Abstract
Guanine-quadruplexes (G4) consist of non-canonical four-stranded helical arrangements of guanine-rich nucleic acid sequences. The bulky and thermodynamically stable features of G4 structures have been shown in many respects to affect normal nucleic acid metabolism. In vivo conversion of G4 structures to single-stranded nucleic acid requires specialized proteins with G4 destabilizing/unwinding activity. RHAU is a human DEAH-box RNA helicase that exhibits G4-RNA binding and resolving activity. In this study, we employed RIP-chip analysis to identify en masse RNAs associated with RHAU in vivo. Approximately 100 RNAs were found to be associated with RHAU and bioinformatics analysis revealed that the majority contained potential G4-forming sequences. Among the most abundant RNAs selectively enriched with RHAU, we identified the human telomerase RNA template TERC as a true target of RHAU. Remarkably, binding of RHAU to TERC depended on the presence of a stable G4 structure in the 5′-region of TERC, both in vivo and in vitro. RHAU was further found to associate with the telomerase holoenzyme via the 5′-region of TERC. Collectively, these results provide the first evidence that intramolecular G4-RNAs serve as physiologically relevant targets for RHAU. Furthermore, our results suggest the existence of alternatively folded forms of TERC in the fully assembled telomerase holoenyzme.
INTRODUCTION
DNA and RNA sequences containing tandem repeats of guanine tracts can form G-quadruplex (G4) structures, non-canonical and thermodynamically stable four-stranded helical arrangements (1,2). The building block of G4 structures is the G-quartet, a square-planar assembly of four guanines held together by Hoogsteen hydrogen bonding. G4 structures result from the consecutive stacking of G-quartets and are further stabilized by alkali metal ions such as Na+ or K+ that position along the helix axis and coordinate the O6 keto oxygens of the tetrad-forming guanines. G4 scaffolds are extremely polymorphic in the relative orientation of strands (parallel, anti-parallel or mixed configurations), length, sequence and conformation of the loops connecting the G-tracts. In addition, G4 structures in vitro can result from the assembly of one (intramolecular G4) or multiple (bi- or tetramolecular-G4) nucleic acid strands.
Biochemical and biophysical studies have shown in vitro that stable G4 structures can form spontaneously from G-rich regions of single-stranded nucleic acid under near physiological conditions. Genome-wide computational surveys have identified more than 300 000 potential intramolecular G4-forming sequences in the human genome (3,4) and revealed a higher prevalence of these sequences in functional genomic regions such as telomeres, promoters (5,6), untranslated regions [UTRs (7,8)] and first introns (9). Taken together, these observations suggest that G4 structures participate in regulating various nucleic acid processes, such as telomere maintenance or the control of gene expression. Nevertheless, because of the lack of evidence that such structures really exist in vivo, G4 structures have often been considered as a structural curiosity without relevance for living organisms. There is now reason to believe that G4 structures are not merely an in vitro artefact, as several recent findings concur with their existence in cells. First, both a G4-specific dye and antibodies raised against telomeric G4-DNA specifically stained telomeres in human and ciliate cells, respectively (10–12). In addition, several potential G4-forming sequences in promoters were shown to form intramolecular G4 structures in vitro and to affect gene expression in vivo (13,14). A possible contribution of G4 to regulating promoter activity was indicated by impairment of the transcriptional activity of several genes by G4-stabilizing ligands (14) or a single-chain antibody specific for intramolecular G4-DNA (15), in a manner correlating with the occurrence of predicted G4 structures in the control regions (16).
Like DNA, RNA can also form G4 structures. Although, to date, G4-RNAs have not attracted as much attention as their DNA counterparts, the formation of G4 structures in RNA is emerging as a plausible regulatory factor in gene expression. RNA is more prone than DNA to form G4 structures due to its single-strandedness, and G4-RNAs have also proved to be more stable than their cognate G4-DNA under physiological conditions (17–19). Bioinformatics analyses of human 5′-UTR sequences revealed potential G4-forming motifs in as many as 3000 different RNAs (7,20). Moreover, the formation of G4 structures in 5′-UTR was shown to impede translation initiation (7,21–23). Given that potential G4 sequences have been identified near splicing and polyadenylation sites (24–26), G4 formation may also affect RNA metabolism at several different stages. Furthermore, formation of parallel G4-RNA structures has also been reported in vitro for telomeric RNA repeats [TERRA, (27–29)] and for the human telomerase template RNA [TERC, (30)], suggesting that G4-RNA formation also plays a part in regulatory processes at telomeres.
The discovery of proteins that positively or negatively stabilize such G4 structures is further indirect evidence for the existence of such structures in vivo. Among the many proteins from various organisms that bind to G4-DNAs in vitro (31), several helicases show ATP-dependent G4-resolving activity (32–36) and have been clearly implicated in the maintenance of genome integrity (37–40). RHAU (alias DHX36 or G4R1), a member of the human DEAH-box family of RNA helicases, exhibits in vitro G4-RNA binding with high affinity for its substrate, and unwinds G4 structures much more efficiently than double-stranded nucleic acid (41,42). Consistent with these biochemical observations, RHAU was also shown to bind to mRNAs in vivo (43) and was identified as the main source of tetramolecular G4-RNA-resolving activity in HeLa cell lysates (42).
Although considerable information is available on the enzymatic activity of RHAU in vitro, almost nothing is known about its biological function as a G4-binding/resolving enzyme in vivo. To address this question, we sought RNAs bound by RHAU in living cells, surmising that the identification of RHAU-bound RNAs and understanding the effects of RHAU should provide important clues to its function in vivo. To this end, we employed high-throughput gene array technologies to identify and quantify the co-purified RNAs on a genome-wide scale. With this screen, we identified about 100 RNAs significantly enriched by RHAU. Computational analysis of RNA sequences for potential intramolecular G4 structures revealed the preferential association of RHAU with transcripts bearing G4-forming motifs, suggesting direct targeting of G4-RNA by RHAU. Amongst the RNAs with the potential to form G4 structures and selectively enriched with RHAU, we identified TERC as a bona fide target of RHAU. Characterization of the RHAU-TERC interaction in vivo and in vitro showed binding of TERC by RHAU to be strictly dependent on the formation of a G4 structure on the 5′-extremity of TERC RNA. Finally, we have demonstrated that RHAU not only interacts with TERC stricto sensu, but is also part of the fully assembled telomerase complex through direct interaction with TERC G4 structure. Taken together, these data demonstrate that intramolecular G4-RNAs are naturally occurring substrates of RHAU in vivo. Moreover, they provide indirect but strong support for the existence of a G4-RNA structure in the telomerase RNP.
MATERIALS AND METHODS
Plasmid constructs, cloning and mutagenesis
The baculoviral expression vector employed for the expression of GST-RHAU and the plasmids used for the expression of the C-terminal FLAG-tagged recombinant RHAU proteins RHAU-FLAG and RHAU(ΔRSM)-FLAG were previously described (43–45). The pIRES.EGFP-myc-N1 vector expressing the C-terminal myc-tagged recombinant RHAU protein was derived from the previously described pIRES.EGFP-FLAG-N1/RHAU expression vector (43) by substituting the FLAG sequence with the myc epitope sequence. Human TERT cDNA (clone: IOH36343, mapped sequence: NM_198253) was obtained from ImaGenes GmbH (Berlin, Germany) and subcloned by PCR amplification into the pSL1-FLAG-N1 mammalian expression vector. The pSL1-FLAG-N1 vector was derived from pEGFP-N1 (Clontech) by replacing the EGFP open reading frame with the FLAG epitope sequence. The human TERC genomic region was PCR-amplified from the genomic DNA of HEK293T cells to yield a 2.1-kb fragment encompassing 1080 bp of the TERC promoter, its coding sequence, and 553 bp of the 3′ flanking genomic region. This fragment was blunt-cloned into pGEM-T Easy vector (Promega) at the HincII/EcoRV sites. The G4 motif sequence of TERC was mutagenized using a variation of the classical QuikChange® (Stratagene) site-directed mutagenesis PCR method (46). To prepare templates for in vitro run-off transcription, the T7 or SP6 phage promoters were inserted upstream of the TERC coding sequence by PCR. The resulting PCR products were cloned into the pSL1-FLAG-N1 vector at the NheI/AgeI sites. Following linearization with NarI or AgeI, in vitro transcription of these templates yielded the TERC (1–71 nt) and full-length TERC (1–451 nt) RNA fragments, respectively. Constructions of all these plasmids were confirmed by sequencing. Sequences of oligonucleotides used in this work and detailed descriptions of the plasmid constructs are available upon request.
Cell culture and transfection
Human cervical carcinoma HeLa and embryonic kidney HEK293T cell lines were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum (FCS) and 2 mM l-glutamine at 37°C in a humidified 5% CO2 incubator. Transient transfections were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Transfected cells were cultured for 24–36 h prior to testing for transgene expression.
