuss our rationale below, and use it to establish structure–function justifications of disease manifestation for the Alpers mutations we assign to Cluster 3.
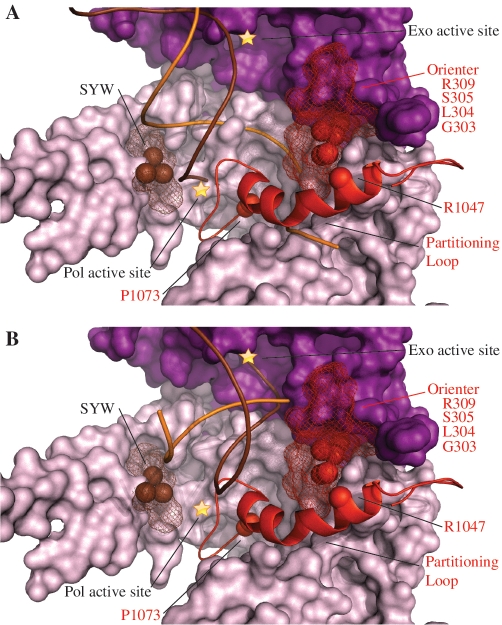
Figure 4.
Alpers Cluster 3 mutations are associated with a novel Pol γ-specific functional module proposed to be involved in primer strand partitioning between the pol and exo sites. Amino acid residues affected by Alpers Cluster 3 mutations in POLG1 are shown as red spheres/mesh adjacent to a novel alpha helix with an associated loop–hairpin (the partitioning loop), also shown in red. The brown spheres and mesh represent the SYW (fly)/ SFW (human) and surrounding residues, respectively, that are described in the text. The pol domain is shown as a surface representation in pink and the exo domain is shown in purple, according to the schematic shown in Figure 1; ptDNA is indicated by orange (template) and brown (primer) strands. (A) The predicted position of the partitioning loop relative to ptDNA in the pol mode, and (B) represents the exo mode. To dock the frayed ptDNA in the exo active site, the exo domain residues 324–518 of Klenow (PDB code 1KLN) were aligned with the exo domain residues 170–440 of Pol γ (PDB code 3IKM) (Supplementary Figure S3).
We propose that the partitioning loop modulates the partitioning of the primer strand between the pol and exo active sites by forming stable contacts with correctly base-paired ptDNA, and destabilizing ptDNA that contains mispairs or lesions. Comprising residues 1050–1095, the atomic structure reveals that the first 10 residues of the partitioning loop adopt an α-helical fold that extends from the fingers subdomain directly into the DNA-binding channel. The beginning of this helix contains a W1049XGG1052 motif, which is strictly conserved in Pol γ, and a similar motif is also present in T7 Pol (W579XAG582). Our Pol γ:DNA model shows that the WXXG motif maps to the same region of the fingers domain in both Pols. In T7 Pol, it is responsible for binding downstream template ssDNA via base-stacking interactions between W579 and the nucleotide base [PDB code 1T8E (25)], and likely performs an equivalent function in Pol γ. The novel functionality that the partitioning loop contributes to Pol γ is achieved by its unusual loop–hairpin component spanning residues 1060–1074 that appears to grip the modeled ptDNA along the major groove as shown in Figure 4A. We propose that it uses a steric exclusion mechanism that confers exquisite specificity by associating closely with the major groove of primer template, such that correctly base-paired substrates are bound stably. In contrast, DNA lesions and mispairs that adopt an altered helical structure clash sterically with the partitioning loop, resulting in exclusion of the primer template from the pol active site. Excluding primer template may facilitate fraying of the duplex, generating ssDNA primer ends that can be bound by and hydrolyzed in the exo active site (Figure 4B).
