Abstract
In many bacteria and archaea, an ancestral pathway is used where asparagine and glutamine are formed from their acidic precursors while covalently linked to tRNAAsn and tRNAGln, respectively. Stable complexes formed by the enzymes of these indirect tRNA aminoacylation pathways are found in several thermophilic organisms, and are called transamidosomes. We describe here a transamidosome forming Gln-tRNAGln in Helicobacter pylori, an ε-proteobacterium pathogenic for humans; this transamidosome displays novel properties that may be characteristic of mesophilic organisms. This ternary complex containing the non-canonical GluRS2 specific for Glu-tRNAGln formation, the tRNA-dependent amidotransferase GatCAB and tRNAGln was characterized by dynamic light scattering. Moreover, we observed by interferometry a weak interaction between GluRS2 and GatCAB (KD = 40 ± 5 µM). The kinetics of Glu-tRNAGln and Gln-tRNAGln formation indicate that conformational shifts inside the transamidosome allow the tRNAGln acceptor stem to interact alternately with GluRS2 and GatCAB despite their common identity elements. The integrity of this dynamic transamidosome depends on a critical concentration of tRNAGln, above which it dissociates into separate GatCAB/tRNAGln and GluRS2/tRNAGln complexes. Ester bond protection assays show that both enzymes display a good affinity for tRNAGln regardless of its aminoacylation state, and support a mechanism where GluRS2 can hydrolyze excess Glu-tRNAGln, ensuring faithful decoding of Gln codons.
INTRODUCTION
Survival in a competitive environment requires a high degree of faithfulness in protein biosynthesis to avoid not only lethal mutations but also those affecting fitness. Decoding of genetic information typically takes place on highly specific enzymes named aminoacyl-tRNA synthetases (aaRSs). Even though there are aaRSs for all 20 canonical amino acids, half of all characterized bacteria and archaea are deprived of asparaginyl-tRNA synthetase (AsnRS). Similarly, glutaminyl-tRNA synthetase (GlnRS) is absent from all archaea and most bacteria (1–3).
The indirect pathway used for the formation of Gln-tRNAGln was discovered in 1968 by Wilcox and Nirenberg (4). The first enzyme in this pathway is a non-discriminating aaRS (ND-aaRS) such as the ND-GluRS of Bacillus subtilis, which glutamylates both tRNAGlu and tRNAGln, or a non-canonical GluRS2 such as those of Helicobacter pylori and Acidithiobacillus ferrooxidans, which glutamylate only tRNAGln (5–7). The second enzyme of this pathway is an aminoacyl-tRNA amidotransferase, such as the trimeric GatCAB of B. subtilis, which transamidates Glu-tRNAGln into Gln-tRNAGln (8). GatCAB also catalyzes the indirect pathway for the formation of Asn-tRNAAsn, when an ND-AspRS is present to provide the Asp-tRNAAsn substrate (9). The formation of these misacylated aa-tRNAs must not lead to translational errors. To this end, several strategies have evolved, such as the proofreading activities of aaRSs (10), and the discrimination by elongation factor EF-Tu against misacylated tRNAs through thermodynamic compensation (11). ND-GluRS and ND-AspRS possess no proofreading activities, but Asp-tRNAAsn and Glu-tRNAGln are deprived of significant affinity for EF-Tu (12,13). Despite this, in vivo experiments have shown that under certain conditions, the Glu residue of Glu-tRNAGln can be incorporated into proteins, suggesting that an additional locking mechanism may be needed to prevent translational errors (6,14). Such a mechanism was shown in Pseudomonas aeruginosa and H. pylori, where the channeling of misacylated aa-tRNAs from the ND-aaRS to the aa-tRNA amidotransferase effectively sequesters them (15), as initially suggested by Schön et al. (16). Sequestration of misacylated aa-tRNAs through the formation of more robust interactions was then confirmed in thermophilic organisms. These were first described in Thermus thermophilus, where ND-AspRS, tRNAAsn and GatCAB form a ternary complex for the biosynthesis of Asn-tRNAAsn (17,18). A ternary complex for the formation of Gln-tRNAGln was then described for Methanothermobacter thermoautotrophicus, involving ND-GluRS, the archaeal amidotransferase GatDE, and tRNAGln (19). Although they differ in their mode of assembly, both complexes were named transamidosomes, in reference to their activity: the archaeal Gln-transamidosome is formed by a tight ND-GluRS/GatDE complex (KD = 100 nM) which then binds tRNAGln, whereas the T. thermophilus Asn-transamidosome assembles through ND-AspRS and GatCAB binding to tRNAAsn without any protein–protein interactions. These transamidosomes are similar to the complex catalyzing Cys-tRNACys biosynthesis in Methanocaldococcus jannaschii (20), suggesting that formation of complexes for tRNA-dependent amino acid biosynthesis is the norm in thermophilic organisms. In agreement with this is the structural data obtained with Thermotoga maritima ND-GluRS, tRNAGln and GatCAB in a ternary complex assisted by a covalent linkage of the GatC subunit to ND-GluRS, which demonstrates that no steric hindrances prevent the simultaneous binding of GatCAB and a GluRS to tRNAGln (21).
In this work, we describe a Gln-tRNAGln synthesizing ternary complex in H. pylori, suggesting that the transamidosome model extends to mesophilic organisms. H. pylori GatCAB and GluRS2 bind to tRNAGln and are further stabilized by a weak interaction directly between GluRS2 and GatCAB. The H. pylori Gln-transamidosome is less stable than those previously described. This fluidity enables conformational changes which allow both enzymes to alternately bind their shared identity elements on the tRNAGln acceptor stem.
MATERIALS AND METHODS
Overproduction and purification of enzymes and tRNAGln
Cells for the overproduction of H. pylori GluRS2 and GatCAB were cultured and lysed as adapted from previous works (6,15). Both enzymes were purified by ion-exchange chromatography on DEAE-cellulose (DE52, Whatman), followed by affinity chromatography on Ni-NTA resin (Novagen). Enzymes used for kinetic measurements were additionally purified by gel-filtration on a Superdex 200 HR 10/30 column (Amersham). The H. pylori tRNAGln gene, cloned into the pES300 plasmid (6), was overexpressed in Escherichia coli DH5α, and tRNAGln was purified as previously described (22); it was eluted between 0.9 and 0.6 M of (NH4)2SO4 on Sepharose-4B, between 20 and 300 mM potassium phosphate on hydroxyapatite (Bio-Rad), and between 0.5 and 1.0 M NaCl on a MonoQ HR10/100 column (GE Healthcare). Fractions of between 40 and 83% pure tRNAGln were obtained.
