Abstract
SimR, a TetR-family transcriptional regulator (TFR), controls the export of simocyclinone, a potent DNA gyrase inhibitor made by Streptomyces antibioticus. Simocyclinone is exported by a specific efflux pump, SimX and the transcription of simX is repressed by SimR, which binds to two operators in the simR-simX intergenic region. The DNA-binding domain of SimR has a classical helix-turn-helix motif, but it also carries an arginine-rich N-terminal extension. Previous structural studies showed that the N-terminal extension is disordered in the absence of DNA. Here, we show that the N-terminal extension is sensitive to protease cleavage, but becomes protease resistant upon binding DNA. We demonstrate by deletion analysis that the extension contributes to DNA binding, and describe the crystal structure of SimR bound to its operator sequence, revealing that the N-terminal extension binds in the minor groove. In addition, SimR makes a number of sequence-specific contacts to the major groove via its helix-turn-helix motif. Bioinformatic analysis shows that an N-terminal extension rich in positively charged residues is a feature of the majority of TFRs. Comparison of the SimR–DNA and SimR–simocyclinone complexes reveals that the conformational changes associated with ligand-mediated derepression result primarily from rigid-body rotation of the subunits about the dimer interface.
INTRODUCTION
The genus Streptomyces accounts for the production of approximately two-thirds of the known antibiotics (1,2). By producing and expelling these compounds into their environment, these bacteria likely acquire a competitive advantage over other organisms inhabiting the same ecological niche. One such antibiotic, simocyclinone, a potent inhibitor of DNA gyrase produced by Streptomyces antibioticus Tü 6040 (3–5), consists of a chlorinated aminocoumarin connected to an angucyclic polyketide at the other end via a tetraene linker and a d-olivose sugar (6,7). Because antibiotics are often potentially lethal to the producing organism, there must be mechanisms to ensure that the machinery responsible for export of the mature antibiotic is in place at the time of biosynthesis. In the case of simocyclinone, such a mechanism is specified by two genes, simR and simX, embedded within the simocyclinone (sim) biosynthetic gene cluster (8–10). The SimR/SimX pair resembles the TetR/TetA repressor–efflux pump pair that confers resistance to clinically important tetracyclines in several human pathogens (11). Simocyclinone is exported from the producing organism by the SimX efflux pump, a member of the major facilitator superfamily. The transcription of simX is repressed by SimR, a TetR-family transcriptional regulator (TFR) that binds to two distinct operators in the intergenic region between the divergently transcribed simR and simX genes (9). Simocyclinone abolishes DNA binding by SimR, inducing expression of the SimX efflux pump, providing a mechanism that couples the biosynthesis of simocyclinone to its export (9).
TFRs are one of the major families of transcriptional regulators in bacteria (12,13). They function as homodimers, with each subunit consisting of two domains: an N-terminal DNA-binding domain (DBD) containing a helix-turn-helix (HTH) motif, and a C-terminal ligand-binding domain (LBD) (12,13). While the LBDs are diverse in amino acid sequence, reflecting the wide range of molecules to which different TFRs respond, the HTH DNA-binding motif is conserved and readily predicted bioinformatically. To date, the structures of only four TFRs bound to cognate DNA have been determined (TetR, DesT, CgmR and QacR), and it is clear that the mode of operator recognition differs from one member of the TFR family to another (14–17). For example, the tetracycline efflux pump repressor, TetR, binds as a dimer to a 15-bp operator and deforms the binding site by 17°, bending away from the protein in order to optimize the position of its HTH for specific base pair interaction (16). In contrast, the multidrug efflux pump repressor from Staphylococcus aureus, QacR, binds its cognate DNA site as a dimer of dimers and bends its operator by just 3°, but widens the major groove to create an optimal DNA environment for a second QacR dimer to bind cooperatively nearby (17).
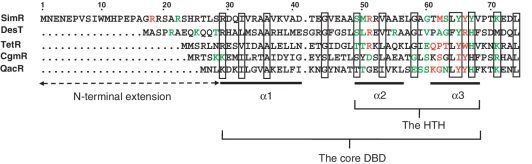
Recently, we determined the structures of apo (unliganded) SimR and SimR in complex with either simocyclinone D8 (SD8) or its biosynthetic intermediate simocyclinone C4 (SC4) (18). These structures revealed the same overall domain architecture for SimR as for other TFRs, including a classical HTH motif. However, SimR possesses an additional arginine-rich N-terminal extension that precedes the core DBD, which is significantly longer than those present in the four TFRs for which protein–DNA crystal structures are available (TetR, DesT, CgmR and QacR) (Figure 1). With the exception of three residues, this 28 amino acid residue extension is disordered in both subunits in the SimR–SD8 structure, and it is only partially ordered in one subunit in the SimR–SC4 structure (18). Consistent with this, the N-terminal extension of SimR is predicted to be disordered in solution.
Figure 1.
Alignment of the amino acid sequence of SimR with the four other TFRs for which protein–DNA crystal structures are available (DesT, TetR, CgmR and QacR), showing the HTH motif, the core DBD and the N-terminal extension present in SimR, herein termed the TFR arm. For each TFR, amino acid residues that interact with the bases of the cognate DNA operator are highlighted in red, and those that contact the phosphate backbone are highlighted in green. Conserved residues are boxed.
Here, we show by deletion analysis that the flexible N-terminal extension of SimR plays an important role in DNA binding, and we present the crystal structure of SimR bound to its operator sequence, which shows that this extension binds in the minor groove adjacent to the major groove occupied by the classical HTH motif. Although the N-terminal extension is hypersensitive to proteolysis in vitro, it becomes protease resistant upon binding cognate DNA. Together these data suggest that the N-terminal extension transitions from a disordered to an ordered state upon DNA binding. Bioinformatic analysis of the entire TetR family shows that an N-terminal extension rich in positively charged residues is a feature of the majority of TFRs. Finally, comparison of the SimR–DNA and SimR–SD8 complexes reveals the conformational changes required to interchange between DNA- and ligand-bound states, which largely involve rigid-body motions of the subunits relative to one another.
MATERIALS AND METHODS
Protein overexpression and purification
The simR gene of Streptomyces antibioticus Tü 6040 encoding a 259 amino acid protein was chemically synthesized with codon optimization (Genscript) for expression in Escherichia coli and was subsequently engineered for expression with a C-terminal hexa-histidine (His6) tag for nickel affinity purification. Construction of the vector for expression of C-terminally tagged protein, pIJ10499, has been described previously (18). This results in a purified protein with an additional 8 amino acid residues at the C-terminus of the native sequence (with sequence LEHHHHHH), giving a total molecular weight of 30 197 Da.
For expression of N-terminally truncated SimR (lacking 10, 15, 22 or 25 amino acid residues from the N-terminus), the gene was amplified by PCR using a downstream primer carrying a XhoI site [R2-full-CtagHis-R: 5′-GATCTCGAGCGCCAGCGCCGGGCGTTCGC-3′] and an upstream primer carrying an NdeI site [R2-M10-trunc-F-NdeI (for SimR-Δ10): GCCCATATGATGCATCCGGAACCGGCCGG; R2-A15-trunc- F-NdeI (for SimR-Δ15): GCCCATATGGCCGGTCGTCGCAGCGCGCG; R2-S22-trunc-F-NdeI (for SimR-Δ22): GCCCATATGAGCCACCGTACCCTGAGCCG; R2-T25-trunc-F-NdeI (for SimR-Δ25): GCCCATATGACCCTGAGCCGCGATCAGATTG]. The amplified DNA fragment was 5′-phosphorylated, cloned into SmaI-cut pUC18 and verified by DNA sequencing. The simR alleles were excised by NdeI/XhoI digestion and cloned into NdeI–XhoI-cut pET20b, giving rise to the overexpression plasmids pIJ10500 (Δ10), pIJ10501 (Δ15), pIJ10502 (Δ22) and pIJ10503 (Δ25). All derivatives of SimR were C-terminally His-tagged and purified as described for wild-type SimR (18).
