Abstract
SWI/SNF is an ATP-dependent remodeler that mobilizes nucleosomes and has important roles in gene regulation. The catalytic subunit of SWI/SNF has an ATP-dependent DNA translocase domain that is essential for remodeling. Besides the DNA translocase domain there are other domains in the catalytic subunit of SWI/SNF that have important roles in mobilizing nucleosomes. One of these domains, termed SnAC (Snf2 ATP Coupling), is conserved in all eukaryotic SWI/SNF complexes and is located between the ATPase and A-T hook domains. Here, we show that the SnAC domain is essential for SWI/SNF activity. The SnAC domain is not required for SWI/SNF complex integrity, efficient nucleosome binding, or recruitment by acidic transcription activators. The SnAC domain is however required in vivo for transcription regulation by SWI/SNF as seen by alternative carbon source growth assays, northern analysis, and genome-wide expression profiling. The ATPase and nucleosome mobilizing activities of SWI/SNF are severely affected when the SnAC domain is removed or mutated. The SnAC domain positively regulates the catalytic activity of the ATPase domain of SWI/SNF to hydrolyze ATP without significantly affecting its affinity for ATP.
INTRODUCTION
Eukaryotic DNA is packaged by wrapping 147 bp of DNA around a histone octamer to form nucleosomes. Nucleosomes are packed into higher order arrays through internucleosomal interactions. Nucleosomal DNA is not readily accessible to many regulatory proteins and therefore nearby regions cannot be easily transcribed, repaired or replicated. Different parts of genomic DNA are accessed at defined periods in cell growth and development by altering the chromatin structure in an ATP-dependent process with proteins called chromatin remodelers. Chromatin remodelers can move nucleosomes to different places on DNA, alter the composition of the nucleosome by exchanging out different histone variants or completely disassemble nucleosomes from DNA (1–3).
In SWI/SNF, there are 12 different subunits that may contribute to its nucleosome remodeling activity (2,4,5). There have been efforts to determine the minimum number of ‘core’ subunits required for SWI/SNF activity. Some subunits, like Swp82 and Snf11, are not required for in vivo SWI/SNF activity, and thus are unlikely to have critical roles in remodeling (6). A minimal yeast SWI/SNF complex consisting of Swi2/Snf2 and Arps 7 and 9 has been found to have activity in vitro comparable to the whole 12 subunit complex; however, the three subunit complex does not displace the H2A–H2B dimer from nucleosomes (7).
A minimal mammalian complex consists of two or four subunits that includes the catalytic subunit BRG1 and the auxiliary subunits BAF155, INI1 and BAF170 (8,9). The BAF155 and BAF170 subunits are homologs of yeast Swi3 and are believed to be the main scaffold subunit(s) of the SWI/SNF complex. INI1 is the homolog of the yeast Snf5 subunit of SWI/SNF. The common factor in these studies is the universally required catalytic subunit. The catalytic subunit alone can remodel chromatin, but does so with lower efficiency than does the 12 subunit holo-complex in part because it binds with a much lower affinity than the holo-complex (9–11). Other data point to the catalytic subunit as part of the holo-complex making most of the critical contacts with nucleosomes. The Swi2/Snf2 catalytic subunit of yeast SWI/SNF was shown to be the primary subunit that contacts nucleosomal DNA and histones by site-directed crosslinking, although Snf5 also appears to be associated with histones (12). Based on these data, it seems likely that the catalytic subunit contains many or most of the domains that interact with nucleosomal DNA and histones by SWI/SNF that are crucial for nucleosome remodeling.
Besides the ATPase domain there are four other known protein domains that are found in the catalytic subunit. Other than the ATPase domain, the two domains most studied in the catalytic subunit of SWI/SNF are the bromodomain and helicase-SANT-associated (HSA) domain. The bromodomain is located near the C terminus of Snf2 and binds to acetylated N-terminal histone tails (13–15) which helps retain SWI/SNF bound to acetylated nucleosomes (16). While the bromodomain does bind to histones it is not required in vitro for nucleosome remodeling with recombinant histones (17) or in vivo for SWI/SNF activity (18,19). The bromodomain probably facilitates in the recruitment of SWI/SNF, but it is not likely to have an essential role in mobilizing nucleosomes. The HSA domain in Swi2/Snf2 and Sth1 is required for binding to the actin related proteins Arp7 and Arp9 (20). Similarly the HSA domain in BRG1 (human SWI/SNF), Eaf1 (histone acetyltransferase NuA4 complex), INO80 and Swr1 associate with other actin related subunits (20,21). In humans, the HSA domain in BRG1was found to be critical for interactions with the BAF250a/ARID1a subunit that is needed for SWI/SNF recruitment by the glucocorticoid-receptor (21). Loss of HSA or the Arp7 and nine subunits had only modest effects on the ATPase and nucleosome mobilizing activities of RSC and did not appear to be critically required for nucleosome remodeling (20).
Two other domains have been identified based on their sequence homology with other known protein motifs or domains. One of these two domains is the two A–T hook motifs located at the N-terminal side of the bromodomain. The other domain is the QLQ domain found between the extreme N-terminus and the HSA domain. The functional significance of these two domains is not well understood. A region between the ATPase domain and the A–T hook domain in BRG1 was recently shown to bind histones and to be required for SWI/SNF remodeling (22). The same general region of BRG1 was also likely to be important in conferring the specific remodeling activity of BRG1 to a chimera with SNF2h in which the ATPase domain of SNF2h was swapped with that of BRG1 (23). Although this region is conserved from yeast to human, it has not been studied before in yeast prior to our current study or identified as a conserved domain of SWI/SNF complexes. We have found this region to have a crucial role in chromatin remodeling for yeast SWI/SNF and refer to it as the Snf2 ATP Coupling or SnAC domain.
MATERIALS AND METHODS
Yeast strains
SNF2 was cloned into yIplac128 plasmid along with C-terminal double FLAG epitope tag with LEU2 marker to generate PSY3. Domain deletion was generated by standard gap repair where two PCR products flanking the domain of interest made with the yIplac128-SNF2-2FLAG-LEU2 plasmid were co-transformed into a Δsnf2 strain (24). Other strains are listed in Table 1.
Table 1.
Yeast strains used in this study
| Strain | Genotype |
|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 |
| PSY1 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 snf6-2FLAG-9 amino acids-LEU2a |
| PSY2 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 snf2-2FLAG-9 amino acids-LEU2a |
| PSY3 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 snf2Δ::kanMX |
| kanMXΔ::snf2Δ1312-1444-2FLAG-9 amino acids-LEU2a | |
| Δsnf2 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 snf2Δ::kanMX |
aThe 9 amino acid insertion has the sequence 5′ gtc gac tct aga gga tcc ccg ggt acc 3′ and translates to VDSRGSPGT.