Gel-filtration
Experiments were conducted as described (22), using an ÄKTA Purifier and a 24-ml Superdex G200 column (GE Healthcare). Na–HEPES buffer (50 mM, pH 7.2) was used, containing 30 mM KCl, 6 mM MgCl2, 0.1 mM Na2-EDTA and 5 mM 2-mercaptoethanol, at 12°C. KD values ([free enzyme] × [free tRNAGln]/[enzyme–tRNAGln complex]) were determined by evaluating the area of enzyme-bound tRNA and free tRNA peaks, the sum of which equals the total tRNA present in the sample. The contaminating non-specific tRNA (~18%) was subtracted from free tRNA. The quantity of free enzyme was determined by subtracting the quantity of enzyme-bound tRNA from the total amount of enzyme present in the sample.
Dynamic light scattering
DLS measurements were performed as described (22), at 20 ± 0.1°C in a Zetasizer NanoS instrument (Malvern, UK). The 20–40 µl samples contained 20 µM protein with or without tRNAGln in the gel-filtration buffer. For the titration experiments, the concentrations of two partners was constant and that of the third (GatCAB or tRNAGln) varied as indicated. Hydrodynamic diameters were corrected for solvent refractive index (n = 1.3353) and absolute viscosity (η = 1.041 mPa.s).
Aminoacylation of tRNA
The reaction mixture contained 100 mM Na–HEPES pH 7.2, 30 mM KCl, 12 mM MgCl2, 2 mM ATP, 20–50 µM [14C]l-Glu (330 cpm pmol−1, GE Healthcare), 0.1 mg ml−1 bovine serum albumin, 10 µM of H. pylori tRNAGln and 0.025–0.5 µM H. pylori GluRS2. A concentration of 200 µM [14C]l-Glu was used to investigate the rate-limiting step. For KM measurements of GluRS2 with or without GatCAB, the tRNAGln concentration varied from 0.1 to 10 µM and that of l-Glu from 50 to 600 µM. Experiments were conducted at 37°C for steady-state measurements, or at 4°C with 10 or 20% glycerol for pre-steady state measurements. The [14C] aa-tRNA formed in 10 or 20 µl aliquots withdrawn at various time intervals was determined as previously described (23).
ATP-PPi exchange
The reaction mixture contained 100 mM Na–HEPES pH 7.2, 10 mM MgCl2, 5 µM tRNAGln, 2 mM l-Glu, 2 mM ATP, 2 mM [32P]PPi (2 cpm/pmol, PerkinElmer) and 0.5 µM GluRS2. The formation of [32P]ATP at 37°C was followed at various time intervals in 40 µl aliquots, as previously described (23).
Gln-tRNAGln formation
Reactions were performed in a mixture containing 50 mM Na–HEPES pH 7.2, 30 mM KCl, 12 mM MgCl2, 2 mM ATP, 1.28 mM l-Gln and 1 mM dithiothreitol. An amount of 50 µM [14C]l-Glu was added to the reaction mixture for glutamylation–transamidation experiments. For experiments measuring only transamidation, GatCAB was used at a concentration of 15 nM, while GluRS2 was used at 6.6 µM when it was in excess. The Glu-tRNAGln substrate was synthesized separately, and purified as described (15). For reactions where glutamylation and transamidation were measured simultaneously, an equimolar concentration of GatCAB and GluRS2 (50 nM), and uncharged tRNAGln at various concentrations were used. Both kinds of experiment were performed at 37°C, with 10 µl reaction aliquots prepared and then analyzed by migration on cellulose thin-layer chromatography plates as previously described (24). Product formation was quantified using radioactivity standards (15).
Stability of Glu-tRNAGln and Gln-tRNAGln
Both aa-tRNAs were purified as described above, with Gln-tRNAGln formed by subjecting Glu-tRNAGln to a transamidation step in the presence of GatCAB before extraction and precipitation. Half-lives were then measured in the transamidation reaction mixture with ATP and Gln excluded, as previously described (15).
Detection of macromolecular interactions by nanopore optical interferometry (NpOI)
These interactions were measured in a Ski Pro NpOI instrument (Silicon Kinetics), using carboxy-functionalized silicon biosensor chips containing covalently-bound GluRS2, GatCAB or GluRS1. These were prepared as previously described (25) using protein concentrations of 0.25, 0.25 and 0.5 mg ml−1, respectively. Assayed against these biosensor chips were target proteins diluted to various concentrations in the buffer used for gel-filtration. To obtain binding data, the target protein was injected into the sample channel of the biosensor chip for 300 s, while the binding buffer was injected into the reference channel. To obtain the dissociation data, binding buffer was injected into the sample channel in addition to the reference channel for at least 100 s to trigger the mass dilution of the target protein. In both cases, the optical path difference of the reference channel was subtracted from that of the sample channel.