RIP-chip assay
Cells were harvested 24–36 h post-transfection, washed with ice-cold PBS and resuspended in lysis buffer {1× PBS, 1% v·v−1 Nonidet P-40, 2 mM EDTA, 2 mM AEBSF [4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride], 1× protease inhibitor cocktail (Complete EDTA-free, Roche), 0.2 U·μl−1 RNasin® Plus (Promega)} for 30 min. All subsequent operations were performed at 4°C. The lysates were cleared by centrifugation (21 000g, 15 min) and mixed with 10 μl of a 50% slurry of anti-FLAG M2-agarose affinity gel (Sigma) that had been equilibrated in lysis buffer. After gentle agitation for 5 h, the resin was recovered by centrifugation, washed 3× with 500 μl lysis buffer, followed by three washes with 500 μl IP-washing buffer [50 mM Tris–HCl (pH 7.5), 300 mM NaCl, 0.1% v·v−1 Nonidet P-40, 5 mM EDTA, 0.4 U·μl−1 RNasin® Plus]. The resin was resuspended in 1 ml TRIzol (Invitrogen) for protein analysis and RNA extraction. For microarray analysis, 100 ng RNA was converted to cRNA according to the manufacturer’s guidelines and the reaction products hybridized to GeneChip® Human Gene 1.0 ST arrays (Affymetrix). The resulting raw expression values were RMA-normalized using R/BioConductor (47) and the Oligo Package [version 1.14.0 (48)]. Probesets were linked to Entrez Gene entries using Affymetrix annotation (NetAffx release 28, 11 March 2009), retaining a single probeset per gene. Genes not clearly detected in input samples [average log2(expression value) < 6.0] were discarded. Differential expression was determined using the Limma package (49), selecting genes with a minimal fold-change of 2.0 and an FDR-adjusted P < 0.01. RHAU targets were identified as transcripts enriched in RHAU-FLAG IP versus RHAU-FLAG input samples but not enriched in control IP versus control RHAU-myc input samples.
G4-RNA structure prediction and bioinformatics analysis
Human RNA sequences were retrieved from Entrez Nucleotide database. Non-overlapping putative intramolecular G4-forming sequences (PQS) and the corresponding G4-score values were computed with the QGRS Mapper algorithm (50) using the default parameters (window size = 30 nt, Min. G-group = 2, loop size = 0–36). For each transcript, the ∑(G4-score) value was calculated as the sum of all non-overlapping G4-scores computed by QGRS Mapper. To obtain a normalized ∑(G4-score) value, the ∑(G4-score) value was divided by the RNA length (kb). Randomization of RNA sequences by single- or dinucleotide shuffling was performed using the Altschul-Erickson algorithm (51).
Protein immunoprecipitation assay
Protein immunoprecipitation experiments were performed under the same conditions employed for the RIP-chip assay. Cleared cell lysates were mixed with rProtein A or Protein G Sepharose™ Fast Flow beads (GE Healthcare) and appropriate antibodies {mouse mAb anti-RHAU [12F33 (43)], rabbit pAb anti-dyskerin (H-300, Santa Cruz Biotechnology)}. After gentle agitation for 5 h, the beads were washed. For telomere repeat amplification protocol (TRAP) assays, beads were resuspended in 40 μl TRAP lysis buffer [10 mM Tris–HCl (pH 8.0), 150 mM NaCl, 1 mM MgCl2, 1 mM EDTA (pH 8.0), 1% v·v−1 Nonidet P-40, 0.25 mM Na-deoxycholate, 10% v·v−1 glycerol, 5 mM 2-mercaptoethanol, 0.1 mM AEBSF]. For protein and RNA analysis, beads were resuspended in 1 ml TRIzol and extraction performed according to the manufacturer’s instructions. Input and co-purified RNA samples were analysed by RT–qPCR. For protein analysis only, beads were directly resuspended in sodium dodecyl sulphate (SDS)-gel loading buffer. Input and immunoprecipitated protein samples were separated by SDS–PAGE and analysed by western blotting.
RNA analysis by quantitative (RT–qPCR) and semi-quantitative RT–PCR
Reverse transcription was performed using the ImProm-II™ Reverse Transcription System (Promega) with oligo(dT)15 or random hexamer primers, according to the manufacturer’s instructions. For monitoring first-strand cDNA synthesis, the reverse transcription reaction was performed in the presence of 20 μCi [α-32P]dATP (3000 Ci·mmol−1). The reaction products were separated by agarose gel electrophoresis and visualized by Phosphor-Imaging. Quantitative and semi-quantitative PCR reactions were performed in technical duplicates using the ABsolute™ QPCR SYBR® Green ROX Mix (Thermo Fisher Scientific), according to the manufacturer’s instructions, on an ABI Prism 7000 Sequence Detection System (SDS) and analysed with the ABI Prism 7000 SDS 1.0 Software (Applied Biosystems). Relative transcript levels were determined using the 2−ΔCt method (52). For each primer pair (Supplementary Table S1), the efficiency of amplification was determined to be equal or superior to 1.8. Control reactions lacking the reverse transcriptase or template RNA confirmed the specificity of the amplification reactions.
TRAP assays
Immunopurified ribonucleoprotein (RNP) complexes were assayed for telomerase activity by the TRAP assay (53). Four microliters of bead-immobilized RNP complexes in TRAP lysis buffer were incubated (30 min, 25°C) in 50 μl TRAP reaction buffer [20 mM Tris–HCl (pH 8.3 at room temperature), 63 mM KCl, 1.5 mM MgCl2, 0.2 mM dNTP mix (50 μM each of dATP, dTTP, dGTP and dCTP), 0.05% v·v−1 Tween®-20, 1 mM EGTA (pH 8.0), 4 ng·μl−1 Cy5-TS primer, 2 ng·μl−1 ACX primer (Supplementary Table S1), 1 U·μl−1 RNasin® Plus, 0.4 mg·ml−1 BSA, 0.04 U·μl−1 Thermo-Start™ Taq DNA Polymerase (Thermo Fisher Scientific)]. The reaction was followed by a 15-min incubation step at 95°C, followed by 15–17 cycles of amplification (30 s at 95°C, 30 s at 52°C, 45 s at 72°C). The reaction products were resolved on a pre-electrophoresed 10% non-denaturing polyacrylamide gel (19:1 acrylamide:bis ratio) in 0.5× TBE at 4°C for 90 min. After electrophoresis, gels were fixed [500 mM NaCl, 50% v·v−1 ethanol, 40 mM Na-acetate (pH 4.2)] for 30 min, scanned on a Typhoon 9400 Imager (GE Healthcare) and analysed with ImageQuant TL software (Nonlinear Dynamics). For immunodepletion experiments of RHAU, the residual telomerase activity in cell extracts was quantified by the quantitative TRAP assay [qTRAP, (53)]. An aliquot of 250 ng protein from immunodepleted HEK293T cell extracts in TRAP lysis buffer was incubated (30 min, 25°C) in 25 μl qTRAP reaction buffer [1× ABsolute™ QPCR SYBR® Green ROX Mix, 1 mM EGTA (pH 8.0), 4 ng·μl−1 TS primer, 4 ng·μl−1 ACX primer (Supplementary Table S1), 0.2 U·μl−1 RNasin® Plus]. The reaction was followed by a 15-min incubation step at 95°C, followed by 40 cycles of amplification (15 s at 95°C, 60 s at 60°C) on an ABI Prism 7000 Sequence Detector. The relative telomerase activity was determined using a standard curve and linear equation model (53).
Expression and purification of recombinant RHAU protein
Recombinant wild-type and ATPase deficient [RHAU(DAIH)] N-terminal GST-tagged RHAU proteins were expressed in Sf9 cells according to the supplier’s instructions (PharMingen) and purified to homogeneity as described previously (45). Purified recombinant GST-RHAU proteins were stored at −80°C. Purity of protein preparations was assessed by SDS–PAGE (Supplementary Figure S1) and protein concentrations determined photometrically at 280 nm using the calculated extinction coefficient ε = 166 615 M−1·cm−1.
In vitro synthesis of 32P-labeled TERC transcripts and intramolecular G4-RNA preparation
Synthetic radio-labeled wild-type and mutant (G4-MT) telomerase RNAs were prepared by in vitro transcription using 50 μCi [α-32P]UTP (3000 Ci·mmol−1) and T7 or SP6 RNA polymerases (Promega), respectively, according to the manufacturer’s instructions. The transcripts were purified by denaturing PAGE, ethanol precipitated and recovered by centrifugation. The purified RNAs were resuspended in potassium- or lithium-based storage buffer [10 mM Li-cacodylate (pH 7.4), 100 mM KCl or LiCl] and annealed by heating at 95°C for 2 min, followed by slow cooling to room temperature. G4-annealed radio-labeled RNAs were stored at −80°C.
RNA electromobility shift assay
Purified recombinant GST-RHAU protein at concentrations from 1 to 320 nM was incubated with 100 pM 32P-labeled G4-RNA in K-Res buffer [50 mM Tris–acetate (pH 7.8), 100 mM KCl, 10 mM NaCl, 3 mM MgCl2, 70 mM glycine, 10% glycerol], supplemented with 10 mM EDTA and 0.2 U·μl−1 SUPERase-In (Ambion) in a 10-μl reaction. The reactions were equilibrated at 22°C for 30 min. RNA–protein complexes were resolved on a pre-electrophoresed 6% non-denaturing polyacrylamide gel (37.5:1 acrylamide:bis ratio) in 0.5× TBE at 4°C for 90 min. After electrophoresis, gels were fixed for 1 h in 10% isopropanol/7% acetic acid. RNA–protein complexes were detected by Phosphor-Imaging, scanned on a Typhoon 9400 Imager (GE Healthcare) and analysed with ImageQuant TL software (Nonlinear Dynamics).
RESULTS
Microarray identification of RHAU-associated RNAs
To identify endogenous RNAs associated with RHAU in vivo on a genome-wide scale, we designed a RIP-chip assay (RNA immunoprecipitation coupled to microarray analysis). Subsets of RHAU target RNAs were isolated by immunoprecipitation (IP) assays under optimized conditions that preserved RNA–protein complexes. Briefly, HeLa cells were transfected with a vector expressing FLAG-tagged or myc-tagged RHAU. Immunoprecipitations of non-cross-linked whole-cell extracts were carried out using anti-FLAG antibodies. Anti-FLAG IP from cells expressing myc-tagged RHAU was employed as an IP control (IPctrl) to assess non-specific interactions that may occur during RIP. Following IP, co-fractionated RNAs were recovered and purified by standard phenol–chloroform extraction and converted to cRNA. Products were subsequently hybridized to human oligonucleotide arrays.
