Abstract
Human polynucleotide kinase/phosphatase (PNKP) is a dual specificity 5′-DNA kinase/3′-DNA phosphatase, with roles in base excision repair, DNA single-strand break repair and non-homologous end joining (NHEJ); yet precisely how PNKP functions in the repair of DNA double strand breaks (DSBs) remains unclear. We demonstrate that PNKP is phosphorylated by the DNA-dependent protein kinase (DNA-PK) and ataxia-telangiectasia mutated (ATM) in vitro. The major phosphorylation site for both kinases was serine 114, with serine 126 being a minor site. Ionizing radiation (IR)-induced phosphorylation of cellular PNKP on S114 was ATM dependent, whereas phosphorylation of PNKP on S126 required both ATM and DNA-PK. Inactivation of DNA-PK and/or ATM led to reduced PNKP at DNA damage sites in vivo. Cells expressing PNKP with alanine or aspartic acid at serines 114 and 126 were modestly radiosensitive and IR enhanced the association of PNKP with XRCC4 and DNA ligase IV; however, this interaction was not affected by mutation of PNKP phosphorylation sites. Purified PNKP protein with mutation of serines 114 and 126 had decreased DNA kinase and DNA phosphatase activities and reduced affinity for DNA in vitro. Together, our results reveal that IR-induced phosphorylation of PNKP by ATM and DNA-PK regulates PNKP function at DSBs.
INTRODUCTION
Exposure of cells to ionizing radiation (IR) results in the generation of complex forms of DNA damage, including damage to bases and cleavage of the DNA backbone to form DNA single strand breaks (SSBs). Two SSBs on opposite DNA strands approximately 6–10 bp apart results in the formation of a DNA double strand break (DSB), which is considered the most lethal form of DNA damage (1). The termini of IR-induced SSBs and DSBs frequently contain 3′-phosphate and 3′-phosphoglycolate groups (2) as well as 5′-hydroxyls (3,4) that must be restored to 3′-hydroxyls and 5′-phosphates before the DNA ends can be ligated. A key enzyme responsible for restoring ligatable end groups to IR-induced DNA strand breaks in mammalian cells is polynucleotide kinase/phosphatase (PNKP), a dual function enzyme with both 3′-DNA phosphatase and 5′-DNA kinase activities (5–8). Cells lacking PNKP are sensitive to IR and other DNA-damaging agents (9,10) and in humans PNKP mutation is associated with microcephaly and seizures (10). PNKP functions in multiple pathways for the repair of IR-induced DNA damage, including base excision repair (BER), DNA SSB repair and non-homologous end joining (NHEJ), making it an attractive therapeutic target (7,11,12).
NHEJ, the major pathway for the repair of IR-induced DSBs in mammalian cells, requires Ku70/80, DNA-dependent protein kinase catalytic subunit (DNA-PKcs), Artemis, XRCC4 (X-ray cross complementing gene 4), XLF (XRCC4-like factor) and DNA-ligase IV (13). PNKP enhances DNA-PK-dependent rejoining of DNA substrates containing 5′-hydroxyls in vitro (14) and interacts with XRCC4 in both a phosphorylation-dependent (15) and a phosphorylation-independent manner (16), providing possible mechanisms to recruit PNKP to DSBs in vivo. Moreover, knock-down of PNKP does not increase the radiation sensitivity of MO59J cells (17), which lack DNA-PKcs and have reduced levels of the related protein kinase ataxia-telangiectasia mutated (ATM) (18,19), suggesting that PNKP functions in the same pathway as DNA-PK and/or ATM. However, precisely how PNKP functions in the repair of IR-induced DSBs is unknown.
Here, we show that both DNA-PK and ATM phosphorylate PNKP in vitro. The major site, serine 114 (S114), is an SQ site located in a flexible linker region that separates the amino-terminal forkhead-associated (FHA) domain from the catalytic DNA phosphatase and DNA kinase domains of PNKP (20,21). A second SQ site, serine 126 (S126) is also phosphorylated in vitro by DNA-PK, but to a lesser extent. Using phosphospecific antibodies and metabolic labeling, we show that both S114 and S126 are phosphorylated in vivo in response to IR. IR-induced phosphorylation of S114 in vivo was ATM dependent, whereas phosphorylation of S126 was reduced by inhibition of ATM and/or DNA-PKcs. Cells expressing PNKP in which serines 114 and 126 were mutated to alanine (to ablate phosphorylation) or aspartic acid (to mimic phosphorylation) were modestly radiation sensitive. Furthermore, inhibition of DNA-PKcs and/or ATM reduced the amount of PNKP detected at DNA damage sites in vivo, suggesting that phosphorylation by ATM and/or DNA-PK regulates PNKP retention or turnover at DSBs. To explore further the effects of DNA-PK and ATM on PNKP function, we examined the effects of mutation of S114 and S126 to alanine or aspartic acid on the biochemical properties of PNKP. Mutation of S114 and S126 reduced PNKP kinase and phosphatase activities as well as the affinity of PNKP for dsDNA. Together, our studies suggest that phosphorylation of the flexible linker region of PNKP by ATM and/or DNA-PK regulates PNKP catalytic activity, its interaction with DNA and turnover and/or retention at sites of DNA damage.
MATERIALS AND METHODS
Cloning and purification of PNKP in bacteria
His-PNKP was expressed as described previously (8). To generate glutathione S transferase (GST)-tagged PNKP, a vector expressing full-length human PNKP with a C-terminal V5 tag (pcDNA3.1-PNK-V5), generously provided by Dr D. Durocher (University of Toronto) (15), was used as a template to amplify full-length human PNKP (GeneBank accession number: AF125807.1) from HeLa cells using primers 5′-BamH1 and 3′-XhoI (see Supplementary Data for primer sequences). The PCR product was cloned into the pGEX-6P-1 vector (GE Healthcare) at BamH1/Xho1 restriction sites to produce an N-terminal GST fusion protein. DNA sequences were verified by the University of Calgary DNA Sequencing Facility. GST-PNKP was expressed and purified in Escherichia coli BL21-DE3 as described previously for XLF (22). Where indicated, the GST tag was removed using PreScission Protease (GE Healthcare) according to the manufacturer's instructions.
In vitro DNA-PK phosphorylation assays
DNA-PKcs and Ku were purified from the nuclear salt wash of unirradiated HeLa cells or from human placenta as described previously (23,24). Unless otherwise indicated, phosphorylation reactions were carried out as described previously (22) and contained 1 µg purified, bacterially expressed PNKP in a final volume of 20 µl. To calculate the stoichiometry of in vitro phosphorylation, reactions were carried out with 0.25 mM ATP containing ~1 µCurie of 32P-γ-ATP. Radioactively labeled bands were excised from Coomassie blue-stained SDS–PAGE gels and radioactivity was determined by Cerenkov counting. The stoichiometry of phosphorylation was calculated from 32P-γ-ATP (in cpm/pmol ATP) and expressed as pmol of phosphate incorporated per pmol protein.
Identification of in vitro PNKP phosphorylation sites
Purified His-PNKP (0.5 µg) was incubated with DNA-PKcs (0.3 µg) and Ku (0.1 µg) and phosphorylated in vitro as described previously (25). Phosphoamino acid analysis and identification of phosphopeptides by mass spectrometry and radiochemical sequencing/Edman degradation was also carried out as described previously (25).
Generation of phosphorylation site mutations in GST-PNKP and PNKP-V5
Serine to alanine mutations at serines 114 and 126 (S114A and S126A, respectively) were generated by site-directed mutagenesis from the pGEX-6P-1-PNKP-wt plasmid using methods described previously (22) and primers S114A and S126A (see Supplementary Data for primer sequences). The double mutant (S114A-S126A) was generated using pGEX-6P-1-PNKP-S126A vector as template with the S114A primer. Alanine to aspartic acid mutations were generated as above using primers A114D and A126D (see Supplementary Data for details). The double mutant was generated using the pGEX-6P-1-PNKP-A114D vector as template with the A126D primer. The same primers were used to generate serine to alanine single and double mutations in the pcDNA3.1-V5 vector for mammalian expression (see below).
Generation of phosphospecific antibodies
A phosphospecific antibody recognizing S114 of PNKP was raised in sheep against the phospho-peptide: RTPESQPDTP, which corresponds to residues 110–119 of human PNKP. The phosphorylated serine is shown in bold and underlined. The peptide was coupled to KLH and BSA and affinity purified as described previously (25). An antibody to S126 was raised in rabbits to the phospho-peptide GTPLVSQDEK, which corresponds to residues 121–130 of human PNKP. Phosphospecific antibodies were purified as described previously (25).
Cell culture and irradiation
HeLa cells were grown in Dulbecco's Modified Eagle Medium (DMEM) containing 5% Hyclone III fetal bovine serum, containing 50 U/ml of penicillin and 50 µg/ml of streptomycin, and maintained at 37°C under an atmosphere of 5% CO2. BT/C3ABR cells were maintained in RPMI media plus 10% Hyclone III fetal bovine serum with antibiotics as described above. Where indicated, irradiation was carried out using a GammaCell 1000 137Cs source (MDS Nordion, Canada) as described previously (22). Cells were pretreated with the ATM inhibitor KU55933 (26) and/or the DNA-PK inhibitor NU7441 (27) at the concentrations indicated prior to irradiation.
In vitro ATM phosphorylation assays
ATM-proficient, human lymphoblastoid cells BT/C3ABR cells were irradiated with 10 Gy IR and whole cell detergent lysis extracts were prepared 30 min after irradiation. ATM was immunoprecipitated and assayed as described previously (28,29). Where indicated, in vitro kinase reactions contained 500 nM KU55933 (26) to inhibit ATM kinase activity.
Transient transfection
PNKP-V5 was transfected into HeLa cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommended conditions, except that half the amount of DNA and one-third the amount of transfection reagent was used. Unless otherwise indicated, cells were harvested 24 h post-transfection.