Genome-wide expression profiling
Expression profiling was performed as described previously (25,26). Briefly, cultures were grown in YPD at 25°C to an A600 of 0.8. Cells were rapidly harvested by centrifugation at room temperature, washed with RNAse-free DEPC treated (diethyl pyrocarbonate) double distilled water and then frozen in liquid nitrogen in less than 15 min to minimize exposure to changes that might affect expression. Total RNA was isolated using the hot-phenol method (27) followed by poly (A+) mRNA purification using Oligo-dT Cellulose (Ambion) according to manufacturer's instructions. Externally generated Bacillus subtilis polyadenylated transcripts (Phe, Lys, Dap, Thr, Trp) were added to the total RNA based on A600 optical density of the culture. These serve as spiking controls. The cDNA generated by reverse transcription using aminoallyl-dUTP was labeled with fluorescent dyes (Cy3 and Cy5). Two micrograms of mRNA was used for hybridizations. Slides were treated with DyeSaver (Genisphere) to preserve signal intensity. Yeast genic arrays were used for the expression analysis. Each of Saccharomyces cerevisiae's ~6200 ORFs were PCR amplified in individual PCR reactions and were printed onto glass slides (Penn State DNA Microarray facility). Microarrays were scanned and quantitated with a GenePix 4000 A scanner and GenePix 4.0 software (Axon Instruments). Raw data are accessible through GEO (Accession # GSE 30675). CLUSTER and TREEVIEW were used for analysis and representation of microarray data (28).
Microarray image files were processed as follows: (i) spots where >25% of the pixels were saturated were eliminated; (ii) the mean foreground signal minus mean background signal had to be greater than the sum of the mean background signal plus one standard deviation of the background intensity. (iii) Ratios of ‘test’ / ‘reference’ were log2 transformed. ‘Test’ samples were ΔSnAC, Δsnf2 and BY4741 against an independent BY4741 reference. SNF2-2FLAG test was referenced against an independent SNF2-2FLAG. (iv) From this ratio, the corresponding ratio of the spiking controls (median) was subtracted. (v) Dye-swapped replicates were averaged.
ChIP-chip
ChIP assays were performed as described previously (29,30). Briefly, a Snf2 TAP-tagged strain (from Open Biosystems, catalog number YSC1178-7502942) and the untagged (BY4741) control were grown at 25°C in 500 ml of YPD to an A600 of 0.8. Cells were cross-linked for 15 min with 1% formaldehyde and quenched with glycine for 5 min. After harvesting, the cells were lysed with zirconium beads. The washed chromatin pellets were sheared by sonication using a Bioruptor generating on average 200- to 300-bp sized DNA fragments. Sheared chromatin was immunoprecipitated with IgG-Sepharose and the ChIP-enriched DNA was then amplified using ligation-mediated PCR (31). The 75–300 bp LM–PCR amplified fragments were gel purified according to the manufacturer's protocol (Qiagen). One hundred nanograms of the gel-purified DNA was amplified by 15 PCR cycles, coupled to fluorescent dyes (Cy5 and Cy3) and co-hybridized to custom low-density tiling arrays. Array design for the low-density tiling microarrays is the same as described previously (29). For each ChIP sample, two biological replicates employing a dye swap for each replicate was performed. Occupancy data were filtered, normalized to corresponding probes in the null (no tag) control, then centered by dividing the dataset by the median normalized probe value for ~300 probes located in intergenic regions between two convergent (T–T) genes, and log2 transformed as described previously (32).
Nucleosome reconstitution, gel shift assays for binding, recruitment and remodeling
Mononucleosomes were reconstituted with 5.2 µg of PCR generated DNA from p-159-1 plasmid that had 29 and 59 bp of DNA flanking either side of the 601 nucleosome positioning sequence (29N59) or with 8 µg of sonicated salmon sperm DNA, 100 fmol 32P labeled 29N59 DNA and 9 µg wild-type Xenopus laevis octamer at 37°C by a rapid salt dilution method (33). The labeled DNA was generated by PCR with an oligonucleotide labeled using Optikinase (USB) and [γ32P] ATP (6000 Ci/mol). Radiolabeled probe for di and trinucleosomes was generated from plasmid DNA as described (34) and reconstituted in presence of salmon sperm DNA by salt dilution method.
SWI/SNF complexes were purified as described previously (35). For standard binding and recruitment gel shift assays, SWI/SNF was incubated with 6.4 nM nucleosomes with or without 25 ng competitor DNA in the presence or absence of 3.2 nM Gal4-VP16, respectively (34). The apparent KD of SWI/SNF for nucleosomes and DNA was determined as described previously by gel shift assay (36). Binding under competitive conditions was performed with 20 nM nucleosomes and 18 nM SWI/SNF in presence of varying amounts of sheared salmon sperm competitor DNA (0, 1.6, 3.2, 6.25, 12.5 and 25 ng). Binding assays with DNA were performed with 7.3 nM 50-bp DNA substrate and increasing amounts of SWI/SNF (0.4, 0.8, 1.6, 3.2, 6.4 and 12.8 nM). Binding with DNA under competitive conditions was performed as with nucleosomes except that DNA was used at 7.3 nM concentration and SWI/SNF at 13.3 nM.
Varying amounts of SWI/SNF was prebound to 29N59 mononucleosomes (6.4 nM), 58N50N23 dinucleosomes (4.7 nM) or 58N50N50N6 trinucleosomes (4.7 nM) containing only PCR-generated DNA for mononucleosomes or salmon sperm and plasmid backbone DNA for di- and trinucleosomes, respectively, at 30°C for 30 min before remodeling for 10 min with 800 µM ATP. The reaction was stopped and SWI/SNF competed off by addition of γ-thio ATP and salmon sperm DNA to a final concentration of 4.5 mM and 0.45 mg ml−1, respectively. The remodeled products were analyzed on 5% native polyacrylamide gels (acrylamide:bisacrylamide = 60:1) at 200 V in 0.5× Tris–Borate–EDTA.
