Abstract
The N-terminal tail of histone H2B is believed to be involved in gene silencing, but how it exerts its function remains elusive. Here, we report the biochemical characterization of p14ARF tumor suppressor as a transcriptional repressor that selectively recognizes the unacetylated H2B tails on nucleosomes. The p14ARF–H2B tail interaction is functional, as the antagonistic effect of p14ARF on chromatin transcription is lost upon deletion or acetylation of H2B tails. Gene expression profiling and chromatin immunoprecipitation studies emphasize the significance of H2B deacetylation and p14ARF recruitment in establishing a repressive environment over the cell cycle regulatory genes. Moreover, HDAC1-mediated H2B deacetylation, especially at K20, constitutes an essential step in tethering p14ARF near target promoters. Our results thus reveal a hitherto unknown role of p14ARF in the regulation of chromatin transcription, as well as molecular mechanisms governing the repressive action of p14ARF.
INTRODUCTION
In eukaryotic cells, DNA is hierarchically packaged into a highly organized structure of chromatin by means of stable association with histones H2A, H2B, H3 and H4. The repeating unit of chromatin is the nucleosome, which comprises 147 bp of DNA spooled around a histone octamer (1,2). The most dynamic parts of the nucleosome are unstructured amino-terminal domains (called histone ‘tails’) of core histones that mediate contacts between adjacent nucleosomes and interact with chromatin-associated proteins (3–5). One H3–H4 tetramer organizes the central part of the nucleosome by interacting with ~80 bp DNA around the dyad, whereas two H2A–H2B dimers each bind to ~30 bp DNA lying at the peripheral part of the nucleosome (2,6,7). Based on this unique structure of the nucleosome, the past decades of studies mainly focused on H3 and H4 tails as a major arbiter of chromatin transcription, with relatively little attention paid to a possible role of H2A and H2B tails. However, there is some experimental evidence suggesting that transcriptional competence of chromatin is also regulated by H2A and H2B tails, and this is particularly true for H2B tails (8). Early studies in yeast demonstrated that the deletion of H2B tails results in a defect in the repression of GAL1 and PHO5 genes (9,10). Additional support for the repressive nature of H2B tails came from yeast genome microarray analysis, showing that the tail deletion of H2B leads to the upregulation of a large number of genes (11). Moreover, acetylation of H2B tails appears to be effective at relieving this H2B tail-derived repression of transcription, as judged by the derepression of more genes in wild-type yeast strain, than was observed in strains containing H2B tails that were mutated at acetylation sites (11).
While these results suggest that the H2B N-terminal tail is a critical regulator of chromatin function, it is currently unclear how the tail domain makes a negative contribution to regulation of gene transcription. One possible mechanism is that H2B tails would serve as an interaction domain to bring specific repressors in close proximity to gene promoters and establish an inactive state of chromatin. An example to support this possibility is photomorphogenesis regulator DET1 in plant cells, which interacts with unacetylated H2B tails of nucleosomes at the promoter region and maintains genes in a repressed state (12). Thus, given that many of the functions of other histone tails involve tail-factor interactions (5,13), H2B tails may employ a similar strategy to play a regulatory role in chromatin transcription.
Related to the current report, p14ARF is a tumor suppressor that controls cellular senescence in response to oncogenic stresses, being often mutated in many types of human cancer (14–16). Much of the tumor suppressor function of p14ARF has been linked to its ability to stabilize p53 by inhibiting Mdm2-dependent ubiquitination and degradation of p53. The resultant stabilization of p53 leads to increased expression of p53-responsive genes, thereby inducing cell cycle arrest and apoptosis (17). However, this paradigm is increasingly challenged by a substantial number of studies demonstrating that p14ARF is capable of regulating cell growth control and apoptosis induction in a p53-independent manner as well (18–21). Mouse models without p53, Mdm2 and p19ARF (mouse homolog of human p14ARF) are much more prone to developing tumors than mice lacking p53 and Mdm2 but retaining p19ARF. The reintroduction of p19ARF into mouse embryo fibroblasts lacking p53, Mdm2 and p19ARF restored the apoptotic response to a level similar to that seen in the wild-type cells (22). The new aspect of p14ARF function was further revealed by the characterization of a wide range of new p14ARF-interacting proteins, such as Tip60, B23/nucleophosmin and ARF-BP1 E3 ubiquitin ligase (23–26). These findings support the idea that p14ARF can exert its function in a p53-independent manner via the interaction with a multitude of different cellular partners.
In the present study, we combine a series of biochemical and cellular techniques to investigate the molecular mechanisms underlying H2B tail-mediated repression of chromatin transcription. We demonstrate that p14ARF participates in establishing and maintaining a transcriptionally repressive chromatin state through its physical interaction with H2B tails. This process is accompanied by HDAC1-mediated deacetylation of H2B–K20, which is critical for the stable localization and prolonged activity of p14ARF in target genes. Thus, our results identify p14ARF as the first human non-histone protein interacting with H2B tail and highlight this physical interaction as a platform to read the chromatin state and establish gene silencing.
MATERIALS AND METHODS
Cell culture, antibodies and constructs
HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). NARF-E6 osteosarcoma cells were maintained in DMEM containing 10% FBS, 150 µg/ml hygromycin and 300 µg/ml G418. For p14ARF expression, NARF-E6 cells were treated with 1 mM isopropyl-β-D-thiogalactoside (IPTG). Further details for antibodies and plasmid constructions are available in the ‘Materials and Methods’ section of Supplementary Data.
Purification and mass spectrometric analysis of nH2BIFs
HeLa cell lines expressing human H2B tails were generated by transfection of pIRES-8xnH2B vector in which Flag-HA–H2B tails are continuously expressed. Colonies were selected with G418 (500 µg/ml) for 2 weeks and tested for their stable expression of H2B tails by immunoblot analysis. Positive cell clones were grown in spinner culture. Nuclear extracts were prepared as described (27,28). Ectopic H2B tails and their interacting proteins were purified from nuclear extracts (300 mg) by sequential immunoprecipitation using anti-Flag M2 and anti-HA antibodies (Sigma) in the precipitation buffer (20 mM Tris–HCl, pH 7.3, 300 mM KCl, 0.2 mM EDTA, 20% Glycerol and 0.1% NP-40). The purified proteins were resolved by 4–20% gradient SDS–PAGE, and stained with Coomassie blue. Protein bands present in the H2B tail lane, but not in the control lane were excised and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) at the Mass Spectrometry and Proteomics Core Facility (University of Southern California).
Preparation of recombinant proteins
Detailed protocols describing the preparation of recombinant proteins are provided in ‘Materials and Methods’ section of Supplementary Data.
Reconstitution of nucleosome arrays and mononucleosomes
For nucleosome array reconstitution, the pG5ML601-280 G containing 280 bp G-less cassettes were digested with EcoRI and HindIII, and the G5ML601 array DNA fragments were gel purified. To reconstitute mononucleosomes, the 207 bp G5ML and 207 bp 601 sequences were polymerase chain reaction (PCR) amplified from the p5GML601-280 G and p601-1 plasmids, respectively. Nucleosome arrays and mononucleosomes were reconstituted by salt gradient dialysis and purified by sedimentation in a 5–30% (v/v) glycerol gradient (29,30).
Immunodepletion of p14ARF from nH2BIFs
Immunodepletion was carried out in a 100 µl reaction containing 10 µg of nH2BIFs and 2 µg of control or p14ARF antibody coupled to 20 μl of protein A/G-PLUS agarose (Santa Cruz Biotechnology). After an overnight incubation at 4°C under constant rotation, the supernatant was recovered and analyzed by SDS–PAGE and immunoblotting.
Transcription and histone acetyl transferase assays
In vitro transcription assays were performed using 100 ng of G5ML601-280 G nucleosome arrays for each reaction. Recombinant p14ARF proteins or nH2BIFs were added before or after p300 (20 ng) and acetyl-CoA (10 µM). The radiolabeled transcripts were resolved on a 5% urea–PAGE and detected by autoradiography. For nucleosome histone acetyl transferase (HAT) assays to examine the effects of lysine mutations, G5ML nucleosomes (200 ng) were pre-incubated with Gal4-VP16 (15 ng) for 20 min, and then with p300 (20 ng) and acetyl-CoA (10 μM) for 60 min. HAT reactions were analyzed by autoradiography or immunoblotting with AcH2A, AcH2B, AcH3 and AcH4 antibodies.