Protein crystallization and cryoprotection
Directly after nickel-affinity purification, fractions of full-length SimR were pooled and concentrated using a Vivaspin 6 30-kDa cut-off concentrator (Vivascience) to 10–12 mg ml−1 (~200 μM SimR dimer). The concentrated protein was exchanged into crystallization buffer [25 mM Tris–HCl (pH 8.4), 300 mM NaCl] using a Zeba desalting micro-column (Thermo Scientific). Complementary pairs of DNA oligonucleotides with different lengths (16–21 bp) and ends (blunt or sticky ends) were ordered from Oligos etc® and DNA duplexes were reconstituted by annealing oligonucleotide pairs overnight in crystallization buffer at a final concentration of 2 mM.
SimR and annealed oligonucleotides were mixed together in the ratio 1 SimR dimer to 1.2 double-stranded oligonucleotide and incubated at 20°C for 10 min before crystallization screening. Crystallization trials of SimR–DNA were set-up in hanging-drop vapour diffusion format with 48-well VDX plates (Hampton Research) using a variety of commercially available screens (Emerald BioSystems and Hampton Research) at a constant temperature of 20°C. Drops consisted of 1 μl SimR–DNA complex solution mixed with 1 μl precipitant solution and the reservoir volume was 150 μl. Improved crystals were subsequently obtained by refining the successful conditions in a hanging-drop format using 24-well VDX plates (Molecular Dimensions) over a reservoir volume of 1 ml.
SimR–DNA crystals were obtained under several different screening conditions, but only with the blunt-ended 17-mer DNA. The best crystals were obtained from solutions containing 10% (w/v) polyethylene glycol 8000, 0.2 M potassium chloride, 0.1 M magnesium acetate in 0.05 M sodium cacodylate (pH 6.5) 2 weeks after set-up. The crystals belonged to the orthorhombic space group P212121. The SimR–17-mer crystals were cryoprotected by a three-step transfer process in which ethylene glycol was added to the drop to a final concentration of 20%.
Structure determination and refinement
All crystals were flash-cooled by plunging into liquid nitrogen and then mounted onto the goniostat at beamline 8.3.1 at the Advanced Light Source (Berkeley, CA, USA). The resultant data were integrated using MOSFLM (19) and subsequently scaled by SCALA (20). Native intensity data were collected from a SimR–17-mer crystal to 2.99 Å resolution. The reflections used to calculate the R-free value were selected in thin resolution shells to avoid bias resulting from the use of non-crystallographic symmetry (NCS) restraints in refinement. The structure of the complex was solved by molecular replacement using the structure of a subunit of C-terminally hexa-histidine-tagged apo SimR (PDB: 2Y2Z) and an idealized B-DNA of the correct sequence as the search models in PHASER (21). SimR–17mer crystals contained two SimR dimer–DNA complexes in the asymmetric unit. The structure of the complex was then rebuilt in COOT (22) and refined using REFMAC5 (23) and PHENIX (24) with NCS restraints. In the final stages, TLS refinement was used with a total of 20 TLS domains, which were defined using the TLS motion determination server (http://skuld.bmsc.washington.edu/~tlsmd/) (25). X-ray data collection and refinement statistics are summarized in Table 1.
Table 1.
Selected crystallographic data
| Data set | SimR–DNA (17-mer) |
|---|---|
| Data collection | |
| Space group | P212121 |
| Cell parameters (Å/°) | a = 85.8, b = 112.6, c = 163.7 |
| Solvent content (%) | 62.5 |
| Wavelength (Å) | 1.11 |
| Resolution rangea (Å) | 92.78–2.99 (3.15–2.99) |
| Unique reflectionsa (#) | 31 030 (4547) |
| Completenessa (%) | 95.2 (96.5) |
| Redundancya | 3.1 (3.1) |
| Rmergeb (%) | 10.0 (59.1) |
| <I>/<σI> | 8.3 (1.8) |
| Wilson B value (Å2) | 53.4 |
| Refinement | |
| Rcrystc (based on 95% of data) | 20.9 |
| Rfreec (based on 5% of data) | 25.1 |
| Coordinate errord (Å) | 0.420 |
| Ramachandran favourede (%) | 98.0 |
| Ramachandran outlierse (%) | 0.22 |
| RMSD bond distances (Å) | 0.010 |
| RMSD bond angles (°) | 1.22 |
| Mean B-value for protein (Å2) | 57.6 |
| Mean B-value for the DNA (Å2) | 54.6 |
| Contents of model | |
| Protein residues in each chain (totals in brackets) | A: 7–241 B: 7–15 and 26–242 C: 7–242 D: 7–15 and 26–242 |
| DNA nucleotides | E and F, G and H : 1–17 |
| PDB accession code | 3ZQL |
aThe figures in brackets indicate the values for highest resolution shell.
bRmerge = ∑hkl ∑i |Ii(hkl) − 〈I(hkl)〉|/∑hkl ∑iIi(hkl), where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
cThe R-factors Rcryst and Rfree are calculated as follows: R = ∑(|Fobs − Fcalc |)/∑| Fobs | × 100, where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively.
dEstimate of the overall coordinate errors calculated in REFMAC5 based on Rfree (23).
eAs calculated using MOLPROBITY (45).
Structural figures were generated using PyMOL (26). The local DNA helical parameters were calculated using Curves+ (27).
Electrophoretic mobility shift assays
The electrophoretic mobility shift assay (EMSA) DNA probe spanning the entire simR–simX intergenic region (138 bp), containing both the OX and OR operators, was amplified by PCR and 5′-end labelled using [γ32-P] ATP and T4 polynucleotide kinase (New England Biolabs). Binding of wild-type or mutated SimR to DNA was carried out in 20 μl EMSA Buffer [20 mM Tris (pH 8.0), 1 μg calf-thymus DNA, 100 mM NaCl, 8% (v/v) glycerol] containing 0.1 nM radiolabelled DNA (~8000 cpm) and varying amounts of SimR. After incubation at 22°C for 10 min, the binding reaction mixtures were loaded on 5% (w/v) native polyacrylamide gels and run in TBE buffer at 100 V for 45 min. EMSA data were collected and analysed on a PhosphoImager (FujiFilm) using Multi Gauge image analysis software (FujiFilm).
DNase I footprinting
Templates for DNase I footprinting were amplified by PCR using one unlabelled primer and one primer 5′-end labelled using [γ32-P] ATP and T4 polynucleotide kinase (New England Biolabs). The primers were the same pair used to generate the simR–simX intergenic region probe for the EMSA experiments. DNase I footprinting assays were performed in 40 μl EMSA buffer containing ~180 000 cpm radiolabelled DNA and varying amounts of SimR. After incubation at 22°C for 10 min, 10 μl DNase I solution (10 U in 10 mM CaCl2) was added and the incubation was continued for a further 60 s. Reactions were stopped by adding 140 μl DNase I stop solution (200 mM unbuffered sodium acetate, 30 mM EDTA, 0.15% SDS and 0.1 mg ml−1 yeast tRNA), the samples were precipitated with ethanol and the pellets were dried and dissolved in 5 μl Sequencing Loading Dye [80% (v/v) formamide, 10 mM NaOH, 1 mM EDTA, 0.1% (w/v) xylene cyanol and 0.1% (w/v) bromophenol blue]. After heating at 80°C for 3 min and cooling on ice, the samples were run on a 6% (w/v) polyacrylamide/8 M urea sequencing gel, which was dried and analysed using a PhosphoImager (FujiFilm). A G+A sequencing ladder was generated from the template DNA by chemical sequencing (28).
Limited proteolysis and protease protection assays
For limited proteolysis assays, 1 nmol of wild-type SimR was incubated with 1 pmol bovine trypsin (Sigma) in a total volume of 100 µl buffer [50 mM Tris (pH 8.0), 20 mM CaCl2 and 150 mM NaCl] at 4°C. For protease protection assays, 1 nmol wild-type SimR was incubated with equimolar amounts of 15, 25 or 31-mer double-stranded oligonucleotide containing the SimR OX operator in a total reaction volume of 100 µl for 5 min at 4°C before addition of 1 pmol bovine trypsin. The 20 µl samples were then taken at 5-min time intervals. Reactions were stopped by adding SDS–PAGE loading buffer, boiled for 5 min, and analysed using SDS–PAGE. Proteins were transferred to PVDF membrane by electroblotting, stained with Coomasie blue and proteolytically resistant species were identified by N-terminal sequencing at the Protein & Nucleic Acid Chemistry Facility, University of Cambridge.