Mutations in the residues composing the partitioning loop, such as the Alpers mutations R1047W and P1073L, as well as the progressive external ophthalmoplegia mutations G1051R and G1076V, have been studied in vivo. Yeast strains homozygous for G1051R Pol γ exhibited a point mutational frequency >10-fold higher than the wild-type strain, and heterozygous strains showed frequencies >2-fold higher relative to homozygous wild-type strains (42). Increased point mutation frequencies were also reported for yeast strains heterozygous and homozygous for the Alpers mutation P1073L and the G1076V mutation associated with progressive external ophthalmoplegia (22,43). Both the P1073L and G1076V mutations cause substitutions in strictly conserved residues located in the loop region that is expected to contact DNA, and the G1051R mutation affects a strictly conserved glycine in the WXXG motif. Interestingly, yeast strains heterozygous and homozygous for L304R, S305R, Q308H, R309L and R309H showed similar increases in point mutation frequency as well as mtDNA depletion (43). These exo domain residues are located in a loop–helix motif adjacent to the partitioning loop, and have been described previously as the orienter module (22) (Figure 4). When S305R and P1073L were present as compound heterozygous, the point mutation frequency increased drastically to >70-fold that of the wild-type strain (44). Non-complementation of these two mutations suggests that both are involved in the same function, and we propose that the role of the orienter module is to position correctly the partitioning loop. L304R, R309H, R309L and W312R were analyzed biochemically by producing and purifying homologous yeast Pol γ variants (22). All variants exhibited reduced DNA-binding affinity and reduced pol activity and in addition, the L304R variant showed a significant increase in exo activity (22). An identical biochemical phenotype was reported previously in the fly Pol γ SYW triple alanine variant, which correspond to thumb residues S799/F800/W801 in human Pol γ (38). As illustrated in Figure 4, the SYW residues are located on the face of the thumb subdomain directly across the DNA-binding channel from the orienter module, and we predict that this region of the thumb plays an equivalent role to the orienter module. Therefore, we define Cluster 3 as mutations affecting residues of the partitioning loop, as well as nearby residues that govern the position and conformation of the partitioning loop. Mutations in Cluster 3 will alter the balance between pol and exo activity and diminish the fidelity of the polymerase. We note the possibility that enhanced fidelity may also be experimentally feasible, introducing the potential to engineer a fine-tuned anti-mutator replicase.
In sum, our justification for the proposed role of the partitioning loop is based on its sequence conservation among Pol γ's from yeast to man, its positioning relative to the specificity loop found in T7 RNA Pol, the clustering of several recessive Alpers mutations in this region, its absence in any other Pol family and the documented high fidelity of Pol γ. We argue that an alternate hypothesis that this structural element would simply rotate away upon DNA binding (7) seems unlikely based on the above considerations and, in particular, on the evolutionary conservation of a novel structural module that is positioned strategically in the DNA-binding channel very near the pol active site. Clearly, validation of either hypothesis warrants future experimentation.
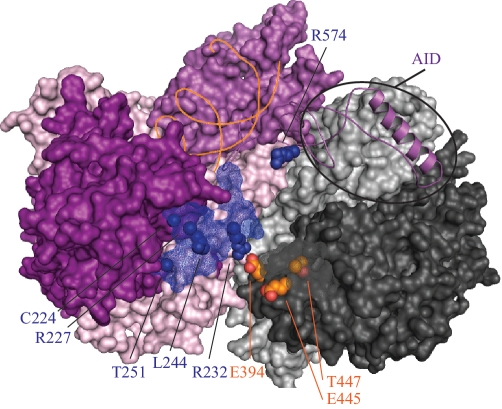
CLUSTER 4: MUTATIONS AFFECTING POL γA INTERACTIONS WITH THE DISTAL POL γB UPON DNA BINDING BY POL γ HOLOENZYME
Pol γA contains independent 5′–3′ DNA polymerase and 3′–5′ exonuclease active sites whose functions are coordinated in proofreading DNA synthesis. The human Pol γ crystal structure showed that its homodimeric Pol γB interacts asymmetrically with Pol γA, such that one Pol γB protomer forms the dominant subunit interface and is designated as the proximal accessory subunit, while the other protomer makes very limited contact with Pol γA and is designated as the distal accessory subunit (7). In fact in the apoezyme structure, the Pol γA interaction with the distal Pol γB is mediated through one ion bond between Pol γA R232 and Pol γB E394 [(7) and Figure 5]. A subsequent biochemical study on the interaction between the catalytic and accessory subunits showed that the proximal Pol γB increases DNA-binding affinity of the holoenzyme, while the distal protomer enhances the polymerization rate of the holoenzyme (45). Again as suggested in the section on Cluster 2, we posit that the holoenzyme likely undergoes substantial conformational rearrangements in the IP and AID subdomains of the spacer upon formation of an active complex with DNA.
Figure 5.
Alpers Cluster 4 mutations affect Pol γA interactions with the distal Pol γB upon DNA binding by Pol γ holoenzyme. Amino acid residues affected by Alpers Cluster 4 mutations in POLG1 are shown as blue spheres/ mesh and are located largely within the exo domain (shown in purple). The helix shown in magenta is the AID (accessory interacting domain) of the spacer region that interacts with the proximal accessory subunit. Other domains are colored according to the schematic shown in Figure 1; duplex DNA is indicated by the orange strand. Orange spheres indicate the positions of the accessory subunit residues described in the text. The proximal and distal accessory subunits are shown as surface representations in light and dark gray, respectively.