ATPase assays
SWI/SNF was incubated with 6.4 nM mononucleosomes (nucleosome stimulated) or 35 ng pUC18 plasmid DNA (DNA stimulated) for 30 min at 30°C. ATPase assays were performed with 8 µM ATP (containing 0.02 mCi γP32 ATP) for 10 min. For ATPase kinetic assays, SWI/SNF was prebound to 29N59 nucleosomes (2.5 nM) containing only PCR-generated DNA at a molar ratio of 3:1 (full-binding conditions) for 15 min at 30°C and an additional 15 min at 25°C. ATP was added to a final concentration of 320 µM for different times and stopped as before. 0.5 µl of the reactions were spotted on PEI-cellulose plates (J.T. Baker) and run with running buffer containing 0.5 M formic acid and 0.5 M lithium chloride. The plates were dried and imaged by autoradiography.
RESULTS
The SnAC domain is required in vivo for gene regulation by SWI/SNF
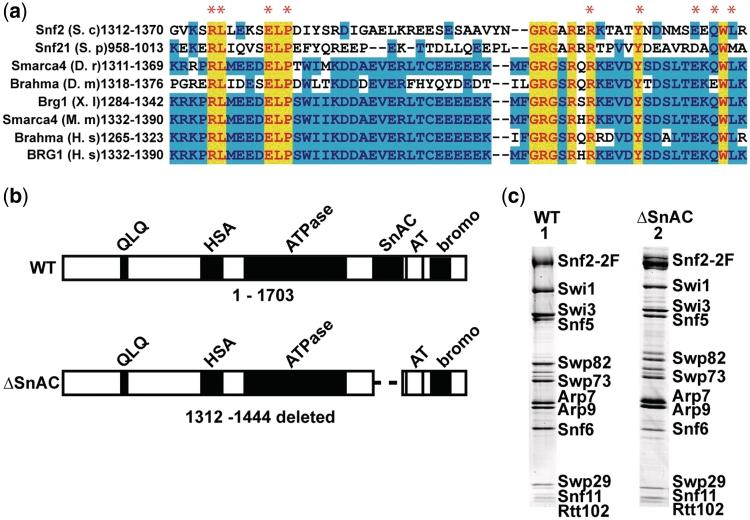
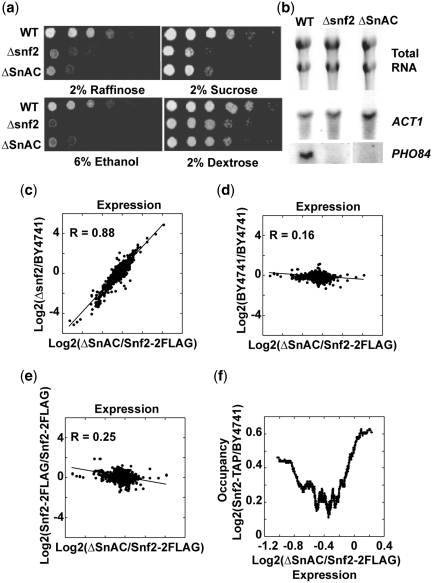
Alignment of yeast Swi2/Snf2 with various homologs including those from higher eukaryotes revealed a ~60 amino acid region located between the ATPase and AT-hook domains that showed remarkable conservation (Figure 1a). This well-conserved region is referred to as the SnAC or Snf2 ATP coupling region because of its ability to facilitate ATP hydrolysis and nucleosome remodeling as will be shown later. A region encompassing the SnAC domain from amino acid 1312 to 1444 was removed from Swi2/Snf2 (ΔSnAC SWI/SNF) to assess the role of the SnAC domain in gene activation and chromatin remodeling by SWI/SNF (Figure 1b). The ΔSnAC strain (PSY3, Table 1) along with Δsnf2 and wild-type strains (BY4741 and PSY2) were analyzed by genome-wide expression profiling with DNA microarrays. Changes in gene expression in the ΔSnAC strain were closely correlated with gene expression changes in the Δsnf2 strain (Figure 2c–e, factor of 0.88), but not with the control strains (correlation factors of 0.16 and 0.25); indicating that the SnAC domain is essential for most of the Swi2/Snf2-mediated gene activation. Northern analysis of SWI/SNF-dependent PHO84 expression and carbon source growth assays confirmed that ΔSnAC behaves like the Δsnf2 strain (Figure 2a and b).
Figure 1.
SnAC domain is a conserved feature of eukaryotic SWI/SNF type remodelers and is not required to maintain complex integrity. (a) Swi2/Snf2 homologs from various eukaryotes are compared in a region between the ATPase and AT-hook domains. Yellow indicates amino acid residues that are strictly conserved across species, whereas blue is slightly less conserved. Residues marked with red asterisk have been mutated to alanine as described. The organisms are S. cerevisiae (S.c), Schizosaccharomyces pombe (S.p), Danio rerio (D.r), Drosophila melanogaster (D.m), Xenopus laevis (X.l), Mus musculus (M.m) and Homo sapiens (H.s). (b) The domain organization of Swi2/Snf2 is shown with domains in black and labeled accordingly. The region from amino acids 1312 to 1444 was removed encompassing the SnAC domain. (c) SWI/SNF was purified from strains with and without the SnAC domain (harboring two FLAG epitopes on Snf2; Snf2-2F) and separated on a 4–20% gradient SDS polyacrylamide gel and stained with Sypro Ruby. Lanes 1 and 2 are SWI/SNF with and without the SnAC domain, respectively. The WT SWI/SNF complex was purified from the PSY2 strain.
Figure 2.
The conserved SnAC domain is required in vivo for Swi2/Snf2-mediated gene activation. (a) Growth spot assays with carbon sources other than dextrose were used to assess the effect on SWI/SNF activity when the SnAC domain was deleted. As a control, a strain missing the entire SWI2/SNF2 gene was tested as well. Plates contained 2% raffinose, 2% sucrose or 6% ethanol. (b) Total RNA was isolated from wild-type (PSY2), Δsnf2 and ΔSnAC (PSY3) strains; blotted and probed for PHO84 expression. Ethidium bromide staining of total RNA in the top panel and detection of the ACT1 mRNA in the middle panel show the extent of RNA recovered from the strains. The bottom panel shows the levels of the PHO84 mRNA transcript detected in these strains. (c–e) Scatter plot comparisons of global changes in gene expression are shown for ΔSnAC compared to Δsnf2 or wild-type strains (BY4741 and PSY2). Correlation coefficients (R) are shown. (f) Wild-type Swi2/Snf2 occupancy levels are plotted as a function of genome-wide changes in gene expression in the ΔSnAC (PSY3) strain.