Stable expression of shRNA-resistant HA-tagged PNKP in the A549-derived PNKP shRNA knockdown cell line, Clone 13
A549 cells containing stable shRNA knock down of PNKP (clone 13) were described previously (9) and were maintained in DMEM/F12 media with 10% Hyclone III fetal bovine serum, 350 µg/ml G418, penicillin and streptomycin as above. To generate shRNA-resistant wt-PNKP, the bases shown in bold type in the shRNA target sequence (1392-1411: AGAGATGACGGACTCCTCT) were mutated using Quick Change II kit (Stratagene), to yield the sequence AGAAATGACCGATTCCTCT, which did not result in any changes in the amino acid sequence of PNKP. The cDNA (in mammalian expression vector pIRESpuro3, Clontech) was transfected into clone 13 cells using Lipofectamine 2000 as above. After 24 h, cells were placed under G418 (0.4 mg/ml) and puromycin (0.8 µg/ml) selection for 14 days. Individual colonies were expanded in selection media and the resultant cell lines were tested for the expression of HA-PNKP by immunoblotting with an antibody to HA (Sigma) following the manufacturer's recommendations. Cells re-expressing shRNA-resistant PNKP were maintained in medium containing 0.4 mg/ml G418 and 0.8 µg/ml puromycin in addition to penicillin and streptomycin as above.
Immunoprecipitation
For immunoprecipitation from HeLa cells transiently transfected with PNKP-V5 vectors, cells were lysed in NP-40 containing buffer (NETN) [150 mM NaCl, 0.2 mM EDTA, 50 mM Tris–HCl, pH 7.5, 1% (v/v) NP-40] containing 1 µM microcystin-LR, 0.2 µM phenylmethylsulfonyl fluoride, 2 µg/ml aprotinin, 2 µg/ml leupeptin and 2 µg/ml pepstatin A, with sonication. Protein concentrations were determined using the Detergent Compatible Protein Assay (BioRad) using BSA as a standard. PNKP was immunoprecipitated from 1 mg of total protein using anti-V5 IgG (Serotec) and Protein G-Sepharose beads (GE Healthcare). Precipitates were washed six times with 1 ml NETN buffer (containing protease inhibitors) per wash, and then analyzed by SDS–PAGE and immunoblot.
For immunoprecipitation from PNKP-depleted cells stably transfected with shRNA-resistant PNKP, cells were harvested and lysed as above, but immunoprecipitated with antibodies to HA (Sigma). Where indicated, immunoblots were also probed with antibodies to XRCC4 (Serotec), DNA ligase IV (Serotec), Mre11 (Novus), ATM (a gift from Dr Martin Lavin, Queensland Institute of Medical Research), Ku80 (in house) and DNA-PKcs (in house) as indicated. Blots were developed with ECL reagent (GE Healthcare), exposed to X-ray film (Fuji) and quantified using ImageQuant (GE Healthcare). In some experiments, immunoblots were imaged and quantified using a Fujifilm LAS-4000 Luminescent Image Analyzer (Fujifilm Corp., Tokyo).
Identification of in vivo phosphorylation sites by mass spectrometry
HeLa cells were transiently transfected with PNKP-V5 and PNKP was immunoprecipitated from 10 mg of total protein. Immunoprecipitates were washed six times with 1 ml of 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer (50 mM HEPES, pH 7.5, 40 mM NaCl, 2 mM EDTA) containing 0.5 M LiCl, 1% (v/v) Triton X-100 plus protease inhibitors as above, followed by a further 10 washes with 1 ml of PBS per wash. Phosphopeptides were identified by WEMB Inc. as described previously (22). Where indicated, beads were treated with 200 U lambda phosphatase (NEB) and incubated at 30°C for 30 min prior to SDS–PAGE and immunoblot.
Metabolic labeling
HeLa cells were transiently transfected with vectors expressing wild-type PNKP with a C-terminal V5 tag (PNKP-V5), or PNKP-V5 containing serine to alanine mutations at S114 or S114 plus S126 (PNKP-S114A-V5 and PNKP-S114A-S126A-V5, respectively) and incubated in phosphate-free media for 2 h. The medium was replaced with fresh phosphate-free media supplemented with 160 µCi 32P inorganic phosphate/ml media, and cells were returned to the incubator for 1 h. The cells were then lysed in NETN buffer containing phosphatase and protease inhibitors and PNKP was immunoprecipitated from 1.5 mg total protein as described above and resolved by SDS–PAGE. The gel was stained with Coomassie Brilliant Blue (Bio-Rad) and exposed to X-ray film (Fuji) for 36 h at −80°C.
Calculation of in vivo stoichiometry of phosphorylation
PNKP was phosphorylated in vitro using purified DNA-PK as above but with non-radioactive ATP. Known amounts of phosphorylated PNKP were run on SDS–PAGE along with PNKP that had been immunoprecipitated from irradiated cells and immunoblots were developed with phospho-specific antibodies to serine 114 and 126 as described above. Phosphorylation of PNKP in vivo was compared with phosphorylation of known amounts of in vitro phosphorylated PNKP after normalization for protein loading (total PNKP by western blot), stoichiometry of phosphorylation in vitro from 32P-labeling experiments (0.34–0.35 pmol phosphate/pmol protein, Supplementary Figure 1) and DNA-PK-mediated phosphorylation of serine 114 and 126 in vitro, by phospho-specific antibody cross-reactivity.
Clonogenic survival assays
Cells were serially diluted and plated at various concentrations ranging from 1 × 102 to 7 × 102 cells/plate. Twenty-four hours after plating, cells were irradiated with 0, 2, 4 or 6 Gy IR and incubated at 37°C for 10–14 days. Colonies were stained with crystal violet and counted on a Bantex 920A colony counter (American Bantex Corp., Burlingame, CA, USA).
Two-photon laser microirradiation
Cells were grown in DMEM/F12 (50/50) plus 10% fetal bovine serum on 35-mm glass bottom culture dishes (MatTek Corp., Ashland, MA, USA). The following day, cells were transfected with a DNA construct (PNKP cDNA cloned into the pCMV6-AC-mRFP vector Origene, Rockville, MD, USA) encoding for PNKP-mRFP expression under the control of the CMV promoter. Twenty-four hours after transfection, cells were incubated with Hoechst 33258 to a final concentration of 2 μg/ml for 20 min and then fed with fresh growth medium for 10 min prior to microirradiation. Where indicated, cells were incubated with 10 µM NU7441 and/or KU55933 for 2 h prior to microirradiation. For laser microirradiation, cells were placed on a 37°C heated stage of a Zeiss LSM510 NLO laser-scanning confocal microscope. Microirradiation was carried out using a near infrared titanium sapphire laser. In order to introduce damage within nuclei of individual cells, a 1.2-μm wide region was pre-defined and subsequently microirradiated with 20 iterations of a 750-nm laser line at 10% power using a Plan-Neofluar ×40/1.3 NA oil immersion objective. Fluorescence imaging of the mRFP moiety was recorded using excitation with a 543-nm He–Ne laser and a 559–634 nm band-pass filter. mRFP-tagged PNKP accumulation at the region of interest was compared with a non-irradiated region, and the accumulation of PNKP at sites of DNA damage was quantified as enrichment relative to unirradiated regions. Cells with low expression levels of PNKP-mRFP were selected and fluorescence intensity at regions of interest was quantified. The intensity was normalized so that the total cell intensity remained constant throughout the experiment. This compensates for photobleaching during acquisition. Images were then realigned using Image J software and subsequently fluorescence signals of the exported Tiff images were quantified using Metamorph software 6.0 (Molecular Devices, Sunnyvale, CA, USA).
In vitro PNKP kinase and phosphatase assays
In vitro PNKP kinase assays were carried out as described previously (16). Briefly, reaction mixtures containing kinase buffer [80 mM succinic acid (pH 5.5), 0.2 nmol of 24 mer ssDNA substrate (HO 24: 5′-GGCGCCCACCACCACTAGCTGGCC-3′) (Integrated DNA Technology), 3.3 pmol of 32P-ATP (Perkin-Elmer), 0.4 nmol unlabeled ATP, 10 mM MgCl2 and 500 ng of enzyme (final volume 50 µl)] were incubated at 37°C. From the reaction mixture, 4 µl samples were taken at several incubation times up to 10 min and mixed with 2 µl of sequencing gel loading dye (Fisher Scientific), boiled and run at 200 V on a 12% polyacrylamide sequencing gel (Bio-Rad) containing 7 M urea (EMD). Gels were scanned on a Typhoon 9400 variable mode imager (GE Healthcare) and the resulting bands were quantified using Image Quant 5.2 software (GE Healthcare).
For in vitro PNKP phosphatase assays, 5′-radiolabeled substrate was prepared by incubation at 37°C for 30 s of 120 pmol 3′-phosphorylated 20-mer DNA oligonucleotide (20P: 5′-ATTACGAATGCCCACACCGC-3′-phosphate) (Integrated DNA Technology) with 30 U of phosphatase-free T4 phage polynucleotide kinase (Roche) and 9.9 pmol γ32P-ATP (Perkin Elmer), in buffer A [50 mM Tris–HCl (pH 8.0), 10 mM MgCl2, 0.1 mM EDTA, 5 mM dithiothreitol, 0.1 mM spermidine] in a total volume of 72 µl. The sample was then heated at 95°C for 5 min to inactivate the T4 phage PNKP. To monitor phosphatase activity, 12 µl of the resulting 32P labeled substrate was mixed with 20 ng of human PNKP in buffer A (total volume 30 µl) and incubated at room temperature. Aliquots (4 µl) were taken at 0, 0.5, 1, 2 and 5 min intervals, mixed with 2 µl sequencing gel loading dye (Fisher Scientific), boiled and analyzed as above.
DNA binding assays
In vitro DNA binding assays were carried out by fluorescence quenching in the presence of 45 bp duplex DNA as described previously (16).