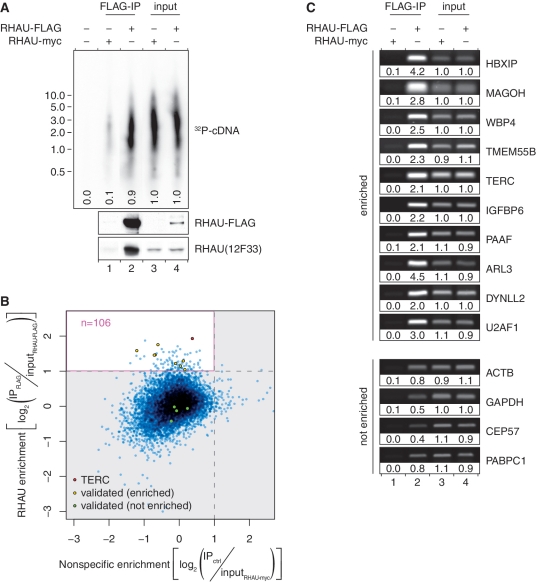
Western blot analysis of immunoprecipitated proteins revealed that RHAU-FLAG, but not RHAU-myc, was efficiently enriched from HeLa cell extracts following IP with anti-FLAG antibody (Figure 1A, bottom). Oligo(dT)-primed reverser-transcription of co-immunoprecipitated RNAs showed the presence of polyadenylated RNAs with sizes similar to the input (Figure 1A, top, compare lane 2 to lanes 3 and 4). With regard to the RHAU-FLAG IP fraction, 10 times less RNA signal was detected in the control immunoprecipitate (lane 1), demonstrating specific co-immunoprecipitation of mRNAs with RHAU. The association between RHAU and target RNAs was deemed specific as the anti-FLAG antibody exhibited no obvious cross-reactivity with other cellular proteins (data not shown).
Figure 1.
Analysis of the RNAs co-immunoprecipitated with RHAU RNPs. (A) RHAU associates with poly(A)+ RNAs in HeLa cells. First strand cDNA synthesis from input (lanes 3 and 4) and co-immunoprecipitated (lanes 1 and 2) RNAs was monitored by incorporation of [α-32P]dATP. The RT–PCR reaction products were separated by agarose gel electrophoresis. An autoradiogram of the gel is shown. The positions and sizes (kb) of marker DNAs are indicated at the left. Signal intensities are expressed relative to the average signal intensity of the input fractions (lanes 3 and 4). Expression and specific enrichment of RHAU-FLAG in input and FLAG-IP fractions were verified by Western blot analysis. (B) Scatter plot representation of the differential enrichment of RHAU-bound versus non-specific RNAs as quantified by microarray analysis. The pink rectangle delineates the area of RNA specifically enriched by RHAU. RNAs whose enrichment was further validated by semi-quantitative RT–PCR are indicated. (C) Validation of novel RHAU-bound target RNAs. The abundance of 10 potential RHAU targets and four non-targeted RNAs in total input RNA (lanes 3 and 4), control IP (lane 1) and RHAU-FLAG IP (lane 2) fractions was monitored by semi-quantitative RT–PCR. Reaction products were separated by agarose gel electrophoresis and visualized by SYBR Green staining. Band intensities are expressed relative to the average signal intensity of the input fractions (lanes 3 and 4).
Microarray analysis of the purified RNAs recovered from input and immunoprecipitated fractions revealed that 9354 (49%) of the 19 089 total genes available on the chip were significantly expressed in HeLa cells overexpressing RHAU (data not shown). In order to maximize the chance of identifying true RNA targets of RHAU as well as to minimize the occurrence of false positives, only those RNAs were chosen that were significantly (adjusted P < 0.01) enriched and were at least 2-fold more abundant in the RHAU-FLAG IP fraction than in the control. Of these potential RHAU targets, we discarded those that were also significantly enriched in the RHAU-myc expressing cells and might thus be RHAU-independent RNAs binding non-specifically to the anti-FLAG antibody matrix.
Thus, of the 9354 genes expressed in HeLa cells, 108 RNAs (1.2%) were found to be significantly enriched in RHAU-FLAG IP fraction compared with total input RNA (Figure 1B). Finally, after subtraction of two non-target RNAs based on the above-mentioned criteria that were associated with the antibody or the beads, 106 RNAs were judged to be specifically enriched by RHAU (Supplementary Table S2). The most abundant transcripts of these potential RHAU targets were selected for further analysis.
Validation of potential RHAU target RNAs
To assess independently the validity and reproducibility of the identified RHAU-associated transcripts, RNA abundance in total input RNA, control IP and RHAU-FLAG IP fractions from fresh whole-cell extracts were analysed by semi-quantitative RT–PCR. As shown in Figure 1C, 10 RNAs randomly selected from the group of plausible RHAU targets were confirmed to be enriched >2-fold in the RHAU-FLAG IP fraction than in the control IP or input fractions. Besides, four non-targeted RNAs were included as negative control to monitor RHAU binding to non-specific RNAs. None of them were found to be enriched in the RHAU-FLAG IP fraction despite their relatively high abundance in the input fraction. Taken together, these independents results validate the previous RIP-chip data. We assume that the remaining RNA species are also part of RHAU RNPs, although additional experiments would be necessary to confirm this supposition.
G4-content analysis for RNAs enriched by RHAU
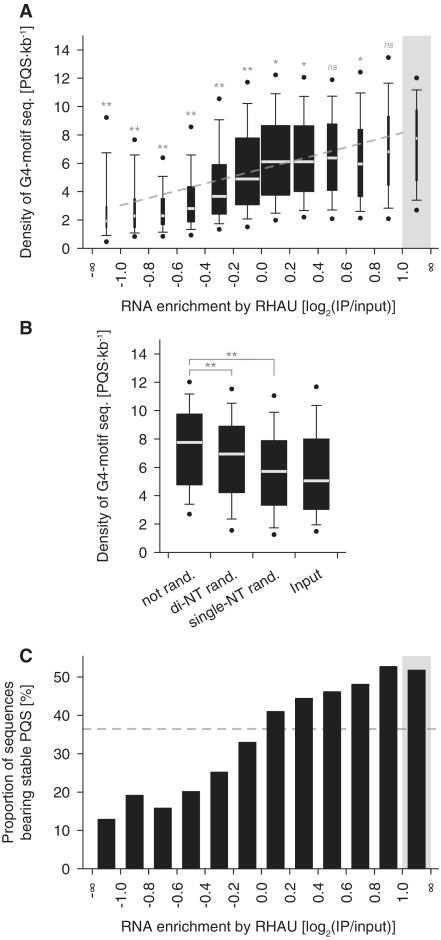
As RHAU shows a high affinity for G4-RNA structures in vitro (42), we next carried out a bioinformatics search for RNAs with potential intramolecular G4 structures. The rationale was that if RHAU binds G4-RNA in vivo, the proportion of potential G4-forming sequences should be higher among RNAs enriched by RHAU than among non-enriched RNAs. Of the various available methods for predicting intramolecular G4 motif sequences, we used QGRS Mapper that identifies and scores each potential G4-forming sequence according to their predicted stabilities (50). Being aware that only limited experimental data so far support the scoring method of QGRS Mapper, we employed the algorithm to identify potential G4-forming sequences, but we also included the G4-score-based analysis as Supplementary Data (Supplementary Figure S2), since the two approaches provided similar results. In fact, with almost eight potential intramolecular G4-forming sequences (PQS) per kilobase, the occurrence of G4 motif sequences was higher in the fraction enriched by RHAU than in any other fraction (Figure 2A and Supplementary Figure S2A). In addition, there was a weak (R2 = 0.10) but significant positive correlation between the predicted G4 motif density per transcript and the magnitude of RNA enrichment by RHAU. Randomization of RHAU target sequences, retaining single- or even dinucleotide frequencies of the original transcripts, significantly reduced the proportion of potential G4-forming sequences (Figure 2B and Supplementary Figure S2B). Thus, the proportion of G4 motif sequences among RHAU-associated transcripts cannot be explained by a mere bias in nucleotide composition.
Figure 2.
Computational analysis of potential intramolecular G4-forming sequences (PQS) among RNAs enriched by RHAU. (A) Box plot representation of the density of potential intramolecular G4-forming sequences per transcript (PQS·kb−1) as a function of their level of enrichment by RHAU. The lower and upper boundaries of the boxes indicate the 25th and 75th percentiles, respectively. The white lines within the boxes mark the median. The width of the boxes is proportional to the number of transcripts found within a group. The error bars indicate the 10th and 90th percentiles and the filled circles the 5th and 95th percentiles. The grey rectangle refers to the group of 106 RNAs specifically enriched by RHAU and serves as a control group for multiple comparison analysis. Statistical significance was determined by Kruskal–Wallis one-way ANOVA on ranks and the Dunn’s test. nsP ≥ 0.05; *P < 0.05; **P < 0.01. The correlation between the two variables was estimated by linear regression analysis (y = 2.56x + 5.60, R2 = 0.10, P < 0.001) and is indicated as a grey line on the graph. (B) PQS analysis among randomized RHAU target sequences. The sequences of the 106 transcripts specifically enriched by RHAU were shuffled, retaining single- or di-nucleotide base composition of the original transcripts. The box plot represents the density of PQS (PQS·kb−1) among the original (not rand.), di-nucleotide shuffled (di-NT rand.) and single-nucleotide shuffled (single-NT rand.) sequences. Statistical significance was determined by one-way repeated measures ANOVA and the Dunnett’s test. (C) Proportions of RNAs bearing stable PQS as a function of their level of enrichment by RHAU. The grey rectangle refers to the group of 106 RNAs specifically enriched by RHAU. The dashed grey line denotes the proportion of sequences showing stable PQS in the input fraction. Significant (P < 0.001) association between the magnitudes of the two variables was estimated by the chi-square-test-for-trend.