Mapping of Alpers mutations within the crystal structure revealed that the majority of the mutations in the exo domain cluster on the protein surface within 13–17 Å from Pol γA residue R232 (Cluster 4 shown in blue in Figures 1 and 5). The functional role of this region can be deduced from biochemical studies on mutations of the neighboring R232 (46) and L244 residues (22,43). Mutations of R232 were shown to decrease pol activity, DNA-binding affinity and processivity of the holoenzyme yet at the same time, to enhance its exonuclease activity, which was also rendered less selective for mismatches (46). These data argue that in the Pol γ:DNA complex, and/or in its ternary complex with dNTP, the distal accessory subunit associates more tightly with the catalytic subunit, thereby enhancing binding of the upstream DNA in the pol mode, and limiting the rate of translocation of the frayed 3′-end of the primer to the exo site (46). Accordingly, an orthologous variant of the L244P human mutation in yeast caused increased mutation frequency (22,43). Previous site-directed mutagenesis of human Pol γB also suggested extensive interaction between the catalytic and distal accessory subunits within the ternary complex (5). Analysis of the Pol γ apoenzyme structure shows that E445 and T447 of Pol γB are located at the tip of the its C-terminal region, and that only in the distal subunit are these residues oriented toward Cluster 4 on the edge of exo domain. A double mutation of these residues (E445A/ T447A) led to a dramatic decrease of the stimulatory effect of Pol γB on the wild-type Pol γA, and a decrease in DNA binding (5). We propose that residues in Cluster 4 of Pol γA contribute to tight interaction between exo domain of catalytic subunit and the distal accessory subunit, resulting in more extensive enclosure of the primer template in the pol mode. We also predict the participation of the AID of the spacer domain in these conformational changes, because it is associated intimately with the accessory subunit. In support of this, biochemical studies performed on the yeast Pol γ core variant of Alpers mutation R574, which is located in the AID subdomain, showed not only reduced DNA-binding affinity, but also reduced exo activity and a severe processivity defect (22,43). To some extent, these interactions would provide structural constraints on the upstream DNA in the direction toward the partitioning loop. Therefore, the partitioning loop, orienter module and thumb in Pol γA, and the interface between its exo domain and the distal accessory subunit might function in a concerted manner to affect switching between the pol and exo modes in Pol γ function.
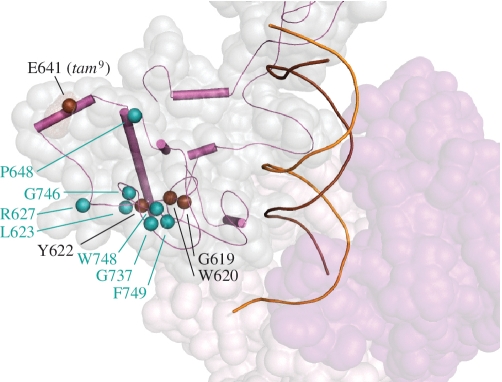
CLUSTER 5: MUTATIONS AFFECTING A REGION OF THE IP SUBDOMAIN THAT IS LIKELY INVOLVED IN REPLISOME CONTACTS
Many reported Alpers mutations (L623W, R627W/Q, P648R, G737R, G746S, W748S and F749S) map to the distal surface of the IP subdomain (Figure 6). We considered these as distinct from the Cluster 2 residues because they are located much further from the DNA-binding channel. In addition, in vitro studies of W748S and R627W/Q variants showed no defects in pol activity, processivity or DNA-binding affinity (47,48). In contrast, biochemical defects were demonstrated earlier in single alanine variants of fly Pol γ within this region of the spacer: a W576A variant was nearly inactive, F578A retained half of wild-type activity, G575A displayed wild-type activity and all three variants showed substantially reduced stimulation by mtSSB (38). The fly G575/W576/F578 residues correspond to residues G619/W620/Y622 in human Pol γ. In the human Pol γ apoenzyme structure, W620 is buried in the hydrophobic core of the IP subdomain, whereas Y622 and G619 are located closer to the surface (7). The combined biochemical data suggest that residues closer to the distal surface of the IP domain are not critical for catalytic function. Cluster 5 mutations map to this surface, and therefore a catalytic defect per se is not expected to be the cause of Alpers manifestation. Rather, we propose that this region is involved in protein–protein contacts, a likely partner being mtSSB as suggested by the GWF fly Pol γ variants (38). Another candidate may be the mitochondrial replicative DNA helicase Twinkle, but such an interaction has not been investigated to date.