RESULTS
DNA-PK and ATM phosphorylate PNKP predominantly on serine 114 in vitro
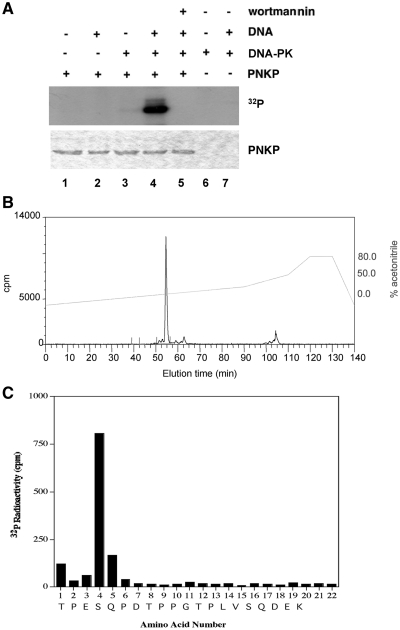
Given that PNKP has been implicated in NHEJ and the catalytic activity of DNA-PK is required for NHEJ (30), we first asked whether PNKP was an in vitro substrate for DNA-PK. His-tagged PNKP was expressed in bacteria, purified, then incubated with purified DNA-PKcs and Ku and phosphorylated under standard assay conditions (Figure 1A). The stoichiometry of phosphorylation was ~0.5 moles of phosphate per mole of PNKP (data not shown). To identify in vitro DNA-PK phosphorylation sites, phosphorylated His-tagged PNKP was extracted from the gel pieces, and, after reduction and alkylation, the protein was digested with trypsin and phosphopeptides were identified using mass spectrometry. A single phosphopeptide was detected which corresponded to S114 in the sequence TPESQPDTPPGTPLVSQDEK (the phosphorylated amino acid is shown in bold and underlined) (Figure 1B and C).
Figure 1.
Identification of DNA-PK phosphorylation sites in PNKP. (A) Purified His-tagged PNKP (0.5 µg) was incubated with or without DNA-PK (400 ng), calf thymus DNA (10 µg/ml) or 10 µM wortmannin, as indicated by the + or − symbols. The reaction was carried out at 30°C for 30 min and stopped by boiling in SDS sample buffer. Proteins were separated by electrophoresis on a 10% SDS polyacrylamide gel stained with Coomassie blue, and dried for autoradiography. (B) Purified PNKP, phosphorylated by DNA-PK, was separated by electrophoresis on an SDS polyacrylamide gel and then digested with trypsin as described in ‘Materials and methods’ section. The resulting 32P-labeled peptides were chromatographed on a Vydac 218TP54 C18 column (Separation Group, Hesperia, CA, USA) equilibrated in 0.1% (v/v) trifluoroacetic acid in water. The column was developed with a linear acetonitrile gradient (dashed diagonal line, right hand Y-axis) at 0.8 ml/min, and fractions of 0.4 ml were collected: >80% of the radioactivity applied to the column was recovered in the fractions. (C) An aliquot of the major 32P-labeled peptide derived from phosphorylated PNKP (Figure 1B) was covalently coupled to a Sequelon acrylamide membrane and analyzed on an Applied Biosystems 476A sequenator. 32P radioactivity was measured after each cycle of Edman degradation. In combination with MALDI-TOF MS, database searching against predicted PNKP tryptic peptides and phosphoamino acid analysis (data not shown) enabled the identification of the site of phosphorylation in the peptide. The putative amino acid sequence of this peptide, deduced from a combination of phosphoamino acid analysis, MS and solid-phase sequencing, is indicated at the bottom of the graph.
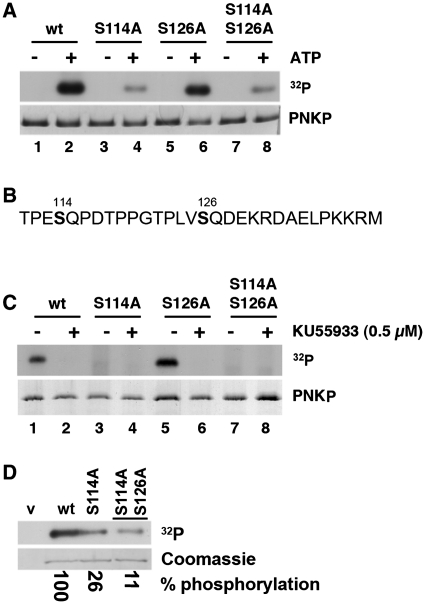
To determine whether S114 represented a major in vitro phosphorylation site, S114 was mutated to alanine (PNKP-S114A) and the protein was again phosphorylated by DNA-PK (Figure 2A). For these experiments, mutations were generated in GST-tagged PNKP and the GST tag was removed prior to phosphorylation. The stoichiometry of phosphorylation of wt-PNKP ranged from 0.34 to 0.35 pmol of phosphate per pmol PNKP (Supplementary Figure S1). Bands corresponding to phosphorylated PNKP were excised and quantified. PNKP in which S114 was replaced by alanine (PNKP-S114A) was phosphorylated to 56 ± 2% of wild-type (WT), confirming mass spectrometry results and indicating that S114 represents a major in vitro phosphorylation site (Figure 2A).
Figure 2.
PNKP is phosphorylated on serines 114 and 126 in vitro and in vivo. (A) Purified recombinant PNKP or PNKP containing serine to alanine mutations at either serines 114, 126 or both was incubated with purified DNA-PK either in the absence (−) or presence (+) of ATP. Reactions were stopped by the addition of SDS sample buffer and samples were run on SDS–PAGE. The top panel shows the resulting autoradiograph. The lower panel is the Coomassie blue-stained gel in the region corresponding to PNKP. To calculate stoichiometry, 32P-labeled bands were excised from the gel and the amount of radioactivity incorporated was determined by Cerenkov counting. 32P incorporation was normalized to total PNKP levels (determined by Coomassie staining) and expressed as percent of wild-type. The experiment was repeated three times and a representative result is shown. Phosphorylation of PNKP mutants S114A, S126A and S114A/S126A were 56 ± 2%, 89 ± 15% and 50 ± 8%, relative to wild-type, respectively. (B) Amino acid sequence of the linker region (amino acids 111–140) between the FHA and catalytic domain of PNKP. Serines 114 and 126 are shown in bold. (C) ATM was immunoprecipitated from irradiated human lymphoblastoid cells and immunoprecipitates were incubated with either wt-PNKP or PNKP containing serine to alanine mutations at serines 114 (S114A) and/or 126 (S126A) as indicated. Where indicated (+), the ATM inhibitor KU55933 was added to reactions prior to addition of ATP. The upper and lower panels correspond to the resulting autoradiogram and Ponceau red-stained nitrocellulose blot, respectively. (D) HeLa cells were transfected with vector alone (v, lane 1), wt-PNKP-V5 (wt, lane 2), PNKP-S114A-V5 (lane 3) or PNKP-S114A-S126A-V5 (lane 4). Cells were labeled with 32P-inorganic phosphate, then PNKP was immunoprecipitated and phosphorylation was determined by autoradiography (top panel) and quantitation after normalization to total PNKP levels (bottom panel). Quantitation as percent phosphorylation compared with wt-PNKP is shown below the panel.
S114 is located in a flexible linker region that connects the FHA domain of PNKP to the catalytic domain (21). Also present in this 35 amino acid linker is a second SQ site, S126 (Figure 2B). To determine whether DNA-PK also phosphorylated this residue in vitro, S126 was mutated to alanine and the mutant protein (PNKP-S126A) was incubated with DNA-PK under standard phosphorylation conditions. Conversion of S126 to alanine decreased DNA-PK mediated phosphorylation of PNKP to 89 ± 15% (Figure 2A), indicating that, under these conditions, S126 represents only ~10% of the total in vitro phosphorylation by DNA-PK. Mutation of both S114 and S126 decreased phosphorylation by ~50% (Figure 2A, lane 8), suggesting the presence of additional in vitro PNKP phosphorylation sites, but these were not pursued further.
To determine whether S114 and S126 were also substrates for ATM, ATM was immunoprecipitated from irradiated human cells and incubated with purified PNKP as substrate. Where indicated, reactions contained the ATM inhibitor KU55933 (26). ATM also phosphorylated PNKP in vitro and again the major site was S114 (Figure 2C).
Serine 114 is a major in vivo PNKP phosphorylation site
To determine whether PNKP was phosphorylated at these sites in vivo, HeLa cells were transiently transfected with a vector encoding PNKP with a C-terminal V5 tag (PNKP-V5) (a kind gift from Drs Durocher and Koch, University of Toronto). Under these conditions, PNKP-V5 was expressed ~12-fold above endogenous PNKP levels (Supplementary Figure 2) and was predominantly nuclear in localization by immunofluoresence staining (data not shown). PNKP was immunoprecipitated from transiently transfected cells and phosphorylation sites were identified by mass spectrometry as described in ‘Materials and Methods’. Both S114 and S126 were detected as in vivo phosphorylation sites (Supplementary Figure 3).
To determine whether S114 and S126 were major sites of in vivo phosphorylation, HeLa cells were transiently transfected with either wild-type PNKP (PNKP-V5), or PNKP-V5 containing serine to alanine mutations at serine 114 (PNKP-S114A-V5) or both serines 114 and 126 (PNKP-S114A-S126A-V5). Cells were then metabolically labeled using 32P-labeled inorganic phosphate as described in ‘Materials and methods’ section and PNKP was immunoprecipitated using antibodies to V5, resolved by SDS–PAGE, and exposed to X-ray film. Metabolic labeling confirmed that PNKP is phosphorylated in vivo under these conditions (Figure 2D). Mutation of S114 to alanine reduced PNKP phosphorylation by ~70%, while mutation of S114 and S126 to alanine reduced phosphorylation by a further 10–15% (Figure 2D). Technical difficulties prevented us from exposing cells to IR during these experiments and therefore the observed phosphorylation likely represents endogenous phosphorylation as well as phosphorylation induced by DNA damage due to incubation with 32P-inorganic phosphate.