It should be mentioned that our prediction is likely to overestimate the number of G4-forming motif sequences per transcript. In fact, we also considered G4 structures consisting of only two successive G-tetrads as these metastable structures have also been demonstrated to have biologically relevant roles (54). However, after discarding these less-stable G4 structures from the analysis, we still observed a significant relationship between the proportion of sequences bearing potentially stable G4-forming sequences (composed of three or more stacked G-quartets) and the magnitude of RNA enrichment by RHAU (Figure 2C). Indeed, with more than half of the sequences presenting potentially stable G4-forming sequences, the group of 106 RNAs identified as true RHAU targets presents one of the highest incidences of potentially stable G4-forming motifs. In summary, the present data are consistent with preferential association of RHAU with transcripts containing potential G4-forming sequences, suggesting direct recognition of G4-RNA structures by RHAU. To further test this hypothesis, we selected one of the identified RHAU targets and addressed the molecular basis of its interaction with RHAU.
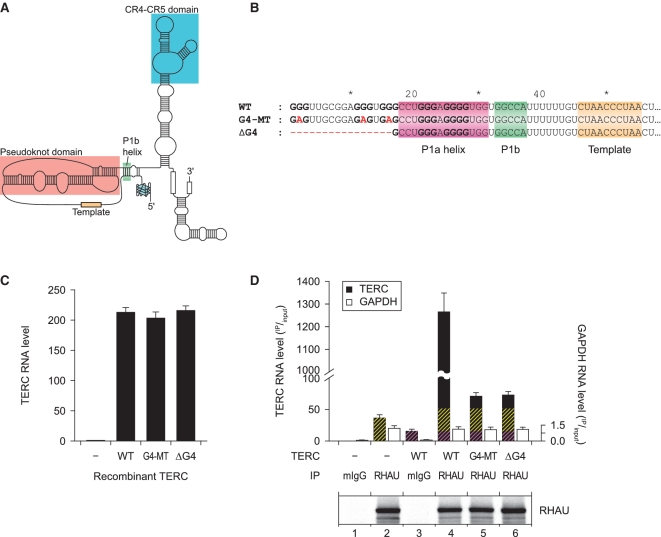
RHAU associates with TERC through its G4 motif sequence
Within the pool of transcripts enriched by RHAU and showing a high potential to form stable G4, TERC is the only RNA reported previously to form a stable and parallel G4 structure in vitro (Figure 3A) (30). TERC was also found to be one of the most abundant RNAs enriched by RHAU in several independent RIP-chip assays using various cell lines (Supplementary Table S2 and unpublished data). To address the role of the TERC G4 structure in its efficient co-immunoprecipitation by RHAU, we cloned the TERC gene, including its promoter and 3′ flanking genomic sequence. The 5′ extremity of the TERC sequence was subsequently mutated (G4-MT) or truncated (ΔG4) to prevent G4 formation (Figure 3B). To avoid effects on the structurally conserved P1 helix by the introduced substitutions, guanine residues that are part of both the predicted G4 structure and the P1a helix region were left intact. The recombinant forms of TERC were transiently transfected into HEK293T cells and the accumulation of stable TERC transcripts was monitored by RT–qPCR (Figure3C). After 24 h, HEK293T cells transiently transfected with the TERC(WT) construct but not with the vector alone showed a substantial (~200-fold) increase in TERC abundance relative to endogenous TERC levels. Importantly, mutations of the G4 motif (G4-MT or ΔG4) did not influence the steady-state level of exogenous TERC expression in these cells since recombinant wild-type, G4-MT and ΔG4 forms of TERC accumulated to comparable levels.
Figure 3.
RHAU associates with TERC through its G4-motif sequence. (A) Schematic representation of the secondary structure of human TERC bearing a parallel G4 structure in the 5′-extremity as described by Mergny et al. (30). (B) Nucleotide sequence of the WT and G4 motif mutant (G4-MT and ΔG4) forms of TERC. Guanine tracts that are predicted to form a stable G4-structure are shown bold. Nucleotide substitutions or deletions in mutant forms of TERC are marked in red. The P1 helix subdomains as well as the template region are indicated. (C) RNA expression levels of endogenous and recombinant WT and G4 motif mutant forms of TERC in HEK293T cells. Expression was quantified by RT–qPCR, normalized to GAPDH expression and endogenous TERC levels set to 1. Data represent the mean ± SEM of three independent experiments. (D) RT–qPCR analysis of the abundance of WT and G4 motif mutant forms of TERC that co-immunoprecipitated with endogenous RHAU protein. RNA levels in IP fractions are represented in function of their respective abundance in the input fraction. Immunoprecipitation with mouse IgGs (mIgG) served as a control to assess non-specific interactions. In lanes 4, 5 and 6, yellow hatches represent the fraction related to endogenous TERC signal and violet hatches represent the fraction of TERC RNA that non-specifically interacts with the antibody matrix or the beads. Data represent the mean ± SEM of five independent experiments. Comparable efficiency of RHAU immunoprecipitation in the various fractions was verified by western blot analysis with anti-RHAU antibodies.
To examine the significance of the G4 motif sequence for co-precipitation of TERC with RHAU, we repeated the RIP assay using HEK293T cells transiently overexpressing the wild-type or G4-MT and ΔG4 mutated forms of TERC. Immunoprecipitations were carried out on endogenous RHAU using a monoclonal antibody against RHAU and TERC RNA abundance was analysed by RT–qPCR (Figure 3D). As already shown for endogenous TERC, a substantial amount of overexpressed TERC(WT) was also recovered with endogenous RHAU, as evidenced by a 25-fold enrichment of TERC transcript over the mock transfection (Figure 3D, compare lanes 4 and 2). In marked contrast, both G4 motif mutated forms of TERC were barely co-immunoprecipitated with RHAU, judged by the 95% reduction in TERC abundance in the corresponding fractions compared with the WT control (Figure 3D, compare lanes 5 and 6 to lane 4). The drastic reduction in immunoprecipitation of the TERC mutants was not due to non-specific RNA degradation in these fractions. Indeed, comparable levels of non-specifically co-immunoprecipitated GAPDH RNA were found to contaminate all of these fractions. Taken together, these results indicate that the intact G4 motif sequence is a prerequisite for TERC association with RHAU in vivo.
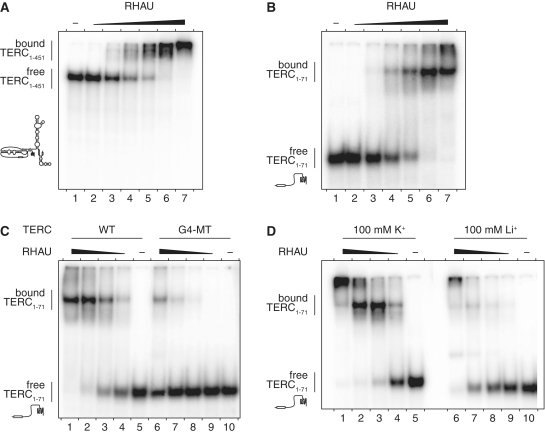
RHAU binds TERC through a G4 structure in the TERC 5′-region
In the absence of Mg·NTPs, RHAU specifically binds to tetramolecular G4-RNA structures with high affinity (42,45). The strict requirement for the G4 motif sequence for effective recovery of TERC by RHAU in vivo suggested that RHAU may also form a stable complex with TERC through direct binding of the G4-RNA forming structure. To address this question in vitro, we performed RNA electromobility shift assays (REMSA) using purified recombinant GST-tagged RHAU protein and in vitro transcribed radio-labeled full-length (1–451 nt) and 5′-end (1–71 nt) TERC fragments. In the absence of protein, the 32P-labeled probes migrated as a single species in the gel (Figure 4A and B). Addition of increasing amounts of RHAU to both full-length and 5′-end TERC fragments resulted in the appearance of a high-affinity (estimated Kd of 10 nM) ribonucleoprotein complex of reduced mobility. In contrast, the stability of the RHAU-TERC interaction was strongly impaired (~20-fold reduction) when formation of the G4 structure was prevented by mutation of the G4 motif sequence (G4-MT, Figure 4C). Similarly, conditions that are unfavourable to G4 stability (substitution of K+ for Li+) impaired the RHAU-TERC interaction to comparable extent (Figure 4D). Replacing K+ by Li+ was indeed shown to strongly reduce the thermodynamic stability of the TERC G4 structure (30). Thus, these results demonstrate a direct and specific interaction between RHAU and TERC dependent on RNA folding into a stable G4 structure. This is consistent with the above finding that RHAU can co-immunoprecipitate the wild-type but not the G4 motif mutated forms of TERC. Together these observations argue that a fraction of TERC RNA forms a G4 structure that can be further bound by RHAU, in vivo.
Figure 4.