Protein–protein and protein–nucleosome interaction studies
Glutathione S-transferase (GST) pull-down assays were carried out using purified recombinant GST or GST-tagged proteins bound to glutathione-Sepharose 4B beads (GE Healthcare) and the indicated recombinants proteins in 400 μl of binding buffer (25 mM HEPES, pH 7.8, 0.2 mM EDTA, 20% glycerol, 150 mM KCl and 0.1% NP-40) for 1.5 h at 4°C. Glutathione beads were washed four times with the binding buffer, and subjected to immunoblot analysis. For nucleosome binding assays, biotinylated nucleosomes (1 µg) were immobilized on Streptavidin agarose beads (Novagen) and incubated with recombinant proteins for 16 h at 4°C. After extensive washing with the binding buffer, the bound proteins were detected by immunoblotting. To immunoprecipitate p14ARF and HDAC1, whole cell lysates (1 mg) from IPTG-treated NARF-E6 cells were incubated with anti-p14ARF and anti-HDAC1 antibodies (2 µg) at 4°C for 16 h. Immune complexes were recovered using protein A/G-PLUS agarose, washed four times with binding buffer and analyzed by immunoblotting.
Microarray and quantitative reverse transcription PCR
Total RNA was isolated from mock- or IPTG-treated NARF-E6 cells using the TRIzol reagent according to the manufacturer’s instructions (Invitrogen). Gene expression microarray experiments were conducted using a whole-genome expression array (Sentrix Human-6 Expression BeadChip version 3, Illumina) at the University of Southern California Epigenome Center. This high density oligonucleotide array chip consists of about 48 000 probe sequences derived from RefSeq genes and the latest Unigene release (http://www.switchtoi.com/annotationfiles.ilmn). Data were processed and analyzed by using the BeadStudio software (Illumina). The quantitative reverse transcription PCR (qRT–PCR) was performed using the iScript cDNA Synthesis Kit (Bio-Rad) and the IQ SYBR Green Supermix (Bio-Rad) with an iCycler IQ5 real time cycler (Bio-Rad). The specificity of the amplification reactions were monitored by melting curve analysis. The threshold cycle (Ct) value for each gene was normalized to the Ct value for GAPDH. All samples were run in triplicate and results were averaged. The primer sets used for qRT–PCR are listed in Supplementary Table S1.
Chromatin immunoprecipitation and RNA interference
Chromatin immunoprecipitation (ChIP) assays with NARF-E6 cells, either treated or not treated with IPTG, were performed as stated in the Agilent mammalian ChIP-on-chip protocol and are explained in detail in the ‘Materials and Methods’ section of Supplementary Data. Antibodies specific for p14ARF, β-catenin, H2B, H2B AcK20, AcH3 and AcH4 were used for immunoprecipitation. The positions of the PCR primers within the promoter regions are shown in Figure 5A. All samples were run in triplicate and results were averaged. For HDAC1 knockdown experiments, a DNA oligonucleotide encoding shRNA specific for HDAC1 mRNA (5′-GCAGATGCAGAGATTCAAC-3′) was subcloned into the pSUPER.puro (OligoEngine). NARF-E6 cells were stably transfected with the HDAC1 shRNA expression construct, and then subjected to qRT–PCR and ChIP analyses. Primers used for ChIP analysis are listed in Supplementary Table S2.
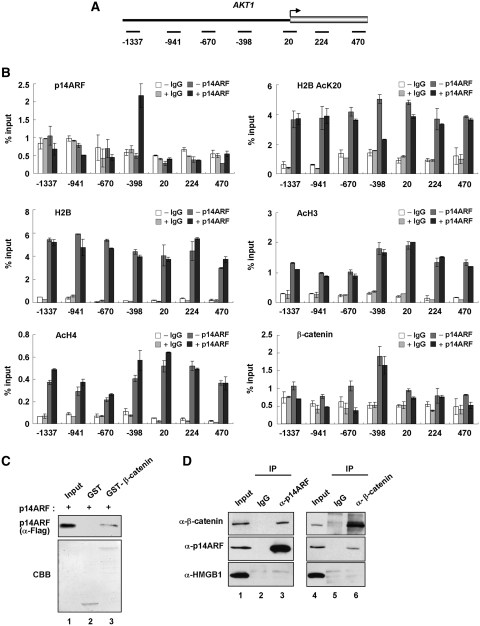
Figure 5.
Reciprocal localization of p14ARF and H2B–K20 acetylation at a target promoter. (A) A schematic diagram showing the PCR amplicons used at the AKT1 gene. The numbers indicate the position of the central base pair of the products relative to the transcription start site. (B) ChIP scanning of p14ARF, H2B AcK20, H2B, AcH3, AcH4 and β-catenin at the AKT1 locus. NARF-E6 cells were mock- or IPTG treated, and analyzed by quantitative chromatin immunoprecipitation (ChIP) using antibodies specifically recognizing p14ARF, H2B AcK20, H2B, AcH3, AcH4 and β-catenin and IgG control. Immunoprecipitated DNA was amplified by quantitative PCR (qPCR) with primers depicted in Figure 5A and listed in Supplementary Table S2. Percentage input is determined as the amount of immunoprecipitated DNA relative to input DNA. Error bars represent standard deviation of three independent experiments. (C) p14ARF interaction with β-catenin in vitro. Flag-tagged p14ARF was tested for binding to GST (lane 2) or GST-β-catenin fusion (lane 3) proteins. p14ARF interaction was determined by immunoblot analysis using anti-Flag antibody. Lane 1 represents 10% of p14ARF used in the binding reactions. (D) p14ARF interaction with β-catenin in vivo. NARF-E6 cells were treated for 48 h with 1 mM IPTG to express p14ARF. Whole cell lysates were prepared and immunoprecipitated with anti-p14ARF (lanes 1–3) or anti-β-catenin (lanes 4–6) antibody, and subjected to immunoblot analysis with anti-p14ARF, anti-β-catenin and anti-HMGB1 antibodies. Input represents 10% of the cell extracts used in immunoprecipitation.
RESULTS
H2B tail-interacting factors repress chromatin transcription
We recently established a protocol that allows purification of factors recognizing N-terminal tails of histones H3 and H4 (28,31). In the present study, we employed the same protocol to search for potential H2B tail-binding proteins. To this end, we generated a cell line that continuously expresses the first 37 amino acids of human H2B fused to the Flag and HA epitopes (Supplementary Figure S1A). This N-terminal domain contains the nuclear localization signal (NLS, amino acids 21–33) that mediates active transport across the nuclear membrane after cytoplasmic translation of the tail domains (Supplementary Figure S1B) (32,33). After two-step purification using anti-Flag and anti-HA immunoaffinity chromatographies (Supplementary Figure S1A), proteins co-purified with the H2B tail were resolved by SDS–PAGE and specific protein bands were subjected to mass spectrometric analysis. As summarized in Figure 1A, our analysis revealed eight proteins that have never been described in association with H2B tails. The mass spectrometry data were further validated by immunoblot analysis using available antibodies (Figure 1B). Since ectopic H2B tails could undergo acetylation, methylation and phosphorylation via actions of cellular modifying activities, we checked the modification status of the ectopic tails by immunoblot analysis. However, our analyses using antibodies specific for acetylations at K5, K12, K15 and K20, phosphorylation at S14 and methylations at K5 and K11 showed no detectable modification of ectopic H2B tails (data not shown).
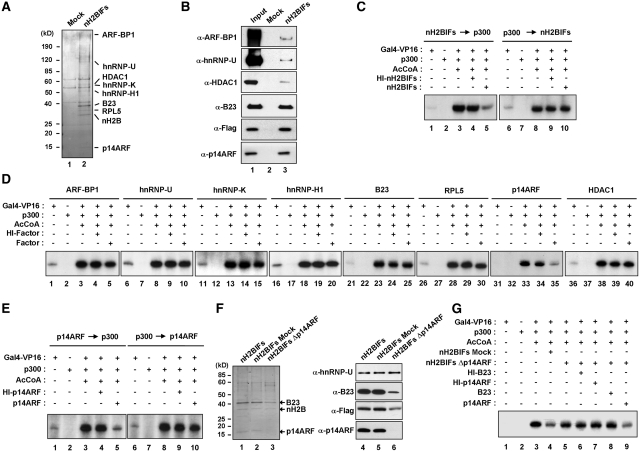
Figure 1.