PNKP is phosphorylated on serine 114 in an ATM-dependent manner in vivo in response to IR
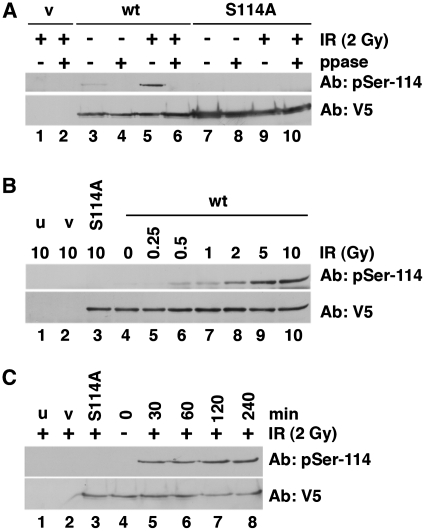
To further characterize the phosphorylation of PNKP in vivo, we generated phospho-specific antibodies to phosphorylated S114 and S126. Both antibodies recognized purified wt-PNKP in a phosphorylation–dependent manner and were specific for their respective phosphorylation sites in vitro and in vivo (Supplementary Figure 4A and B). HeLa cells were transiently transfected with PNKP-V5 or PNKP-S114A-V5, then, after 24 h, cells were irradiated (2 Gy) and 30 min later, PNKP was immunoprecipitated as described above. Immunoprecipitates were either run directly on SDS–PAGE (Figure 3A, odd numbered lanes) or incubated in the presence of lambda phosphatase (ppase) prior to SDS–PAGE (Figure 3A, even numbered lanes). After SDS–PAGE, immunoblots were probed with either the phospho-specific antibody to S114 (Ab: pSer-114) or an antibody to V5 (to detect total transfected PNKP) (Figure 3A). The results confirm that PNKP is phosphorylated on S114 in vivo in response to IR. Phosphorylation of S114 was detected at 0.5 Gy and the amount of phosphorylation increased up to 10 Gy (Figure 3B). Phosphorylation was detected as early as 30 min and persisted up to 4 h post IR (Figure 3C).
Figure 3.
IR induces phosphorylation of PNKP on S114. (A) HeLa cells were transfected with vector alone (v, lanes 1 and 2), wt-PNKP (wt, lanes 3–6) or PNKP-S114A (S114A, lanes 7–10). Where indicated (+), cells were irradiated with 2 Gy and after 0.5 h, cells were lysed and PNKP immunoprecipitated as described in ‘Materials and Methods’ section. Where indicated (even numbered lanes), samples were incubated with lambda phosphatase (ppase) prior to SDS–PAGE. Immunoblots were probed either with the phospho-specific antibody to S114 (upper panel) or V5 (lower panel) for total PNKP protein. (B) Cells were either untreated (u, lane 1), or transfected with vector alone (v, lane 2), PNKP-S114A-V5 (lane 3) or wt-PNKP-V5 (lanes 4–10) as indicated. Cells were irradiated and harvested after 1 h. Samples in lanes 1–3 and 10 received 10 Gy; the sample in lane 4 was unirradiated; samples in lanes 5–9 received 0.25, 0.5, 1, 2, or 5 Gy as indicated. Immunoblots were probed for phosphorylation of PNKP on S114 or total PNKP (V5) as indicated. (C) Cells were either untreated (u, lane 1), or transfected with vector alone (v, lane 2), PNKP-S114A-V5 (lane 3) or wt-PNKP-V5 (lanes 4–10) as in (B). Cells were either unirradiated (lane 4), or irradiated at 2 Gy and harvested after 4 h (lanes 1–3 and 8) or after 0.5, 1 or 2 h (lanes 5–7). PNKP was immunoprecipitated and analyzed by SDS–PAGE and immunoblot as described above.
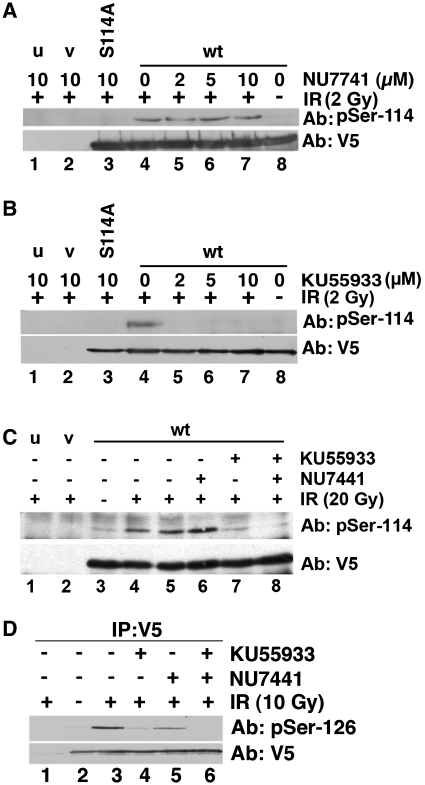
Given that both DNA-PK and ATM phosphorylate S114 in vitro, we next asked whether phosphorylation of PNKP on S114 required DNA-PK or ATM in vivo. Cells were transfected with either vector alone, PNKP-V5, or PNKP-S114A-V5 as described above, but prior to irradiation, cells were pretreated with either the DNA-PK inhibitor NU7441 (27) or the ATM inhibitor KU55933 (26) at doses experimentally determined to be specific for the respective kinase (Supplementary Figure 5). Significantly, pre-treatment with the DNA-PK inhibitor NU7441 had no effect on IR-induced phosphorylation of PNKP on S114 (Figure 4A), while pre-treatment with the ATM inhibitor KU55933 virtually abolished phosphorylation (Figure 4B).
Figure 4.
IR-induced phosphorylation on serine 114 is ATM dependent, while phosphorylation at serine 126 is both ATM and DNA-PK dependent. (A) Cells were either untransfected (u, lane 1), or transfected with vector (v, lane 2), PNKP-S114A-V5 (lane 3) or wt-PNKP-V5 (lanes 4–8). Twenty-four hours after transfection, cells were pretreated with 10 µM NU7441 (DNA-PK inhibitor) (lanes 1, 2, 3 and 7), DMSO control (lane 4), 2 µM NU7441 (lane 5) or 5 µM NU7441 (lane 6) for 2 h prior to irradiation (2 Gy, lanes 1–7). The sample in lane 8 was not irradiated. Thirty minutes after irradiation, PNKP was immunoprecipitated and analyzed by western blot using the phospho-specific antibody to S114 or V5 for total PNKP as above. (B) Cells were pre-treated with the indicated concentrations of the ATM inhibitor KU55933 for 2 h, then irradiated (2 Gy) and analyzed exactly as described above. PNKP was immunoprecipitated and observed by western blotting with either the phospho-specific antibody to S114 or V5 for total PNKP. As above, u and v represent untransfected (lane 1) and vector (lane 2). S114A refers to the S114A mutant of PNKP (lane 3). (C) Cells were pre-treated with either 10 µM NU7441 or 5 µM KU55933 and either unirradiated (lane 3) or irradiated at 20 Gy (lanes 1, 2, 4–8) and harvested after 1 h (lane 4) or 4 h (lanes 1, 2, 5–8). PNKP was immunoprecipitated and analyzed by western blotting with either the phosphospecific antibody to S114 or V5 for total PNKP as above. (D) Cells were pre-treated with NU7441 and/or KU55933 as in (C), then irradiated (10 Gy). PNKP was immunoprecipitated after 30 min and analyzed by western blot as described, but using a phospho-specific antibody to serine 126.
Although, these results eliminated a role for DNA-PK under these experimental conditions, we considered it possible that DNA-PK might play a role at higher doses of IR or at longer times after irradiation. Indeed, we have previously observed that IR-induced phosphorylation of the PIKK target Artemis is largely ATM dependent at low doses of IR (<2 Gy) and short times (<2 h) after IR, while at higher doses (10–20 Gy) and longer times (4 h), phosphorylation becomes in part DNA-PK dependent (R. Ye, S. Hiebert and S.P. Lees-Miller, unpublished data). However, phosphorylation of PNKP on S114 remained highly ATM dependent 4 h post 20 Gy IR (Figure 4C).
We next examined phosphorylation of S126 in response to IR. HeLa cells were transiently transfected with PNKP-V5, pretreated with NU7441 or KU55933 then irradiated with 10 Gy. Phosphorylation at S126 was also IR inducible (Figure 4D, lane 3) and pre-incubation with the ATM inhibitor KU55933 reduced phosphorylation by ~90% (Figure 4D, lane 4). However, pre-treatment with the DNA-PK inhibitor reduced phosphorylation by ~65% (Figure 4D, lane 5), while pre-treatment with both inhibitors abolished phosphorylation (Figure 4D, lane 6). Thus, both ATM and DNA-PK contribute to IR-induced phosphorylation of PNKP on S126.