Global bioinformatic analysis of TFRs
We searched the PFAM database (http://pfam.sanger.ac.uk) for proteins that match the Hidden Markov Model profile PF00440, identifying 23 137 TFR candidates. Protein sequences longer than 300 amino acid residues were removed to eliminate false positives, and highly similar orthologous TFRs were removed using Jalview with a threshold of 99% identity (29), resulting in a non-redundant set of 12 715 TFRs.
The non-redundant set of TFRs was divided into clusters of 200 sequences using USEARCH and UCLUST (30). The amino acid sequences of the TFRs in each cluster were then aligned using MUSCLE (31) to identify their N-terminal extensions, which were defined as the amino-acid sequences preceding the conserved core DBDs (Figure 1). The globular body of the TFRs was defined by excluding the N-terminal extension from the whole protein sequence. In-house Perl scripts were used to quantify the length of the N-terminal extension and the fractions of R+K or D+E residues within these extensions. The sequences of the N-terminal extensions were concatenated together and submitted to the Regional Order Neural Network (RONN) programme (32) to predict the disorder probability for each residue. QtiPlot (http://soft.proindependent.com/qtiplot.html) was used to produce histograms.
RESULTS AND DISCUSSION
N-terminally truncated SimR derivatives bind DNA with reduced affinity
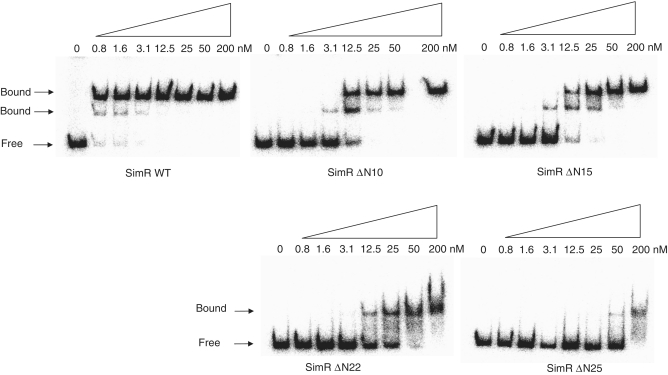
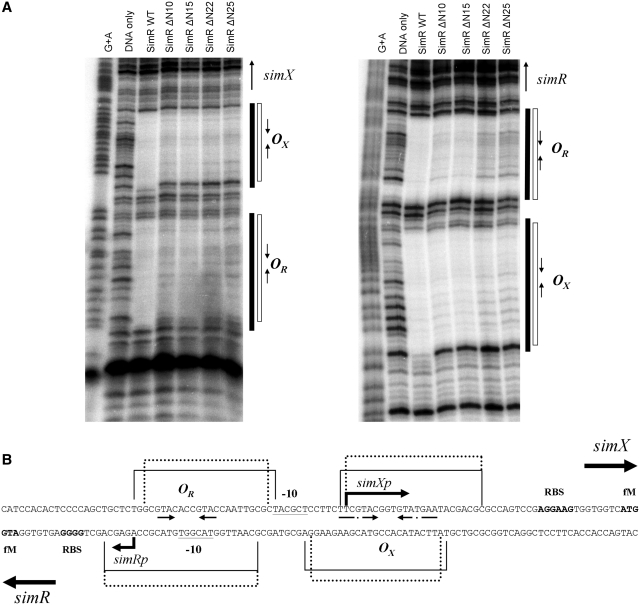
SimR possesses a 28-residue N-terminal extension that precedes the core DBD, herein termed the TFR arm (Figure 1), which carries four arginine residues at positions 18, 19, 22 and 25. This TFR arm is significantly longer than those in DesT, TetR, CgmR and QacR (Figure 1), the four TFRs for which DNA–protein crystal structures are available (14–17). To determine if the TFR arm of SimR is involved in DNA binding, we made C-terminally His-tagged SimR derivatives with progressively shorter N-terminal extensions and tested them for binding to the simR–simX intergenic region by EMSA. Wild-type SimR and SimR derivatives with 10, 15, 22 or 25 amino acid residues deleted from the N-terminus were overexpressed and purified (Supplementary Figure S1). Increasing concentrations of protein were incubated with a DNA probe spanning the simR–simX intergenic region and the complexes were resolved on native polyacrylamide gels (Figure 2). The simR–simX intergenic region contains two SimR operators: OR closer to simR, and a higher affinity binding site, OX, closer to simX (9). The lower and upper sets of shifted protein–DNA complexes seen in Figure 2 correspond, respectively, to single and double occupancy of these two SimR-binding sites (9). SimR DNA-binding affinity was reduced ~30-fold when 10 or 15 amino acid residues were deleted from the N-terminus, and was reduced by at least 120-fold when 22 or 25 amino acid residues were removed (Figure 2). These results suggested the TFR arm plays a role in DNA binding.
Figure 2.
Electrophoretic mobility shift assay (EMSA) showing the binding of purified wild-type and N-terminally truncated derivatives of SimR to the simR–simX intergenic region. Bands correspond to SimR–DNA complexes (Bound) and free DNA (Free) are indicated. The final concentration of SimR is indicated above each lane.
The TFR arm becomes protease resistant upon DNA binding
The 28-amino acid TFR arm of SimR has a high proportion of disorder-promoting amino acids and is predicted by the Proteins Disorder Prediction System (PrDOS; http://prdos.hgc.jp/cgi-bin/top.cgi) and by the Regional Order Neural Network (RONN; http://www.strubi.ox.ac.uk/RONN) servers to be disordered in solution (Supplementary Figure S2). Additionally, with the exception of three residues (residues 8–10, here termed the anchor string), this extension is disordered in both monomers in the SimR–SD8 structure, and it is only partially ordered in one monomer in the SimR–SC4 structure (18). The TFR arm is ordered in the SimR–apo structure, but its structure is the likely result of crystal packing (Supplementary Figure S3).
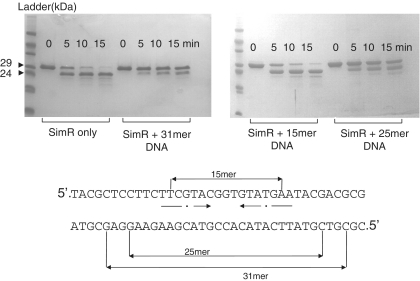
Because disordered regions are often hypersensitive to proteolysis (33), we examined the sensitivity of SimR to trypsin. The TFR arm was rapidly digested, leaving a much more stable product with a N-terminus at either residue Ser20 or Ser23 (Figure 3 and Supplementary Figure S4). Taken together, these observations suggest that the TFR arm is solvent exposed and displays conformational flexibility in solution in the absence of cognate DNA.
Figure 3.
Limited tryptic proteolysis of SimR in the presence or absence of DNA. SimR was incubated either alone or with the OX operator DNA duplexes indicated, before the addition of trypsin. Note that the 15-mer DNA duplex does not bind SimR (Supplementary Figure S5A). After SDS–PAGE, SimR species were visualized by Coomassie blue staining. The major product of tryptic digestion (arrowed) was shown by Edman sequencing to have an N-terminus corresponding to Ser20 or Ser23 of wild-type SimR.
Since many unstructured regions exhibit increased resistance to proteolysis on binding of a partner (33,34), we determined the effect of DNA binding on the sensitivity of the TFR arm to trypsin. Addition of 25- or 31-bp DNA duplexes spanning the OX operator substantially decreased the rate of SimR proteolysis, suggesting that DNA binding renders the TFR arm more resistant to trypsin (Figure 3). Consistent with this interpretation, proteolysis was not inhibited when a 15-bp OX DNA duplex that is unable to bind to SimR was incubated with SimR (Figure 3 and Supplementary Figure S5A). In total, these experiments suggest that the TFR arm transitions from a disordered or conformationally flexible state to a more ordered, rigid state upon DNA binding.