Those genes shown to be physically associated with SWI/SNF by chromatin immunoprecipitation (ChIP) tended to be in one of two expression groups that were either positively or negatively affected when the SWI2/SNF2 gene was deleted. Both sets of genes were equally affected when the SnAC domain was absent (Figures 2f) and suggest that the SnAC domain is important for both gene activation and repression by Swi2/Snf2.
The SnAC domain is not required for proper assembly of SWI/SNF, efficient SWI/SNF binding to nucleosomes or recruitment by Gal4-VP16
ΔSnAC SWI/SNF was purified by immunoaffinity chromatography to determine the role of the SnAC domain in complex integrity/assembly, binding, nucleosome and DNA-dependent stimulation of ATP hydrolysis and nucleosome mobilization. SDS–PAGE analysis of the purified protein showed that in the absence of the SnAC domain, Swi2/Snf2 was properly assembled into a complete SWI/SNF complex with all the other 11 subunits (Figure 1c). The stoichiometry of most of the 12 subunits remained the same in ΔSnAC as wild-type (WT) SWI/SNF except for Snf6. Snf6 has two copies per Swi2/Snf2 in the complex and was half that in ΔSnAC as compared to WT SWI/SNF, suggesting that one copy of this subunit has been lost.
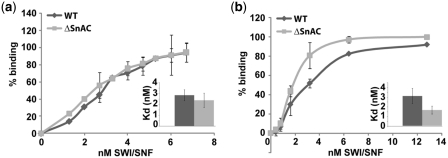
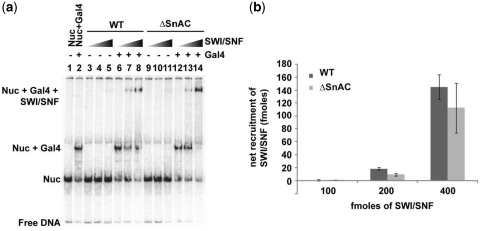
The affinity of ΔSnAC SWI/SNF for nucleosomes was determined using native gel shift assays. The amount of SWI/SNF was varied with a constant amount of reconstituted mononucleosomes. The SnAC domain was not required for efficient binding of SWI/SNF to nucleosomes and the apparent KD of WT and ΔSnAC complex was 2.85 ± 0.5 and 2.38 ± 0.7 nM, respectively (Figure 3a). Another approach was taken to determine the relative binding affinities of WT and ΔSnAC SWI/SNF to nucleosomes. Increasing amounts of competitor DNA was added to nucleosomes saturated with WT and ΔSnAC complexes and the percentage of bound complex left estimated (Supplementary Figure S1a and b). The apparent KD values and the competitor DNA titration revealed similar binding affinities of WT and ΔSnAC to nucleosomes. Similar approaches were used to determine that ΔSnAC has a slightly higher affinity for 50 bp of DNA than WT SWI/SNF (Figure 3b, Supplementary Figure S1c and d). The apparent KD for DNA binding was 3.1 ± 0.8 and 1.6 ± 0.4 nM for WT and ΔSnAC complex, respectively. SWI/SNF however is recruited by transcription factors in order to activate select genes in vivo and the SnAC domain could be essential for SWI/SNF recruitment by transcription activators. Three subunits in the SWI/SNF complex (Snf5, Swi1 and Swi2/Snf2) have been implicated to interact with activators such as Gal4-VP16, Hap4, Gcn4, Pho4 and Swi5 (37–39). The N-terminus of Snf5 and a region encompassing the ARID domain of Swi1 have been shown to interact with these activators directly (38). Investigators have been unable to assign a particular region in Swi2/Snf2 that could be involved in recruitment. In our assays, recruitment of ΔSnAC SWI/SNF was observed with nucleosomes having Gal4-VP16 bound to its cognate site in the extranucleosomal DNA region under stringent binding conditions with competitor DNA. The efficiency of Gal4-VP16 recruitment of WT and ΔSnAC SWI/SNF was measured based on the amount of nucleosomes bound under recruitment conditions and compared to those without Gal4-VP16. Three different concentrations of SWI/SNF were used to discriminate the effect of the deletion of the SnAC domain on Gal4-VP16 recruitment. SWI/SNF efficiently bound nucleosomes in the presence of 25 ng of competitor DNA only when Gal4-VP16 was present (Figure 4a, lanes 3–5 and 6–8). ΔSnAC SWI/SNF behaved the same as WT SWI/SNF suggesting that this region is not involved in interacting with transcription activators (Figure 4b).
Figure 3.
The SnAC domain is not required for efficient binding of SWI/SNF to nucleosomes or DNA. (a) Different amounts of SWI/SNF ranging from 1.3 to 6.7 nM were incubated with 6.4 nM nucleosomes. The percent of nucleosome bound was determined by gel shift and plotted. The apparent KD (concentration of SWI/SNF at which half of the nucleosomes were bound) was determined and shown in the inset. (b) Increasing amounts of SWI/SNF ranging from 0.4 to 12.8 nM was incubated with 7.3 nM 50 bp DNA and the percent of bound DNA determined and plotted. The apparent KD for DNA binding is shown in the inset.
Figure 4.
The SnAC domain is dispensable for Gal4-VP16-mediated recruitment of SWI/SNF. (a) Recruitment of SWI/SNF by Gal4-VP16 was monitored by binding SWI/SNF (6.4, 13 and 26 nM) to 29N59 mononucleosomes (6.4 nM) either in the presence or absence of Gal4-VP16 (3.2 nM) and competitor DNA (25 ng). Bound complexes were resolved by 4% native PAGE. (b) The amount of SWI/SNF recruited by Gal4-VP16 was determined by subtracting the amount of SWI/SNF bound in the absence from that in the presence of Gal4-VP16. The net recruitment with 6.4, 12.8 and 26 nM SWI/SNF for WT and ΔSnAC SWI/SNF is shown in the bar graph.