Gel mobility shift assay for TERC G4 binding by RHAU. (A) Radio-labeled full-length (1–451 nt) and (B) 5′-end (1–71 nt) TERC fragments at a concentration of 100 pM were incubated without protein (−) or with increasing amounts (1, 3.2, 10, 32, 100 and 320 nM) of GST-tagged RHAU in the absence of ATP. The reaction mixtures were analysed by non-denaturing PAGE. An autoradiogram of the gel is shown. The positions of the free RNA substrate and the protein–RNA complex are indicated on the left. At concentrations up to 320 nM, GST protein alone had no effect on TERC mobility (data not shown). (C) RNA-binding assay with WT and G4 motif mutant (G4-MT) forms of TERC 5′-end (1–71 nt) fragments. Radio-labeled TERC fragments were incubated without protein (−) or with increasing amounts (3.2, 10, 32 and 100 nM) of GST-tagged RHAU. (D) Cation dependency of RHAU interaction with TERC. The 5′-end WT TERC fragment was incubated under standard (100 mM K+) or lithium-based (100 mM Li+) REMSA conditions without protein (−) or with increasing amounts (10, 32 100 and 320 nM) of GST-tagged RHAU.
RHAU associates with telomerase RNPs by direct interaction with TERC
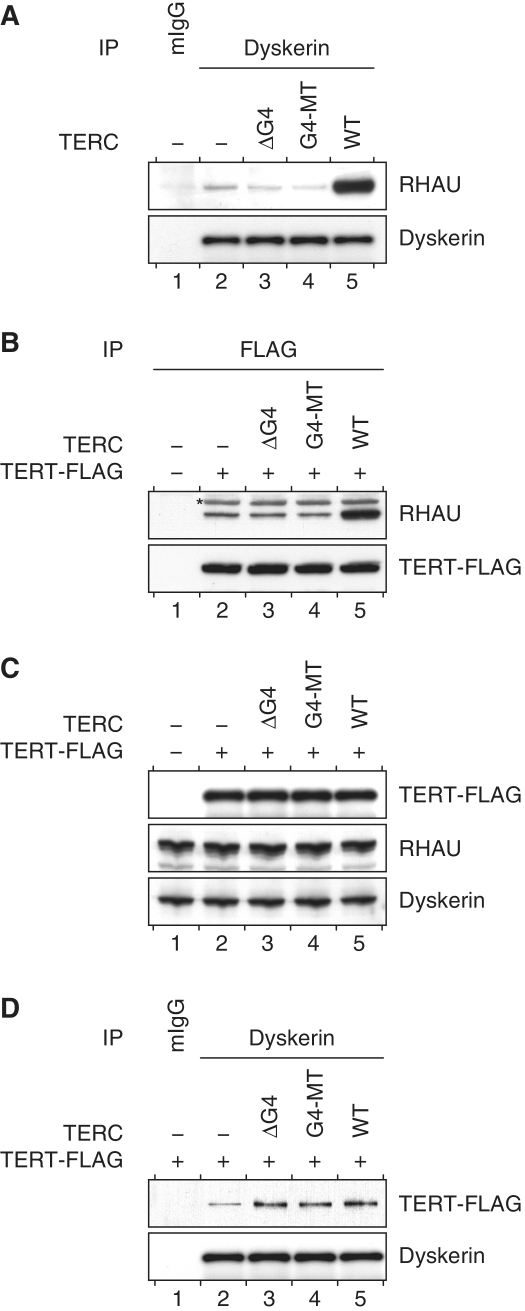
In cells, the biogenesis of the telomerase holoenzyme follows a stepwise RNP assembly. Upon transcription, nascent TERC is first bound by proteins dyskerin, NHP2 and NOP10 and subsequently joined by GAR1 (55). The co-transcriptional binding of this RNP complex is essential for the processing and accumulation of TERC transcripts. Finally, the catalytic protein component telomerase reverse transcriptase (TERT) assembles with the processed telomerase RNPs and forms the active telomerase holoenzyme. To determine whether RHAU was exclusively interacting with TERC during the transcriptional process or whether it was also part of the telomerase RNP, we performed co-IP assays and examined whether endogenous RHAU co-fractionated with endogenous dyskerin or FLAG-tagged TERT proteins. Immunoblot analysis showed that RHAU was present in both dyskerin and TERT immunoprecipitated fractions but absent in the control IP (Figure 5A and B, compare lanes 1 and 2). The association of RHAU with components of the telomerase holoenzyme proved to be strictly dependent on the ability of RHAU to bind the 5′-end of TERC, since overexpression of wild-type but not the G4 motif mutated forms of TERC resulted in extensive enrichment of co-precipitating RHAU protein (Figure 5A and B, compare lane 2 to lanes 3, 4 and 5). Although mutations of the TERC G4 motif sequence severely impinged on the recruitment of RHAU to the telomerase, they had no apparent repercussions on the steady-state expression level (Figure 5C) or on the assembly of the core components of telomerase. Indeed, as shown in Figure 5D (compare lane 2 to lanes 3, 4 and 5), similar levels of TERT protein co-fractionated with dyskerin following transient overexpression of either wild-type or G4 motif mutated forms of TERC. These results suggest that RHAU does not strictly bind TERC in the course of its biogenesis but that a fraction of RHAU also associates with the fully assembled telomerase holoenzyme through direct interaction with the G4 motif sequence of TERC.
Figure 5.
Association of RHAU with telomerase holoenzyme subunits. (A) TERC G4 motif dependent association of RHAU with dyskerin. Proteins from whole HEK293T cell lysates of either mock-transfected (−) cells or transiently expressing WT or G4 motif mutant (G4-MT, ΔG4) forms of TERC were immunoprecipitated with either control mouse IgGs (mIgG) or anti-dyskerin antibodies. Immunopurified RNP complexes were separated by SDS–PAGE and probed with anti-RHAU or anti-dyskerin antibodies. (B) TERC G4 motif-dependent association of RHAU with TERT. Protein immunoprecipitation experiments with anti-FLAG antibodies were performed with whole cells lysates of HEK293T cells transiently expressing TERT–FLAG protein together with WT or G4 motif mutant (G4-MT, ΔG4) forms of TERC. Immunopurified RNP complexes were separated by SDS–PAGE and probed with anti-RHAU or anti-FLAG antibodies. The asterisk denotes immunoprecipitated TERT–FLAG protein that cross-reacted unspecifically with the horsearadish peroxidase-conjugated secondary antibody. (C) Western blot analysis of comparable protein expression in input HEK293T cell lysates. (D) Protein immunoprecipitation experiments with anti-dyskerin antibodies were performed with whole cells lysates of HEK293T cells transiently expressing TERT–FLAG protein together with WT or G4 motif mutant (G4-MT, ΔG4) forms of TERC. Immunopurified RNP complexes were separated by SDS–PAGE and probed with anti-FLAG or anti-dyskerin antibodies.
RHAU associates with telomerase activity
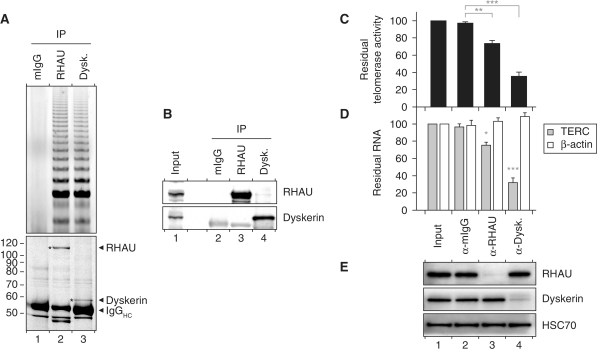
As RHAU binding to TERC requires a G4 structure, the finding that RHAU co-immunoprecipitated with components of the telomerase complex suggested the existence of an alternatively folded form of TERC bearing a G4 structure in a fraction of telomerase RNP. To address this further, immunopurified RHAU RNP complexes were analysed for telomerase activity by TRAP assay. In all the subsequent experiments, parallel immunopurification of dyskerin served as a positive control for telomerase activity (56). In agreement with previous observations, TRAP assays of antibody-bound complexes confirmed that substantial telomerase activity was recovered with RHAU but not with control IgGs (Figure 6A, top). Coomassie staining demonstrated similar yields of immunoprecipitated RHAU and dyskerin proteins (Figure 6A, bottom) and protein identity was subsequently verified by Western blotting (Figure 6B). To further examine the apparent significance of G4 binding by RHAU for its recruitment to the telomerase holoenzyme, we transiently expressed a FLAG-tagged mutant version of RHAU (ΔRSM, Supplementary Figure S3A) that was unable to bind to G4 structures (45). Western blot analysis confirmed that the wild-type and G4-binding-deficient (ΔRSM) forms of RHAU were expressed and recovered to similar levels (Supplementary Figure S3B). Immunopurified RHAU fractions were further assayed for telomerase activity and enrichment of TERC (Supplementary Figure S3C and D). As previously observed for endogenous RHAU, immunopurified FLAG-tagged RHAU(WT) efficiently recovered telomerase activity and co-precipitated TERC. In contrast, negligible telomerase activity and TERC signal were retrieved from the FLAG-tagged RHAU(ΔRSM) mutant, showing that G4 binding by RHAU is a prerequisite for RHAU binding to TERC. Altogether, these findings provide persuasive evidence that a fraction of RHAU associates with a subpopulation of the telomerase holoenzyme in vivo through direct interaction with the G4-motif sequence of TERC.
Figure 6.