The structure of SimR bound to its DNA operator
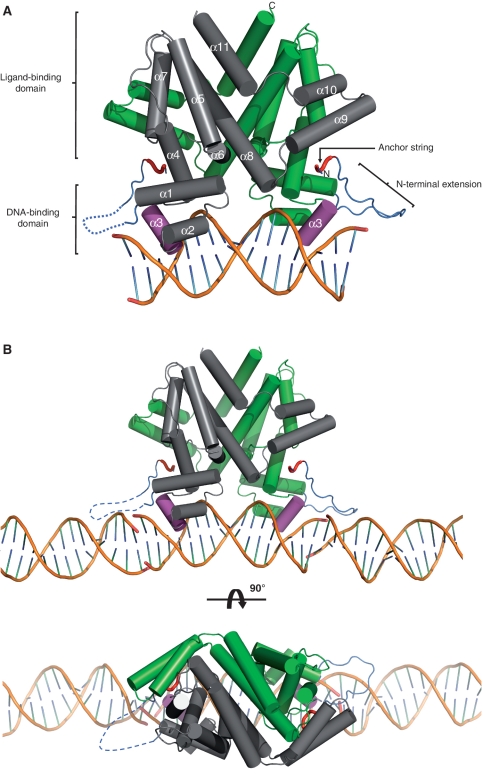
To understand how SimR binds to its operator sequence and to shed light on the role of the TFR arm in DNA binding, we crystallized SimR in complex with DNA. We tested DNA duplexes from 17 to 21 bp in length and found that only the minimal, blunt-ended 17-bp duplex crystallized in complex with SimR. The 17-bp DNA duplex used was the OX operator (5′-TTCGTACGGTGTATGAA-3′), but carrying 2 bp changes to generate a near perfect inverted repeat (5′-TTCGTACGGCGTACGAA-3′), which bound SimR at least as tightly as the wild-type 17-bp DNA duplex (Supplementary Figure S5B). We solved the structure of full-length SimR (residues 1–259) in complex with this 17-bp DNA duplex to 2.99 Å resolution (Figure 4A). X-ray data collection and refinement statistics are summarized in Table 1.
Figure 4.
Structure of the SimR–17-mer complex (A) in isolation or (B) showing the adjacent DNA duplexes in the crystal. A cylindrical helix representation is used to highlight the secondary structure of SimR with key features labelled in (A). One subunit of the biological-relevant dimer is shown in grey and one in green. The recognition helix α3 is shown in magenta, the TFR arm is shown in blue and the N- and C-termini are labelled. The anchor string of the TFR arm (residues 8–11) is shown as a red tube cartoon. The dotted blue line represents the disordered TFR arm in the left-hand SimR subunit. In (B) only the DNA components of the adjacent symmetry complexes are shown in order to highlight the pseudo-continuous DNA filament running through the crystal (See also Supplementary Figure S6 and Figure 7).
The asymmetric unit contained two SimR dimers, each bound to a 17-bp DNA duplex. The two SimR dimer–DNA complexes are essentially identical [root mean square deviation (RMSD) between complexes for the Cα backbone = 0.15 Å], and thus only one complex is discussed throughout (Figure 4A). The conformation of bound DNA is mostly regular B-form but is bent away from the SimR dimer by ~15° (see below and Supplementary Figure S10A). The bases at the end of adjacent DNA duplexes stack and interact to form a pseudo-continuous double-helical DNA filament running through the crystal (Figure 4B and Supplementary Figure S6).
Interactions between the HTH motif and the major groove
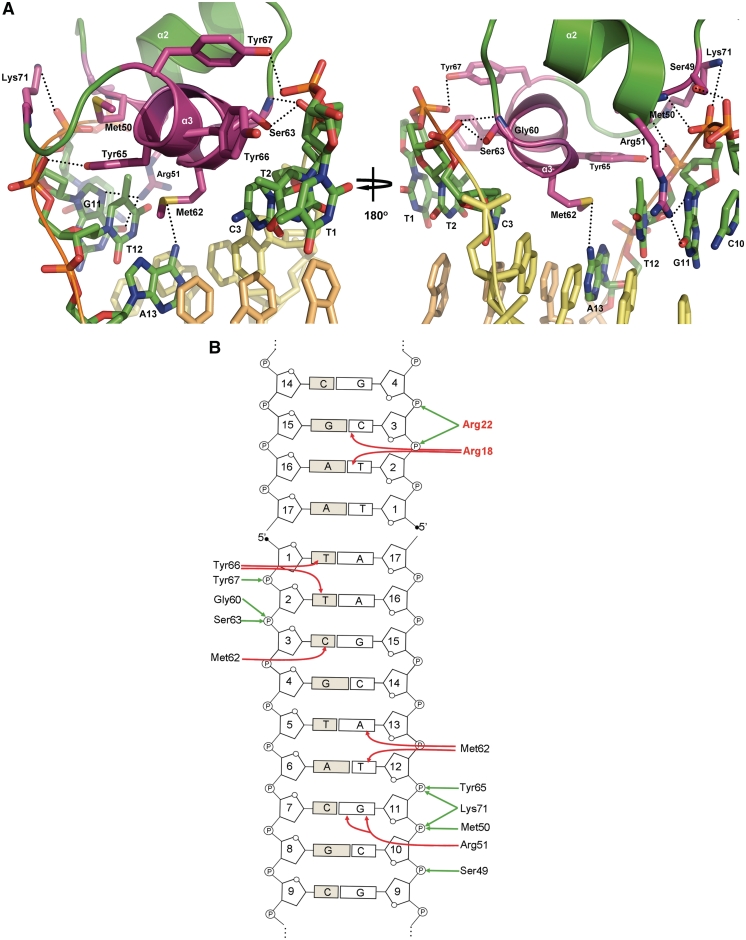
The core DBD is composed of helices α1–α3 (residues 29–67). Helix α2 (residues 49–58) and the recognition helix α3 (residues 61–67) form the HTH motif which packs against α1 for stabilization (Figure 4A). Surprisingly, the recognition helix makes no canonical hydrogen bonds with the bases. However, the side chain of Met62 makes a series of contacts to three different bases including van der Waals to C3 (Cβ to C5), and an uncommon electrostatic interaction between the S atom and the face of the base of T12, which is analogous to S stacking over the aromatic side chains of tryptophan, histidine and phenylalanine (35) (Figure 5). This interaction is buttressed by van der Waals contacts to the C7 methyl group of T12. The S atom of Met62 also accepts a hydrogen bond from the N6 hydrogen bond donor of A13. Another key interaction involved in the DNA sequence recognition mechanism of SimR is the stacking of the side chain of residue Tyr66 with the C7 exocyclic methyl groups of T1 and T2. This interaction explains in great part why SimR has a higher affinity for the OX operator, which has this pair of thymines, than for OR, which has a pair of guanines at these positions (9). The dominant recognition helix interactions are with the phosphate backbone. For each operator half-site, there are hydrogen bonds between the hydroxyl group of Ser63 and the phosphate group of C3, between the hydroxyl group of Tyr65 and the phosphate group of T12 and between Tyr67 and the phosphate group of T2 (Figure 5). Just outside helix α3, the backbone NH group of Gly60 hydrogen bonds with the phosphate group of C3. On binding DNA, the recognition helix adopts a 310 helical conformation, in contrast to the canonical α-helical conformation seen in the structures of SimR–apo and SimR–simocyclinone complexes (9). This conformational alteration in the recognition helix on DNA binding is also observed in TetR, and is believed to facilitate intimate interaction with the DNA (16).
Figure 5.