Repression of chromatin transcription by p14ARF. (A) Affinity purification of nH2BIFs. Flag-HA-tagged H2B tails were stably expressed in HeLa cells and subjected to sequential immunoprecipitations utilizing anti-Flag and anti-HA antibodies. The co-purified proteins were separated by 4–20% gradient SDS–PAGE and identified by liquid chromatography–tandem mass spectrometry (LC–MS/MS). The positions of the molecular mass markers (in kDa) are indicated on the left. Lane 1, mock-purified control; lane 2, nH2BIFs (H2B tail-interacting factors). (B) Immunoblot analysis of the purified nH2BIFs. The purified nH2BIFs were separated by 4–20% gradient SDS–PAGE and analyzed by immunoblotting with the indicated antibodies. Lane 1, nuclear extract input; lane 2, mock-purified control; lane 3, nH2BIFs. (C) Effects of the purified nH2BIFs on chromatin transcription. Reconstituted nucleosome arrays were transcribed in the presence of Gal4-VP16 (15 ng), p300 (20 ng), acetyl-CoA (10 µM) and/or nH2BIFs (15 ng) as summarized in Supplementary Figure S2A. The purified nH2BIFs were added before or after p300-mediated chromatin acetylation as indicated. Radiolabeled transcripts were resolved on a 5% PAGE containing 7 M urea and detected by autoradiography. Heat-inactivated tail-interacting factors (HI-nH2BIFs) were used in control reactions, and the results shown are representative of three independent experiments. (D) Effects of the recombinant nH2BIFs on chromatin transcription. Transcription reactions with G5ML601 nucleosome arrays were essentially as described in Figure 1C, but nH2BIFs were replaced by the indicated recombinant factors. Each of recombinant nH2BIFs was added prior to p300. (E) Loss of p14ARF activity upon pre-acetylation of chromatin. Transcription assays were performed under the conditions described in the legend to Figure 1D, but p14ARF was added before (lanes 1–5) or after (lanes 6–10) p300 acetylation reactions. (F) Depletion of p14ARF from the purified nH2BIFs. The purified nH2BIFs were immunodepleted of p14ARF as described in the ‘Materials and Methods’ section. To confirm that the purified factors had been depleted of p14ARF, mock- and p14ARF-depletion reactions were analyzed by Coomassie blue staining (lanes 1–3) and immunoblotting (lanes 4–6). (G) Effects of the p14ARF-depleted nH2BIFs on chromatin transcription. In vitro transcription was conducted as described in Figure 1D, except that mock-depleted (lane 4) or p14ARF-depleted (lanes 5–9) nH2BIFs were used as indicated. The p14ARF-depleted nH2BIFs were supplemented with either recombinant B23 (lane 8) or p14ARF (lane 9). Heat-inactivated (HI) B23 and p14ARF were used in control reactions (lanes 6 and 7).
To functionally characterize H2B tail-interacting factors (nH2BIFs), we next assessed the ability of the purified factors to regulate transcription from chromatin. Nucleosome arrays were reconstituted onto the linear G5ML601 DNA fragment containing Gal4 binding sites linked to the adenovirus major late core promoter proceeding 280 bp G-less cassette and, on both sides, seven direct repeats of the 207 bp 601 nucleosome positioning sequence (Supplementary Figure S2A). The successful reconstitution of nucleosome arrays using bacterially expressed histone octamers was confirmed by partial micrococcal nuclease (MNase) digestion as previously described (Supplementary Figure S2B) (34). Transcription assays with nucleosome arrays were carried out with Gal4-VP16 activator and p300 HAT as summarized in Supplementary Figure S2A. Given the fact that, the addition or removal of acetyl groups on histones influences the ability of factors to bind to the nucleosome (5,28), we assessed the effect of p14ARF on chromatin transcription before and after p300 acetylation reactions. As previously reported (35,36), transcription from nucleosome arrays was completely dependent on Gal4-VP16, p300, and acetyl-CoA (Figure 1C, lanes 1–3 and 6–8). When the effects of nH2BIFs were examined, chromatin transcription was significantly repressed by the addition of the factors prior to p300 (lane 5). In contrast, no detectable change in chromatin transcription was observed upon the addition of the purified factors after p300 (lane 10). Due to addition of the same amount of heat-inactivated factors (HI-nH2BIFs) to our reactions had no effect on chromatin transcription (lanes 4 and 9), the observed repression is unlikely from non-specific interference in the transcription reaction.
p14ARF among nH2BIFs is responsible for the repression
To define the key factors directly involved in this repression process, we decided to evaluate the effects of individual factors on chromatin transcription by using recombinant proteins. Thus, nH2BIFs were expressed in insect cells (for ARF-BP1) or in bacteria (for hnRNP-U, HDAC1, hnRNP-K, hnRNP-H1, B23, RPL5 and p14ARF) as described in the ‘Materials and Methods’ section. After affinity tag-based purification of the recombinant factors, their purity was confirmed by SDS–PAGE analysis (Supplementary Figure S3). When transcription assays were performed with each of these factors, no detectable repression in chromatin transcription was observed by the addition of ARF-BP1, hnRNP-U, HDAC1, hnRNP-K, hnRNP-H1, B23 or RPL5 prior to p300 (Figure 1D, lanes 5, 10, 15, 20, 25, 30 and 40). However, the addition of p14ARF prior to p300 resulted in a dramatically lower level of transcription (lane 35), strongly supporting that p14ARF is mainly responsible for the repressive effects of nH2BIFs. As expected, no measurable effect of p14ARF was observed when p14ARF was added to transcription reactions after p300 (Figure 1E, lane 5 versus lane 10), suggesting that pre-acetylation of chromatin mediated by p300 prevents the repressive action of p14ARF. It is worth noting that, the advantage of using recombinant histones over their native counterparts in chromatin reconstitution is to exclude the effects of pre-existing acetylation of native histones (Supplementary Figure S4A); chromatin templates reconstituted with native histones were immune to inhibitory effects of p14ARF (Supplementary Figure S4B, lane 5 versus lane 10).
To further confirm the above results, the fraction containing p14ARF was removed from the purified nH2BIFs by treatment with anti-p14ARF antibody. The SDS–PAGE analysis of p14ARF antibody-treated factors showed almost complete disappearance of the 14 kDa band (Figure 1F, lane 3 versus lanes 1 and 2), indicating a specific depletion of p14ARF from the unfractionated nH2BIFs. A minor reduction of B23 in the depleted nH2BIFs was also observed after p14ARF depletion, possibly due to its partial interaction and thus co-depletion with p14ARF (lanes 1–3). Immunoblot analysis further confirmed that p14ARF was efficiently removed from the purified nH2BIFs (Figure 1F, lanes 4–6). As expected, when the p14ARF-depleted nH2BIFs were tested in chromatin transcription, no apparent repression of transcription was observed (Figure 1G, lane 5 versus lane 3). The addition of recombinant p14ARF, but not its heat-inactivated form, to the depleted nH2BIFs resulted in the repression of chromatin transcription (Figure 1G, lane 9 versus lane 7). In contrast, the depleted nH2BIFs supplemented with recombinant B23 failed to show any change in transcription (Figure 1G, lane 8), further confirming that the p14ARF among nH2BIFs plays a major role in repressing chromatin transcription.