RESULTS
Helicobacter pylori GluRS2 and GatCAB have comparable affinities for tRNAGln
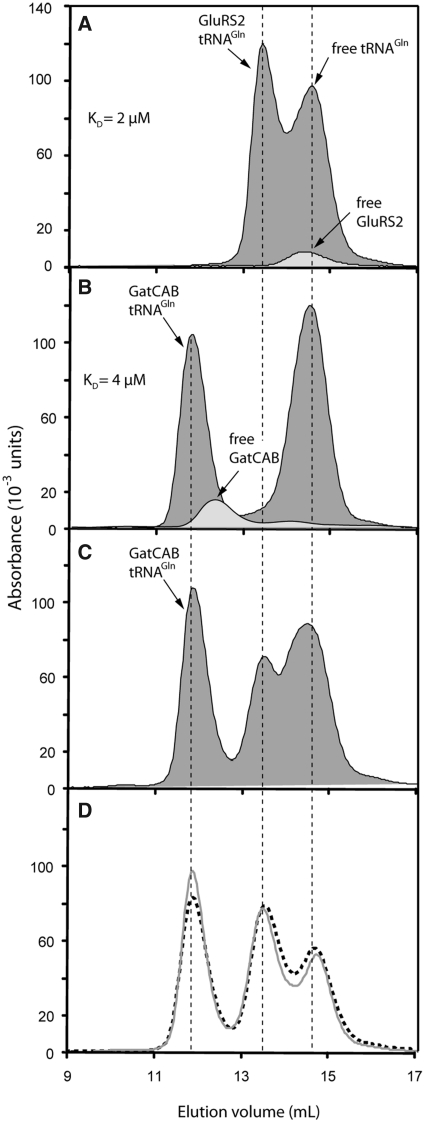
An important characteristic of previously described transamidosomes is that the GatCAB and the aaRS can both bind tRNAGln or tRNAAsn (17,19). We therefore used gel-filtration to investigate the capacity of GluRS2 and GatCAB to form binary complexes with tRNAGln. GluRS2 binds to Hp tRNAGln with a KD value estimated at 2 ± 0.6 µM (Figure 1A). In accordance with previous observations, GatCAB also made a complex with uncharged tRNAGln with a KD value of 4 ± 1 µM (Figure 1B), close to that measured for GluRS2. No ternary complex was detected, either using tRNAGln or Glu-tRNAGln, whether the latter was simultaneously formed, or prepared in advance (Figure 1C and D). Direct protein–protein interactions were also undetectable by gel-filtration, as seen when comparing elution volumes of GluRS2 and GatCAB (Figure 1A, B and C). As the free and bound forms of these macromolecules are not at equilibrium during gel-filtration, these results do not rule out the existence of a ternary complex, nor of a direct GluRS2/GatCAB interaction, and they imply that the KD values obtained are upper limits.
Figure 1.
Binding experiments with tRNAGln, GluRS2 and GatCAB as shown by size-exclusion chromatography. (A) Comparison of isolated GluRS2 and GluRS2/tRNAGln complex profiles. The association enabled us to estimate a KD value for GluRS2 and tRNAGln binding (see ‘Materials and Methods’ section). (B) Comparison between free and tRNAGln-complexed GatCAB. (C) Gel-filtration of a solution containing the three partners. (D) The same experiment as described in (C) was performed with preformed Glu-tRNAGln (grey line) or after incubation of the three partners within an aminoacylation medium containing free Glu and ATP, to provide endogenous Glu-tRNAGln (dotted line). Proteins and tRNAGln were added at concentrations of 20 µM each for the experiments shown in all panels.
Characterization of GluRS2/GatCAB interactions by nanopore optical interferometry (NpOI)
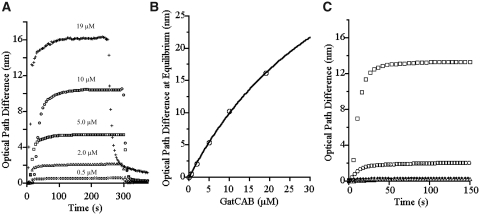
Gel-filtration experiments indicated that if a Gln-transamidosome exists in H. pylori, it would be more unstable than those previously described. Nevertheless, we attempted to determine if protein–protein interactions occur, as is the case for the M. thermoautotrophicus Gln-transamidosome (19). Using NpOI, we measured the quantity of GatCAB bound by a GluRS2 biosensor chip, for increasing GatCAB concentrations in a mobile phase (Figure 2A). Dissociation of the GatCAB from the biosensor chip was measured by injecting buffer. Using the detected amounts of GluRS2-bound GatCAB at equilibrium, we determined a KD of 40 ±5 µM for the GluRS2/GatCAB complex (Figure 2B) (26). Using the dissociation equation for the reversible binding of two molecules (27), we calculated a koff of 0.12 s−1 for the dissociation of this complex, which together with the KD value, yields a kon of 0.003 s−1 µM−1. Control experiments using GatCAB, GluRS2, and the discriminating H. pylori GluRS1 in various combinations revealed minimal binding between GluRS1 and GluRS2, between GluRS1 and GatCAB and between GluRS1 and GluRS1 itself (Figure 2C). Although we obtained data for the binding of GluRS2 to fixed GatCAB, the ability of GluRS2 to bind to itself (Figure 2C) interfered with data interpretation.
Figure 2.
GluRS2–GatCAB interactions characterized by optical interferometry. (A) Kinetics of association and dissociation of GatCAB to fixed GluRS2. GatCAB concentrations of 0.5, 2, 5, 10 and 19 µM yielded average optical path differences of 0.6, 2.2, 5.4, 10.4 and 16.2 nm at each plateau. The standard deviation of these averages was <1%. (B) Curve of GatCAB bound to GluRS2 at equilibrium (from A) versus the concentration of GatCAB in the mobile phase gives a KD of 40 ± 5 µM (standard error). The curve was obtained using the equation Y = L/(KD + L)·Ymax, where Y is the GatCAB bound at equilibrium, L is its concentration in the mobile phase and Ymax the maximum amount of GatCAB bound by the GluRS2 biosensor (26). (C) As controls, H. pylori GluRS1 was assayed against chip-bound GatCAB (open circle), chip-bound GluRS2 (filled diamond) and chip-bound GluRS1 (open triangle). GluRS1 concentrations of 10, 10 and 40 µM, respectively, were used as the target protein. Binding of GluRS1 with itself or with chip-bound GluRS2 was negligible relative to the ability of GluRS2 to bind to itself (open square). A small amount of GluRS1/GatCAB binding was detected (open circle) but this remained limited compared to the ability of GluRS2 to bind GatCAB (A).
Characterization of a ternary complex of GluRS2/tRNAGln/GatCAB by dynamic light scattering (DLS)
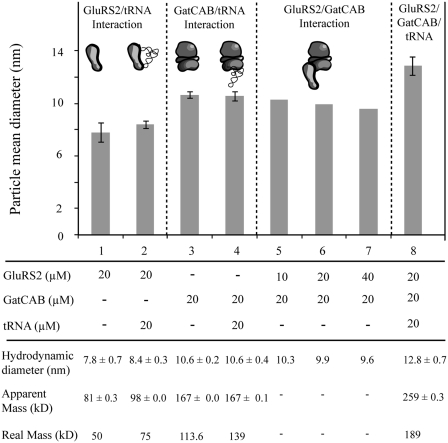
The GluRS2/tRNAGln, GatCAB/tRNAGln and GluRS2/GatCAB binary complexes detected by gel-filtration (Figure 1) and NPoI (Figure 2B) did not show a significant increase in apparent mean hydrodynamic diameter over those of the free proteins (Figure 3, lanes 1–7). The particle size measured for GluRS2/GatCAB decreased when the concentration of GluRS2 was increased, confirming that this complex was very labile. Finally, we tested the effect of tRNAGln addition: when the three partners were mixed, a new particle of significantly larger size was formed (Figure 3, lane 8). This shows that while weak GluRS2/GatCAB interactions occur, the strongest interactions in the ternary complex are those between both enzymes and tRNAGln. The apparent mass (259 kDa) deduced from the particle size (12.8 nm) is roughly equal to the sum of those measured for GluRS2/tRNAGln and GatCAB, or for GatCAB/tRNAGln and GluRS2.