(A) Interactions between the HTH motif and the major groove. Stick representations of the interacting residues are shown in magenta. The Cα backbone of recognition helix α3 is shown in magenta and that of helix α2 is shown in green. Hydrogen bonds are represented by dotted black lines. The interacting bases are labelled and only the ring frames are shown for non-interacting bases. (B) Schematic representation of SimR–DNA contacts. For simplicity, only a recognition half-site and the first 4 bp of an adjacent duplex are shown. Interactions between amino acid residues and the bases of the cognate DNA operator are indicated by red arrows, and those between amino acid residues and the phosphate backbone are represented by green arrows. Amino acid residues belonging to the TFR arm are shown in red.
Three residues in helix α2 contribute to DNA binding, with the side chain hydroxyl group of Ser49 forming a hydrogen bond with the phosphate backbone of C10 and the backbone NH group of Met50 forming a hydrogen bond with the phosphate backbone of G11 (Figure 5). The guanidinium group of Arg51 is involved in direct base recognition by bifurcated hydrogen bonds from the Nη2 atom to the O6 and N7 acceptors of G11. Other interactions between SimR and the major groove are hydrogen bonds between the amino group of Lys71 and the phosphate group of G11, and between the backbone NH group of Lys71 and the phosphate group of T12. Lys71 lies at the N-terminus of helix α4 at the very beginning of the LBD, just outside the core HTH motif of the DBD. This residue is highly conserved among TFRs and the equivalent lysine in TetR also forms a hydrogen bond with the phosphate backbone (16).
TFRs frequently rely on phosphate backbone contacts to mediate interaction with the DNA. In an extreme case, the DesT–DNA interface involves 11 phosphate backbone contacts but only two specific interactions with a pair of guanine bases within each half site (15). In contrast, TetR and QacR make extensive direct hydrogen bond contacts with the bases (16,17). In this sense, SimR is perhaps more similar to DesT than to TetR or QacR in its DNA sequence recognition mechanism. Thus, although the overall structure of the DBD in TFRs is conserved, it is clear that the mode of operator recognition differs from one member of the TFR family to another (14–17). TFRs recycle conserved residues and inventively employ non-conserved ones within the DBD for either base-specific hydrogen bond formation or for phosphate backbone contacts (Figure 1). It seems that there is no deterministic set of rules for TFR–DNA recognition.
Interactions between the TFR arm and the minor groove
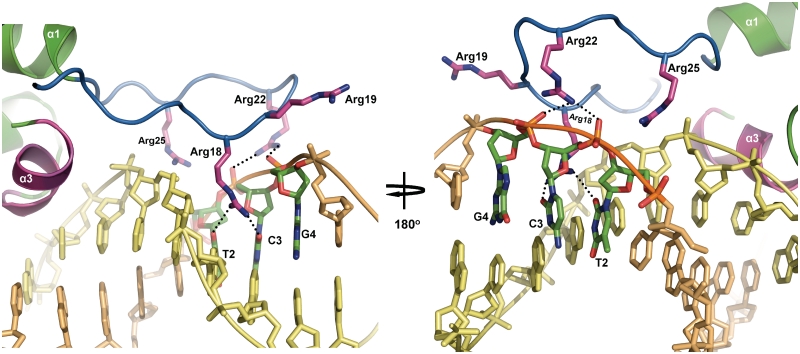
If the structure of a single SimR–DNA complex is viewed in isolation, it can be seen that the TFR arm does not make contact with the cognate DNA duplex (Figure 4A). Instead, the TFR arm binds the minor groove of the adjacent DNA duplex in the pseudo-filament (Figure 4B). This binding to the minor groove is mediated through arginine residues that sit at the tip of the TFR arm (Figures 5B and 6). Specifically, the Nη2 atom of the guanidinium group of Arg18 forms a hydrogen bond with the O2 of C3, while the Nη1 atom interacts with the O2 of T2. In addition, the guanidinium group of Arg22 forms two salt bridges to the phosphate backbone of C3 and G4 (Figures 5B and 6). The electropositive side chain of Arg18 is deeply buried in this minor groove (Figure 6), where the electronegative potential of the phosphate backbone is focused (36,37). This helps anchor the tip of the TFR arm in the minor groove. A third arginine in the flexible TFR arm, Arg19, does not contact DNA in the structure reported here (Figure 6). However, given the non-covalent nature of the DNA pseudo-filament, we considered the possibility that Arg19 might be involved in DNA binding in truly continuous double-stranded DNA. To examine this possibility, we mutagenized Arg19 to alanine and assayed the resulting protein for its ability to bind to the simR–simX intergenic region by EMSA. SimR R19A-bound DNA with an affinity equal to that of wild-type SimR (Supplementary Figure S7), suggesting Arg19 does not contribute to DNA binding. In contrast, when we constructed SimR R18A and SimR R22A variants, we found that each exhibited an approximate 15-fold reduction in binding affinity (Supplementary Figure S7), consistent with roles for R18 and R22 in DNA binding, as suggested by the structure of the SimR–DNA complex.
Figure 6.
Interactions between the TFR arm and the minor groove. The Cα backbone of the TFR arm is shown in blue and stick representations of arginine residues Arg18, Arg19, Arg22 and Arg25 are shown in magenta. Hydrogen bonds are represented by dotted black lines. The interacting bases are labelled and only the ring frames are shown for non-interacting bases.
Initially, it was difficult to understand why SimR variants lacking just 10 or 15 N-terminal amino acid residues should have reduced DNA-binding affinity, given that they retain the interacting arginine residues. In the previously solved structures of apo–SimR and SimR–ligand complexes, although the TFR arm is mostly disordered, residues 8–10, herein termed the anchor string, are always visible in electron density maps (18), probably because this string of amino acid residues is stabilized by van der Waals interactions with the cleft between the LBD and the DBD. It therefore seems likely that this short segment, highlighted in red in Figure 4, serves as an anchoring point for the TFR arm to loop back onto the body of SimR. This arrangement may be important for restricting the flexibility of the TFR arm, so that it is poised appropriately to interact with the minor groove. Deleting 10 or 15 amino acids from the N-terminus would remove this anchor point, destabilizing loop formation and reducing DNA-binding affinity. The more severe deletions, removing 22 or 25 amino acids, further reduce binding affinity because they remove the interacting arginine residues themselves.
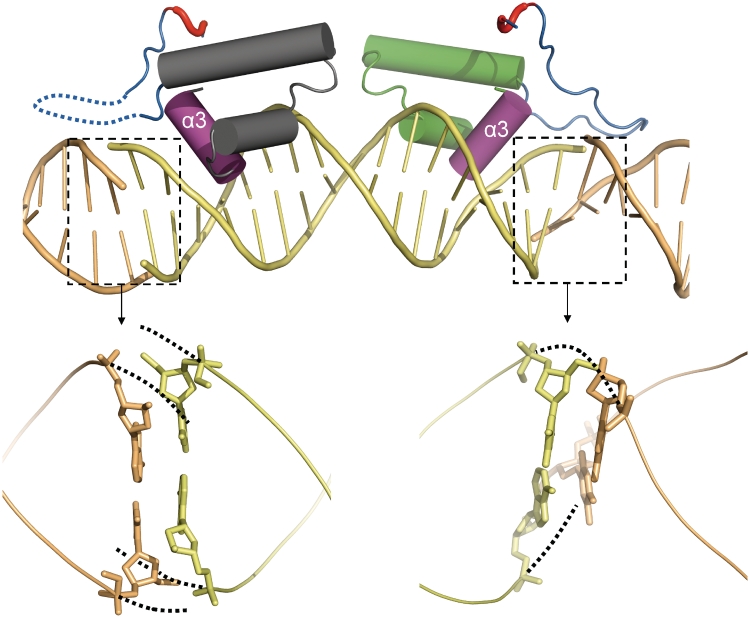
In the crystal structure of the SimR–DNA complex, the TFR arm is seen in one SimR subunit but is disordered in the other subunit (Figure 4). From an inspection of the end-to-end base stacking between adjacent DNA duplexes within the crystal, it is clear that the two ends are not equivalent. The stacking at the right-hand end (as viewed in Figure 7) allows the neighbouring DNA strands to transit smoothly across the gap, producing a relatively normal minor groove. However, on the left-hand end the strands veer away to avoid a steric clash while maintaining base pair stacking, producing a much wider minor groove (Figure 7). It seems likely that the TFR arm is unable to interact with this ‘abnormal’ minor groove and is therefore disordered in the crystal. In the structure of the SimR–17-mer duplex, apart from the interaction of the anchor string with the body of SimR, the only contacts made by the TFR arm are with the minor groove of DNA (Figure 4 and Supplementary Figure S6). Based on the crystal structure of the SimR–DNA complex and the results of the proteolysis protection assays, we propose that the TFR arm transitions from a disordered or conformationally flexible state to a more ordered state upon binding to its cognate DNA.