The interaction between p14ARF and H2B tail is required for transcriptional repression
As p14ARF was co-purified with free H2B tails, the transcription results we obtained thus far, suggest that p14ARF inhibits chromatin transcription by interacting with unacetylated H2B tails associated with promoter-encompassing nucleosomes. To gain support for this idea, we examined the interaction of Flag-p14ARF with equimolar amounts of GST-histone tail fusion proteins immobilized on glutathione-Sepharose beads. As confirmed by immunoblot analysis of the binding reactions, p14ARF interaction was highly selective for GST-H2B tails (Figure 2A). In similar binding experiments with truncated versions of H2B tails, p14ARF efficiently binds to three H2B tail deletion mutants lacking the first 3, 9 or 15 amino acids, but not to another tail mutant lacking the first 26 amino acids (Figure 2B). To further characterize the interaction between H2B tail and p14ARF, we reconstituted nucleosomes containing wild-type H2B or H2B N-terminal deletion mutants on a 601 nucleosome positioning sequence (Supplementary Figure S5A) and checked the binding of p14ARF. In agreement with the interactions observed with GST-H2B tails, we detected a remarkable binding preference of p14ARF for the immobilized nucleosome containing intact H2B or H2B mutant lacking the first 15 amino acids over the nucleosome containing H2B mutant lacking the first 26 amino acids (Figure 2C). To confirm these results, we repeated in vitro binding experiments using GST-nH2B Δ16–26 and nucleosomes containing H2B Δ16–26. Our results show that the ability of p14ARF to bind H2B tails and nucleosomes is significantly compromised when amino acids 16–26 of H2B tail domain were deleted (Supplementary Figure S6). Additionally, in mapping the interaction region of p14ARF, we found that N-terminal domain (residues 1–64) of p14ARF retained strong affinity for GST-H2B tail, whereas, no apparent interaction was observed with the remainder (residues 65–132) of the protein (Figure 2E and Supplementary Figure S5B). Similar results were obtained when nucleosomes were reconstituted on the 601 nucleosome positioning sequence and used to study the binding properties of p14ARF deletion mutants (Figure 2F). These results argue strongly against the possibility that the observed binding of p14ARF to H2B tail is due to their non-specific associations.
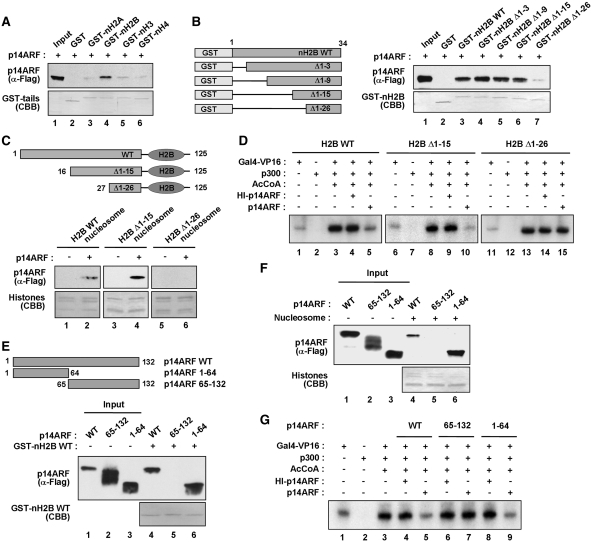
Figure 2.
Mapping of the H2B and p14ARF interaction domains. (A) Preferential binding of p14ARF to H2B tail. Interaction of 14ARF with histone tails was examined by GST pull-down assays using GST (lane 2) or GST–histone tail fusions (lanes 3–6), and the binding reactions were analyzed by immunoblotting and Coomassie brilliant blue (CBB) staining. Lane 1 contains 50% of the input p14ARF. (B) p14ARF interaction with H2B tail deletion mutants. The left panel shows the schematic illustration of H2B tail and its deletion mutants. Numbers indicate amino acid residues. The right panel shows the detection of Flag-p14ARF in GST-pull down assays with GST (lane 2) or GST-H2B tail deletion mutants (lanes 3–7). (C) p14ARF interaction with H2B tail-deleted nucleosomes. Nucleosomes containing wild-type or tailless H2B were reconstituted on biotinylated 207 bp 601 fragments and immobilized on Streptavidin agarose beads. The binding assays were performed with Flag-p14ARF, and the presence of p14ARF in the beads was analyzed by immunoblotting with anti-Flag antibody. (D) Impairment of p14ARF-induced chromatin repression by H2B tail deletions. In vitro transcription experiments were performed as described in Figure 1D, but with chromatin templates containing H2B N-terminal deletion mutants. (E) H2B tail interaction with p14ARF deletion mutants. The top panel shows the schematic illustration of p14ARF deletion mutants. The bottom panel shows the detection of p14ARF deletion mutants in GST pull-down assay with GST–H2B tail. Numbers indicate amino acid residues. Input lanes 1–3 represent 25% of p14ARF used in the binding reactions. (F) Nucleosome interaction with p14ARF deletion mutants. Nucleosomes containing wild-type H2B were reconstituted on biotinylated 207 bp 601 sequences and immobilized on Streptavidin agarose beads. The binding assays were performed with Flag-p14ARF deletion mutants, and the presence of p14ARF deletion mutants in the beads was analyzed by immunoblotting with anti-Flag antibody. (G) Effects of N- and C-terminal regions of p14ARF on chromatin transcription. Transcription reactions were essentially as described in Figure 1D, but contained p14ARF deletion mutants. Heat-treated (HI) p14ARF proteins were also tested in control reactions.
Having found that residues 16–26 of H2B interact with N-terminal domain of p14ARF, we next assessed whether this interaction is critical for p14ARF-induced transcriptional repression. The use of recombinant histones for chromatin reconstitution allowed us to make deletions or point mutations in the histone tail regions that could influence p14ARF-induced repression. Consistent with the previous finding that H3 and H4 tails are sufficient for p300-mediated chromatin transcription (37), deletion of the first 26 amino acids of H2B showed minimal effects on transcription stimulatory activity of p300 (Figure 2D, lane 13 versus lane 3). In parallel transcription experiments with p14ARF, deletion of the first 15 amino acids of H2B did not cause any change in the ability of p14ARF to repress chromatin transcription (lane 10 versus lane 5). However, further truncation of H2B up to amino acid 26 overtly interfered with the repressive action of p14ARF on chromatin transcription (lane 15 versus lane 5). These results confirm that p14ARF-mediated repression of chromatin transcription is highly dependent on a small subdomain in the H2B tail, comprised of residues 16–26. In checking the effects of N-terminal domain (residues 1–64) and the remainder (residues 65–132) of p14ARF on transcription reactions, we found that the N-terminal domain can repress chromatin transcription to the same extent as the full-length p14ARF (Figure 2G, lane 9 versus lane 5). In contrast, chromatin transcription was not repressed when the C-terminal fragment of p14ARF was added to the reaction (lane 7 versus lane 5).
p14ARF suppresses multiple cell cycle regulatory genes
The observation that p14ARF–H2B tail interaction is required for chromatin repression, suggests that this might be responsible for the p53-independent action of p14ARF. To identify genes whose expression is regulated by this mechanism, we examined gene expression profiles using NARF-E6, a p53-null osteosarcoma cell line, which expresses p14ARF from an IPTG-inducible promoter (Supplementary Figure S7A) (38). Western blotting of cytoplasmic and nuclear fractions confirmed that the major fraction of p14ARF is accumulated in the cell nucleus after its expression (Supplementary Figure S7C). RNA was extracted and labeled from cells grown in the absence or presence of IPTG for 48 h, and was hybridized to the Illumina microarrays. Statistical analysis identified a total of 330 genes to be downregulated and 20 genes to be upregulated with a 2-fold change cutoff in response to p14ARF expression (Figure 3A and Supplementary Table S3). Among the genes downregulated by p14ARF, the most significant repression was observed for genes that are involved in cell cycle control and apoptosis. qRT–PCR was performed on the five genes that were repressed (AKT1, Cyclin E1, CHD5 and eIF4E3) or unchanged (RUNX2) in our microarray studies and the results were consistent with the microarray results (Figure 3B).
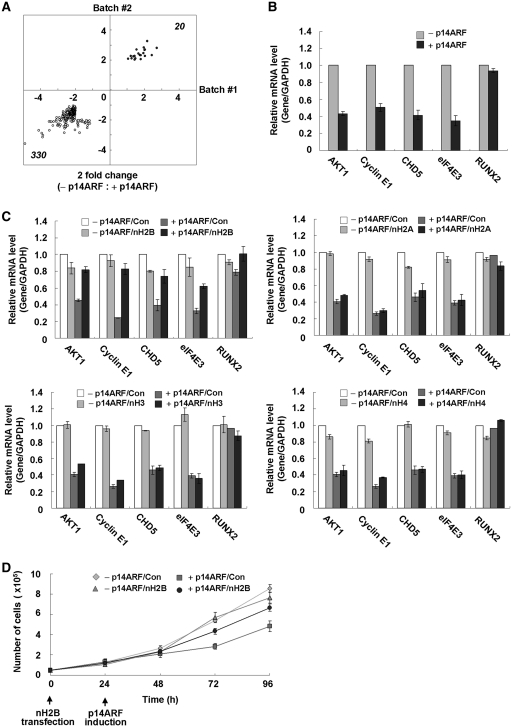
Figure 3.