Figure 6.
Alpers Cluster 5 mutations are proposed to affect replisome interactions. Amino acid residues affected by Alpers Cluster 5 mutations in POLG1 are shown as cyan spheres. Spacer domain SSEs within the IP (intrinsic processivity subdomain) are shown in magenta; ptDNA is indicated by orange (template) and brown (primer) strands. The spacer domain is also shown as a transparent surface representation in pale gray and the exo domain is shown in purple. Brown spheres indicate the positions of the IP residues described in the text.
PROSPECTS
Toward a diagnostic tool to assess new human mitochondrial disease mutations
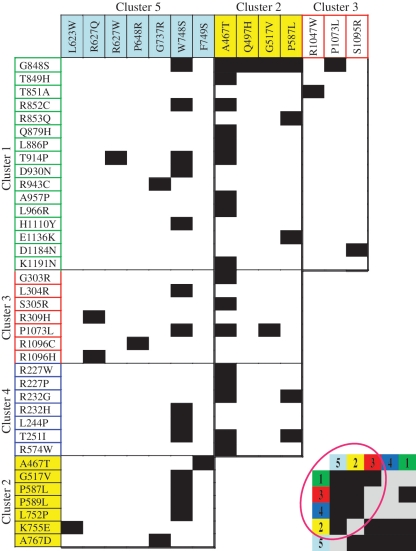
We show that although the reported Alpers mutations scatter across the entire POLG1 gene, they cluster to distinct functional regions in the Pol γ tertiary structure. Table 1 summarizes the structure–function predictions for each cluster and also displays the cluster combinations that have been reported in compound heterozygous patients with Alpers syndrome. These combinations do not occur randomly; rather, multiple occurrences of specific cluster combinations is evident whereas others are absent, suggesting that the latter combinations are not tolerated. Cluster 1 mutations locate in the pol active site region and will invariably cause reduced pol activity, potentially combined with reduced DNA binding. Cluster 2 mutations locate in the upstream DNA-binding channel, resulting in reduced DNA-binding affinity, but are too distant from the catalytic site to affect directly pol activity to the extent of Cluster 1 mutations. Therefore, Cluster 2 mutations will be recessive, whereas Cluster 1 mutations may be either dominant or recessive, depending on the severity of the biochemical defect. Cluster 3 mutations locate to the partitioning loop or its environs, such as the orienter module, and we predict that they will alter the balance of polymerase and exonuclease activities of mutant Pol γ. Cluster 4 mutations locate on the surface of the exo domain along the interface of the distal accessory subunit. These mutations are predicted to reduce the stimulation affected by the distal accessory subunit, which has been documented to enhance pol rate and to reduce exo activity (45). Both Clusters 3 and 4 mutations will likely cause increased mutagenesis in vivo, a phenotype that has been observed in yeast models to be associated with amino acid alterations within these clusters (43). Cluster 5 mutations locate on the distal surface of the intrinsic processivity (IP) subdomain, removed from the pol active site, DNA-binding channel and accessory subunit interface. Their distant location and lack of biochemical defects prompt us to speculate that this region is involved in replisome contacts. This hypothesis, in particular, highlights the need for future experimentation on both the physical and functional interactions among the key proteins at the mtDNA replication fork.
Table 1.
Structural and functional features of the five proposed Alpers Clusters
| Cluster | Structural location | Predicted primary biochemical defect | Predicted phenotype (primary, secondary) | Causes Alpers when found in trans with Cluster |
|---|---|---|---|---|
| 1 | Pol active site and environs | Pol activity |
|
2, 3 or 5 |
| 2 | Upstream DNA-binding channel | DNA-binding affinity |
|
1, 3, 4 or 5 |
| 3 | Partitioning loop | Partitioning of primer strand between pol and exo active sites |
|
1, 2 or 5 |
| 4 | Exo-IP interaction (Stabilized by distal Pol γB) | Stabilization of ternary complex |
|
2 or 5 |
| 5 | Periphery of IP domain | Protein–protein interactions |
|
1, 2, 3 or 4 |
POLG1 shows significant polymorphic variation in the human population, and a constant challenge in DNA diagnosis is to distinguish pathogenic mutations from neutral variants. We suggest that the remarkable clustering of Alpers mutations into specific functional regions (Figure 1) enables the use of our Pol γ:DNA model as a diagnosis-supporting tool for evaluating pathogenic potential of new sequence variants. For example, Q1102 is located within the Cluster 1 region, and mutations affecting this amino acid can be predicted to cause catalytic defects. This prediction is consistent with biochemical studies on the Q1102 counterpart in bacteriophage T7 Pol (Q615), which was shown to be essential for the fidelity of nucleotide incorporation and for ptDNA binding (25). Functional predictions can be made in the absence of biochemical data as well; for example, the tam9 mutant allele, carrying an E595V mutation in the catalytic subunit of Pol γ, causes mtDNA depletion in the fly (49,50). E595V corresponds to E641V in human Pol γ, which maps to Cluster 5 (Figure 6). Our model predicts that the tam9-associated mtDNA depletion is caused by deficient replisome interactions. Consequently, we would predict that both Q1102 and E641 are potential candidates for causing recessive POLG syndrome with mtDNA depletion.