The SnAC domain is required for DNA and nucleosome-stimulation of the ATPase activity of SWI/SNF
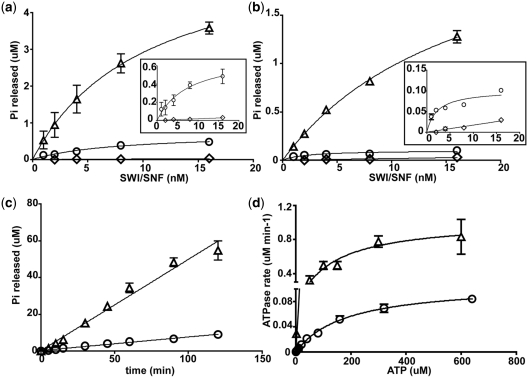
The roles of the SnAC domain in the enzymatic activity of SWI/SNF were determined by comparing the ATPase activity of WT and ΔSnAC SWI/SNF with nucleosomes and free DNA. Previously, SWI/SNF has been shown to have its ATPase activity equally stimulated by free DNA or nucleosomes (18,37,40). The concentration of nucleosomes/DNA and ATP remained constant along with the incubation time while varying the amounts of SWI/SNF (Figure 5a and b). The nucleosome stimulated ATPase activity of ΔSnAC SWI/SNF was ~8-fold less than WT SWI/SNF (Figure 5a and inset). The ATPase activity of ΔSnAC SWI/SNF when stimulated by free DNA was also 12- to 13-fold down as compared to WT SWI/SNF (Figure 5b and inset). Besides titrating with SWI/SNF, the rate of ATP hydrolysis was also measured for both WT and ΔSnAC SWI/SNF with higher concentrations of ATP (320 µM) so as to not be limiting and was with nucleosomes as substrate (Figure 5c). The difference in rates between WT and ΔSnAC SWI/SNF was ~7-fold (8.2 versus 1.2 nM ATP s−1). These data show that the SnAC domain has an important role in stimulating the ATPase activity of SWI/SNF in the presence of either nucleosomes or free DNA.
Figure 5.
The SnAC domain is necessary for efficient stimulation of the ATPase activity of SWI/SNF by nucleosomes or free DNA. (a) ATPase assays were done with fixed concentrations of γ-[32P]-ATP (8 µM) and 29N59 nucleosomes (8 nM). The reactions were incubated for 10 min and inorganic phosphate separated from ATP by thin layer chromatography on PEI-cellulose (33). SWI/SNF concentrations ranged from 1 to 16 nM. Reactions contained WT (open triangle) or ΔSnAC SWI/SNF (open circle) or WT SWI/SNF with no nucleosomes added (open diamond). Inset shows only ΔSnAC SWI/SNF with nucleosomes (open circle) and WT SWI/SNF with no nucleosomes added (open diamond) with an expanded y-axis. (b) Similar ATPase assays were done as in (a) except they contained 35 ng pUC18 DNA instead of 29N59 nucleosomes. Reactions contained WT (open triangle) or ΔSnAC SWI/SNF (open circle) or WT SWI/SNF with no DNA added (open diamond). Inset shows only ΔSnAC SWI/SNF with DNA (open circle) and WT SWI/SNF with no DNA added (open diamond) with an expanded y-axis. (c) The rate of ATP hydrolysis of WT (open triangle) and ΔSnAC SWI/SNF (open circle) with 320 µM ATP was measured with excess SWI/SNF (7.5 nM) relative to nucleosomes (2.5 nM). (d) The Km and Kcat values for ATP hydrolysis of WT and ΔSnAC SWI/SNF were found using 8 nM nucleosomes, 1.6 nM SWI/SNF, and ATP ranging from 0.2 to 640 µM. The Km and Kcat values were obtained by graphing initial rates of ATP hydrolysis versus ATP concentration as shown and performing non-linear regression analysis using GraphPad Prism.
How does the SnAC domain regulate the activity of the ATPase domain? The affinity for ATP (Km) and the catalytic activity (Kcat) of WT and ΔSnAC SWI/SNF was determined with nucleosomes in excess to SWI/SNF (5:1 molar ratio of nucleosome to SWI/SNF) and different concentrations of ATP (Figure 5d). The parameters obtained for the WT complex are similar to those reported earlier (41). In comparison, the ΔSnAC SWI/SNF had only a 1.6-fold increase in Km while there was a ~10-fold decrease in Kcat (Table 2). Due to the similar Km values, it appears the SnAC domain does not facilitate binding ATP, but instead affects the hydrolysis of the gamma phosphate. Since the SnAC domain is not directly part of the two lobes making up the ATPase domain it may be that the SnAC domain regulates the orientation and positioning of the two lobes in order to promote catalysis. Other studies have shown that the position of the lobe bound to ATP relative to the other lobe containing an arginine finger is critical for efficient hydrolysis of ATP by DNA-dependent ATPases (42–44).
Table 2.
Kinetic parameters obtained from Michaelis–Menten analysis
| WT | ΔSnAC | R1316A, L1317A | E1322A, P1324A | R1354A | Y1359A | E1366A, Q1368A | L1370A | |
|---|---|---|---|---|---|---|---|---|
| Km (µM) | 110 ± 30 | 180 ± 21 | 300 ± 80 | ND | 200 ± 41 | 66 ± 19 | 130 ± 22 | 77 ± 10 |
| Kcat (min−1) | 670 ± 76 | 68 ± 3.1 | 140 ± 21 | ND | 110 ± 11 | 79 ± 7.5 | 73 ± 2.3 | 48 ± 2.7 |
| Kcat /Km (min−1 µM−1) | 6.1 | 0.38 | 0.47 | ND | 0.55 | 0.84 | 0.56 | 0.62 |
ND, not determined.
The SnAC domain is required for mobilizing nucleosomes
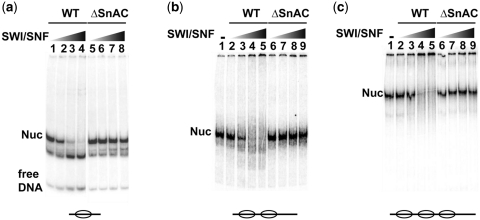
The remodeling efficiency of ΔSnAC SWI/SNF was examined using gel shift assays with mono-, di- or trinucleosomes and different concentrations of SWI/SNF (Figure 6a–c). SWI/SNF characteristically mobilizes asymmetrically positioned 29N59 mononucleosomes (the numbers refer to the length of extranucleosomal DNA) to positions slightly off the DNA ends causing the nucleosome to migrate faster [Figure 6a lanes 1–4 and reference (45)]. Nucleosomes are moved towards each other in dinucleosomes with an H2A–H2B dimer being displaced first and later an entire nucleosome being evicted (34). At the highest concentration of SWI/SNF, WT SWI/SNF shifted most of the nucleosomes as seen by changes in their electrophoretic mobilities, but were not moved with ΔSnAC SWI/SNF at comparable concentrations and reaction conditions (Figure 6a, lanes 5–8, Figure 6b and c, lanes 6–9). Different preparations of ΔSnAC SWI/SNF all had the same inability to mobilize nucleosomes and showed that the SnAC domain has a vital role in mobilizing nucleosomes.
Figure 6.