Suppression of multiple genes by p14ARF. (A) Global analysis of p14ARF-dependent gene regulation. Dots in the two-dimensional scatter-plot represent expression values for the genes with a change >2-fold in either of two independent analyses (Batch #1 and Batch #2). These analyses identified 330 genes that were downregulated (indicated by white dots) and 20 genes that were upregulated (indicated by black dots) in the IPTG-treated cells, compared with the mock-treated cells. (B) Validation of microarray data. The RNA samples from mock-treated (-p14ARF) or IPTG-treated (+p14ARF) NARF-E6 cells were subjected to qRT–PCR to examine the expression levels of AKT1, Cyclin E1, CHD5, eIF4E3 and RUNX2 genes. All expression levels were normalized to that of GAPDH, and average and standard deviation of three independent experiments are shown. (C) Inhibition of p14ARF-mediated repression by ectopic H2B tails. NARF-E6 cells were transfected with an empty vector (Con) or vectors expressing H2B tails (nH2B), H2A tail (nH2A), H3 tails (nH3) or H4 tail (nH4) for 24 h and were further mock-treated (-p14ARF) or treated with 1 mM IPTG (+p14ARF) for 48 h. Changes in transcription of AKT1, Cyclin E1, CHD5, eIF4E3 and RUNX2 genes were measured by qRT–PCR (right panel). (D) Attenuation of p14ARF-enhanced cell proliferation by ectopic H2B tails. NARF-E6 cells were treated as in Figure 3C, and cell viability was measured by trypan blue staining analysis. All reactions were performed in triplicate.
As p14ARF selectively interacts with H2B tails protruding from the nucleosome, it is possible that free H2B tails might influence the repressive activities of p14ARF on its target nucleosomes through competing for binding with p14ARF. To test this, we expressed the N-terminal tail domains of human histones and checked their effects on transcription of the five selected genes after IPTG treatment. As expected, our western blot analysis confirmed that all ectopic histone tails are predominantly located in the cell nucleus after their expression (Supplementary Figure S7B and S7C). Expression of the H2B tail led to a dramatic enhancement in transcription of the four target genes (AKT1, Cyclin E1, CHD5 and eIF4E3), but only a modest increase in transcription of the control gene (RUNX2) (Figure 3C, +p14ARF/nH2B versus +p14ARF/Con). The observed effects of ectopic H2B tails were specific, since other histone tails produced a slight or no detectable change in transcription of the selected genes (Figure 3C). The inability of the H2B tails to affect transcription in the absence of p14ARF further confirmed that the observed enhancement was due to the sequestration of p14ARF activity (−p14ARF/nH2B versus −p14ARF/Con). To gain more support for the competition results above, we also checked whether H2B tail expression targeting p14ARF is directly attributable to alteration in cell proliferation. As summarized in Figure 3D, overexpression of H2B tails resulted in a sharp increment in the proliferation of p14ARF-expressing cells, but no detectable change in control cells.
H2B–K20 acetylation interferes with the repressive action of p14ARF
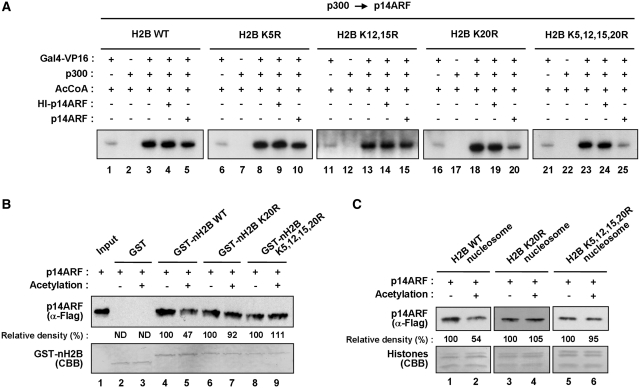
One of the key observations made in our experiments so far is that p300-mediated pre-acetylation of H2B tails eliminates the action of p14ARF as a negative regulator of chromatin transcription (Figure 1E). To determine which of the acetylation sites in H2B is important in blocking the p14ARF activity, we prepared a series of chromatin templates in which four major acetylation sites (K5, K12, K15 and K20) of H2B were mutated individually or in combination (Supplementary Figure S8A). As expected, no detectable change in chromatin transcription was observed by the addition of p14ARF after p300-mediated acetylation of chromatin (Figure 4A, lane 5). Similarly, neither the individual mutation of K5, nor the simultaneous mutation of K12 and K15 in H2B prevented p300-mediated acetylation from blocking the action of p14ARF (lanes 10 and 15). Surprisingly, however, p14ARF was capable of repressing transcription from chromatin-carrying K20-mutated H2B, when p14ARF was added after the p300-mediated acetylation reaction (lane 20). Likewise, the repressive effect of p14ARF on pre-acetylated chromatin was also rescued by simultaneous mutation of all four acetylation sites (K5, K12, K15 and K20) of H2B (lane 25), further underscoring the prominent role of K20 acetylation in antagonizing the repressive action of p14ARF.
Figure 4.
Regulation of p14ARF function by H2B–K20 acetylation. (A) Antagonistic effects of H2B–K20 acetylation on p14ARF transcriptional repression. Transcription assays were performed as in Figure 1D, but p300 was added to the reactions before p14ARF. Chromatin templates contain wild-type (WT) (lanes 1–5) or lysine-mutated (K5R, K12, 15R, K20R and K5, 12, 15, 20R) (lanes 6–25) H2B proteins. (B) Antagonistic effects of K20 acetylation on H2B tail–p14ARF interaction. Flag-tagged p14ARF was tested for binding to GST (lanes 2 and 3) or GST-fused wild-type (WT) or lysine-mutated (K5R, K12,15R, K20R and K5,12,15,20R) H2B tails (lanes 4–9). Lane 1 shows 25% of p14ARF used in the binding reactions. Experiments were repeated three times with comparable results. Data were quantitated by phosphoimager analysis. (C) Antagonistic effects of H2B–K20 acetylation on nucleosome-p14ARF interaction. Mononucleosomes containing wild-type (WT) or mutant (K20R and K5,12,15,20R) H2B were reconstituted on biotinylated 207 bp G5ML fragments and immobilized on Streptavidin agarose beads. Flag-tagged p14ARF was incubated with the nucleosomes containing wild-type (lanes 1 and 2) or mutant (lanes 3–6) H2B, and p14ARF binding was determined by immunoblot analysis using anti-Flag antibody. Data were quantitated by phosphoimager analysis, and similar results were obtained in two independent binding experiments.
To explore the molecular basis for the transcription results described above, we conducted in vitro binding assays with pre-acetylated GST fusion proteins containing wild-type or lysine-mutated forms of H2B tails. As expected, pre-acetylation of wild-type H2B tails by p300 reduced p14ARF binding (Figure 4B, lane 5 versus lane 4). However, parallel binding experiments with K20- or K5-, 12-, 15-, 20-mutated H2B tails showed a similar binding with p14ARF regardless of pre-incubation with p300 and acetyl-CoA (lanes 6–9). We also found that p300-mediated pre-acetylation reduced the binding affinity of p14ARF to nucleosomes containing wild-type H2B, but did not change p14ARF interaction with nucleosomes containing K20- or all four acetylation site-mutated H2B (Figure 4C and Supplementary Figure S8B). Hence, these results establish H2B–K20 acetylation as a major regulatory mark, and fit well with the transcription data showing that pre-acetylation of H2B–K20 is sufficient to block the inhibitory action of p14ARF in chromatin transcription.
p14ARF occupancy at AKT1 promoter coincides with H2B–K20 deacetylation
Among the p14ARF-regulated genes identified through microarray analysis, AKT1 is of particular interest, as the overexpression of this gene was recently found to be an early event in carcinogenesis (39). To elucidate a possible role of p14ARF in the regulation of AKT1 transcription, we established a high resolution ChIP protocol that allows us to map factor occupancy and histone modification on the AKT1 locus. As depicted in Figure 5A, the four different promoter regions (amplicons −1337, −941, −670 and −398), transcription start site (amplicon 20) and two different coding regions (amplicons 224 and 470) of the AKT1 gene were amplified for quantification of the ChIP-enriched DNA. In confirmation of our microarray results, the IPTG-induced expression of p14ARF led to a stable localization of p14ARF at amplicon −398 of the AKT1 (Figure 5B, p14ARF). Also, consistent with our results indicating a selective association of p14ARF with K20-deacetylated H2B, parallel ChIP experiments showed a dramatic reduction of H2B–K20 acetylation at −398 of the AKT1 promoter (H2B AcK20). As there were little or no change in H3 and H4 acetylation (AcH3 and AcH4) at all regions, these results constitute a powerful argument that the observed repression of AKT1 transcription by p14ARF is mainly contributed by deacetylated H2B–K20. Furthermore, a very similar distribution of H2B was seen in control and p14ARF-expressing cells (H2B), making it highly unlikely that the decrease in K20 acetylation is a result of H2B dissociation from nucleosomes. To explore the possible effect of p14ARF on other target genes, we repeated ChIP experiments at the Cycline E1 locus; similar results were obtained from these parallel experiments (Supplementary Figure S9).