Predicting the consequences of compound heterozygosity via Alpers cluster analysis
Alpers syndrome is typically caused by compound heterozygosity of two mutations occurring along the entire length of the POLG1 gene. In contrast, homozygosity for a single mutation often is associated with a somewhat milder disorder (2,51). Utilizing the Human DNA Polymerase γ Database, we compiled data to show all combinations of mutations that have been reported to trigger Alpers disease (Figure 7). Alpers patients typically do not show a combination of two mutations from the same cluster. In addition, Cluster 4 mutations only manifest as Alpers disease when combined with Clusters 2 or 5 mutations. These trends provide merit for our structure-guided assignment of the mutations into the five proposed clusters, and suggest unique functional roles inherent to each. With the rapid development of massive parallel sequencing methodologies, the number of new Pol γA variants is likely to increase significantly, emphasizing the importance of bioinformatic tools to evaluate the pathogenic role of identified variants. Furthermore, the identification of synergistic functional relationships between the characterized clusters provides novel insight into the mechanism of Alpers disease manifestation, and will serve as a framework for future structure–function studies of the mitochondrial DNA replicase.
Figure 7.
Combinations of mutations found in Alpers patients. Individual Alpers mutations are grouped by cluster as shown in Figure 1, and black blocks represent Alpers manifesting combinations. The inset in the lower right presents a simplified version of the table by reducing the axes to the five clusters only, where gray blocks represent cluster combinations that have not been found in Alpers patients. The tabulated data suggest that two mutations from the same cluster do not typically manifest as early-onset Alpers disease. Furthermore, the data indicate that Cluster 4 (blue) mutations manifest as Alpers disease only when in combination with Cluster 2 (yellow) or Cluster 5 (cyan) mutations. These trends support the existence of unique functional relationships in Pol γA that are inherent to each cluster.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (GM45295 to L.S.K.); Academy of Finland, University of Helsinki, Jane, and Aatos Erkko Foundation and Sigrid Juselius Foundation (to A.S.). Funding for open access charge: National Institutes of Health (GM45295 to L.S.K.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Dr Li Fan (University of California, Riverside) and the members of their labs for critical reading of the manuscript and many helpful discussions. G.F. thanks Professor Robert L. Laduca for his enthusiastic mentoring, scientific inspiration and exceptional training in undergraduate research.