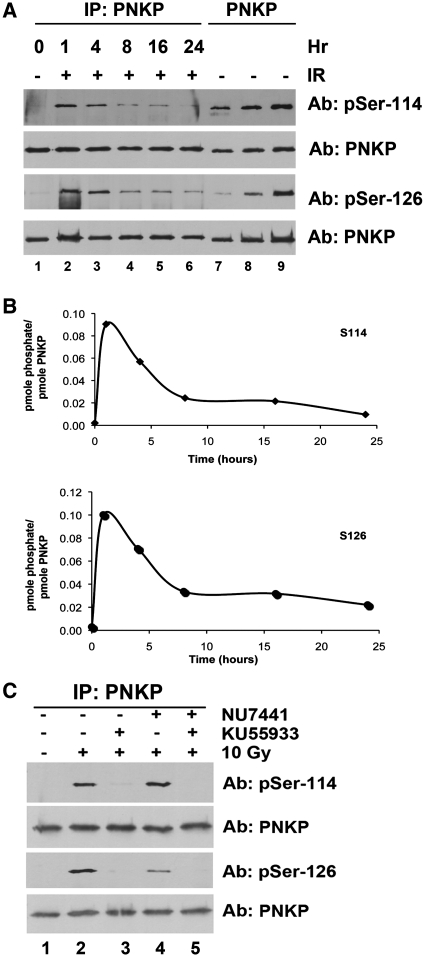
The experiments described above were carried out in cells that had been transiently transfected with wt-PNKP. We have previously observed that transient transfection can promote DNA-PK-dependent phosphorylation of some substrates in the absence of additional DNA damage (22). To eliminate any possible effects of transient transfection, we examined PNKP phosphorylation in PNKP-depleted cells (clone 13) into which shRNA-resistant HA-tagged wt-PNKP had been stably introduced (see ‘Materials and methods’ section). In these cells, PNKP was expressed at approximately 5-fold higher levels than endogenous PNKP in the parental A549 cell line (Supplementary Figure S6A), and PNKP expression was predominantly nuclear (Supplementary Figure S6B). Cells stably expressing wt-PNKP were irradiated (10 Gy) and PNKP was immunoprecipitated 1, 4, 8, 16 and 24 h later. Immunoprecipitates were run on SDS–PAGE and immunoblots were analysed for phosphorylation at serine 114, serine 126 and total PNKP (Figure 5A). As found for cells transiently transfected with PNKP, IR induced phosphorylation of PNKP at serines 114 and 126 within 1 h. Interestingly, phosphorylation at both sites started to diminish by 4 h post IR and returned to close to background levels by 24 h (Figure 5A and B).
Figure 5.
IR-induced phosphorylation of PNKP at S114 is transient and ATM dependent while phosphorylation of S126 is transient and requires both DNA-PK and ATM. (A) PNKP knockdown cells (C13) stably expressing shRNA-resistant wt-PNKP were unirradiated (lane 1) or irradiated (10 Gy) and harvested 1, 4, 8, 16 or 24 h post IR. PNKP was immunoprecipitated and immunoblots were probed for phosphorylation at S114, S126 or total PNKP as indicated. (B) Quantitation of immunoblots shown in (A). (C) PNKP knockdown cells (C13) stably expressing shRNA-resistant wt-PNKP were pre-treated with 10 µM KU55933, 10 µM NU7441 or both for 60 min prior to treatment with 10 Gy IR. Cells were harvested after 60 min and PNKP was immunoprecipitated using an antibody to HA, resolved by SDS–PAGE and blotted for phosphoserine 114, phosphoserine 126 or total PNKP as indicated.
To estimate the stoichiometry of in vivo phosphorylation, known amounts of purified PNKP that had been phosphorylated in vitro by DNA-PK were run on the same gels (Figure 5A, lanes 7–9). The stoichiometry of phosphorylation at both sites at the time of maximum phosphorylation (1 h) was ~0.1 pmol of phosphate/pmol of protein indicating that ~10% of the total PNKP is phosphorylated in response to DNA damage (Figure 5A and B).
To determine whether ATM or DNA-PK was responsible for IR-induced phosphorylation in these cells, cells were pre-incubated with either NU7441 or KU5593 or both inhibitors together. As found for phosphorylation of PNKP after transient transfection, PNKP phosphorylation at both S114 and S126 was virtually abolished by pre-incubation with either the ATM inhibitor alone or both the ATM and DNA-PK inhibitors together (Figure 5C, lanes 3 and 5), while incubation with the DNA-PK inhibitor alone reduced phosphorylation of S126 by ~70% (Figure 5C), consistent with phosphorylation of S114 being ATM dependent, while both ATM and DNA-PK contribute to phosphorylation of S126.
Mutation of S114 and S126 to phospho-mutants or phospho-mimics imparts modest radiosensitivity to cells
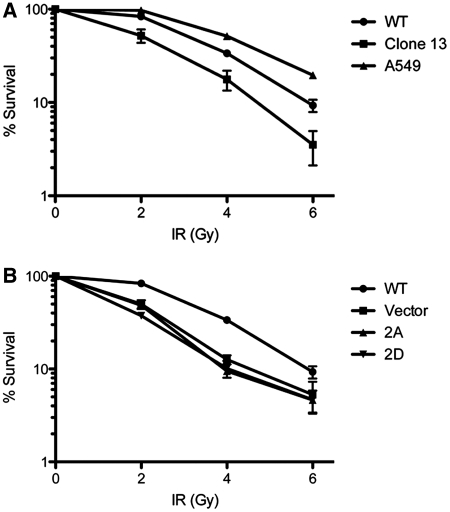
To further examine the effects of PNKP phosphorylation on the cellular response to IR, shRNA-resistant PNKP containing A or D mutations at both S114 and S126 (PNKP-2A and PNKP-2D, respectively) was re-introduced into the shRNA-depleted cell line (clone 13) and stable cell lines were isolated. Sensitivity to IR was determined using clonogenic/colony formation assays. Cells lacking PNKP protein were radiosensitive as reported previously (9) and stable expression of wt-PNKP partially restored radioresistance (Figure 6A). However, expression of PNKP in which both S114 and S126 had either been ablated (PNKP-2A) or mutated to the phospho-mimic aspartic acid (PNKP-2D) did not restore radiation resistance and were as radiosensitive as the PNKP-depleted cells (Figure 6B).
Figure 6.
Mutation of S114 and S126 to either alanine or aspartic acid induces a radiosensitive phenotype. (A) A549 cells (triangles), A549 cells with stable downregulation of PNKP (clone 13 cells, squares) and cells containing stably re-expressed shRNA-resistant wt-HA-PNKP (circles) were plated in triplicate and allowed to adhere for 24 h. Cells were then treated with 0, 2, 4, or 6 Gy IR and incubated for 10–14 days. Colonies were stained with crystal violet and counted. The surviving fraction and the SEM were calculated. Data are representative of three separate experiments. (B) Cells containing stably re-expressed wt-HA-PNKP (circles), HA-PNKP-S114A-S126A (2A, triangles), HA-PNKP-S114D-S126D (2D, inverted triangles) or a stably introduced vector control (squares) were plated and analyzed as above.
Mutation of S114 or S126 to a phospho-mutant or a phospho-mimic does not alter the interaction of PNKP with XRCC4
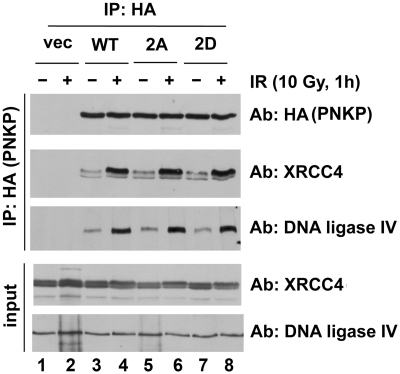
PNKP interacts with XRCC4, which in turn interacts with DNA ligase IV (31), providing a possible mechanism for recruitment of PNKP to DSBs in NHEJ. To test whether phosphorylation might disrupt the interaction of PNKP with XRCC4, cells stably expressing PNKP-WT, PNKP-2A or PNKP-2D were irradiated, PNKP was immunoprecipitated and immunoblots were probed for the presence of XRCC4 and DNA ligase IV. Although the total amount of immunoprecipitated PNKP remained constant, the amount of XRCC4 and DNA ligase IV that interacted with PNKP increased significantly after IR (Figure 7, lanes 4–9 and Supplementary Figure S7). However, mutation of the two PNKP phosphorylation sites did not affect the IR-enhanced interaction of PNKP with XRCC4 and DNA-ligase IV (Figure 7 and Supplementary Figure S7), suggesting that the enhanced interaction of PNKP and XRCC4 after IR does not require phosphorylation at these sites. Immunoblots were also probed for the presence of ATM, DNA-PKcs and Ku, but these proteins did not co-immunoprecipitate with PNKP under these conditions (Supplementary Figure S7).
Figure 7.
IR enhances the interaction between PNKP, XRCC4 and DNA ligase IV. C13 PNKP knockdown cells stably re-expressing vector (vec, lanes 1 and 2), shRNA-resistant wt HA-PNKP (lanes 3 and 4), HA-PNKP-S114A-S126A (2A, lanes 5 and 6) or HA-PNKP-S14D-S126D (2D, lanes 7 and 8) were either unirradiated (−) or irradiated 10 Gy (+) and harvested after 60 min. HA-PNKP was immunoprecipitated and resolved by SDS–PAGE as described above. Immunoblots were probed for HA (for PNKP), XRCC4 or DNA Ligase IV as indicated. Whole cell extract of 50 µg (input) was also analyzed for XRCC4 and DNA ligase IV levels (bottom two panels). See also Supplementary Figure S7.