Since β-catenin has a regulatory function in AKT1 gene transactivation (40), promoter localization of β-catenin was also determined. β-Catenin showed a similar pattern of localization at the AKT1 promoter regions compared to p14ARF (Figure 5B; compare β-catenin with p14ARF). However, because β-catenin showed the same occupancy profiles without p14ARF expression, p14ARF does not seem to affect β-catenin localization at the AKT1 promoter. Rather, the fact that p14ARF has no distinguishable DNA binding motif suggests that p14ARF may be initially recruited by β-catenin. To check this possibility, Flag-p14ARF was incubated with GST-β-catenin immobilized on glutathione-Sepharose beads, and p14ARF binding was analyzed by immunoblotting. p14ARF was precipitated from the reaction by GST-β-catenin, but GST control failed to show any detectable precipitation of p14ARF under the same conditions (Figure 5C). The interaction of p14ARF with β-catenin in physiological conditions was further tested by immunoblot analysis of anti-p14ARF immunoprecipitates from lysates of p14ARF-induced NARF-E6 cells. As shown in Figure 5D, β-catenin co-immunoprecipitated with p14ARF. A reverse co-immunoprecipitation experiment in which immunoprecipitation was carried out using anti-β-catenin antibody also showed that p14ARF co-immunoprecipitated with β-catenin (Figure 5D). The interaction between p14ARF and β-catenin is specific, as they showed no detectable binding to HMGB1 control in the precipitated complex (α-HMGB1). It thus seems that, although other factors may be involved, the initial recruitment of p14ARF at the AKT1 promoter reflects its direct interaction with β-catenin.
p14ARF cooperates with HDAC1 to repress transcription
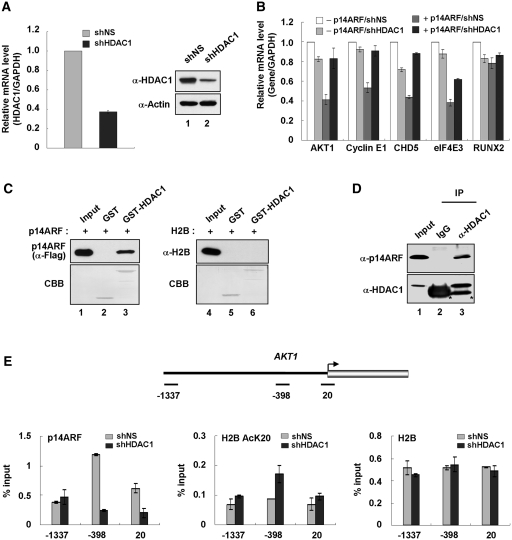
Although the inhibitory action of p14ARF in chromatin transcription reflects its direct interaction with unacetylated H2B tails, it is not clear which factors are mainly responsible for establishing and maintaining the unacetylated state of H2B tails. Given the fact that HDAC1 is co-purified with ectopic H2B tails (Figure 1A and B), we next sought to explore whether HDAC1 is functionally connected to the H2B tail-dependent action of p14ARF. As first confirmed by immunoblot analysis, transfection of NARF-E6 cells with shRNA targeting HDAC1 efficiently depleted HDAC1 (Figure 6A). In checking the transcription of the five selected genes by qRT–PCR, we found that HDAC1 knockdown gave a substantial reactivation of the four target genes (AKT1, Cyclin E1, CHD5 and eIF4E3) in p14ARF-expressing cells, but little or no effect on the control gene (RUNX2) (Figure 6B). Due to the cooperative action of HDAC1 at p14ARF, target genes might reflect its direct association with p14ARF or unacetylated H2B, we conducted a series of interaction studies. In vitro experiments with GST-HDAC1 and Flag-p14ARF reveal that p14ARF directly interacts with HDAC1 (Figure 6C, lane 3). In contrast, similar binding experiments with free H2B failed to show any detectable binding to GST-HDAC1 (lane 6). Consistent with in vitro binding data, immunoblot analysis of HDAC1 immunoprecipitates from extracts of the IPTG-treated NARF-E6 cells, showed a strong interaction with p14ARF (Figure 6D, lane 3). Since HDAC1 can directly associate with p14ARF, a possible explanation for the higher level of transcription after HDAC1 knockdown could be the increased acetylation of H2B–K20 at promoter regions, upon which p14ARF exerts minimal repressive effects on transcription. Predictably, when ChIP experiments were performed at the AKT1 promoter region in p14ARF-expressing cells, HDAC1 knockdown led to a dramatic increase in H2B–K20 acetylation and a near complete loss of p14ARF at amplicon −398 of the AKT1 promoter (Figure 6E, p14ARF and H2B AcK20). There was a comparable distribution of H2B over the AKT1 promoter (H2B) in mock and HDAC1-depleted cells, indicating that ChIP results were not due to non-specific crosslinking or nucleosome aggregation.
Figure 6.
Cooperative action of p14ARF and HDAC1 in gene repression. (A) Stable depletion of HDAC1. NARF-E6 cells were stably transfected with control shRNA (lane 1) or HDAC1 shRNA (lane 2), and the depletion of HDAC1 was analyzed by qRT–PCR (left panel) and immunoblotting (right panel). (B) Requirement of HDAC1 for p14ARF-mediated repression. Mock-depleted or HDAC1-depleted cells were treated with 1 mM IPTG for 48 h, and mRNA levels of AKT1, Cyclin E1, CHD5, eIF4E3 and RUNX2 were analyzed by qRT–PCR. All reactions were performed in triplicate. (C) HDAC1 interaction with p14ARF in vitro. GST or GST-fused HDAC1 was incubated with Flag-tagged p14ARF or untagged H2B, and bound proteins were analyzed by immunoblotting using anti-Flag and anti-H2B antibodies as indicated. Lanes 1 and 4 represent 50% of p14ARF and 20% of H2B used in the binding reactions, respectively. (D) HDAC1 interaction with p14ARF in vivo. Whole cell extracts were prepared from IPTG-treated NARF-E6 cells and immunoprecipitated with anti-HDAC1 antibody. The precipitates were analyzed by immunoblotting with HDAC1 and p14ARF antibodies as indicated. Asterisks indicate non-specific bands containing IgG heavy chain (lanes 2 and 3). Lane 1 represents 10% of the input. (E) HDAC1-dependent localization of p14ARF at the AKT1 promoter. Mock-depleted and HDAC1-depleted cells were treated with 1 mM IPTG to express p14ARF as in Figure 6B, and ChIP assays of the AKT1 gene were performed using antibodies against p14ARF, H2B AcK20 and H2B. Input DNA and immunoprecipitated DNA were quantified by qPCR analysis as described in Figure 5A.
DISCUSSION
The regulatory effects of chromatin have been studied with much greater attention to H3 and H4 N-terminal tails, but H2A and H2B tails are also implicated in the regulation of chromatin function (8). Genome-wide expression profiling of yeast strains indicates that the deletion of H2B tails upregulates a large number of genes (11), suggesting a general repressive effect of the H2B tail domain in transcription. Despite this distinct role of H2B tails in transcription, the molecular mechanism underlying this process has been unclear, partly because of a lack of understanding of upstream regulatory factors that contribute to the observed trans-repression potential of H2B tails. In this report, we demonstrate that p14ARF, a critical human tumor suppressor, binds to the H2B tails and functions as a negative regulator of gene transcription.