REFERENCES
- 1.Kaguni LS. DNA polymerase gamma, the mitochondrial replicase. Ann. Rev. Biochem. 2004;73:293–320. doi: 10.1146/annurev.biochem.72.121801.161455. [DOI] [PubMed] [Google Scholar]
- 2.Graziewicz MA, Longley MJ, Copeland WC. DNA polymerase gamma in mitochondrial DNA replication and repair. Chem. Rev. 2006;106:383–405. doi: 10.1021/cr040463d. [DOI] [PubMed] [Google Scholar]
- 3.Yakubovskaya E, Chen Z, Carrodeguas JA, Kisker C, Bogenhagen DF. Functional human mitochondrial DNA polymerase gamma forms a heterotrimer. J. Biol. Chem. 2006;281:374–382. doi: 10.1074/jbc.M509730200. [DOI] [PubMed] [Google Scholar]
- 4.Longley MJ, Prasad R, Srivastava DK, Wilson SH, Copeland WC. Identification of 5′-deoxyribose phosphate lyase activity in human DNA polymerase gamma and its role in mitochondrial base excision repair in vitro. Proc. Natl Acad. Sci. USA. 1998;95:12244–12248. doi: 10.1073/pnas.95.21.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fan L, Kim S, Farr CL, Schaefer KT, Randolph KM, Tainer JA, Kaguni LS. A novel processive mechanism for DNA synthesis revealed by structure, modeling and mutagenesis of the accessory subunit of human mitochondrial DNA polymerase. J. Mol. Biol. 2006;358:1229–1243. doi: 10.1016/j.jmb.2006.02.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim SE, Longley MJ, Copeland WC. The mitochondrial p55 accessory subunit of human DNA polymerase gamma enhances DNA binding, promotes processive DNA synthesis, and confers N-ethylmaleimide resistance. J. Biol. Chem. 1999;274:38197–38203. doi: 10.1074/jbc.274.53.38197. [DOI] [PubMed] [Google Scholar]
- 7.Lee YS, Kennedy WD, Yin YW. Structural insight into processive human mitochondrial DNA synthesis and disease-related polymerase mutations. Cell. 2009;139:312–324. doi: 10.1016/j.cell.2009.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suomalainen A, Majander A, Haltia M, Somer H, Lonnqvist J, Savontaus ML, Peltonen L. Multiple deletions of mitochondrial DNA in several tissues of a patient with severe retarded depression and familial progressive external ophthalmoplegia. J. Clin. Invest. 1992;90:61–66. doi: 10.1172/JCI115856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeviani M, Servidei S, Gellera C, Bertini E, DiMauro S, DiDonato S. An autosomal dominant disorder with multiple deletions of mitochondrial DNA starting at the D-loop region. Nature. 1989;339:309–311. doi: 10.1038/339309a0. [DOI] [PubMed] [Google Scholar]
- 10.Hakonen AH, Heiskanen S, Juvonen V, Lappalainen I, Luoma PT, Rantamaki M, Goethem GV, Lofgren A, Hackman P, Paetau A, et al. Mitochondrial DNA polymerase W748S mutation: a common cause of autosomal recessive ataxia with ancient European origin. Am. J. Hum. Genet. 2005;77:430–441. doi: 10.1086/444548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Goethem G, Luoma P, Rantamaki M, Al Memar A, Kaakkola S, Hackman P, Krahe R, Lofgren A, Martin JJ, De Jonghe P, et al. POLG mutations in neurodegenerative disorders with ataxia but no muscle involvement. Neurology. 2004;63:1251–1257. doi: 10.1212/01.wnl.0000140494.58732.83. [DOI] [PubMed] [Google Scholar]
- 12.Winterthun S, Ferrari G, He L, Taylor RW, Zeviani M, Turnbull DM, Engelsen BA, Moen G, Bindoff LA. Autosomal recessive mitochondrial ataxic syndrome due to mitochondrial polymerase gamma mutations. Neurology. 2005;64:1204–1208. doi: 10.1212/01.WNL.0000156516.77696.5A. [DOI] [PubMed] [Google Scholar]
- 13.Naviaux RK, Nguyen KV. POLG mutations associated with Alpers' syndrome and mitochondrial DNA depletion. Ann. Neurol. 2004;55:706–712. doi: 10.1002/ana.20079. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen KV, Ostergaard E, Ravn SH, Balslev T, Danielsen ER, Vardag A, McKiernan PJ, Gray G, Naviaux RK. POLG mutations in Alpers syndrome. Neurology. 2005;65:1493–1495. doi: 10.1212/01.wnl.0000182814.55361.70. [DOI] [PubMed] [Google Scholar]