Figure 3.
Characterization of the ternary complex of glutaminylation by dynamic light scattering. The error bars represent the standard deviation of triplicate experiments. Attempts at detecting GluRS2–GatCAB interactions were performed once, for each of three distinct GluRS2 concentrations.
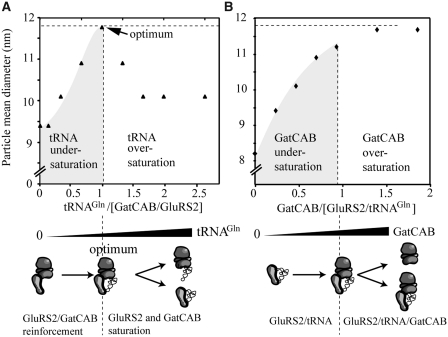
In order to confirm that this complex is a GluRS2/tRNAGln/GatCAB transamidosome, we performed a titration of an equimolar mix of GluRS2 and GatCAB with free tRNAGln, increasing the molar ratio from 0.3 to 2.5 (Figure 4A, upper panel), and a titration of a equimolar mix of GluRS2 and tRNAGln with free GatCAB, with molar ratios of 0 to 2 (Figure 4B). Titration with tRNA led to a nearly symmetrical bell-shaped curve, indicating that increasing tRNAGln concentrations first led to the formation of a GluRS2/tRNAGln/GatCAB ternary complex with a 1/1/1 stoichiometry, and then to separate saturation of both GluRS2 and GatCAB with free tRNAGln (Figure 4A, lower panel), disrupting the ternary complex. Titration with GatCAB led to a hyperbolic saturation curve (Figure 4B), also indicating a stoichiometry of 1/1/1. The titration of a GatCAB/tRNAGln stoichiometric mix with GluRS2 gave strong dispersion in hydrodynamic parameters, probably due to the ability of this aaRS to bind to itself (Figure 2C).
Figure 4.
Stoichiometry of the GluRS2/tRNAGln/GatCAB complex. (A) Titration curve of a 1/1 mix of GluRS2 and GatCAB (20 µM each) with tRNAGln in molar ratios of 0.2–2.7. (B) Titration of a 1/1 mix of GluRS2 and tRNAGln (20 µM each) with free GatCAB in molar ratios of 0.3–2.0.
Kinetic experiments reveal a model for Gln-tRNAGln formation in the H. pylori glutamine transamidosome
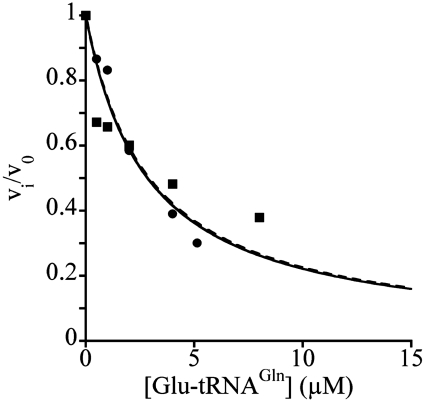
We found that the KM for Glu and the kcat of the non-canonical GluRS2 (140 ± 26 µM; 0.16 ± 0.012 s−1) are comparable to those of the B. subtilis ND-GluRS (28). Because of this high KM value, GluRS2 aminoacylation kinetic values for tRNAGln were obtained with a non-saturating concentration of Glu. Each of the two protein partners involved in Gln-tRNAGln formation altered the kinetic constants of the other. The presence of 6.6 µM GatCAB increased the KM of GluRS2 for tRNAGln by a factor of 2.1 (Table 1). Similarly, the presence of 6.6 µM GluRS2 increased the KM of GatCAB for Glu-tRNAGln nearly 4-fold (Table 1). This is consistent with the fact that GluRS2 and GatCAB have a similar affinity for tRNAGln (Figure 1). It indicates that both GluRS2 and GatCAB can sequester the acceptor stem away from the other, leading to a kinetic competition, as suggested by the 3D structural data for the T. maritima Gln-transamidosome (21). Additional support for this competition model is the fact that Glu-tRNAGln was an efficient competitive inhibitor of GluRS2, with a KI value of 1.32 µM (Figure 5) close to the KM for uncharged tRNAGln (0.87 µM, Table 1). A concentration of GatCAB equal to that of GluRS2/tRNAGln allowed formation of a ternary complex, which was not disrupted by the addition of excess GatCAB (Figure 4). This confirms that the effect of an excess of GatCAB on the KM of GluRS2 for tRNAGln is not caused by a sequestration of the entire tRNAGln molecule away from GluRS2, but by competition for the tRNAGln acceptor stem inside the full-size ternary complex.
Table 1.
Kinetic constants of GluRS2 for tRNAGln glutamylation and of GatCAB for Glu-tRNAGln transamidation in the absence or in the presence of the second protein
| KM (µM) | kcat (s−1) | |
|---|---|---|
| GluRS2 | 0.87 ± 0.13 | 0.094 ± 0.004 |
| GluRS2 + GatCAB | 1.86 ± 0.19 | 0.24 ± 0.009 |
| GluRS2 + GatCAB | 0.69 ± 0.15 | 0.23 ± 0.015 |
| GatCAB | 0.42 ± 0.11 | 0.21 ± 0.011 |
| GatCAB + GluRS2 | 1.34 ± 0.38 | 0.21 ± 0.014 |
| GatCAB + GluRS2 | 0.97 ± 0.30 | 0.23 ± 0.019 |
Constants were measured using the assay for the first enzyme indicated. When present as the second enzyme, GatCAB and GluRS2 were in equimolar amounts (normal font), or in excess 330- and 440-fold, respectively (in bold). Reported KM values are for tRNAGln in all experiments except GatCAB alone and with an excess of GluRS2, in which they were for Glu-tRNAGln. Errors shown are the standard errors for non-linear curve-fitting of data obtained from at least two experiments.