A key question that this study presages is whether gene regulation by H2B tails is through the binding of specific chromatin factors to H2B tails. Our initial purification and transcription studies indicate that H2B tails are physically associated with eight factors, which can significantly repress chromatin transcription. In screening factors responsible for the observed repression, p14ARF is found to antagonize transcription from chromatin, when it is added to chromatin prior to p300 HAT. Furthermore, much to our surprise, our results reveal a direct link between the unacetylated state of H2B–K20 and the repressive property of p14ARF. As a potential mechanism explaining why deacetylation of H2B–K20 is critical for p14ARF action, we demonstrate that the p14ARF N-terminal domain selectively interacts with K20-unacetylated H2B tails. These results point to a previously unrecognized contribution of H2B–K20 to the control of gene transcription, through its effect on p14ARF-induced changes in chromatin competence. Gene regulatory factors that affect transcription are generally unable to access their target nucleosomes, so a local remodeling of nucleosomes is a prerequisite for their activity (41,42). However, p14ARF does not appear to require prior disruption of the nucleosome, as we have observed that p14ARF can directly access unacetylated H2B tails in the nucleosome. Thus, apart from our demonstration of repressor activity of p14ARF, these results bear an important implication on the accessibility of H2B tails for the establishment of repressed state of chromatin. In many cases, distinct tertiary structures are known to be involved in protein–histone tail interactions, and histone tails acquire distinct features of recognition as a consequence of post-translational modifications (4,43). However, our sequence alignment and motif prediction fail to detect any known histone tail binding motifs in p14ARF. Whether H2B tails recognize a previously undocumented binding motif in p14ARF is an intriguing question that needs to be addressed in future studies.
In accord with our in vitro results, our gene expression profiling revealed that p14ARF downregulates 330 genes, many of which are involved in cell cycle control and apoptosis. The repressive role of p14ARF is further underscored by ChIP and RNAi analyses demonstrating that p14ARF is specifically localized at promoter nucleosomes containing K20-hypoacetylated H2B to block transcription. As HDAC1 is co-purified with ectopic H2B tails, we reasoned that HDAC1 might contribute to promoter localization of p14ARF by removing and/or preventing H2B acetylation. Indeed, HDAC1 depletion increased H2B–K20 acetylation and decreased p14ARF occupancy at the promoter, placing HDAC1 as a key regulator of p14ARF activity. Interestingly, however, our ChIP analysis failed to show continuous localization of HDAC1 over promoter-proximal nucleosomes (data not shown), implicating transient action of HDAC1 at AKT1 promoter. In this case, HDAC1-mediated deacetylation of H2B should be categorized as an initial regulator for p14ARF interaction with nucleosome. Mechanistically, these aspects of functional interactions between p14ARF and HDAC1 appear to be critical for H2B tail-mediated repression, as misregulation of H2B–K20 deacetylation and thus H2B tail–p14ARF interaction can lead to inappropriate gene repression. Since our analyses have been restricted to the effect of H2B acetylation, future studies should determine the extent to which the p14ARF–H2B interaction is regulated by other histone modifications such as H2B–S14 phosphorylation (44).
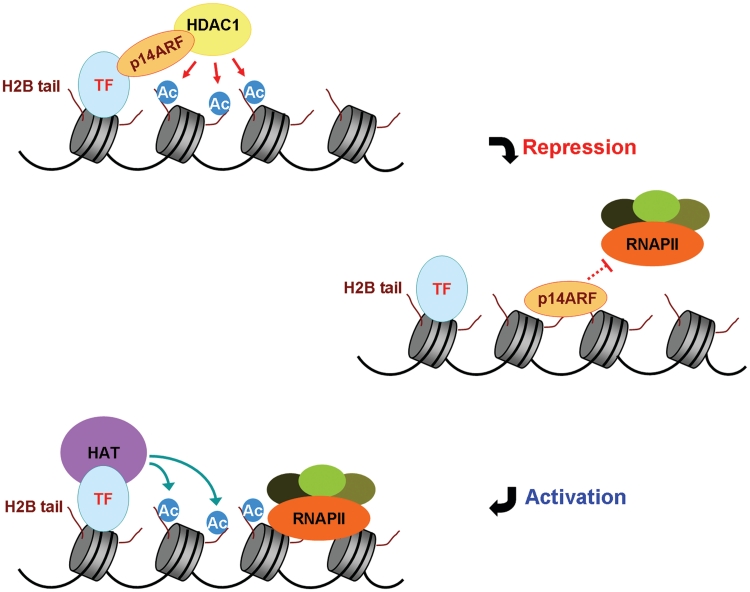
Based on the functional relationship and biochemical characteristics, we have established for H2B tail, p14ARF and HDAC1, we propose the following model of transcriptional repression (Figure 7D). The nuclear p14ARF–HDAC1 complex is brought to target genes by interaction with specific transcription factors (TFs) at the initial stage of gene repression. After their localization at promoter regions, HDAC1 removes acetylation of H2B–K20, mostly over promoter-proximal nucleosomes. This unacetylated H2B–K20 serves as a binding platform for p14ARF, thereby enabling p14ARF to maintain a repressive state of nucleosomes at its target genes. This model implies that HDAC1 recruitment and H2B deacetylation are the rate-limiting steps for the repressive action of p14ARF. This model also has implications for H2B tail-dependent transcriptional activation, because H2B acetylation will antagonize tethering of p14ARF to the promoter.
Figure 7.
Model for the role of p14ARF in chromatin silencing. p14ARF recognizes unacetylated H2B tails at target promoters, and acts as a molecular rheostat to block transcription. For the most efficient repression, p14ARF cooperates with HDAC1 to remove H2B acetylation. Upon p14ARF dissociation, HAT acetylates H2B tails to establish active chromatin environment and to achieve transcription initiation. See the ‘Discussion’ section for further details.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes Health Grant R01GM84209; ACS Research Scholar Grant DMC-1005001; Department of Defense Grant W81XWH0910330 (awarded to W.A.). Funding for open access charge: National Institutes of Health (R01GM84209).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Francisco Baralle for pET11a-hnRNP-H1; Gideon Dreyfuss for pGEM4-hnRNP-U; Petra Jakob for pSG5-His-hnRNP-K; Karolin Luger for histone expression vectors; Gordon Peters for NARF-E6 cells; Daniela Rhodes for p601-7 and p601-1; Michael Stallcup for pGEX4T1-β-catenin; Xiaodong Wang for His-ARF-BP1 Baculovirus; and Wendell Yarbrough for pcDNA-Myc-p14ARF.