- 15.Suomalainen A, Isohanni P. Mitochondrial DNA depletion syndromes–many genes, common mechanisms. Neuromuscul. Disord. 2010;20:429–437. doi: 10.1016/j.nmd.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 16.Wong LJ, Naviaux RK, Brunetti-Pierri N, Zhang Q, Schmitt ES, Truong C, Milone M, Cohen BH, Wical B, Ganesh J, et al. Molecular and clinical genetics of mitochondrial diseases due to POLG mutations. Hum. Mutat. 2008;29:E150–172. doi: 10.1002/humu.20824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stumpf JD, Copeland WC. Mitochondrial DNA replication and disease: insights from DNA polymerase gamma mutations. Cell Mol. Life Sci. 2011;68:219–233. doi: 10.1007/s00018-010-0530-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atanassova N, Fuste JM, Wanrooij S, Macao B, Goffart S, Backstrom S, Farge G, Khvorostov I, Larsson NG, Spelbrink JN, et al. Sequence-specific stalling of DNA polymerase {gamma} and the effects of mutations causing progressive ophthalmoplegia. Hum. Mol. Genet. 2011;20:1212–1223. doi: 10.1093/hmg/ddq565. [DOI] [PubMed] [Google Scholar]
- 19.Chan SS, Longley MJ, Copeland WC. The common A467T mutation in the human mitochondrial DNA polymerase (POLG) compromises catalytic efficiency and interaction with the accessory subunit. J. Biol. Chem. 2005;280:31341–31346. doi: 10.1074/jbc.M506762200. [DOI] [PubMed] [Google Scholar]
- 20.Kasiviswanathan R, Longley MJ, Chan SS, Copeland WC. Disease mutations in the human mitochondrial DNA polymerase thumb subdomain impart severe defects in mitochondrial DNA replication. J. Biol. Chem. 2009;284:19501–19510. doi: 10.1074/jbc.M109.011940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luoma PT, Luo N, Loscher WN, Farr CL, Horvath R, Wanschitz J, Kiechl S, Kaguni LS, Suomalainen A. Functional defects due to spacer-region mutations of human mitochondrial DNA polymerase in a family with an ataxia-myopathy syndrome. Hum. Mol. Genet. 2005;14:1907–1920. doi: 10.1093/hmg/ddi196. [DOI] [PubMed] [Google Scholar]
- 22.Szczepanowska K, Foury F. A cluster of pathogenic mutations in the 3′-5′ exonuclease domain of DNA polymerase gamma defines a novel module coupling DNA synthesis and degradation. Hum. Mol. Genet. 2010;19:3516–3529. doi: 10.1093/hmg/ddq267. [DOI] [PubMed] [Google Scholar]
- 23.Chan SS, Copeland WC. DNA polymerase gamma and mitochondrial disease: understanding the consequence of POLG mutations. Biochim. Biophys. Acta. 2009;1787:312–319. doi: 10.1016/j.bbabio.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brieba LG, Eichman BF, Kokoska RJ, Doublie S, Kunkel TA, Ellenberger T. Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. EMBO J. 2004;23:3452–3461. doi: 10.1038/sj.emboj.7600354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berdis AJ. Mechanisms of DNA polymerases. Chem. Rev. 2009;109:2862–2879. doi: 10.1021/cr800530b. [DOI] [PubMed] [Google Scholar]
- 28.Johnson KA. The kinetic and chemical mechanism of high-fidelity DNA polymerases. Biochim. Biophys. Acta. 2010;1804:1041–1048. doi: 10.1016/j.bbapap.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee HR, Helquist SA, Kool ET, Johnson KA. Base pair hydrogen bonds are essential for proofreading selectivity by the human mitochondrial DNA polymerase. J. Biol. Chem. 2008;283:14411–14416. doi: 10.1074/jbc.M705006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee HR, Helquist SA, Kool ET, Johnson KA. Importance of hydrogen bonding for efficiency and specificity of the human mitochondrial DNA polymerase. J. Biol. Chem. 2008;283:14402–14410. doi: 10.1074/jbc.M705007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polesky AH, Dahlberg ME, Benkovic SJ, Grindley ND, Joyce CM. Side chains involved in catalysis of the polymerase reaction of DNA polymerase I from Escherichia coli. J. Biol. Chem. 1992;267:8417–8428. [PubMed] [Google Scholar]
- 32.Polesky AH, Steitz TA, Grindley ND, Joyce CM. Identification of residues critical for the polymerase activity of the Klenow fragment of DNA polymerase I from Escherichia coli. J. Biol. Chem. 1990;265:14579–14591. [PubMed] [Google Scholar]
- 33.Graziewicz MA, Longley MJ, Bienstock RJ, Zeviani M, Copeland WC. Structure-function defects of human mitochondrial DNA polymerase in autosomal dominant progressive external ophthalmoplegia. Nat. Struct. Mol. Biol. 2004;11:770–776. doi: 10.1038/nsmb805. [DOI] [PubMed] [Google Scholar]
- 34.Van Goethem G, Dermaut B, Lofgren A, Martin JJ, Van Broeckhoven C. Mutation of POLG is associated with progressive external ophthalmoplegia characterized by mtDNA deletions. Nat. Genet. 2001;28:211–212. doi: 10.1038/90034. [DOI] [PubMed] [Google Scholar]