Association of RHAU with telomerase activity. (A) TRAP assay of immunopurified endogenous RHAU and dyskerin RNP complexes. RHAU or dyskerin RNP complexes were enriched by immunoprecipitation and a fraction of the immunopurified RNP was essayed for telomerase activity by the TRAP assay. Mouse IgGs (mIgGs) served as a control to assess non-specific interactions to the antibody matrix. A fraction of the immunopurified RNP was separated by SDS–PAGE. The Coomassie stained gel was scanned on a LI-COR Odyssey infrared imaging system. Positions of the immunoprecipitated proteins as well as the immunoglobulin heavy chains (IgGHC) are indicated at the right. Positions and sizes (kDa) of marker proteins are shown at the left. (B) Western blot analysis of the immunopurified RNP complexes assayed for telomerase activity. (C) qTRAP analysis of the residual telomerase activity present in immunodepleted HEK293T cell lysates. Data represent the mean ± SEM of three independent experiments. Statistical significance was determined by one-way ANOVA and the Bonferroni t-test. *P < 0.05; **P < 0.01; ***P < 0.001. (D) RT–qPCR analysis of the residual levels of TERC and β-actin RNAs in immunodepleted HEK293T cell lysates. The RNA signal was normalized to GAPDH. Data represent the mean ± SEM of three independent experiments. Statistical significance was determined by one-way ANOVA and the Bonferroni t-test. (E) Western blot analysis of input and immunodepleted HEK293T cell lysates. Heat shock protein 70 cognate (HSC70) was used as a loading control.
Finally, to determine the proportion of telomerase activity associated with RHAU, we carried out immunodepletion studies of RHAU in HEK293T cell lysates and subsequently quantified the residual TERC level and telomerase activity within these fractions (Figure 6C and D). Depletion with RHAU antibody significantly ablated telomerase activity in an aliquot of the supernatant by 25%, while ~70% of the telomerase activity of the input fraction was lost following depletion of endogenous dyskerin. As a control, immunodepletion with control mouse IgGs produced no significant reduction in telomerase activity. Western blot analysis of the supernatants confirmed that both RHAU and dyskerin proteins were effectively depleted from the corresponding fractions (Figure 6E). Furthermore, the quantification by RT–PCR of RNA levels in the immunodepleted fractions was consistent with the above findings. Indeed, approximately one quarter of input TERC, but not unrelated RNAs such as β-actin mRNA, vanished following depletion of endogenous RHAU. These results not only corroborate the association of RHAU with telomerase, but also show that the fraction of telomerase interacting with RHAU accounts for merely 25% of the total telomerase activity. Nevertheless, these data provide indirect but explicit evidence of the existence of an alternatively folded TERC structure bearing a G4 scaffold in the fully assembled telomerase holoenzyme.
ATPase-dependent interaction of RHAU with TERC
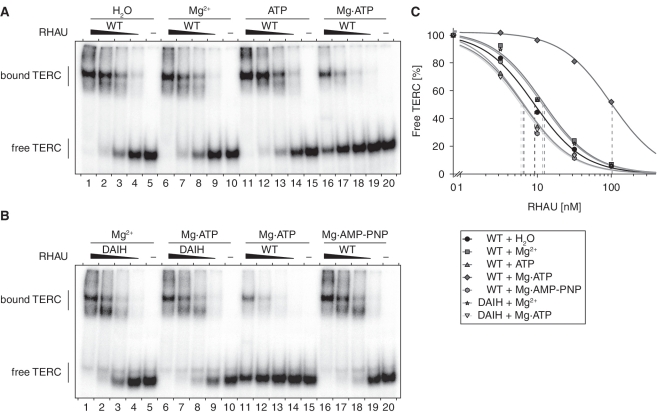
The fact that only one quarter of telomerase activity is linked to RHAU may reflect a dynamic interaction between RHAU and TERC. Indeed, most of the DEAH-box RNA helicases only transiently interact with nucleic acid, because they do not remain bound after the ATPase-dependent remodelling of their substrate (57–60). To gain insight into the role of ATP-hydrolysis by RHAU in its interaction with TERC, we repeated gel mobility shift assays with 5′ TERC fragments in the presence of Mg2+ or ATP or both. As shown in Figure 7A, ATP or Mg2+ alone had little effect on the binding affinity of RHAU for TERC. In contrast, under conditions supporting ATP hydrolysis (Mg·ATP), RHAU displayed reduced binding affinity for TERC, as evidenced by a 10-fold increase in the apparent equilibrium Kd value (Figure 7C). Such a reduction in the affinity of RHAU for TERC was not observed when Mg·ATP was substituted with the non-hydrolysable ATP analogue Mg·AMP-PNP (Figure 7B and C). Thus, the present data suggest that intrinsic RHAU ATP hydrolysis rather than ATP binding is essential for disrupting the interaction of RHAU with TERC. To further validate this, we turned to the previously described RHAU(DAIH) mutant in which the E335A amino-acid substitution within the Walker B site abolishes RHAU ATPase activity (45). Unlike wild-type RHAU, the binding affinity of RHAU(DAIH) mutant for TERC was similar in the absence and presence of Mg·ATP (Figure 7B and C). The absence of RHAU helicase activity as a consequence of the loss of ATPase activity probably prevents RHAU dissociation from its substrate. In fact, a similar reduction in in vivo RNA binding dynamics after suppression of RHAU intrinsic ATPase activity was reported previously in studies of the shuttling of RHAU in cytoplasmic stress granules (43). Taken together, these data corroborate the idea that RHAU is not permanently associated with TERC but dissociates upon ATP hydrolysis.
Figure 7.
Role of Mg·ATP on the interaction of RHAU with TERC. (A) Gel mobility shift assay for TERC G4 binding by RHAU. Radio-labeled 5′-end (1–71 nt) TERC fragment at a concentration of 100 pM was incubated without protein (−) or with increasing amounts (3.2, 10, 32 and 100 nM) of purified recombinant GST-tagged RHAU in the presence of 1 mM Mg2+, ATP or Mg·ATP (as indicated). The reaction mixtures were analysed by non-denaturing PAGE. An autoradiogram of the gel is shown. (B) Gel mobility shift assay of TERC G4 binding by the ATPase deficient RHAU(DAIH) mutant. Radio-labeled 5′-end TERC fragment was incubated without protein (−) or with increasing amounts (3.2, 10, 32 and 100 nM) of purified recombinant WT or DAIH mutant GST-tagged RHAU in the presence of 1 mM Mg2+, Mg·ATP or Mg·AMP-PNP (as indicated). (C) Quantification of gel mobility shift assays of WT and RHAU(DAIH) mutant binding to TERC 5′-end (1–71 nt) fragment. The data represent the mean of three independent experiments. Error bars for SEM were omitted for clarity.
Apart from a high affinity for G4 structures, RHAU has also been shown to couple ATP hydrolysis with tetramolecular G4-RNA resolving activity (42,45). These biochemical properties prompted us to test whether RHAU could catalyse the conversion of TERC G4 structure to ssRNA form. However, the faster rate of refolding of intramolecular G4 structures with regard to tetramolecular G4-RNAs made it technically difficult to monitor the G4-resolving activity of RHAU using standard non-denaturing gel electrophoresis.
DISCUSSION
The propensity of nucleic acid guanine-rich sequences to self-assemble into G4 structures in vitro has been recognized for several decades. Although RNA is also prone to form such structures, G4-RNA has not attracted as much attention as G4-DNA. Nevertheless, a growing body of evidence indicates a significant role for G4 structure formation during RNA metabolism (20,61). As a consequence, G4 structures emerged as a plausible post-transcriptional means of regulating the function of coding and non-coding RNAs. However, little is known about the mechanisms by which G4-RNA formation is regulated in the cells. Indeed, only a few proteins have been reported so far to interact with G4-RNAs in vitro (62–66) and, apart from FMRP and FMR2P, experimental data on their biological activity subsequent to their interaction with G4-RNAs is scarce. Of these G4-RNA binding proteins, RHAU is the only protein that exhibits robust in vitro ATPase-dependent G4-RNA resolving activity, in addition to a high affinity and specificity for its target RNAs (42). These particular biochemical features promote RHAU as an ideal candidate for regulating G4-dependent RNA metabolism, although its in vivo RNA targets have not yet been determined. The present study aimed to identify naturally occurring RHAU targets with the emphasis on RNAs containing potential G4-forming sequences. By RIP-chip assays, 106 RNAs were found to be significantly enriched with RHAU. Importantly, more than half of these RNAs contained G-rich sequences with potential to form stable G4 structures. In addition, there was a weak but significant correlation between the predicted G4 motif density per transcript and the magnitude of RNA enrichment by RHAU. Nevertheless, because RHAU appears to show significant affinity for RNAs not bearing G4-forming sequences, we do not exclude the possibility that RHAU has affinity to other RNA structural features.
In-depth studies revealed that TERC, one of the identified RNAs, was a bona fide target of RHAU. Several independent RIP-chip assays using various cell lines further showed TERC to be one of the most abundant RNAs enriched by RHAU. TERC bears a 5′ G-rich sequence that was previously shown to adopt a stable intramolecular G4 structure in vitro (30). Further investigations dissecting the basis of the interaction between RHAU and TERC showed RHAU binding to be strictly dependent on G4 structure formation in the 5′ region of TERC and to require the N-terminal RSM domain of RHAU, an ancillary domain necessary for specific recognition of G4 by RHAU (45). Moreover, RHAU was found to interact transiently with TERC in a manner dependent on its own ATPase activity. Together, these data provide the first evidence of a specific and direct interaction between a G4-resolvase enzyme and a potentially relevant intramolecular G4-RNA substrate. Furthermore, in agreement with our previous reports (42,45), these data attest that G4-RNAs are naturally occurring substrates of RHAU in vivo.