The SnAC domain is crucial for SWI/SNF-mediated nucleosome movement. (a) The nucleosome mobilization activity of ΔSnAC SWI/SNF was compared to WT SWI/SNF by gel shift on a 5% native PAGE. The reactions contained 6.4 nM 29N59 mononucleosomes and 1.6 (lanes 1 and 5), 3.2 (lanes 2 and 6), 6.4 (lanes 3 and 7) or 13 nM SWI/SNF (lanes 4 and 8) and were incubated for 10 min with 800 µM ATP. Reactions were stopped and SWI/SNF competed off as described in ‘Materials and Methods’ section. (b) Same as in (a) except 4.7 nM dinucleosomes were used as substrate with 2.4 (lanes 2 and 6), 4.8 (lanes 3 and 7), 9.6 (lanes 4 and 8) and 19 nM SWI/SNF (lanes 5 and 9). Lane 1 contained 4.7 nM dinucleosome with no SWI/SNF added. (c) Same as in (b) except 4.7 nM trinucleosomes were used.
Amino acid substitution mutations in the SnAC domain disrupt the ATPase and remodeling activities of SWI/SNF
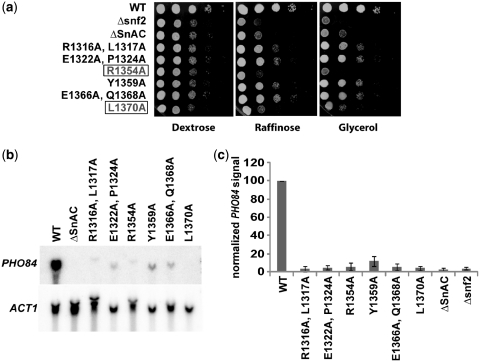
Sequence alignment of the SnAC domain from yeast Swi2/Snf2 with various Swi2/Snf2 homologs in other eukaryotes revealed several invariant amino acids (Figure 1a, yellow boxes). Single and double amino acid substitutions to alanine were made in this region to generate six individual mutants—(i) R1316A, L1317A, (ii) E1322A, P1324A, (iii) R1354A, (iv) Y1359A, (v) E1366A, Q1368A and (vi) L1370A (Figure 1a, asterisks). Strains were grown on alternative carbon sources to screen for growth defects characteristic of the lack of SWI/SNF with the WT (PSY2), Δsnf2, ΔSnAC (PSY3) and the various point mutant strains (Figure 7a). The R1354A and L1370A strains were the most strongly affected of the point mutant strains and had growth defects most like Δsnf2. Northern blot assays with the PHO84 gene seemed to be more sensitive than the growth assays since all of the point mutant strains had a considerably lower level of PHO84 expression than the wild-type strain (Figure 7b and c).
Figure 7.
Mutations of the conserved residues in the SnAC domain negatively affect SWI/SNF mediated function in vivo. (a) Six strains containing substitution mutations within the SnAC domain were grown on alternative carbon media containing either 2% raffinose or 2% glycerol to assay for growth defects. (b–c) The same strains from (a) were assayed for PHO84 gene expression by northern blotting with SWI/SNF-independent ACT1 control. The right panel shows percentage of PHO84 expression relative to WT after normalizing relative to the ACT1 signal.
Next, SWI/SNF was immunoaffinity purified for all the mutants in the SnAC domain and characterized biochemically. Complex integrity was maintained for all the SWI/SNF complexes containing the different point mutations in the SnAC domain and there was no reduction in the amount of Snf6 (Supplementary Figure S2). Mutations in the SnAC domain adversely affected ATP hydrolysis by SWI/SNF when stimulated by mononucleosomes similar to that observed for ΔSnAC SWI/SNF (Table 2). Most of the point mutations in the SnAC domain negatively affected the Kcat of ATP hydrolysis and were 5–14 times lower than WT SWI/SNF. The affinity of SWI/SNF for ATP for most of the SnAC mutants was comparable to WT SWI/SNF as reflected by their KM values. The R1316A/ L1317A mutant reduced the affinity of SWI/SNF for ATP slightly more than most of the other mutations. The E1322A and P1324A mutant was particularly deficient in nucleosome stimulated ATPase activity which made it too difficult to measure its Kcat and KM values.
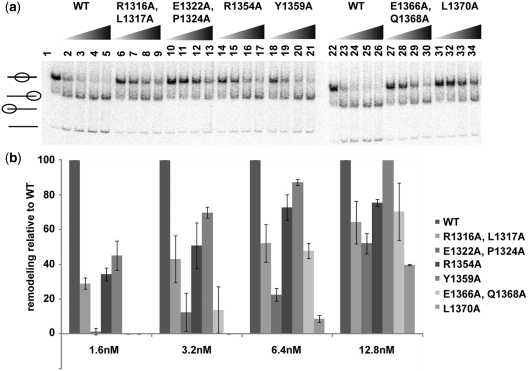
The nucleosome mobilizing activity of SWI/SNF with mutations in the SnAC domain were assessed by measuring the amount of nucleosomes moved with different SWI/SNF concentrations by native gel shift assay (Figure 8a). At the lowest SWI/SNF concentration three of the six mutants moved approximately two to three times less nucleosomes than WT SWI/SNF (Figure 8b, 1.6 nM SWI/SNF). The other three SnAC mutant SWI/SNF had much less nucleosome remodeling activity which could not be readily detected until higher concentrations of SWI/SNF were used. At the highest SWI/SNF concentration there are significant amounts of nucleosomes moved by all six SnAC domain mutants. While the point mutations in the SnAC domain do interfere with the nucleosome mobilizing activity of SWI/SNF, they do not generally seem to be as defective as the complete deletion of the SnAC region (compare Figures 6a and 8a). The reduction in the ATPase and nucleosome mobilizing activities when the conserved residues in the SnAC domain are changed shows the functional importance of the SnAC domain consistent with that shown by deletion of this region.
Figure 8.
Mutations of the conserved residues in the SnAC domain inhibit nucleosome mobilization by SWI/SNF. (a) Varying amounts of WT and mutant SWI/SNF (1.6, 3.2, 6.4 and 12.8 nM) was added to 6.4 nM mononucleosomes and remodeled for 10 min with 800 µM ATP. (b) The amount of nucleosomes remodeled relative to WT SWI/SNF was determined and plotted for each concentration of mutant and WT SWI/SNF.