- 35.Johnson SJ, Taylor JS, Beese LS. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc. Natl Acad. Sci. USA. 2003;100:3895–3900. doi: 10.1073/pnas.0630532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin Z, Johnson KA. Role of a GAG hinge in the nucleotide-induced conformational change governing nucleotide specificity by T7 DNA polymerase. J. Biol. Chem. 2011;286:1312–1322. doi: 10.1074/jbc.M110.156737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baruffini E, Horvath R, Dallabona C, Czermin B, Lamantea E, Bindoff L, Invernizzi F, Ferrero I, Zeviani M, Lodi T. Predicting the contribution of novel POLG mutations to human disease through analysis in yeast model. Mitochondrion. 2011;11:182–190. doi: 10.1016/j.mito.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 38.Luo N, Kaguni LS. Mutations in the spacer region of Drosophila mitochondrial DNA polymerase affect DNA binding, processivity, and the balance between Pol and Exo function. J. Biol. Chem. 2005;280:2491–2497. doi: 10.1074/jbc.M411447200. [DOI] [PubMed] [Google Scholar]
- 39.Murakami KS, Davydova EK, Rothman-Denes LB. X-ray crystal structure of the polymerase domain of the bacteriophage N4 virion RNA polymerase. Proc. Natl Acad. Sci. USA. 2008;105:5046–5051. doi: 10.1073/pnas.0712325105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeruzalmi D, Steitz TA. Structure of T7 RNA polymerase complexed to the transcriptional inhibitor T7 lysozyme. EMBO J. 1998;17:4101–4113. doi: 10.1093/emboj/17.14.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gleghorn ML, Davydova EK, Rothman-Denes LB, Murakami KS. Structural basis for DNA-hairpin promoter recognition by the bacteriophage N4 virion RNA polymerase. Mol. Cell. 2008;32:707–717. doi: 10.1016/j.molcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baruffini E, Ferrero I, Foury F. Mitochondrial DNA defects in Saccharomyces cerevisiae caused by functional interactions between DNA polymerase gamma mutations associated with disease in human. Biochim. Biophys. Acta. 2007;1772:1225–1235. doi: 10.1016/j.bbadis.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 43.Stumpf JD, Bailey CM, Spell D, Stillwagon M, Anderson KS, Copeland WC. mip1 containing mutations associated with mitochondrial disease causes mutagenesis and depletion of mtDNA in Saccharomyces cerevisiae. Hum. Mol. Genet. 2010;19:2123–2133. doi: 10.1093/hmg/ddq089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baruffini E, Lodi T. Construction and validation of a yeast model system for studying in vivo the susceptibility to nucleoside analogues of DNA polymerase gamma allelic variants. Mitochondrion. 2010;10:183–187. doi: 10.1016/j.mito.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 45.Lee YS, Lee S, Demeler B, Molineux IJ, Johnson KA, Yin YW. Each monomer of the dimeric accessory protein for human mitochondrial DNA polymerase has a distinct role in conferring processivity. J. Biol. Chem. 2010;285:1490–1499. doi: 10.1074/jbc.M109.062752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee YS, Johnson KA, Molineux IJ, Yin YW. A single mutation in human mitochondrial DNA polymerase Pol gammaA affects both polymerization and proofreading activities of only the holoenzyme. J. Biol. Chem. 2010;285:28105–28116. doi: 10.1074/jbc.M110.122283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan SS, Longley MJ, Copeland WC. Modulation of the W748S mutation in DNA polymerase gamma by the E1143G polymorphismin mitochondrial disorders. Hum. Mol. Genet. 2006;15:3473–3483. doi: 10.1093/hmg/ddl424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palin EJ, Lesonen A, Farr CL, Euro L, Suomalainen A, Kaguni LS. Functional analysis of H. sapiens DNA polymerase gamma spacer mutation W748S with and without common variant E1143G. Biochim. Biophys. Acta. 2010;1802:545–551. doi: 10.1016/j.bbadis.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iyengar B, Roote J, Campos AR. The tamas gene, identified as a mutation that disrupts larval behavior in Drosophila melanogaster, codes for the mitochondrial DNA polymerase catalytic subunit (DNApol-gamma125) Genetics. 1999;153:1809–1824. doi: 10.1093/genetics/153.4.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baqri RM, Turner BA, Rheuben MB, Hammond BD, Kaguni LS, Miller KE. Disruption of mitochondrial DNA replication in Drosophila increases mitochondrial fast axonal transport in vivo. PLoS ONE. 2009;4:e7874. doi: 10.1371/journal.pone.0007874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naviaux RK, Nguyen KV. POLG mutations associated with Alpers syndrome and mitochondrial DNA depletion. Ann. Neurol. 2005;58:491. doi: 10.1002/ana.20544. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.