Figure 5.
Glu-tRNAGln is an inhibitor of the aminoacylation reaction catalyzed by GluRS2. Glu-tRNAGln was formed as described in ‘Materials and Methods’ section and added to the GluRS2 reaction mixture at concentrations varying from 0 to 7 µM. Vi/V0 is equal to 2/[2 + (I/Ki)] when the substrate concentration is equal to its KM (15). Two independent experiments were conducted (filled square, filled circle) with 1 µM uncharged tRNAGln as the substrate, yielding an average Ki of 1.3 ± 0.2 µM.
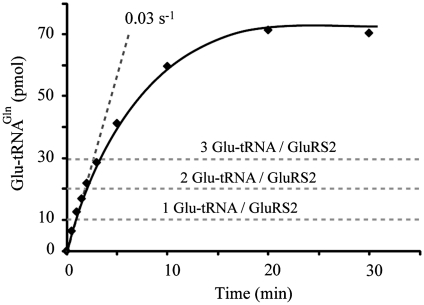
Furthermore, GatCAB increased 2.6-fold the aminoacylation kcat of GluRS2 despite the increase in the KM for tRNAGln (Table 1). Since dissociation of Glu-tRNAGln was not rate-limiting (Figure 6), this stimulation by GatCAB can be explained by an effect on the activation of Glu or on its transfer to tRNAGln in the GluRS2/GatCAB complex. The latter is suggested by the fact that the kcat of the ATP-PPi exchange reaction monitoring the tRNA-dependent activation of Glu by GluRS2 was one order of magnitude higher (1.5 s−1) than the kcat of the transfer step (0.16 s−1).
Figure 6.
Pre-steady state measurements of Glu-tRNAGln formation by GluRS2. Aminoacylation reactions were performed at 4°C with 10 µM tRNAGln, and 1 µM GluRS2, taking 10 µl aliquots to measure product formation. No burst indicative of a limiting product-release step was observed.
Finally, assays were performed measuring glutamylation and transamidation simultaneously using uncharged tRNAGln and equal concentrations of GluRS2 and GatCAB. Glutamylation occurred with a KM of 0.69 µM for tRNAGln, and kcat of 0.23 s−1, while transamidation occurred with values of 0.97 µM for tRNAGln and 0.23 s−1 (Table 1). Therefore, an equimolar concentration of both enzymes offers the best possible overall rate of Gln-tRNAGln formation with KM values nearer to those obtained in the absence of acceptor stem competition. This competition may be the result of inefficient switching between glutamylation and transamidation tRNAGln binding modes inside the Gln-transamidosome. The overall ternary complex formation could be regulated by the concentration of tRNAGln, for which there would be an optimal cellular concentration for maximum efficiency.
Stability of Glu-tRNAGln and Gln-tRNAGln
GluRS2 and GatCAB had opposite effects on the stability of the aminoacyl-tRNA link of both of these molecules. GluRS2 reduced 2-fold the half-life of both aa-tRNAs, while GatCAB increased their half-life nearly 5- and 8-fold, respectively (Table 2). Therefore, both enzymes appeared to interact with tRNAGln, regardless of its aminoacylation state, which is consistent with a ternary complex inside which potential competition dictates that neither enzyme should have a greater affinity than the other for the shared substrate. When GluRS2 and GatCAB were both present, no overall deacylation effect was observed (Table 2).
Table 2.
Stability of Glu-tRNAGln and Gln-tRNAGln
| Half-life (min) |
||
|---|---|---|
| Glu-tRNAGln | Gln-tRNAGln | |
| No enzyme | 54 ± 1 | 57 ± 3 |
| GatCAB | 245 ± 43 | 444 ± 111 |
| GluRS2 | 26 ± 3 | 26 ± 2 |
| GluRS2 + GatCAB | 50 ± 1 | 55 ± 6 |
Half-lives for both aa-tRNAs were measured by incubating purified Glu-tRNAGln or Gln-tRNAGln in the standard reaction mixture for transamidation, from which Gln and ATP were excluded, at 37°C. When added, proteins were at a concentration of 20 µM. GatCAB protected the ester bond of both aa-tRNAs, while GluRS2 showed a deacylating activity.
DISCUSSION
The dynamic H. pylori glutamine transamidosome
In archaea, Cys-tRNACys and Gln-tRNAGln are synthesized by stable complexes between a misacylating aaRS and a second enzyme providing the final, cognate aa-tRNA (19–21). Our results show that in H. pylori, the non-canonical GluRS2 and GatCAB also interact but in a very unstable complex (KD of 40 ± 5 µM). This is in contrast to the stable complex between ND-GluRS and the archaeal GatDE amidotransferase in Methanothermobacter thermautotrophicus (KD of 100 nM). The weaker complex observed in H. pylori may be a result of the absence of an AspRS-like domain in the GatB subunit (19). However, DLS and kinetic experiments have shown that tRNAGln acts as a scaffold molecule transiently reinforcing the interaction between both GluRS2 and GatCAB which stabilizes the Gln-transamidosome through independent binding of each protein (Figures 3 and 4). The mode of assembly for the H. pylori Gln-transamidosome is therefore a hybrid of the protein–RNA mechanism of the T. thermophilus Asn-transamidosome, and of the protein–protein mechanism of the Gln-transamidosome of M. thermoautotrophicus. The recent structure of the Thermotoga maritima Gln-transamidosome reveals that its ND-GluRS and GatCAB can bind simultaneously to a single tRNAGln molecule (21). The ND-GluRS binds most of the tRNAGln, with the acceptor end within the active site, while the GatB subunit of GatCAB interacts through its C-terminal region with the elbow domain of tRNAGln. Additionally, GluRS2/GatCAB interactions can be seen in the N-terminal regions of both proteins. Consequently, it is possible that GluRS2/tRNAGln or GatCAB/tRNAGln complexes can bind the missing partner with a better affinity than either free enzyme, as suggested by the DLS titration experiments (Figure 4). This effect would be similar to the one observed for the binding of aa-AMP or bi-modular inhibitors to aaRSs which are typically more effective than the sum of their parts (29,30).