The 5′ extremity of TERC folds into an intramolecular G4 structure in vivo
In agreement with the observations presented here, Mergny and co-workers reported previously the formation in vitro of a G4 structure in the 5′ region of human TERC (30). However, as for the great majority of nucleic acid structures, direct experimental demonstration of G4 structures in vivo has proven very difficult. As such, our results do not directly prove the existence of G4 structures in vivo, but strongly support the notion that TERC can form a G4 structure in the cells. Notably, RHAU can only bind TERC provided that all conditions necessary for the formation of an intramolecular G4 structure (RNA sequence and ionic conditions) are met. Modifying any one of the conditions is sufficient to reduce substantially the affinity of RHAU for TERC. In addition, our data argue that TERC G4 structure can also be formed in a fraction of telomerase holoenzyme, since RHAU associates with TERT and dyskerin proteins and significant amounts of telomerase activity were recovered with RHAU. Based on the fraction of telomerase activity co-precipitating with RHAU, we can estimate that at least 25% of TERC in the human telomerase holoenzyme bears a 5′ G4 structure. Moreover, we predict that formation of a G4 structure in TERC is not exclusive to human cells, since the majority of the mammalian orthologues of TERC also harbours a potential 5′ G4-forming sequence (Supplementary Figure S4). Insofar as the G4-RNA binding and resolving activities of RHAU are conserved in higher eukaryotes (45), it would not be surprising to find that RHAU is part of the telomerase holoenzymes of other mammals.
As reported previously (30), folding of the 5′ region of TERC into a G4 structure is likely to interfere with the widely adopted secondary structure of human TERC (Supplementary Figure S5). Indeed, according to the standard model of TERC secondary structure (67), two of the guanine tracts that are part of the intramolecular G4 also form the P1a helix. As a consequence, the P1a helix and G4 represent two mutually exclusive structures and may correspond to different functional states of TERC. Insofar as helix P1 is required for template boundary definition in mammalian telomerase (68), formation of a G4 structure in TERC may be detrimental to telomerase activity. On the other hand, G4 structure formation may protect TERC from degradation during telomerase biogenesis. Such a scenario was suggested quite recently by Collins and co-workers, who found that nucleotide substitutions within the G4-forming sequence markedly reduced accumulation of the mature form of TERC, thereby causing telomere shortening (69). Similar to our data, they also found that RHAU associates with the G4-forming sequence of TERC in vivo, but in contrast to our finding, detected negligible telomerase activity with immunopurified RHAU fraction. Thus, further experiments are necessary to clarify the functional impact of G4 formation in TERC on telomerase biogenesis and activity.
In cells, the P1 duplex- and G4-folded forms of TERC are likely to coexist in dynamic equilibrium. However, under physiological conditions, restoration of helix P1 base pairing from the G4-folded conformation of TERC is likely to require a catalyst due to the high thermodynamic stability of the quadruplex (30). The biochemical properties and substrate specificity of RHAU make it a likely candidate for catalysing such a reaction. Quite apart from its high affinity for TERC, RHAU manifests robust tetramolecular G4-RNA resolving activity (42,45) and was also shown to unwind various intramolecular G4-DNA structures in vitro (70). These data suggest that RHAU can resolve G4 structures irrespective of the strand stoichiometry of the G4 stem. Although unwinding of TERC G4 by RHAU has not yet been demonstrated experimentally, our observation that RHAU dissociates from TERC upon ATP-hydrolysis, but not upon ATP binding, supports the idea that RHAU couples ATP-hydrolysis to conformational changes of TERC, resulting in the resolution of the G4 and further release of RHAU from its target RNA.
RHAU may target other RNAs containing G4 structures
Apart from TERC, 54 further RNAs found to be associated with RHAU are predicted to form stable G4 structures (Supplementary Table S2). The determination of the modalities and the significance of these interactions are essential issues to be addressed in the future. Nonetheless, the finding that RHAU associates preferentially with transcripts bearing potential G4-forming sequences strongly suggests that RHAU targets G4-RNA structures in cells. Although nearly three-quarters of these G4-forming sequences are located in 5′ or 3′ UTRs of mRNAs (Supplementary Table S2), it is important to stress that, to date, we have not observed any suppressive action of RHAU on translational repression by G4 formation in 5′-UTRs. Therefore, RHAU is unlikely to function as a translational activator but may instead intervene in other aspects of mRNA metabolism, such as pre-mRNA processing or mRNA turnover. Indeed, RHAU localizes predominantly in the nucleus, where it concentrates in nuclear speckles (71). These are sites of high transcriptional activity and mRNA splicing. Furthermore, RHAU relocalizes to cytoplasmic stress granules in response to various cellular stresses (43). Although recruitment of RHAU to stress granules is mediated by interaction with RNA, we have not yet examined whether this phenomenon depends upon binding of RHAU to G4-RNA structures. However, considering that RSM-domain mutated forms of RHAU deficient in G4-RNA binding also show reduced relocalization to stress granules, it is likely that a fraction of RHAU binds G4 structures in stress granules.
Together with our previous findings that RHAU binds and exhibits robust resolvase activity on various types of G4 structures (41,42,70), the data presented here bring forth the idea that G4-RNAs and especially intramolecular G4-RNAs serve as physiologically relevant targets for RHAU. Identification of naturally occurring substrates of RNA helicases is a prerequisite to any further investigation of their biochemical properties in vitro. However, although most of the RNA helicases achieve highly specific tasks in vivo, they often show only little or even no nucleic acid binding specificity in vitro. Currently, our major limitation towards understanding the mechanisms, whereby RNA helicases melt nucleic acid structures stem from the difficulty of identifying such physiologically relevant substrates. Hence, few detailed molecular models available for studying RNA helicases in vitro. However, with the finding that the 5′ region of TERC constitutes a biologically relevant substrate, RHAU emerges as a novel and promising prototype of DEAH-box protein and deserves more investigations to explore its functions as a G4 resolving enzyme.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Novartis Research Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Stéphane Thiry for excellent technical assistance and Susan Gasser, Patrick King, Janice Ching Lai and Sandra Pauli for critical comments on the article.
REFERENCES
- 1.König S, Evans A, Huppert J. Seven essential questions on G-quadruplexes. BioMolecular Concepts. 2010;1:197–213. doi: 10.1515/bmc.2010.011. [DOI] [PubMed] [Google Scholar]
- 2.Lane AN, Chaires JB, Gray RD, Trent JO. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hershman SG, Chen Q, Lee JY, Kozak ML, Yue P, Wang LS, Johnson FB. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36:144–156. doi: 10.1093/nar/gkm986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5' UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huppert JL, Bugaut A, Kumari S, Balasubramanian S. G-quadruplexes: the beginning and end of UTRs. Nucleic Acids Res. 2008;36:6260–6268. doi: 10.1093/nar/gkn511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eddy J, Maizels N. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2008;36:1321–1333. doi: 10.1093/nar/gkm1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang CC, Chu JF, Kao FJ, Chiu YC, Lou PJ, Chen HC, Chang TC. Verification of antiparallel G-quadruplex structure in human telomeres by using two-photon excitation fluorescence lifetime imaging microscopy of the 3,6-Bis(1-methyl-4-vinylpyridinium)carbazole diiodide molecule. Anal. Chem. 2006;78:2810–2815. doi: 10.1021/ac052218f. [DOI] [PubMed] [Google Scholar]
- 11.Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl Acad. Sci. USA. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 13.Grand CL, Han H, Munoz RM, Weitman S, Von Hoff DD, Hurley LH, Bearss DJ. The cationic porphyrin TMPyP4 down-regulates c-MYC and human telomerase reverse transcriptase expression and inhibits tumor growth in vivo. Mol. Cancer Ther. 2002;1:565–573. [PubMed] [Google Scholar]
- 14.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernando H, Rodriguez R, Balasubramanian S. Selective recognition of a DNA G-quadruplex by an engineered antibody. Biochemistry. 2008;47:9365–9371. doi: 10.1021/bi800983u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernando H, Sewitz S, Darot J, Tavare S, Huppert JL, Balasubramanian S. Genome-wide analysis of a G-quadruplex-specific single-chain antibody that regulates gene expression. Nucleic Acids Res. 2009;37:6716–6722. doi: 10.1093/nar/gkp740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joachimi A, Benz A, Hartig JS. A comparison of DNA and RNA quadruplex structures and stabilities. Bioorg. Med. Chem. 2009;17:6811–6815. doi: 10.1016/j.bmc.2009.08.043. [DOI] [PubMed] [Google Scholar]
- 18.Arora A, Maiti S. Differential biophysical behavior of human telomeric RNA and DNA quadruplex. J. Phys. Chem. B. 2009;113:10515–10520. doi: 10.1021/jp810638n. [DOI] [PubMed] [Google Scholar]