DISCUSSION
In this study, a new domain has been found in the catalytic subunit of yeast SWI/SNF (Swi2/Snf2) that has a critical role in regulating the nucleosome mobilization and ATPase activities of SWI/SNF independent of binding or recruitment. The SnAC (Snf2 ATP Coupling) domain is well conserved throughout the different members of the SWI/SNF subfamily and is not present in other ATP-dependent remodeler families. While there have been two reports pointing to a region between the A–T hook and ATPase domain being important for BRG1 remodeling activity, it has not been recognized before to be a universal domain in all SWI/SNF complexes and its functional role in remodeling had not been previously characterized. Kingston and colleagues (23) when swapping ATPase domains between BRG1 and SNF2h included a segment of 136 amino acids on the C-terminal side of the ATPase domain from residue 1250 to 1386 with the ATPase domain to ensure its functionality. The SnAC domain identified in our study from S. cerevisiae corresponds to a region from amino acids 1332 to 1390 in BRG1 based on sequence homology (Figure 1) and is likely the functional element needed for BRG1 activity. We find for the first time that the pivotal role of the SnAC domain is not for complex assembly or to contribute to the affinity of SWI/SNF for nucleosomes, but rather to regulate the activity of the ATPase domain. The KM and Kcat values for nucleosome-stimulated ATP hydrolysis revealed that the SnAC domain is important for efficiently hydrolyzing ATP or releasing ADP and not for binding ATP. Muchardt and colleagues (22) also identified a region in BRG1 from residue 1223 to 1420 that was important for remodeling and selectively bound histone H3 which includes the SnAC domain. Although the region they examined is larger than the SnAC domain, it is most likely that the activities they attributed to this region are those of the SnAC domain. It may be that interactions of the SnAC domain with histones have a positive feedback on the ATPase domain. We have confirmed that it is the conserved 58 amino acid region that is responsible for these activites and not other adjacent regions by making point mutations in the most highly conserved positions. The SnAC domain is not found in other ATP-dependent chromatin remodelers outside of the SWI/SNF family and therefore has a role in remodeling unique to the SWI/SNF subfamily of remodelers that could be connected to the ability of SWI/SNF complexes to disassemble nucleosomes.
Regulation of the ATPase activity by the SnAC domain could be through interactions with the ATPase domain (direct) or by interacting with some other target that influences the ATPase domain (indirect). The ATPase domain contains two RecA like lobes, one of which binds to ATP and the other promotes the cleavage of the ß–γ phosphodiester bond of ATP. The orientation of the two lobes is a critical factor in determining the efficiency of ATP hydrolysis. The SnAC domain can have a direct effect on the ATPase efficiency by interacting with and orienting the two lobes of the ATPase domain. The SnAC domain could indirectly stimulate the ATPase activity by binding to DNA or nucleosomes thereby helping to tether the ATPase domain to its substrate. This however does not seem to be the case since the affinity of SWI/SNF for DNA or nucleosomes is not adversely affected when the SnAC domain is absent. Another indirect mode may be that the SnAC domain interacts with another part of the catalytic subunit or other SWI/SNF subunit that in its turn stimulates the activity of the ATPase domain. The reduction in the Snf6 subunit observed when the region containing the SnAC domain is deleted may support this idea; however, the point mutations in the SnAC domain did not cause any reduction in Snf6 associated with the complex but still adversely affected ATP hydrolysis and remodeling. Since the activities of the ΔSnAC and point mutant SWI/SNF complexes are so similar, the reduced level of Snf6 likely does not influence the activity of SWI/SNF. Both halves of the conserved SnAC domain are required for enhancing the ATPase activity of SWI/SNF since mutations in either side of the SnAC domain inhibit the ATPase activity of SWI/SNF. The extent to which the SnAC domain positively regulates the ATPase domain is more than that previously observed for other domains outside of the ATPase domain. Deletion of the HSA domain caused a change of <2-fold in the ATPase activity of RSC, whereas deletion or mutation of the SnAC domain reduced the ATPase activity of SWI/SNF as much as ~14-fold (WT versus L1370A).
The SnAC domain is also essential for the in vivo activity of SWI/SNF. Deletion of the SnAC domain had the same effect as the complete deletion of the SWI2/SNF2 gene when analyzed at a genomic level by monitoring mRNA expression with DNA microarrays and is equivalent to the complete loss of SWI/SNF. These effects were also seen by measuring the expression of the PHO84 gene by northern analysis when the SnAC domain was deleted or its conserved residues changed to alanine. The simpler carbon source growth assays were consistent with these data, but the quantitative differences did not correspond to those found when measuring biochemically the activities of SWI/SNF with mutant SnAC domains. The SnAC domain has an important role in SWI/SNF remodeling as shown both in vitro and in vivo for promoting nucleosome movement and ATP hydrolysis. In the future it will be important to find the interaction partners of the SnAC domain and how they together have such a critical role in controlling SWI/SNF activity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: National Institutes of Health (GM48413 to B.B.; GM59055 to B.F.P.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Jerry Workman for the yIplac128 plasmid, Judy Davie for the yeast knockout strains, Geena Skariah for help generating the SnAC deletion, Bhawana Uprety for help in purifying the SnAC mutants and all BB lab members for critical reading of the article.