PNKP recruitment to DSBs is impaired in cells deficient for DNA-PKcs and ATM
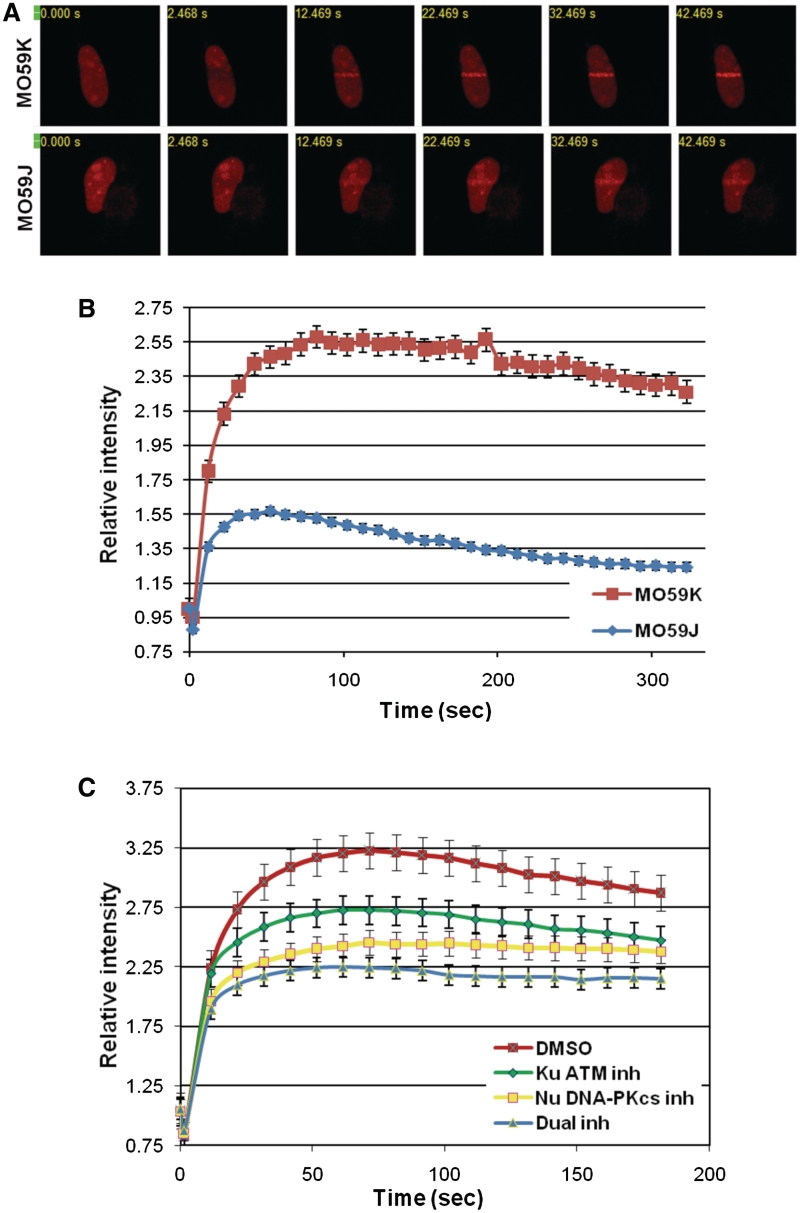
We have previously shown that depletion of PNKP in the DNA-PKcs/ATM-defective cell line MO59J (18,19) does not increase radiation sensitivity, suggesting an epistatic relationship between PNKP and DNA-PKcs and/or ATM (17). To explore further the relationship between PNKP, DNA-PKcs and ATM, RFP-tagged PNKP was expressed in MO59J cells or the DNA-PKcs/ATM proficient cell line MO59K (isolated from the same tumor specimen as MO59J), and DNA damage was introduced using two-photon laser microirradiation. PNKP was recruited to sites of DNA damage rapidly (<1 min) and with similar kinetics in both MO59J and MO59K cells, suggesting that DNA-PKcs and ATM are not required for actual recruitment of PNKP to sites of DNA damage in vivo. However, considerably less PNKP was recruited in MO59J cells, suggesting that DNA-PKcs and/or ATM might play a role in the retention of PNKP at sites of damage (Figure 8A and B). To explore this possibility further, RFP-tagged PNKP was expressed in HeLa cells and cells were pre-treated with either KU55933, NU7441 or both inhibitors together prior to laser microirradiation. Inhibition of either DNA-PKcs or ATM or both together significantly reduced the amount of PNKP at laser microirradiation-induced sites of damage without altering the initial time of recruitment (Figure 8C), suggesting that phosphorylation of PNKP by ATM and DNA-PK regulates the retention or turnover of PNKP at DNA damage sites in vivo.
Figure 8.
Recruitment of PNKP to sites of DNA damage induced by two-photon laser microirradiation is impaired in DNA-PK/ATM-deficient cells. (A) MO59K and MO59J glioma cells transiently expressing PNKP-mRFP were subjected to laser microirradiation (as described in ‘Materials and Methods’ section). Recruitment kinetics of PNKP-mRFP at laser-induced DNA damage sites in individual cells was monitored in real time. (B) The amount of the fluorescently tagged protein at tracks of DNA damage was quantified (n = 11 cells for each data set; error bars represent ± SEM). The red line represents MO59K and the blue line represents MO59J cells. (C) HeLa cells expressing PNKP-mRFP were incubated with DMSO (red squares), the ATM inhibitor KU55933, 10 µM (green diamonds), the DNA-PK inhibitor NU7741, 10 µM (yellow squares) or both inhibitors together, 10 µM (blue triangles) for 2 h prior to microirradiation. Samples were quantified as in (A) and (B).
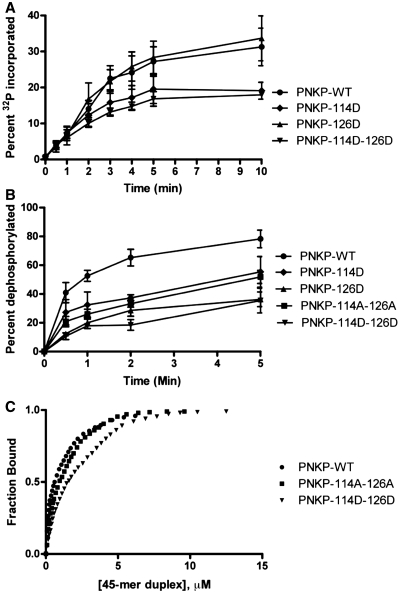
Both ablation and mimicry of S114 and S126 reduce PNKP activity in vitro
To investigate whether phosphorylation of PNKP on S114 and S126 might affect its biochemical properties, wild-type (wt) PNKP or PNKP-containing alanine (to ablate phosphorylation) or aspartic acid (to mimic phosphorylation) at serines 114 and 126 were expressed and purified from E. coli and PNKP DNA kinase and DNA phosphatase activities were determined as described in ‘Materials and Methods’ section. Mutation of either S114 or S126 to A had no effect on PNKP kinase activity (data not shown). Similarly, mutation of S126 to D (phosphomimic) had no effect on PNKP kinase activity, whereas mutation of S114 to D (S114D) or mutation of both S114 and S126 to D (PNKP-114D-126D) reduced by ~40% the ability of PNKP to phosphorylate the 5′-end of ssDNA (Figure 9A). Thus, replacement of S114 with a phosphomimic negatively affects PNKP kinase activity, suggesting that phosphorylation at S114 may regulate PNKP kinase activity in vitro (see also ‘Discussion’ section). Mutation of S114 to D also reduced PNKP phosphatase activity toward 3′-phosphorylated ssDNA by ~25% (Figure 9B, triangles). Mutation of S126 to D had a greater effect on PNKP phosphatase activity, reducing activity by 50% (Figure 9B, inverted triangles), while mutation of both S114 and S126 to A or D reduced PNKP phosphatase activity by 25 and 50%, respectively (Figure 9B). Thus, mutations that result in either ablation or phosphomimicry of S114 and S126 negatively affect PNKP phosphatase activity, with replacement of both S114 and S126 by aspartic acid having the most significant effect.
Figure 9.
Effect of mutation of PNKP S114 and S126 on in vitro PNKP kinase, phosphatase and DNA binding activities. Wild-type (wt) PNKP or PNKP with serine to aspartic acid (phosphomimic) or alanine (to ablate phosphorylation) mutations at positions 114 and/or 126 were expressed and purified from E. coli and assayed for DNA kinase (A) or DNA phosphatase (B) activity as described in ‘Materials and Methods’ section. Circles, wt PNKP; diamonds, PNKP-S114D; triangles, S126D; inverted triangles, S114D–S126D (2D). The concentrations of PNKP and DNA in the kinase assays (A) were 0.175 and 4 µM, respectively, and in the phosphatase assays (B), 11.6 nM and 0.667 µM, respectively. (C) wt PNKP and PNKP containing S to A or S to D mutations at serines 114 and/or 126 were prepared and purified from E. coli as in (A) and (B) and assayed for binding to 45 bp duplex DNA by fluorescence quenching as described in ‘Materials and Methods’ section. Results for wt (circles), 2A (squares) and 2D (inverted triangles) are shown.
We also asked whether mutation of serines 114 and 126 affected the ability of PNKP to interact with dsDNA. Wt-PNKP bound DNA with a Kd of 0.55 ± 0.05 µM (Figure 9C). Kd values for PNKP with S114A, S114D, S126A and S126D mutations were 0.75 ± 0.05, 0.9 ± 0.05, 1.2 ± 0.1 and 1.10 ± 0.1 µM, respectively (data not shown). PNKP with mutation of both 114 and 126 to either A or D had Kd values of 1.0 ± 0.1 and 1.8 ± 0.1 µM, respectively (Figure 9C). Thus, ablation or phosphomimicry of PNKP phosphorylation sites adversely affected the affinity of PNKP for DNA, with the double phosphomimic mutation (PNKP-S114D-S126D) having the strongest effect on DNA binding.
DISCUSSION
PNKP interacts with XRCC4 (15) and enhances DNA-PK-dependent rejoining of NHEJ substrates containing 5′-hydroxyl groups in vitro (14). Moreover, PNKP-deficient cells are radiosensitive (9). These observations, combined with its enzymatic properties, have suggested a role for PNKP in NHEJ; yet precisely how PNKP functions in DSB repair has not been determined. Here we show that both DNA-PK and ATM phosphorylate PNKP on SQ sites located in a 35 amino acid flexible linker region, which tethers the FHA domain to the catalytic phosphatase and kinase domains (20). This linker, which corresponds to amino acids 111–145 of human PNKP, was disordered in the crystal structure (21) and its function is unknown. The major in vitro PNKP phosphorylation site for both DNA-PK and ATM was S114, with S126 being a minor site. Our results also suggest that DNA-PK may phosphorylate additional sites in PNKP, but these were not identified in this study (Figure 2A).