Figure 7.
Non-equivalent stacking between adjacent DNA duplexes in the crystal pseudo-filament creates two different minor grooves. Only the DBD of SimR is shown. At the right-hand end of the central DNA duplex the base stacking allows the DNA phosphate backbone to transit smoothly (dotted lines) between adjacent duplexes, creating a relatively normal minor groove. At the left-hand end of the central DNA duplex the base stacking causes the phosphate backbone to veer away to avoid a steric clash (dotted lines), producing an abnormal minor groove. Adjacent DNA duplexes are shown in contrasting colours.
N-terminally truncated SimR derivatives have a smaller footprint on DNA than wild-type SimR
We used DNase I protection to compare the footprints of wild-type SimR and the N-terminally truncated SimRs on the OX and OR operators in the simR–simX intergenic region (Figure 8A). In each case, saturating amounts of SimR protein were used to ensure complete protection of the binding sites. The footprint for wild-type SimR was comparable with that reported previously (9). In contrast, in the footprints generated using the N-terminally truncated SimR proteins, the edge of the protected region retracted at both ends of the footprint when compared to the footprint of full-length SimR (Figure 8). Specifically, when N-terminally truncated proteins were used, on the upper DNA strand the OR footprint retracted by two base pairs at the left edge and by one base pair at the right edge (Figure 8). No retraction of the OR footprint was apparent on the lower DNA strand. When N-terminally truncated proteins were used, on the upper DNA strand the left edge of the OX footprint retracted by 1 bp, while no retraction was apparent at the right edge (Figure 8). On the lower DNA strand, the OX footprint receded by 1 bp at both ends. These observations indicate that the TFR arm sterically hinders DNase I, protecting additional phosphodiester bonds from cleavage by the nuclease. Each SimR mutant protein produced the same footprint, regardless of whether 10, 15, 22 or 25 amino acids had been deleted from the N-terminus, consistent with the idea that residues 8–10, (i.e. the anchor string), are needed for the TFR arm to be fully functional, as discussed above. Note that the retraction of the footprint occurs at both ends of the operator, suggesting that the TFR arms of both monomers in the SimR dimer function in solution.
Figure 8.
(A) DNase I footprinting analysis of the binding of wild-type and N-terminally truncated derivatives of SimR to the simR–simX intergenic region. A DNA fragment containing the simR–simX intergenic region, 5′-end labelled on either the upper strand (left panel) or the lower strand (right panel), was exposed to DNase I in the presence of saturating concentrations of SimR protein (200 nM for wild-type SimR, SimRΔN10 and SimRΔN15; 400 nM for SimRΔN22 and SimRΔN25). The sequencing ladders were generated by subjecting the probes to Maxam-Gilbert G+A chemical sequencing. Regions protected from DNase I cleavage (operators OX and OR) by wild-type SimR are indicated by solid vertical bars, and those protected by the N-terminally truncated SimR derivatives are indicated by open bars. Inverted repeats within the DNase I protected regions are indicated by convergent arrows. (B) Sequence of the simR–simX intergenic region summarizing the DNase I footprinting data. Regions protected by wild-type SimR are indicated by solid lines, and those protected by the N-terminally truncated SimR derivatives are indicated by dotted lines. Also indicated are the simRp and simXp transcription start points and putative −10 sequences, the simR and simX ribosome-binding sites (RBS), and the imperfect inverted repeats within the footprints.
We also performed a complementary experiment to determine the binding affinity of wild-type SimR to three DNA duplexes of different lengths (15, 17 and 23 bp) spanning the OX inverted repeat sequence. The 23-bp duplex bound SimR more strongly than the minimal 17-bp duplex, showing that DNA flanking the core 17-bp inverted repeat contributes to SimR binding (Supplementary Figure S5A). The 15-bp duplex failed to bind SimR (Supplementary Figure S5A). In addition, although the minimal 17-bp duplex binds to SimR relatively well (Supplementary Figure S5A), it is unable to protect the TFR arm of SimR from tryptic digestion, while a 23-mer reduced the rate of proteolysis considerably (Supplementary Figure S8). Taken together, these observations suggest that, in solution, the TFR arm interacts with DNA outside the core 17-bp OX operator, consistent with the SimR–DNA structure, which shows dimer–DNA interactions spanning 21 bp.
Among the five TFRs for which protein–DNA crystal structures are available (DesT, TetR, CgmR, QacR and SimR; Figure 1), only SimR possesses a flexible TFR arm that undergoes a transition to an ordered state upon DNA binding. DesT has a 12 amino acid residue N-terminal extension (Figure 1) but it is not disordered, instead forming part of an extended helix α1. Residues Arg5 and Lys9 of this short N-terminal extension in DesT nevertheless contribute to DNA binding (15), which is unusual because the main role of helix α1 is in stabilizing the HTH motif (α2– α3). Residues N-terminal to the core DBD in two other TFRs, Neisseria gonorrhoeae MtrR (11 amino acids) and Streptomyces coelicolor ActR (32 amino acids) have also been suggested to be involved in DNA binding (38,39), implying a possible common role for TFR N-terminal extensions when present (see also the global TFR bioinformatic analysis presented below). Similar kinds of extensions have been identified in at least two other families of DNA-binding proteins. For example, members of the eukaryotic Hox family recognize nearly identical major groove sequences through the recognition helix of their homeodomain but use an extended arm to insert into the minor groove to enhance binding specificity (40). A related example is phage lambda repressor, which has a conventional HTH motif and an additional N-terminal extension that promotes DNA binding, in this case by interacting with the major groove (41). A comprehensive analysis of all available protein–DNA structures has shown that the binding of arginine residues to narrow minor grooves is a widely used mechanism in protein–DNA recognition. This readout mechanism exploits the fact that narrow minor grooves, often associated with A-tracts, strongly enhance the negative electrostatic potential of the DNA (36,37). However, it should be noted that the minor groove bound by the TFR arm of SimR is not associated with an A-tract, and has a slightly enlarged width with respect to canonical B DNA (Supplementary Figure S10C).
The arginine- and lysine-rich TFR arm is likely to be a common feature of TetR family members
We searched the PFAM database (http://pfam.sanger.ac.uk/) for proteins that match the Hidden Markov Model profile PF00440, identifying 12 715 non-redundant TFRs (see Materials and methods for further details). The amino acid sequences of these TFRs were then aligned using MUSCLE (31) to identify the core DBD and any N-terminal extension. Twenty-eight per cent had N-terminal extensions of less than 10 amino acids, 44% had N-terminal extensions of 11–20 amino acids, 17% had N-terminal extensions of 21–30 amino acids and 11% had N-terminal extensions >31 amino acids. Further, the fraction of Arg and Lys residues in these N-terminal extensions (mean value = 20.5%) was almost double the frequency found in the globular body of the TFRs (mean value = 11.4%) (Supplementary Figure S9A). Finally, the RONN server predicts that the majority of these N-terminal extensions are likely to be disordered in solution (Supplementary Figure S9B). It therefore seems likely that a conformationally malleable, DNA-binding N-terminal extension is a common feature of TFRs.