Our data further refines the T. maritima Gln-transamidosome model by experimentally confirming that Glu-tRNAGln cannot directly flip between both active sites without a rearrangement in the ternary complex structure, but the mechanism we describe differs slightly. The existence of this complex in H. pylori depends on a critical concentration of tRNAGln (Figure 4A) above which the formation of separate binary complexes is favoured, which is not the case for T. maritima where a 2:1:1 ratio for tRNAGln/ND-GluRS/GatCAB allowed the formation of a ternary complex detectable by gel-shift assay [(21) and supplementary information therein]. Excess of the first enzyme over the second increases the KM of the second enzyme for tRNAGln (Table 1). An excess of GatCAB does not disrupt the ternary complex (Figure 4B), and therefore induces a conformational change in the complex to the tRNA-binding mode specific to its activity. This may also occur when an excess of GluRS2 is present, but could not be detected by DLS (see above). Such conformational change would be unnecessary if the tRNA acceptor stem was able to flip directly between the catalytic pockets of GluRS2 and GatB.
GluRS2 can hydrolyze excess Glu-tRNAGln
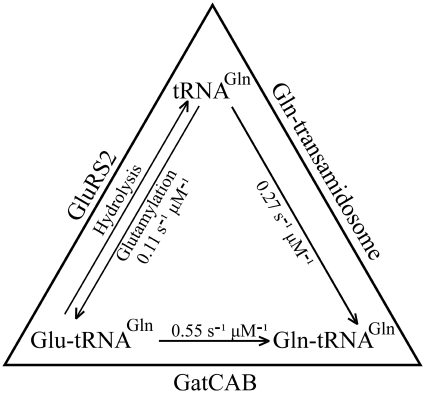
Optimally, upon glutamylation within the Gln-transamidosome, GluRS2 releases the Glu-tRNAGln acceptor stem towards GatCAB for transamidation before leaving the complex. When this fails to occur, Glu-tRNAGln may be released without being transamidated and is susceptible to retroactive binding and deacylation by GluRS2 (Figure 5 and Table 2), or could bind to a free GatCAB. Either sequence is possible since Glu-tRNAGln release is not the rate-limiting step for glutamylation (Figure 6). This mechanism bears a resemblance to the binding and hydrolysis, or resampling, of free Tyr-tRNAPhe by the E. coli PheRS which can erroneously synthesize it (31). The GluRS2 resampling-like mechanism (Figure 7) is supported by the fact that aaRSs and their cognate tRNAs are present in similar concentrations within the cell, near the KM value (32).
Figure 7.
A dynamic system for Gln-tRNAGln synthesis, which limits free Glu-tRNAGln in H. pylori. The constants shown are the kcat/KM values calculated from the data in Table 2, for GluRS2 and GatCAB as free enzymes, and for the stoechiometric mix forming the Gln-transamidosome. Starting with tRNAGln, the Gln-transamidosome pathway is favoured, with a kcat/KM value greater than that of the GluRS2 pathway. Glu-tRNAGln which is formed outside the Gln-transamidosome is susceptible to deacylation by GluRS2, or to an efficient transamidation by free GatCAB. The entire system therefore regulates against the presence of free Glu-tRNAGln.
The aa-tRNA ester bonds of Glu-tRNAGln and of Gln-tRNAGln can be broken by GluRS2, but are protected by GatCAB (Table 2). No net effect on the stability of these aa-tRNAs was observed when both enzymes were present. Asp and Glu are very similar, as are their amidated counterparts, and GatCAB interacts with tRNAGln and tRNAAsn through the same identity elements. Therefore, these results provide an intriguing contrast to those obtained for the Asn-tRNAAsn forming pathways in H. pylori and in T. thermophilus, where the individual enzymes (ND-AspRS or AspRS2 and GatCAB) had negligible effects and where the presence of both enzymes had a protective effect on Asn-tRNAAsn(15,17).
Stability of the GluRS2/GatCAB complex
KD values of several protein–protein interactions are temperature-dependent (33,34). The transamidosomes of thermophilic organisms previously described, and of H. pylori, were observed at temperatures between 4°C and room temperature. For the mesophilic H. pylori complex, experimental conditions were therefore much less efficient in slowing the normal dynamics.
This lesser stability may also be necessary to leave enough free GatCAB to allow the formation of an Asn-transamidosome: the amidotransferases of both stable transamidosomes described so far, those of T. thermophilus and M. thermoautotrophicus, catalyze only one activity. On the other hand, the T. maritima and H. pylori GatCABs fill both transamidation roles in the cell, and may therefore only form unstable Gln-transamidosomes which would facilitate the formation of Asn-transamidosomes. This may be why covalent linkage between ND-GluRS and GatC was needed to obtain structural data for T. maritima (21).
A ‘loose’ association may also be what allows the rearrangements in the complex which are necessary for movement of the acceptor stem. Structural data obtained for GatCAB indicates that the tail domain of GatB responsible for the binding of the D-loop of tRNAGln is very mobile in relation to the catalytic cradle domain of GatB, which binds the acceptor stem identity element (35). Inside the complex, this flexibility might be what allows contact to be maintained with the D-loop when the cradle domain loses the acceptor stem to GluRS2.
Attempts to characterize the Gln-transamidosome in vivo
Attempts were made to detect the complex in vivo using the TAP-tag method (36) under conditions where the GluRS2 is overexpressed in H. pylori in the presence of physiological concentrations of GatCAB and of tRNAGln. No complex was detected, making it unlikely that an unidentified protein stabilizes the transamidosome. This result is not surprising, given the stringency of the method, which requires that any complexes remain stable over the course of two affinity chromatographies, in order to be detected. The two transamidosomes which have been detected in vivo thus far are assembled with affinities of 0.1–0.6 µM (17,19). These experiments required overexpression or high concentrations of all the protein partners in order to isolate the complex. In vitro detection of the H. pylori transamidosome, as well as of the T. maritima transamidosome (21), required protein concentrations one to two orders of magnitude greater than the affinities above. This suggests that further work is needed to establish the existence of such GluRS/tRNAGln/GatCAB complexes under physiological conditions, without relying on abnormally high concentrations of some or all partners.