In vivo, IR-induced phosphorylation of PNKP on S114 was largely ATM dependent, suggesting that ATM may directly phosphorylate S114 in vivo in response to IR. S126 was also phosphorylated in vivo in response to IR and with similar kinetics to S114. Using the highly selective ATM and DNA-PK inhibitors KU55933 and NU7441, we show that the major S126 kinase was again ATM; however, NU7441 also reduced PNKP S126 phosphorylation, suggesting that both ATM and DNA-PK contribute to phosphorylation at this site in irradiated cells. Indeed, our results suggest that ATM kinase activity may be required for DNA-PK to phosphorylate PNKP at S126 (Figures 4C and 5C), suggesting cross-talk between ATM and DNA-PKcs in response to IR.
The stoichiometry of phosphorylation of PNKP on S114 and 126 in response to IR was ~0.1 pmol phosphate/pmol protein, or ~10% (Figure 5). We have calculated that a similar fraction of DNA-PKcs is phosphorylated in response to IR (32), consistent with only a fraction of the total population of NHEJ proteins recruited to and phosphorylated at DSBs. IR-induced phosphorylation of PNKP at both S114 and S126 was rapid, reaching a maximum within 1 h but transient, diminishing to 50% within ~5 h and to pre-irradiation levels by 24 h (Figure 5B). Since the total amount of PNKP was unchanged, these data suggest that IR-induced phosphorylation of PNKP is regulated by protein phosphatases.
Recent phosphoproteomics studies have also identified serines 114 and 126 as IR- (33,34) and UV- (35) inducible sites of phosphorylation. Our study confirms and extends these results, showing that phosphorylation at S114 is ATM dependent after IR while both ATM and DNA-PK contribute to phosphorylation of S126. Metabolic labeling experiments revealed that mutation of both serine 114 and 126 to alanine reduced phosphorylation by only 85%, suggesting that PNKP is phosphorylated on additional sites in vivo. Our preliminary mass spectrometry analysis provided evidence for phosphorylation of T39 and T111 (data not shown) and T111 was independently identified as a UV-inducible phosphorylation site in a proteomics screen (35). Moreover, T118 and T122 have also been identified as DNA damage-inducible in vivo phosphorylation sites in phosphoproteomics screens (33–35) (see also PhosphoSitePlus®, www.phosphosite.org, search term PNKP); thus, remarkably, all five of the phosphorylatable amino acids (Ser or Thr) in the 35 amino acid flexible linker of PNKP (Figure 2B), can be phosphorylated in response to DNA damage in vivo. It is interesting to note that the related protein, Aprataxin-PNKP-like factor (APLF), which has also been implicated in NHEJ, is phosphorylated by ATM on an SQ site located between the FHA module and the carboxy-terminal portion of the protein (36). Together, these studies suggest that the flexible linker region plays important roles in regulating the function of the PNKP family of proteins.
We show that PNKP is recruited to sites of microlaser-induced DNA damage rapidly (within 20s), consistent with its role in the rapid repair of modified DNA termini (Figure 8). Initial recruitment of PNKP to sites of DNA damage did not require DNA-PKcs or ATM; however, the amount of PNKP at DNA damage sites was reduced in cells lacking DNA-PKcs and ATM protein or in which their activity had been inhibited, suggesting that phosphorylation of PNKP by ATM and DNA-PK could play a role in the retention or turnover of PNKP at sites of DNA damage.
Consistent with previous reports, shRNA PNKP-depleted cells were modestly radiosensitive (9). shRNA PNKP-depleted cells stably expressing shRNA-resistant wt-PNKP partially restored radiation resistance, while expression of PNKP in which serines 114 and 126 had been replaced by alanine did not. Although this result is consistent with phosphorylation of PNKP at S114 and S126 being required for survival after IR, cells expressing PNKP with aspartic acid substitutions (PNKP-2D) were as radiation sensitive as PNKP-depleted cells. It is possible that aspartic acid at these sites may be a poor phosphomimic or that amino acid substitution within the linker region might affect PNKP function, perhaps by altering its conformation. Alternatively, the finding that IR-induced phosphorylation of PNKP is transient (Figure 5) suggests that PNKP phosphorylation may play a regulatory role required for the correct coordination of PNKP function in DNA repair. It is therefore possible that constitutive phosphorylation, as mimicked by the PNKP-2D mutant, could have deleterious consequences for the cell, preventing complementation.
Our experiments also reveal that IR significantly enhances the interaction of XRCC4 and DNA ligase IV with PNKP. However, interaction of XRCC4 with PNKP-2A and PNKP-2D was also enhanced after IR, indicating that phosphorylation at these sites is not required for the enhanced interaction of PNKP and XRCC4 after IR, and the mechanism for this enhanced interaction is under investigation. Our results do not exclude the possibility that PNKP phosphorylation at these sites may regulate the interaction with other proteins after DNA damage and this possibility is also under investigation.
PNKP protein containing mutations at S114 and S126 was also assayed for the effects on PNKP kinase, phosphatase and DNA binding activities in vitro. Although mutation of S114 or S126 to aspartic acid had little effect on the initial rate of PNKP kinase activity, mutation of S114 to aspartic acid either in the context of PNKP-S114D or PNKP-2D, resulted in a clear plateau in PNKP activity at later times (Figure 9A). This suggests that the S114D mutation, which mimics phosphorylation at S114, may increase product inhibition of PNKP kinase activity, which in turn suggests that phosphorylation at S114 may increase the affinity of PNKP for 5′-phosphorylated termini. We speculate that this increased affinity could serve to protect the exposed ends of a partially repaired DSB from degradation, analogous to the ‘passing the baton’ model for BER (37). This effect was specific for S114, suggesting that phosphorylation at S114 and S126 might have distinct effects on PNKP kinase activity. In contrast, mutation of either S114 or S126 to D, as well as mutation of both sites to either alanine or aspartic acid (PNKP-2A and PNKP-2D, respectively), reduced phosphatase activity suggesting different effects of phosphorylation on PNKP kinase and phosphatase activities.
It is also interesting to note that mutation of both the identified phosphorylation sites to aspartic acid (PNKP-2D) had significant effects on multiple PNKP activities in vitro, reducing the ability of PNKP to either remove a phosphate group from the 3′-end of the DNA substrate or to add a phosphate group to the 5′-OH termini of DNA and reducing the affinity of PNKP for DNA approximately 4-fold. Although the linker region does not appear to be in close contact with the two active sites of PNKP (21), it is possible that the mutation of the two serines to aspartic acid residues causes sufficient conformational change to the protein to reduce both of its enzymatic activities. On the other hand, ablation of the two phosphorylation sites by mutation to alanine reduced PNKP phosphatase activity but not kinase activity. We speculate that because mutation of serine to alanine can be considered more benign than mutation to the negatively charged aspartic acid, any conformational change may only affect the phosphatase domain of PNKP to which the linker is directly attached, rather than the more distal kinase domain.
Together, our results suggest that PNKP is recruited to sites of DNA damage presumably through its interactions with XRCC4 and that IR-induced phosphorylation of PNKP by ATM and/or DNA-PK may promote the retention of PNKP at sites of DNA damage. In addition, our in vitro experiments suggest that modification of the flexible linker region of PNKP has significant effects on PNKP activity, and that phosphorylation of the linker may be important for PNKP function.
While this article was under review, Shiloh and colleagues reported ATM-dependent phosphorylation of PNKP on S114 and S126 in response to the radiomimetic, neocarzinostatin (NCS) (38). The authors showed that PNKP-depleted cells were modestly sensitive to DNA-damaging agents and that re-introduction of PNKP lacking S114 and/or S126 only partially restored resistance to X-rays and NCS. Cells unable to undergo phosphorylation at S114 or S126 were also defective in DSB repair and showed impaired phosphatase activity in cell extracts. Our results confirm a role for ATM in phosphorylation of PNKP at S114 and extend these studies to show that DNA-PK is also involved in the phosphorylation of S126. Our study also reveals that phosphorylation of both sites after IR is transient, suggesting regulation by protein phosphatases. The ability of phosphorylation mutants to restore resistance to DNA damage was modest in the study by Segal-Raz et al. (38) and the difference between our results and theirs may reflect differences in the cell lines used. Together, our studies reveal new roles for ATM and DNA-PK in the regulation of PNKP function in human cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institutes of Health Research (grant numbers 13639 to S.P.L.-M. and 15385 to M.W.); Alberta Cancer Foundation (grant number 23124 to M.W. and S.P.L.-M.). Funding for open access charge: Alberta Cancer Foundation grant 23124.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Rulin Zhang of WEMB Inc. for mass spectrometry analysis of in vivo phosphorylation sites, Drs Dan Durocher and Anne Koch, University of Toronto for the PNKP-V5 vector, Dr Graeme Smith, KuDOs Pharmaceuticals Inc. for providing ATM and DNA-PK inhibitors; Hillary McLaughlin, University of Dundee for phosphopeptide synthesis and generation of phospho-specific antibodies to serine 114, ProSci Inc. for generation of phospho-specific antibodies to serine 126 and Dr Sun, Cross Cancer Institute, Edmonton for assistance with two-photon microscopy experiments. M.J.H. is an Alberta Innovates—Health Solutions Research Senior Scholar. S.P.L.-M. is a Research Scientist of Alberta Innovates—Health Solutions and holds the Engineered Air Chair in Cancer Research at the Southern Alberta Cancer Research Institute.