REFERENCES
- 1.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 2.Gangaraju VK, Bartholomew B. Mechanisms of ATP dependent chromatin remodeling. Mutat. Res. 2007;618:3–17. doi: 10.1016/j.mrfmmm.2006.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith CL, Horowitz-Scherer R, Flanagan JF, Woodcock CL, Peterson CL. Structural analysis of the yeast SWI/SNF chromatin remodeling complex. Nat. Struct. Biol. 2003;10:141–145. doi: 10.1038/nsb888. [DOI] [PubMed] [Google Scholar]
- 6.Treich I, Cairns BR, de los Santos T, Brewster E, Carlson M. SNF11, a new component of the yeast SNF-SWI complex that interacts with a conserved region of SNF2. Mol. Cell. Biol. 1995;15:4240–4248. doi: 10.1128/mcb.15.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang X, Zaurin R, Beato M, Peterson CL. Swi3p controls SWI/SNF assembly and ATP-dependent H2A-H2B displacement. Nat. Struct. Mol. Biol. 2007;14:540–547. doi: 10.1038/nsmb1238. [DOI] [PubMed] [Google Scholar]
- 8.Kadam S, McAlpine GS, Phelan ML, Kingston RE, Jones KA, Emerson BM. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev. 2000;14:2441–2451. doi: 10.1101/gad.828000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 10.Phelan ML, Schnitzler GR, Kingston RE. Octamer transfer and creation of stably remodeled nucleosomes by human SWI-SNF and its isolated ATPases. Mol. Cell. Biol. 2000;20:6380–6389. doi: 10.1128/mcb.20.17.6380-6389.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 2002;16:2120–2134. doi: 10.1101/gad.995002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dechassa ML, Zhang B, Horowitz-Scherer R, Persinger J, Woodcock CL, Peterson CL, Bartholomew B. Architecture of the SWI/SNF-nucleosome complex. Mol. Cell. Biol. 2008 doi: 10.1128/MCB.00693-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Owen DJ, Ornaghi P, Yang JC, Lowe N, Evans PR, Ballario P, Neuhaus D, Filetici P, Travers AA. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 15.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 16.Hassan AH, Neely KE, Workman JL. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell. 2001;104:817–827. doi: 10.1016/s0092-8674(01)00279-3. [DOI] [PubMed] [Google Scholar]
- 17.Awad S, Hassan AH. The Swi2/Snf2 bromodomain is important for the full binding and remodeling activity of the SWI/SNF complex on H3- and H4-acetylated nucleosomes. Ann. NY Acad. Sci. 2008;1138:366–375. doi: 10.1196/annals.1414.038. [DOI] [PubMed] [Google Scholar]
- 18.Laurent BC, Treich I, Carlson M. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 1993;7:583–591. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- 19.Laurent BC, Treitel MA, Carlson M. Functional interdependence of the yeast SNF2, SNF5, and SNF6 proteins in transcriptional activation. Proc. Natl Acad. Sci. USA. 1991;88:2687–2691. doi: 10.1073/pnas.88.7.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szerlong H, Hinata K, Viswanathan R, Erdjument-Bromage H, Tempst P, Cairns BR. The HSA domain binds nuclear actin-related proteins to regulate chromatin-remodeling ATPases. Nat. Struct. Mol. Biol. 2008;15:469–476. doi: 10.1038/nsmb.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trotter KW, Fan HY, Ivey ML, Kingston RE, Archer TK. The HSA domain of BRG1 mediates critical interactions required for glucocorticoid receptor-dependent transcriptional activation in vivo. Mol. Cell. Biol. 2008;28:1413–1426. doi: 10.1128/MCB.01301-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavigne M, Eskeland R, Azebi S, Saint-Andre V, Jang SM, Batsche E, Fan HY, Kingston RE, Imhof A, Muchardt C. Interaction of HP1 and Brg1/Brm with the globular domain of histone H3 is required for HP1-mediated repression. PLoS Genet. 2009;5:e1000769. doi: 10.1371/journal.pgen.1000769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan HY, Trotter KW, Archer TK, Kingston RE. Swapping function of two chromatin remodeling complexes. Mol. Cell. 2005;17:805–815. doi: 10.1016/j.molcel.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 24.Johnston M, Riles L, Hegemann JH. Gene disruption. Methods Enzymol. 2002;350:290–315. doi: 10.1016/s0076-6879(02)50970-8. [DOI] [PubMed] [Google Scholar]
- 25.Chitikila C, Huisinga KL, Irvin JD, Basehoar AD, Pugh BF. Interplay of TBP inhibitors in global transcriptional control. Mol. Cell. 2002;10:871–882. doi: 10.1016/s1097-2765(02)00683-4. [DOI] [PubMed] [Google Scholar]
- 26.Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- 27.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 28.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venters BJ, Pugh BF. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 2009;19:360–371. doi: 10.1101/gr.084970.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zanton SJ, Pugh BF. Changes in genomewide occupancy of core transcriptional regulators during heat stress. Proc. Natl Acad. Sci. USA. 2004;101:16843–16848. doi: 10.1073/pnas.0404988101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zanton SJ, Pugh BF. Full and partial genome-wide assembly and disassembly of the yeast transcription machinery in response to heat shock. Genes Dev. 2006;20:2250–2265. doi: 10.1101/gad.1437506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kassabov SR, Henry NM, Zofall M, Tsukiyama T, Bartholomew B. High-resolution mapping of changes in histone-DNA contacts of nucleosomes remodeled by ISW2. Mol. Cell. Biol. 2002;22:7524–7534. doi: 10.1128/MCB.22.21.7524-7534.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dechassa ML, Sabri A, Pondugula S, Kassabov SR, Chatterjee N, Kladde MP, Bartholomew B. SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes. Mol. Cell. 38:590–602. doi: 10.1016/j.molcel.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dechassa ML, Zhang B, Horowitz-Scherer R, Persinger J, Woodcock CL, Peterson CL, Bartholomew B. Architecture of the SWI/SNF-nucleosome complex. Mol. Cell. Biol. 2008;28:6010–6021. doi: 10.1128/MCB.00693-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kagalwala MN, Glaus BJ, Dang W, Zofall M, Bartholomew B. Topography of the ISW2-nucleosome complex: insights into nucleosome spacing and chromatin remodeling. EMBO J. 2004;23:2092–2104. doi: 10.1038/sj.emboj.7600220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cote J, Quinn J, Workman JL, Peterson CL. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 38.Prochasson P, Neely KE, Hassan AH, Li B, Workman JL. Targeting activity is required for SWI/SNF function in vivo and is accomplished through two partially redundant activator-interaction domains. Mol. Cell. 2003;12:983–990. doi: 10.1016/s1097-2765(03)00366-6. [DOI] [PubMed] [Google Scholar]
- 39.Neely KE, Hassan AH, Brown CE, Howe L, Workman JL. Transcription activator interactions with multiple SWI/SNF subunits. Mol. Cell. Biol. 2002;22:1615–1625. doi: 10.1128/MCB.22.6.1615-1625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cairns BR, Kim YJ, Sayre MH, Laurent BC, Kornberg RD. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc. Natl Acad. Sci. USA. 1994;91:1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith CL, Peterson CL. A conserved Swi2/Snf2 ATPase motif couples ATP hydrolysis to chromatin remodeling. Mol. Cell. Biol. 2005;25:5880–5892. doi: 10.1128/MCB.25.14.5880-5892.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 43.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 44.Ogura T, Whiteheart SW, Wilkinson AJ. Conserved arginine residues implicated in ATP hydrolysis, nucleotide-sensing, and inter-subunit interactions in AAA and AAA+ ATPases. J. Struct. Biol. 2004;146:106–112. doi: 10.1016/j.jsb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 45.Kassabov SR, Zhang B, Persinger J, Bartholomew B. SWI/SNF unwraps, slides, and rewraps the nucleosome. Mol. Cell. 2003;11:391–403. doi: 10.1016/s1097-2765(03)00039-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.