Insights into evolution
In H. pylori, translation-targeted Gln is synthesized on a tRNAGln matrix using the indirect aminoacylation pathway. Such pathways may be remnants of ancient routes for aa-tRNA formation, as suggested by the presence of GatCAB and GluRS in the ‘paleome’ (37), proposed to include ancestral proteins. EF-Tu can discriminate against mischarged aa-tRNA, such as Asp-tRNAAsn, Glu-tRNAGln or Ser-tRNASec through a thermodynamic compensation mechanism by which aa-tRNAs are most strongly recognized either by their amino acid moiety or their tRNA moiety (11,13). It is probable that this mechanism appeared very early to prevent the use of misacylated tRNAs in protein biosynthesis. Owing to the ancient origin of all aaRSs, it is possible that this property of EF-Tu already existed before GatCAB appeared, and that today's Asp-tRNAAsn and Glu-tRNAGln escaped early translation, as is the case with some tRNAGly isoacceptors in S. aureus (38). The appearance of Asn- and Gln-transamidosomes provided new aa-tRNAs suitable for EF-Tu binding via the strongly recognized amidated amino acids, leading to a new specification for the present day tRNAGln (UUG and CUG anticodons) and tRNAAsn (GUU anticodon). Thus, GatCAB, beyond its role in matrix-assisted modifications for amidated aa-tRNA biosynthesis, could also be considered as a codon reassignment enzyme, that led to the present-day genetic code.
FUNDING
Natural Sciences and Engineering Research Council of Canada (9597-2010 to J.L.); Fond de Recherche sur la Nature et les Technologies du Québec (PR-105093 to J.L. and graduate fellowship to J.L.H.); Regroupement québecois de recherche sur la fonction, la structure et l’ingénierie des proteines (PROTEO) (graduate fellowship to J.L.H.); Ministère de l’Éducation Nationale, de la Recherche et de la Technologie (graduate fellowship to F.F.); Université de Strasbourg, the Centre National de la Recherche Scientifique and the Association pour la Recherche sur le Cancer (to D.K.); National Institutes of Health (R01GM071480 to T.L.H.). Funding for open access charge: Natural Sciences and Engineering Research Council of Canada (9597-2010).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Prof. E. Westhof, Dr R. Giegé and Prof. H.D. Becker (IBMC) for support, M. Brayé (IBMC) for technical assistance and Dr H. De Reuse and J. Gallaud (Institut Pasteur, Paris) for performing the TAP-tag experiments.
REFERENCES
- 1.Feng L, Sheppard K, Tumbula-Hansen D, Söll D. Gln-tRNAGln formation from Glu-tRNAGln requires cooperation of an asparaginase and a Glu-tRNAGln kinase. J. Biol. Chem. 2005;280:8150–8155. doi: 10.1074/jbc.M411098200. [DOI] [PubMed] [Google Scholar]
- 2.Kern D, Roy H, Becker HD. In: The Aminoacyl-tRNA Synthetases. Ibba M, Francklyn C, Cusack S, editors. Georgetown, TX: Landes Bioscience; 2005. pp. 193–209. [Google Scholar]
- 3.Sheppard K, Söll D. On the evolution of the tRNA-dependent amidotransferases, GatCAB and GatDE. J. Mol. Biol. 2008;377:831–844. doi: 10.1016/j.jmb.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilcox M, Nirenberg M. Transfer RNA as a cofactor coupling amino acid synthesis with that of protein. Proc. Natl Acad. Sci. USA. 1968;61:229–236. doi: 10.1073/pnas.61.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lapointe J, Duplain L, Proulx M. A single glutamyl-tRNA synthetase aminoacylates tRNAGlu and tRNAGln in Bacillus subtilis and efficiently misacylates Escherichia coli tRNAGln1 in vitro. J. Bacteriol. 1986;165:88–93. doi: 10.1128/jb.165.1.88-93.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skouloubris S, Ribas de Pouplana L, De Reuse H, Hendrickson TL. A noncognate aminoacyl-tRNA synthetase that may resolve a missing link in protein evolution. Proc. Natl Acad. Sci. USA. 2003;100:11297–11302. doi: 10.1073/pnas.1932482100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salazar JC, Ahel I, Orellana O, Tumbula-Hansen D, Krieger R, Daniels L, Söll D. Coevolution of an aminoacyl-tRNA synthetase with its tRNA substrates. Proc. Natl Acad. Sci. USA. 2003;100:13863–13868. doi: 10.1073/pnas.1936123100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curnow AW, Hong K, Yuan R, Kim S, Martins O, Winkler W, Henkin TM, Söll D. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc. Natl Acad. Sci. USA. 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curnow AW, Tumbula DL, Pelaschier JT, Min B, Söll D. Glutamyl-tRNAGln amidotransferase in Deinococcus radiodurans may be confined to asparagine biosynthesis. Proc. Natl Acad. Sci. USA. 1998;95:12838–12843. doi: 10.1073/pnas.95.22.12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ling J, Reynolds N, Ibba M. Aminoacyl-tRNA synthesis and translational quality control. Annu. Rev. Microbiol. 2009;63:61–78. doi: 10.1146/annurev.micro.091208.073210. [DOI] [PubMed] [Google Scholar]
- 11.LaRiviere FJ, Wolfson AD, Uhlenbeck OC. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science. 2001;294:165–168. doi: 10.1126/science.1064242. [DOI] [PubMed] [Google Scholar]
- 12.Stanzel M, Schön A, Sprinzl M. Discrimination against misacylated tRNA by chloroplast elongation factor Tu. Eur. J. Biochem. 1994;219:435–439. doi: 10.1111/j.1432-1033.1994.tb19956.x. [DOI] [PubMed] [Google Scholar]
- 13.Roy H, Becker HD, Mazauric MH, Kern D. Structural elements defining elongation factor Tu mediated suppression of codon ambiguity. Nucleic Acids Res. 2007;35:3420–3430. doi: 10.1093/nar/gkm211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruan B, Palioura S, Sabina J, Marvin-Guy L, Kochhar S, Larossa RA, Söll D. Quality control despite mistranslation caused by an ambiguous genetic code. Proc. Natl Acad. Sci. USA. 2008;105:16502–16507. doi: 10.1073/pnas.0809179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huot JL, Balg C, Jahn D, Moser J, Emond A, Blais SP, Chênevert R, Lapointe J. Mechanism of a GatCAB amidotransferase: aspartyl-tRNA synthetase increases its affinity for Asp-tRNAAsn and novel aminoacyl-tRNA analogues are competitive inhibitors. Biochemistry. 2007;46:13190–13198. doi: 10.1021/bi700602n. [DOI] [PubMed] [Google Scholar]