REFERENCES
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2 edn. Washington, DC: ASM Press; 2005. [Google Scholar]
- 2.Henner WD, Rodriguez LO, Hecht SM, Haseltine WA. gamma Ray induced deoxyribonucleic acid strand breaks. 3′ Glycolate termini. J. Biol. Chem. 1983;258:711–713. [PubMed] [Google Scholar]
- 3.Lennartz M, Coquerelle T, Bopp A, Hagen U. Oxygen–effect on strand breaks and specific end-groups in DNA of irradiated thymocytes. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1975;27:577–587. doi: 10.1080/09553007514550611. [DOI] [PubMed] [Google Scholar]
- 4.Buchko GW, Weinfeld M. Influence of nitrogen, oxygen, and nitroimidazole radiosensitizers on DNA damage induced by ionizing radiation. Biochemistry. 1993;32:2186–2193. doi: 10.1021/bi00060a009. [DOI] [PubMed] [Google Scholar]
- 5.Jilani A, Ramotar D, Slack C, Ong C, Yang XM, Scherer SW, Lasko DD. Molecular cloning of the human gene, PNKP, encoding a polynucleotide kinase 3′-phosphatase and evidence for its role in repair of DNA strand breaks caused by oxidative damage. J. Biol. Chem. 1999;274:24176–24186. doi: 10.1074/jbc.274.34.24176. [DOI] [PubMed] [Google Scholar]
- 6.Karimi-Busheri F, Weinfeld M. Purification and substrate specificity of polydeoxyribonucleotide kinases isolated from calf thymus and rat liver. J. Cell. Biochem. 1997;64:258–272. [PubMed] [Google Scholar]
- 7.Weinfeld M, Mani RS, Abdou I, Aceytuno RD, Glover JN. Tidying up loose ends: the role of polynucleotide kinase/phosphatase in DNA strand break repair. Trends Biochem. Sci. 2011;36:262–271. doi: 10.1016/j.tibs.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karimi-Busheri F, Daly G, Robins P, Canas B, Pappin DJ, Sgouros J, Miller GG, Fakhrai H, Davis EM, Le Beau MM, et al. Molecular characterization of a human DNA kinase. J. Biol. Chem. 1999;274:24187–24194. doi: 10.1074/jbc.274.34.24187. [DOI] [PubMed] [Google Scholar]
- 9.Rasouli-Nia A, Karimi-Busheri F, Weinfeld M. Stable down-regulation of human polynucleotide kinase enhances spontaneous mutation frequency and sensitizes cells to genotoxic agents. Proc. Natl Acad. Sci. USA. 2004;101:6905–6910. doi: 10.1073/pnas.0400099101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen J, Gilmore EC, Marshall CA, Haddadin M, Reynolds JJ, Eyaid W, Bodell A, Barry B, Gleason D, Allen K, et al. Mutations in PNKP cause microcephaly, seizures and defects in DNA repair. Nat. Genet. 2010;42:245–249. doi: 10.1038/ng.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allinson SL. DNA end-processing enzyme polynucleotide kinase as a potential target in the treatment of cancer. Future Oncol. 2010;6:1031–1042. doi: 10.2217/fon.10.40. [DOI] [PubMed] [Google Scholar]
- 12.Bernstein NK, Karimi-Busheri F, Rasouli-Nia A, Mani R, Dianov G, Glover JN, Weinfeld M. Polynucleotide kinase as a potential target for enhancing cytotoxicity by ionizing radiation and topoisomerase I inhibitors. Anticancer Agents Med. Chem. 2008;8:358–367. doi: 10.2174/187152008784220311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem. J. 2009;417:639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chappell C, Hanakahi LA, Karimi-Busheri F, Weinfeld M, West SC. Involvement of human polynucleotide kinase in double-strand break repair by non-homologous end joining. EMBO J. 2002;21:2827–2832. doi: 10.1093/emboj/21.11.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koch CA, Agyei R, Galicia S, Metalnikov P, O'Donnell P, Starostine A, Weinfeld M, Durocher D. Xrcc4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. EMBO J. 2004;23:3874–3885. doi: 10.1038/sj.emboj.7600375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mani RS, Yu Y, Fang S, Lu M, Fanta M, Zolner AE, Tahbaz N, Ramsden DA, Litchfield DW, Lees-Miller SP, et al. Dual modes of interaction between XRCC4 and polynucleotide kinase/phosphatase: implications for nonhomologous end-joining. J. Biol. Chem. 2010;285:37619–37629. doi: 10.1074/jbc.M109.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karimi-Busheri F, Rasouli-Nia A, Allalunis-Turner J, Weinfeld M. Human polynucleotide kinase participates in repair of DNA double-strand breaks by nonhomologous end joining but not homologous recombination. Cancer Res. 2007;67:6619–6625. doi: 10.1158/0008-5472.CAN-07-0480. [DOI] [PubMed] [Google Scholar]
- 18.Lees-Miller SP, Godbout R, Chan DW, Weinfeld M, Day RS, III, Barron GM, Allalunis-Turner J. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science. 1995;267:1183–1185. doi: 10.1126/science.7855602. [DOI] [PubMed] [Google Scholar]
- 19.Chan DW, Gately DP, Urban S, Galloway AM, Lees-Miller SP, Yen T, Allalunis-Turner J. Lack of correlation between ATM protein expression and tumour cell radiosensitivity. Int. J. Radiat. Biol. 1998;74:217–224. doi: 10.1080/095530098141591. [DOI] [PubMed] [Google Scholar]
- 20.Bernstein NK, Hammel M, Mani RS, Weinfeld M, Pelikan M, Tainer JA, Glover JN. Mechanism of DNA substrate recognition by the mammalian DNA repair enzyme, polynucleotide kinase. Nucleic Acids Res. 2009;37:6161–6173. doi: 10.1093/nar/gkp597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernstein NK, Williams RS, Rakovszky ML, Cui D, Green R, Karimi-Busheri F, Mani RS, Galicia S, Koch CA, Cass CE, et al. The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase. Mol. Cell. 2005;17:657–670. doi: 10.1016/j.molcel.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 22.Yu Y, Mahaney BL, Yano K, Ye R, Fang S, Douglas P, Chen DJ, Lees-Miller SP. DNA-PK and ATM phosphorylation sites in XLF/Cernunnos are not required for repair of DNA double strand breaks. DNA Repair. 2008;7:1680–1692. doi: 10.1016/j.dnarep.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan DW, Mody CH, Ting NS, Lees-Miller SP. Purification and characterization of the double-stranded DNA-activated protein kinase, DNA-PK, from human placenta. Biochem. Cell. Biol. 1996;74:67–73. doi: 10.1139/o96-007. [DOI] [PubMed] [Google Scholar]
- 24.Goodarzi AA, Lees-Miller SP. Biochemical characterization of the ataxia-telangiectasia mutated (ATM) protein from human cells. DNA Repair. 2004;3:753–767. doi: 10.1016/j.dnarep.2004.03.041. [DOI] [PubMed] [Google Scholar]
- 25.Douglas P, Sapkota GP, Morrice N, Yu Y, Goodarzi AA, Merkle D, Meek K, Alessi DR, Lees-Miller SP. Identification of in vitro and in vivo phosphorylation sites in the catalytic subunit of the DNA-dependent protein kinase. Biochem. J. 2002;368:243–251. doi: 10.1042/BJ20020973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 27.Zhao Y, Thomas HD, Batey MA, Cowell IG, Richardson CJ, Griffin RJ, Calvert AH, Newell DR, Smith GC, Curtin NJ. Preclinical evaluation of a potent novel DNA-dependent protein kinase inhibitor NU7441. Cancer Res. 2006;66:5354–5362. doi: 10.1158/0008-5472.CAN-05-4275. [DOI] [PubMed] [Google Scholar]
- 28.Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 29.Ye R, Bodero A, Zhou BB, Khanna KK, Lavin MF, Lees-Miller SP. The plant isoflavenoid genistein activates p53 and Chk2 in an ATM-dependent manner. J. Biol. Chem. 2001;276:4828–4833. doi: 10.1074/jbc.M004894200. [DOI] [PubMed] [Google Scholar]
- 30.Kurimasa A, Kumano S, Boubnov NV, Story MD, Tung CS, Peterson SR, Chen DJ. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol. Cell. Biol. 1999;19:3877–3884. doi: 10.1128/mcb.19.5.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Critchlow SE, Bowater RP, Jackson SP. Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr. Biol. 1997;7:588–598. doi: 10.1016/s0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- 32.Douglas P, Cui X, Block WD, Yu Y, Gupta S, Ding Q, Ye R, Morrice N, Lees-Miller SP, Meek K. The DNA-dependent protein kinase catalytic subunit (DNA-PKcs) is phosphorylated in vivo on threonine 3950, a highly conserved amino acid in the protein kinase domain. Mol. Cell. Biol. 2007;27:1581–1591. doi: 10.1128/MCB.01962-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bennetzen MV, Larsen DH, Bunkenborg J, Bartek J, Lukas J, Andersen JS. Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response. Mol. Cell. Proteomics. 2010;9:1314–1323. doi: 10.1074/mcp.M900616-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 35.Stokes MP, Rush J, Macneill J, Ren JM, Sprott K, Nardone J, Yang V, Beausoleil SA, Gygi SP, Livingstone M, et al. Profiling of UV-induced ATM/ATR signaling pathways. Proc. Natl Acad. Sci. USA. 2007;104:19855–19860. doi: 10.1073/pnas.0707579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macrae CJ, McCulloch RD, Ylanko J, Durocher D, Koch CA. APLF (C2orf13) facilitates nonhomologous end-joining and undergoes ATM-dependent hyperphosphorylation following ionizing radiation. DNA Repair. 2008;7:292–302. doi: 10.1016/j.dnarep.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 37.Prasad R, Shock DD, Beard WA, Wilson SH. Substrate channeling in mammalian base excision repair pathways: passing the baton. J. Biol. Chem. 2010;285:40479–40488. doi: 10.1074/jbc.M110.155267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segal-Raz H, Mass G, Baranes-Bachar K, Lerenthal Y, Wang SY, Chung YM, Ziv-Lehrman S, Strom CE, Helleday T, Hu MC, et al. ATM-mediated phosphorylation of polynucleotide kinase/phosphatase is required for effective DNA double-strand break repair. EMBO Rep. 2011;12:713–719. doi: 10.1038/embor.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.