DNA bending induced by SimR binding
DNA helical parameters were analysed using the Curves+ programme (27). The overall conformation of the 17-bp duplex is B-DNA, with an average helical twist of 33.7° (compared to a helical twist value of 36.0° for an idealized B-form DNA). It should be noted that individual steps might show significant deviations from the average value. The global bending of DNA is ~15° (Supplementary Figure S10A). Since bending is most affected by the base step roll and twist angles (42), we plotted the roll and twist angles against the base steps to pinpoint the source of bending (Supplementary Figure S10B). There are two significant positive rolls (10–10.7°) centred around base steps 6–7–8 in the operator half-site and symmetrically around steps 9–10–11 of the opposite half-site (Supplementary Figure S10B). The increase in roll angle coincides with the decrease in twist angle (26.7– 26.9°) (Supplementary Figure S10B). The average global roll and twist angles are 2.9° and 33.4°, respectively. Thus local kinks around those base steps produce a global bend in the DNA, rather than a smooth bending. Moreover, there is a significant increase in the width of the minor groove from base step 6 through to base step 12, while the major groove width is just below the value for an idealized B-form DNA (Supplementary Figure S10C). Since the average distance between the two recognition helices in the SimR–DNA complex is 36.8 Å [assessed as the distance between the Cα atom of Tyr65 in each subunit (13)], greater than the distance between two consecutive major grooves in idealized B-DNA (34 Å), it is likely that the bending and the unwinding of the central DNA steps might be necessary for optimal positioning of the HTH motifs in adjacent major grooves. Lastly, although the sequence of the 17-bp duplex used in this study is a perfect inverted repeat with the exception of the central GC base pair, the groove width and roll parameters are not symmetrical across this central base pair. This reflects the non-equivalent end-to-end interactions between neighbouring DNA duplexes described above (Figure 7).
Comparison of the SimR–DNA and SimR–simocyclinone complexes suggests the mechanism of derepression
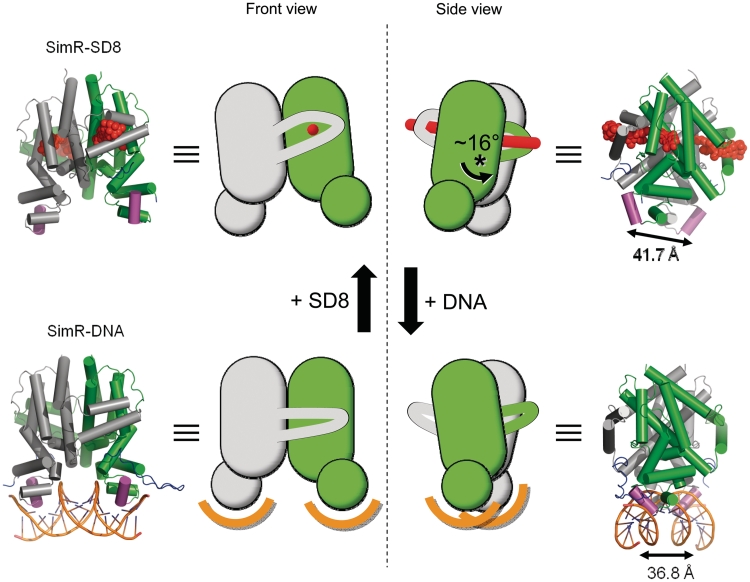
In a previous report, we speculated about the mechanism of simocyclinone-mediated derepression, based on a comparison of the structures of SimR–apo and the SimR–SD8 complex (18). However, it was apparent that SimR–apo had not crystallized in its DNA-binding form, since the distance between its recognition helices was 42.3 Å, a spacing incompatible with binding to two consecutive major grooves (18). Moreover, this spacing was comparable to the corresponding value of 41.7 Å obtained for the SimR–SD8 complex. Indeed, TFR apo-proteins in general do not crystallize in their DNA-binding form (13). The helix separation obtained for SimR–DNA was significantly shorter at 36.8 Å (averaged over the two complexes in the asymmetric unit), this value lying within the range of 34.7–38.8 Å observed in other TFR–DNA complexes (13,15). The major structural differences between the repressed, DNA-bound conformation of SimR and the derepressed, SD8-bound conformation, result from a 16° rotation of the subunits relative to one another roughly about the centre of the dimer interface (Figure 9 and Supplementary Figure S11). This re-defines many of the inter-subunit contacts, although the interface areas remain similar at 2795 and 2640 Å2 for SimR–SD8 and SimR–DNA, respectively [as calculated by the Protein Interactions, Surfaces and Assemblies server (PISA, http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html) (43)]. However, five reciprocated inter-subunit hydrogen bonds (i.e. 10 in total) are preserved between the two conformational states. These link the C-terminal end of α8 and the α9–α10 wrapping arm to the LBD of the adjacent subunit. As a consequence, when the subunits rotate, the α9–α10 wrapping arm moves with the adjacent subunit and the C-terminal end of α8 bends (Supplementary Figure S11). Pair-wise superpositions of individual subunits taken from the SimR–SD8 and SimR–DNA structures based on the subunit cores (i.e., inclusive of residues 29–168 plus 222–247 and exclusive of the TFR arm, the C-terminal end of α8 and the α9–α10 wrapping arm) gave RMSD values in the range 0.85–0.96 Å, indicating that the cores move essentially as rigid bodies at the protein backbone level and, importantly, there is no significant re-orientation of the DBD with respect to the LBD, in contrast to the ‘pendulum-like’ motion seen in TetR (Supplementary Figure S12) (12,16). Nevertheless, the crystal structures do not convey the dynamic behaviour of the system and, as has been illustrated for other TFRs (13,44), in the absence of ligands or DNA, the protein is generally highly flexible and capable of sampling a variety of conformations, presumably including those akin to both the ligand- and DNA-bound states. The binding of SD8, a relatively hydrophobic molecule, contributes to the hydrophobic core of the SimR dimer; this will have a stabilizing effect on the overall structure, locking it into a relatively rigid, low-energy state. Moreover, the combination of the threading of the ligand through both subunits and the projection of the side chain of Arg122 into the opposing subunit contribute to the rigidification of the system (18). The flexibility of the apo form is important to enable the TFR arms and the recognition helices to engage optimally with the DNA. The resulting favourable protein–DNA interactions will have a stabilizing effect on this conformation of SimR. Moreover, in the DNA binding conformation, the repositioning of the C-terminal end of helix α8 appropriately places it to make salt bridges to the DBD of the same subunit and to that of the opposing subunit, specifically between Arg179 and Glu46, and between Arg180 and Glu72, respectively. These interactions, not present in the SD8-bound form, will further stabilize the DNA-bound conformation of SimR.
Figure 9.
Structures of SimR–simocyclinone and SimR–DNA together with schematic representations illustrating the rigid-body rotation of the subunits relative to one another. In order to emphasize the subunit rotation, the grey coloured subunits are shown fixed in the same relative orientations. This can be clearly seen in the side view where the green subunit rotates by ~16° relative to the grey subunit; the approximate pivot point is indicated by the asterisk (see also Supplementary Figures S11 and S12). The distances separating the recognition helices α3 and α3′ in the two structures are indicated.
ACCESSION NUMBER
Coordinates and structure factors for the SimR–DNA structure described herein have been deposited in the Protein Data Bank with accession number 3ZQL.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
John Innes Centre Rotation Studentship (to T.B.K.L.); Short Term EMBO Fellowship ASTF: 290-2010 (to T.B.K.L.); Korner Travelling Fellowship (to T.B.K.L.); BBSRC grant BB/I002197/1 (to M.J.B. and D.M.L.); BBSRC Core Strategic Grant to the John Innes Centre; R.A. Welch Foundation grant G-0040 (to R.G.B.); M.D. Anderson Trust Fellowship (to M.A.S.). Funding for open access charge: BBSRC grant BB/I002197/1 (to M.J.B. and D.M.L.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Clare Stevenson for assistance with X-ray crystallography, to Richard Little and Mahmoud Al-Bassam for advice on limited proteolysis experiments and to beamline scientists at the Advanced Light Source (Berkeley, CA, USA) and the Diamond Light Source (Harwell, UK) for assistance with X-ray data collection.