REFERENCES
- 1.Harp JM, Hanson BL, Timm DE, Bunick GJ. Asymmetries in the nucleosome core particle at 2.5 A resolution. Acta Crystallogr. D Biol. Crystallogr. 2000;56:1513–1534. doi: 10.1107/s0907444900011847. [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Cosgrove MS, Wolberger C. How does the histone code work? Biochem. Cell Biol. 2005;83:468–476. doi: 10.1139/o05-137. [DOI] [PubMed] [Google Scholar]
- 4.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Oohara I, Wada A. Spectroscopic studies on histone-DNA interactions. I. The interaction of histone (H2A, H2B) dimer with DNA: DNA sequence dependence. J. Mol. Biol. 1987;196:389–397. doi: 10.1016/0022-2836(87)90699-1. [DOI] [PubMed] [Google Scholar]
- 7.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 8.Wyrick JJ, Parra MA. The role of histone H2A and H2B post-translational modifications in transcription: a genomic perspective. Biochim. Biophys. Acta. 2009;1789:37–44. doi: 10.1016/j.bbagrm.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Lenfant F, Mann RK, Thomsen B, Ling X, Grunstein M. All four core histone N-termini contain sequences required for the repression of basal transcription in yeast. EMBO J. 1996;15:3974–3985. [PMC free article] [PubMed] [Google Scholar]
- 10.Nag R, Kyriss M, Smerdon JW, Wyrick JJ, Smerdon MJ. A cassette of N-terminal amino acids of histone H2B are required for efficient cell survival, DNA repair and Swi/Snf binding in UV irradiated yeast. Nucleic Acids Res. 2010;38:1450–1460. doi: 10.1093/nar/gkp1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parra MA, Kerr D, Fahy D, Pouchnik DJ, Wyrick JJ. Deciphering the roles of the histone H2B N-terminal domain in genome-wide transcription. Mol. Cell. Biol. 2006;26:3842–3852. doi: 10.1128/MCB.26.10.3842-3852.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benvenuto G, Formiggini F, Laflamme P, Malakhov M, Bowler C. The photomorphogenesis regulator DET1 binds the amino-terminal tail of histone H2B in a nucleosome context. Curr. Biol. 2002;12:1529–1534. doi: 10.1016/s0960-9822(02)01105-3. [DOI] [PubMed] [Google Scholar]
- 13.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harland M, Taylor CF, Chambers PA, Kukalizch K, Randerson-Moor JA, Gruis NA, de Snoo FA, ter Huurne JA, Goldstein AM, Tucker MA, et al. A mutation hotspot at the p14ARF splice site. Oncogene. 2005;24:4604–4608. doi: 10.1038/sj.onc.1208678. [DOI] [PubMed] [Google Scholar]
- 15.Dominguez-Brauer C, Brauer PM, Chen YJ, Pimkina J, Raychaudhuri P. Tumor suppression by ARF: gatekeeper and caretaker. Cell Cycle. 2010;9:86–89. doi: 10.4161/cc.9.1.10350. [DOI] [PubMed] [Google Scholar]
- 16.Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc. Natl Acad. Sci. USA. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat. Rev. Cancer. 2006;6:663–673. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- 18.Hemmati PG, Gillissen B, von Haefen C, Wendt J, Starck L, Guner D, Dorken B, Daniel PT. Adenovirus-mediated overexpression of p14(ARF) induces p53 and Bax-independent apoptosis. Oncogene. 2002;21:3149–3161. doi: 10.1038/sj.onc.1205458. [DOI] [PubMed] [Google Scholar]
- 19.Yarbrough WG, Bessho M, Zanation A, Bisi JE, Xiong Y. Human tumor suppressor ARF impedes S-phase progression independent of p53. Cancer Res. 2002;62:1171–1177. [PubMed] [Google Scholar]
- 20.Eymin B, Leduc C, Coll JL, Brambilla E, Gazzeri S. p14ARF induces G2 arrest and apoptosis independently of p53 leading to regression of tumours established in nude mice. Oncogene. 2003;22:1822–1835. doi: 10.1038/sj.onc.1206303. [DOI] [PubMed] [Google Scholar]
- 21.Ha L, Ichikawa T, Anver M, Dickins R, Lowe S, Sharpless NE, Krimpenfort P, Depinho RA, Bennett DC, Sviderskaya EV, et al. ARF functions as a melanoma tumor suppressor by inducing p53-independent senescence. Proc. Natl Acad. Sci. USA. 2007;104:10968–10973. doi: 10.1073/pnas.0611638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber JD, Jeffers JR, Rehg JE, Randle DH, Lozano G, Roussel MF, Sherr CJ, Zambetti GP. p53-independent functions of the p19(ARF) tumor suppressor. Genes Dev. 2000;14:2358–2365. doi: 10.1101/gad.827300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertwistle D, Sugimoto M, Sherr CJ. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol. Cell. Biol. 2004;24:985–996. doi: 10.1128/MCB.24.3.985-996.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eymin B, Claverie P, Salon C, Leduc C, Col E, Brambilla E, Khochbin S, Gazzeri S. p14ARF activates a Tip60-dependent and p53-independent ATM/ATR/CHK pathway in response to genotoxic stress. Mol. Cell. Biol. 2006;26:4339–4350. doi: 10.1128/MCB.02240-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Itahana K, Bhat KP, Jin A, Itahana Y, Hawke D, Kobayashi R, Zhang Y. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol. Cell. 2003;12:1151–1164. doi: 10.1016/s1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- 26.Chen D, Kon N, Li M, Zhang W, Qin J, Gu W. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 27.Malik S, Roeder RG. Isolation and functional characterization of the TRAP/mediator complex. Methods Enzymol. 2003;364:257–284. doi: 10.1016/s0076-6879(03)64015-2. [DOI] [PubMed] [Google Scholar]
- 28.Choi J, Kim B, Heo K, Kim K, Kim H, Zhan Y, Ranish JA, An W. Purification and characterization of cellular proteins associated with histone H4 tails. J. Biol. Chem. 2007;282:21024–21031. doi: 10.1074/jbc.M703883200. [DOI] [PubMed] [Google Scholar]
- 29.Robinson PJ, Fairall L, Huynh VA, Rhodes D. EM measurements define the dimensions of the “30-nm” chromatin fiber: evidence for a compact, interdigitated structure. Proc. Natl Acad. Sci. USA. 2006;103:6506–6511. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaskelioff M, Gavin IM, Peterson CL, Logie C. SWI-SNF-mediated nucleosome remodeling: role of histone octamer mobility in the persistence of the remodeled state. Mol. Cell. Biol. 2000;20:3058–3068. doi: 10.1128/mcb.20.9.3058-3068.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heo K, Kim B, Kim K, Choi J, Kim H, Zhan Y, Ranish JA, An W. Isolation and characterization of proteins associated with histone H3 tails in vivo. J. Biol. Chem. 2007;282:15476–15483. doi: 10.1074/jbc.M610270200. [DOI] [PubMed] [Google Scholar]
- 32.Mosammaparast N, Jackson KR, Guo Y, Brame CJ, Shabanowitz J, Hunt DF, Pemberton LF. Nuclear import of histone H2A and H2B is mediated by a network of karyopherins. J. Cell Biol. 2001;153:251–262. doi: 10.1083/jcb.153.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreland RB, Langevin GL, Singer RH, Garcea RL, Hereford LM. Amino acid sequences that determine the nuclear localization of yeast histone 2B. Mol. Cell. Biol. 1987;7:4048–4057. doi: 10.1128/mcb.7.11.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Kundu TK, Palhan VB, Wang Z, An W, Cole PA, Roeder RG. Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol. Cell. 2000;6:551–561. doi: 10.1016/s1097-2765(00)00054-x. [DOI] [PubMed] [Google Scholar]
- 36.Black JC, Choi JE, Lombardo SR, Carey M. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol. Cell. 2006;23:809–818. doi: 10.1016/j.molcel.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 37.An W, Palhan VB, Karymov MA, Leuba SH, Roeder RG. Selective requirements for histone H3 and H4 N termini in p300-dependent transcriptional activation from chromatin. Mol. Cell. 2002;9:811–821. doi: 10.1016/s1097-2765(02)00497-5. [DOI] [PubMed] [Google Scholar]
- 38.Stott FJ, Bates S, James MC, McConnell BB, Starborg M, Brookes S, Palmero I, Ryan K, Hara E, Vousden KH, et al. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roy HK, Olusola BF, Clemens DL, Karolski WJ, Ratashak A, Lynch HT, Smyrk TC. AKT proto-oncogene overexpression is an early event during sporadic colon carcinogenesis. Carcinogenesis. 2002;23:201–205. doi: 10.1093/carcin/23.1.201. [DOI] [PubMed] [Google Scholar]
- 40.Dihlmann S, Kloor M, Fallsehr C, von Knebel Doeberitz M. Regulation of AKT1 expression by beta-catenin/Tcf/Lef signaling in colorectal cancer cells. Carcinogenesis. 2005;26:1503–1512. doi: 10.1093/carcin/bgi120. [DOI] [PubMed] [Google Scholar]
- 41.Luger K, Hansen JC. Nucleosome and chromatin fiber dynamics. Curr. Opin. Struct. Biol. 2005;15:188–196. doi: 10.1016/j.sbi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 42.Lorch Y, Maier-Davis B, Kornberg RD. Chromatin remodeling by nucleosome disassembly in vitro. Proc. Natl Acad. Sci. USA. 2006;103:3090–3093. doi: 10.1073/pnas.0511050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Latham JA, Dent SY. Cross-regulation of histone modifications. Nat. Struct. Mol. Biol. 2007;14:1017–1024. doi: 10.1038/nsmb1307. [DOI] [PubMed] [Google Scholar]
- 44.Cheung WL, Ajiro K, Samejima K, Kloc M, Cheung P, Mizzen CA, Beeser A, Etkin LD, Chernoff J, Earnshaw WC, et al. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell. 2003;113:507–517. doi: 10.1016/s0092-8674(03)00355-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.