- 19.Mergny JL, De Cian A, Ghelab A, Sacca B, Lacroix L. Kinetics of tetramolecular quadruplexes. Nucleic Acids Res. 2005;33:81–94. doi: 10.1093/nar/gki148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beaudoin JD, Perreault JP. 5'-UTR G-quadruplex structures acting as translational repressors. Nucleic Acids Res. 2010;38:7022–7036. doi: 10.1093/nar/gkq557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arora A, Dutkiewicz M, Scaria V, Hariharan M, Maiti S, Kurreck J. Inhibition of translation in living eukaryotic cells by an RNA G-quadruplex motif. RNA. 2008;14:1290–1296. doi: 10.1261/rna.1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris MJ, Basu S. An unusually stable G-quadruplex within the 5'-UTR of the MT3 matrix metalloproteinase mRNA represses translation in eukaryotic cells. Biochemistry. 2009;48:5313–5319. doi: 10.1021/bi900498z. [DOI] [PubMed] [Google Scholar]
- 23.Gomez D, Guedin A, Mergny JL, Salles B, Riou JF, Teulade-Fichou MP, Calsou P. A G-quadruplex structure within the 5'-UTR of TRF2 mRNA represses translation in human cells. Nucleic Acids Res. 2010;38:7187–7198. doi: 10.1093/nar/gkq563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gomez D, Lemarteleur T, Lacroix L, Mailliet P, Mergny JL, Riou JF. Telomerase downregulation induced by the G-quadruplex ligand 12459 in A549 cells is mediated by hTERT RNA alternative splicing. Nucleic Acids Res. 2004;32:371–379. doi: 10.1093/nar/gkh181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kostadinov R, Malhotra N, Viotti M, Shine R, D'Antonio L, Bagga P. GRSDB: a database of quadruplex forming G-rich sequences in alternatively processed mammalian pre-mRNA sequences. Nucleic Acids Res. 2006;34:D119–D124. doi: 10.1093/nar/gkj073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Didiot MC, Tian Z, Schaeffer C, Subramanian M, Mandel JL, Moine H. The G-quartet containing FMRP binding site in FMR1 mRNA is a potent exonic splicing enhancer. Nucleic Acids Res. 2008;36:4902–4912. doi: 10.1093/nar/gkn472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Randall A, Griffith JD. Structure of long telomeric RNA transcripts: the G-rich RNA forms a compact repeating structure containing G-quartets. J. Biol. Chem. 2009;284:13980–13986. doi: 10.1074/jbc.M900631200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martadinata H, Phan AT. Structure of propeller-type parallel-stranded RNA G-quadruplexes, formed by human telomeric RNA sequences in K+ solution. J. Am. Chem. Soc. 2009;131:2570–2578. doi: 10.1021/ja806592z. [DOI] [PubMed] [Google Scholar]
- 29.Xu Y, Kaminaga K, Komiyama M. G-quadruplex formation by human telomeric repeats-containing RNA in Na+ solution. J. Am. Chem. Soc. 2008;130:11179–11184. doi: 10.1021/ja8031532. [DOI] [PubMed] [Google Scholar]
- 30.Gros J, Guedin A, Mergny JL, Lacroix L. G-Quadruplex formation interferes with P1 helix formation in the RNA component of telomerase hTERC. Chembiochem. 2008;9:2075–2079. doi: 10.1002/cbic.200800300. [DOI] [PubMed] [Google Scholar]
- 31.Fry M. Tetraplex DNA and its interacting proteins. Front Biosci. 2007;12:4336–4351. doi: 10.2741/2391. [DOI] [PubMed] [Google Scholar]
- 32.Huber MD, Lee DC, Maizels N. G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 2002;30:3954–3961. doi: 10.1093/nar/gkf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun H, Karow JK, Hickson ID, Maizels N. The Bloom's syndrome helicase unwinds G4 DNA. J. Biol. Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 34.Fry M, Loeb LA. Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem. 1999;274:12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- 35.Wu Y, Shin-ya K, Brosh RM., Jr FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell. Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanders CM. Human Pif1 helicase is a G-quadruplex DNA binding protein with G-quadruplex DNA unwinding activity. Biochem. J. 2010;430:119–128. doi: 10.1042/BJ20100612. [DOI] [PubMed] [Google Scholar]
- 37.Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat. Rev. Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 38.Wu Y, Suhasini AN, Brosh RM., Jr Welcome the family of FANCJ-like helicases to the block of genome stability maintenance proteins. Cell Mol. Life Sci. 2009;66:1209–1222. doi: 10.1007/s00018-008-8580-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat. Rev. Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 40.Bochman ML, Sabouri N, Zakian VA. Unwinding the functions of the Pif1 family helicases. DNA Repair. 2010;9:237–249. doi: 10.1016/j.dnarep.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaughn JP, Creacy SD, Routh ED, Joyner-Butt C, Jenkins GS, Pauli S, Nagamine Y, Akman SA. The DEXH protein product of the DHX36 gene is the major source of tetramolecular quadruplex G4-DNA resolving activity in HeLa cell lysates. J. Biol. Chem. 2005;280:38117–38120. doi: 10.1074/jbc.C500348200. [DOI] [PubMed] [Google Scholar]
- 42.Creacy SD, Routh ED, Iwamoto F, Nagamine Y, Akman SA, Vaughn JP. G4 resolvase 1 binds both DNA and RNA tetramolecular quadruplex with high affinity and is the major source of tetramolecular quadruplex G4-DNA and G4-RNA resolving activity in HeLa cell lysates. J. Biol. Chem. 2008;283:34626–34634. doi: 10.1074/jbc.M806277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chalupnikova K, Lattmann S, Selak N, Iwamoto F, Fujiki Y, Nagamine Y. Recruitment of the RNA helicase RHAU to stress granules via a unique RNA-binding domain. J. Biol. Chem. 2008;283:35186–35198. doi: 10.1074/jbc.M804857200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tran H, Schilling M, Wirbelauer C, Hess D, Nagamine Y. Facilitation of mRNA deadenylation and decay by the exosome-bound, DExH protein RHAU. Mol. Cell. 2004;13:101–111. doi: 10.1016/s1097-2765(03)00481-7. [DOI] [PubMed] [Google Scholar]
- 45.Lattmann S, Giri B, Vaughn JP, Akman SA, Nagamine Y. Role of the amino terminal RHAU-specific motif in the recognition and resolution of guanine quadruplex-RNA by the DEAH-box RNA helicase RHAU. Nucleic Acids Res. 2010;38:6219–6233. doi: 10.1093/nar/gkq372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 50.Kikin O, D'Antonio L, Bagga PS. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:W676–W682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altschul SF, Erickson BW. Significance of nucleotide sequence alignments: a method for random sequence permutation that preserves dinucleotide and codon usage. Mol. Biol. Evol. 1985;2:526–538. doi: 10.1093/oxfordjournals.molbev.a040370. [DOI] [PubMed] [Google Scholar]
- 52.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 53.Herbert BS, Hochreiter AE, Wright WE, Shay JW. Nonradioactive detection of telomerase activity using the telomeric repeat amplification protocol. Nat. Protoc. 2006;1:1583–1590. doi: 10.1038/nprot.2006.239. [DOI] [PubMed] [Google Scholar]
- 54.Wieland M, Hartig JS. RNA quadruplex-based modulation of gene expression. Chem. Biol. 2007;14:757–763. doi: 10.1016/j.chembiol.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 55.Collins K. Physiological assembly and activity of human telomerase complexes. Mech. Ageing Dev. 2008;129:91–98. doi: 10.1016/j.mad.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 57.Schwer B, Guthrie C. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature. 1991;349:494–499. doi: 10.1038/349494a0. [DOI] [PubMed] [Google Scholar]
- 58.Teigelkamp S, McGarvey M, Plumpton M, Beggs JD. The splicing factor PRP2, a putative RNA helicase, interacts directly with pre-mRNA. EMBO J. 1994;13:888–897. doi: 10.1002/j.1460-2075.1994.tb06332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwer B, Gross CH. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 1998;17:2086–2094. doi: 10.1093/emboj/17.7.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bohnsack MT, Martin R, Granneman S, Ruprecht M, Schleiff E, Tollervey D. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol. Cell. 2009;36:583–592. doi: 10.1016/j.molcel.2009.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Halder K, Wieland M, Hartig JS. Predictable suppression of gene expression by 5'-UTR-based RNA quadruplexes. Nucleic Acids Res. 2009;37:6811–6817. doi: 10.1093/nar/gkp696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bashkirov VI, Scherthan H, Solinger JA, Buerstedde JM, Heyer WD. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol. 1997;136:761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 64.Khateb S, Weisman-Shomer P, Hershco I, Loeb LA, Fry M. Destabilization of tetraplex structures of the fragile X repeat sequence (CGG)n is mediated by homolog-conserved domains in three members of the hnRNP family. Nucleic Acids Res. 2004;32:4145–4154. doi: 10.1093/nar/gkh745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Davidovic L, Bechara E, Gravel M, Jaglin XH, Tremblay S, Sik A, Bardoni B, Khandjian EW. The nuclear microspherule protein 58 is a novel RNA-binding protein that interacts with fragile X mental retardation protein in polyribosomal mRNPs from neurons. Hum. Mol. Genet. 2006;15:1525–1538. doi: 10.1093/hmg/ddl074. [DOI] [PubMed] [Google Scholar]
- 66.Bensaid M, Melko M, Bechara EG, Davidovic L, Berretta A, Catania MV, Gecz J, Lalli E, Bardoni B. FRAXE-associated mental retardation protein (FMR2) is an RNA-binding protein with high affinity for G-quartet RNA forming structure. Nucleic Acids Res. 2009;37:1269–1279. doi: 10.1093/nar/gkn1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen JL, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- 68.Chen JL, Greider CW. Template boundary definition in mammalian telomerase. Genes Dev. 2003;17:2747–2752. doi: 10.1101/gad.1140303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sexton AN, Collins K. The 5' guanosine tracts of human telomerase RNA are recognized by the G-quadruplex binding domain of the RNA helicase DHX36 and function to increase RNA accumulation. Mol. Cell. Biol. 2011;31:736–743. doi: 10.1128/MCB.01033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Giri B, Smaldino PJ, Thys RG, Creacy SD, Routh ED, Hantgan RR, Lattmann S, Nagamine Y, Akman SA, Vaughn JP. G4 Resolvase 1 tightly binds and unwinds unimolecular G4-DNA. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr234. doi:10.1093/nar/gkr234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Iwamoto F, Stadler M, Chalupnikova K, Oakeley E, Nagamine Y. Transcription-dependent nucleolar cap localization and possible nuclear function of DExH RNA helicase RHAU. Exp. Cell Res. 2008;314:1378–1391. doi: 10.1016/j.yexcr.2008.01.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.