REFERENCES
- 1.Berdy J. Bioactive microbial metabolites. J. Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 2.Walsh C. Antibiotics: Actions, Origins, Resistance. Washington, DC: ASM Press; 2003. [Google Scholar]
- 3.Edwards MJ, Flatman RH, Mitchenall LA, Stevenson CE, Le TB, Clarke TA, McKay AR, Fiedler HP, Buttner MJ, Lawson DM, et al. A crystal structure of the bifunctional antibiotic simocyclinone D8, bound to DNA gyrase. Science. 2009;326:1415–1418. doi: 10.1126/science.1179123. [DOI] [PubMed] [Google Scholar]
- 4.Flatman RH, Howells AJ, Heide L, Fiedler HP, Maxwell A. Simocyclinone D8, an inhibitor of DNA gyrase with a novel mode of action. Antimicrob. Agents Chemother. 2005;49:1093–1100. doi: 10.1128/AAC.49.3.1093-1100.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oppegard LM, Hamann BL, Streck KR, Ellis KC, Fiedler HP, Khodursky AB, Hiasa H. In vivo and in vitro patterns of the activity of simocyclinone D8, an angucyclinone antibiotic from Streptomyces antibioticus. Antimicrob. Agents Chemother. 2009;53:2110–2119. doi: 10.1128/AAC.01440-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holzenkämpfer M, Walker M, Zeeck A, Schimana J, Fiedler HP. Simocyclinones, novel cytostatic angucyclinone antibiotics produced by Streptomyces antibioticus Tu 6040 II. Structure elucidation and biosynthesis. J. Antibiot. 2002;55:301–307. doi: 10.7164/antibiotics.55.301. [DOI] [PubMed] [Google Scholar]
- 7.Schimana J, Fiedler HP, Groth I, Süssmuth R, Beil W, Walker M, Zeeck A. Simocyclinones, novel cytostatic angucyclinone antibiotics produced by Streptomyces antibioticus Tu 6040. I. Taxonomy, fermentation, isolation and biological activities. J. Antibiot. 2000;53:779–787. doi: 10.7164/antibiotics.53.779. [DOI] [PubMed] [Google Scholar]
- 8.Galm U, Schimana J, Fiedler HP, Schmidt J, Li SM, Heide L. Cloning and analysis of the simocyclinone biosynthetic gene cluster of Streptomyces antibioticus Tu 6040. Arch. Microbiol. 2002;178:102–114. doi: 10.1007/s00203-002-0429-z. [DOI] [PubMed] [Google Scholar]
- 9.Le TB, Fiedler HP, den Hengst CD, Ahn SK, Maxwell A, Buttner MJ. Coupling of the biosynthesis and export of the DNA gyrase inhibitor simocyclinone in Streptomyces antibioticus. Mol. Microbiol. 2009;72:1462–1474. doi: 10.1111/j.1365-2958.2009.06735.x. [DOI] [PubMed] [Google Scholar]
- 10.Trefzer A, Pelzer S, Schimana J, Stockert S, Bihlmaier C, Fiedler HP, Welzel K, Vente A, Bechthold A. Biosynthetic gene cluster of simocyclinone, a natural multihybrid antibiotic. Antimicrob. Agents Chemother. 2002;46:1174–1182. doi: 10.1128/AAC.46.5.1174-1182.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001;65:232–260. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramos JL, Martinez-Bueno M, Molina-Henares AJ, Teran W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 2005;69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu Z, Reichheld SE, Savchenko A, Parkinson J, Davidson AR. A comprehensive analysis of structural and sequence conservation in the TetR family transcriptional regulators. J. Mol. Biol. 2010;400:847–864. doi: 10.1016/j.jmb.2010.05.062. [DOI] [PubMed] [Google Scholar]
- 14.Itou H, Watanabe N, Yao M, Shirakihara Y, Tanaka I. Crystal structures of the multidrug binding repressor Corynebacterium glutamicum CgmR in complex with inducers and with an operator. J. Mol. Biol. 2010;403:174–184. doi: 10.1016/j.jmb.2010.07.042. [DOI] [PubMed] [Google Scholar]
- 15.Miller DJ, Zhang YM, Subramanian C, Rock CO, White SW. Structural basis for the transcriptional regulation of membrane lipid homeostasis. Nat. Struct. Mol. Biol. 2010;17:971–975. doi: 10.1038/nsmb.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orth P, Schnappinger D, Hillen W, Saenger W, Hinrichs W. Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nat. Struct. Biol. 2000;7:215–219. doi: 10.1038/73324. [DOI] [PubMed] [Google Scholar]
- 17.Schumacher MA, Miller MC, Grkovic S, Brown MH, Skurray RA, Brennan RG. Structural basis for cooperative DNA binding by two dimers of the multidrug-binding protein QacR. EMBO J. 2002;21:1210–1218. doi: 10.1093/emboj/21.5.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le TB, Stevenson CE, Fiedler HP, Maxwell A, Lawson DM, Buttner MJ. Structures of the TetR-like simocyclinone efflux pump repressor, SimR, and the mechanism of ligand-mediated derepression. J. Mol. Biol. 2011;408:40–56. doi: 10.1016/j.jmb.2011.02.035. [DOI] [PubMed] [Google Scholar]
- 19.Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 20.Evans P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 21.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 23.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 24.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 25.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 26.DeLano WL. The PyMol User’s Manual. San Carlos, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- 27.Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K. Conformational analysis of nucleic acids revisited: curves+ Nucleic Acids Res. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 29.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 31.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang ZR, Thomson R, McNeil P, Esnouf RM. RONN: the bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics. 2005;21:3369–3376. doi: 10.1093/bioinformatics/bti534. [DOI] [PubMed] [Google Scholar]
- 33.Receveur-Brechot V, Bourhis JM, Uversky VN, Canard B, Longhi S. Assessing protein disorder and induced folding. Proteins. 2006;62:24–45. doi: 10.1002/prot.20750. [DOI] [PubMed] [Google Scholar]
- 34.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 35.Pal D, Chakrabarti P. Non-hydrogen bond interactions involving the methionine sulfur atom. J. Biomol. Struct. Dyn. 2001;19:115–128. doi: 10.1080/07391102.2001.10506725. [DOI] [PubMed] [Google Scholar]
- 36.Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS. Origins of specificity in protein-DNA recognition. Annu. Rev. Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffmann KM, Williams D, Shafer WM, Brennan RG. Characterization of the multiple transferable resistance repressor, MtrR, from Neisseria gonorrhoeae. J. Bacteriol. 2005;187:5008–5012. doi: 10.1128/JB.187.14.5008-5012.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willems AR, Tahlan K, Taguchi T, Zhang K, Lee ZZ, Ichinose K, Junop MS, Nodwell JR. Crystal structures of the Streptomyces coelicolor TetR-like protein ActR alone and in complex with actinorhodin or the actinorhodin biosynthetic precursor (S)-DNPA. J. Mol. Biol. 2008;376:1377–1387. doi: 10.1016/j.jmb.2007.12.061. [DOI] [PubMed] [Google Scholar]
- 40.Joshi R, Passner JM, Rohs R, Jain R, Sosinsky A, Crickmore MA, Jacob V, Aggarwal AK, Honig B, Mann RS. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell. 2007;131:530–543. doi: 10.1016/j.cell.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beamer LJ, Pabo CO. Refined 1.8 A crystal structure of the lambda repressor-operator complex. J. Mol. Biol. 1992;227:177–196. doi: 10.1016/0022-2836(92)90690-l. [DOI] [PubMed] [Google Scholar]
- 42.Dickerson RE. DNA bending: the prevalence of kinkiness and the virtues of normality. Nucleic Acids Res. 1998;26:1906–1926. doi: 10.1093/nar/26.8.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krissinel E, Henrick K. Detection of protein assemblies in crystals. In: Berthold MR, Glen RC, Diederichs K, Kohlbacher O, Fischer I, editors. Computational Life Sciences. Vol. 3695. Berlin Heidelberg: Springer-Verlag; 2005. pp. 163–174. [Google Scholar]
- 44.Reichheld SE, Yu Z, Davidson AR. The induction of folding cooperativity by ligand binding drives the allosteric response of tetracycline repressor. Proc. Natl Acad. Sci. USA. 2009;106:22263–22268. doi: 10.1073/pnas.0911566106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.