- 16.Schön A, Kannangara CG, Gough S, Söll D. Protein biosynthesis in organelles requires misaminoacylation of tRNA. Nature. 1988;331:187–190. doi: 10.1038/331187a0. [DOI] [PubMed] [Google Scholar]
- 17.Bailly M, Blaise M, Lorber B, Becker HD, Kern D. The transamidosome: a dynamic ribonucleoprotein particle dedicated to prokaryotic tRNA-dependent asparagine biosynthesis. Mol. Cell. 2007;28:228–239. doi: 10.1016/j.molcel.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 18.Blaise M, Bailly M, Frechin M, Behrens MA, Fischer F, Oliveira CL, Becker HD, Pedersen JS, Thirup S, Kern D. Crystal structure of a transfer-ribonucleoprotein particle that promotes asparagine formation. EMBO J. 2010;29:3118–3129. doi: 10.1038/emboj.2010.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rampias T, Sheppard K, Söll D. The archaeal transamidosome for RNA-dependent glutamine biosynthesis. Nucleic Acids Res. 2010;38:5774–5783. doi: 10.1093/nar/gkq336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang CM, Liu C, Slater S, Hou YM. Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNACys. Nat. Struct. Mol. Biol. 2008;15:507–514. doi: 10.1038/nsmb.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito T, Yokoyama S. Two enzymes bound to one transfer RNA assume alternative conformations for consecutive reactions. Nature. 2010;467:612–616. doi: 10.1038/nature09411. [DOI] [PubMed] [Google Scholar]
- 22.Bailly M, Blaise M, Roy H, Deniziak M, Lorber B, Birck C, Becker HD, Kern D. tRNA-dependent asparagine formation in prokaryotes: characterization, isolation and structural and functional analysis of a ribonucleoprotein particle generating Asn-tRNAAsn. Methods. 2008;44:146–163. doi: 10.1016/j.ymeth.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 23.Kern D, Lapointe J. The twenty aminoacyl-tRNA synthetases from Escherichia coli. General separation procedure, and comparison of the influence of pH and divalent cations on their catalytic activities. Biochimie. 1979;61:1257–1272. doi: 10.1016/s0300-9084(80)80285-9. [DOI] [PubMed] [Google Scholar]
- 24.Bailly M, Giannouli S, Blaise M, Stathopoulos C, Kern D, Becker HD. A single tRNA base pair mediates bacterial tRNA-dependent biosynthesis of asparagine. Nucleic Acids Res. 2006;34:6083–6094. doi: 10.1093/nar/gkl622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Latterich M, Corbeil J. Label-free detection of biomolecular interactions in real time with a nano-porous silicon-based detection method. Proteome Sci. 2008;6:31. doi: 10.1186/1477-5956-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serdyuk I, Zaccai NR, Zaccai J. Methods in Molecular Biophysics: Structure, Dynamics, Function. New York: Cambridge University Press; 2007. pp. 240–242. [Google Scholar]
- 27.Johnson KA. Transient-state kinetic analysis of enzyme reaction pathways. In: Sigman DS, editor. The Enzymes. Vol. XX. New York: Academic Press; 1992. pp. 1–61. [Google Scholar]
- 28.Proulx M, Duplain L, Lacoste L, Yaguchi M, Lapointe J. The monomeric glutamyl-tRNA synthetase from Bacillus subtilis 168 and its regulatory factor. Their purification, characterization, and the study of their interaction. J. Biol. Chem. 1983;258:753–759. [PubMed] [Google Scholar]
- 29.Lin SX, Baltzinger M, Remy P. Fast kinetic study of yeast phenylalanyl-tRNA synthetase: an efficient discrimination between tyrosine and phenylalanine at the level of the aminoacyladenylate-enzyme complex. Biochemistry. 1983;22:681–689. doi: 10.1021/bi00272a024. [DOI] [PubMed] [Google Scholar]
- 30.Balg C, Blais SP, Bernier S, Huot JL, Couture M, Lapointe J, Chênevert R. Synthesis of beta-ketophosphonate analogs of glutamyl and glutaminyl adenylate, and selective inhibition of the corresponding bacterial aminoacyl-tRNA synthetases. Bioorg. Med. Chem. 2007;15:295–304. doi: 10.1016/j.bmc.2006.09.056. [DOI] [PubMed] [Google Scholar]
- 31.Ling J, So BR, Yadavalli SS, Roy H, Shoji S, Fredrick K, Musier-Forsyth K, Ibba M. Resampling and editing of mischarged tRNA prior to translation elongation. Mol. Cell. 2009;33:654–660. doi: 10.1016/j.molcel.2009.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jakubowski H, Goldman E. Quantities of individual aminoacyl-tRNA families and their turnover in Escherichia coli. J. Bacteriol. 1984;158:769–776. doi: 10.1128/jb.158.3.769-776.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banerjee A, Hu J, Goss DJ. Thermodynamics of protein-protein interactions of cMyc, Max, and Mad: effect of polyions on protein dimerization. Biochemistry. 2006;45:2333–2338. doi: 10.1021/bi0522551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roos H, Karlsson R, Nilshans H, Persson A. Thermodynamic analysis of protein interactions with biosensor technology. J. Mol. Recognit. 1998;11:204–210. doi: 10.1002/(SICI)1099-1352(199812)11:1/6<204::AID-JMR424>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 35.Nakamura A, Sheppard K, Yamane J, Yao M, Söll D, Tanaka I. Two distinct regions in Staphylococcus aureus GatCAB guarantee accurate tRNA recognition. Nucleic Acids Res. 2010;38:672–682. doi: 10.1093/nar/gkp955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 37.Danchin A, Fang G, Noria S. The extant core bacterial proteome is an archive of the origin of life. Proteomics. 2007;7:875–889. doi: 10.1002/pmic.200600442. [DOI] [PubMed] [Google Scholar]
- 38.Giannouli S, Kyritsis A, Malissovas N, Becker HD, Stathopoulos C. On the role of an unusual tRNAGly isoacceptor in Staphylococcus aureus. Biochimie. 2009;91:344–351. doi: 10.1016/j.biochi.2008.10.009. [DOI] [PubMed] [Google Scholar]