Abstract
Zinc-finger recombinases (ZFRs) are chimaeric proteins comprising a serine recombinase catalytic domain linked to a zinc-finger DNA binding domain. ZFRs can be tailored to promote site-specific recombination at diverse ‘Z-sites’, which each comprise a central core sequence flanked by zinc-finger domain-binding motifs. Here, we show that purified ZFRs catalyse efficient high-specificity reciprocal recombination between pairs of Z-sites in vitro. No off-site activity was detected. Under different reaction conditions, ZFRs can catalyse Z-site-specific double-strand DNA cleavage. ZFR recombination activity in Escherichia coli and in vitro is highly dependent on the length of the Z-site core sequence. We show that this length effect is manifested at reaction steps prior to formation of recombinants (binding, synapsis and DNA cleavage). The design of the ZFR protein itself is also a crucial variable affecting activity. A ZFR with a very short (2 amino acids) peptide linkage between the catalytic and zinc-finger domains has high activity in vitro, whereas a ZFR with a very long linker was less recombination-proficient and less sensitive to variations in Z-site length. We discuss the causes of these phenomena, and their implications for practical applications of ZFRs.
INTRODUCTION
Site-specific recombinases are enzymes that promote programmed DNA rearrangements by breaking and rejoining DNA strands at specific sequence loci. Many site-specific recombinases belong to one of two large groups of enzymes called the serine recombinases and the tyrosine recombinases. These two groups are structurally unrelated and promote recombination by different mechanisms (1). Site-specific recombinases have become essential tools for the experimental manipulation of transgenic DNA sequences within the cells of living organisms. The bacteriophage tyrosine recombinase Cre, the yeast tyrosine recombinase FLP and the bacteriophage ϕC31 serine integrase have been the enzymes most commonly used for these purposes to date, but many other microbial recombinases have been shown to have potential utility (2). The recombination sites for these enzymes (typically ~30–40 bp) are normally incorporated into the design of a transgenic construct which is introduced into the organism being studied. This need for exogenous recombination sites restricts the current applications of site-specific recombination to genetically modified organisms. There has therefore been much interest in the development of methods to reprogramme recombinases to act on natural genomic sequences (3).
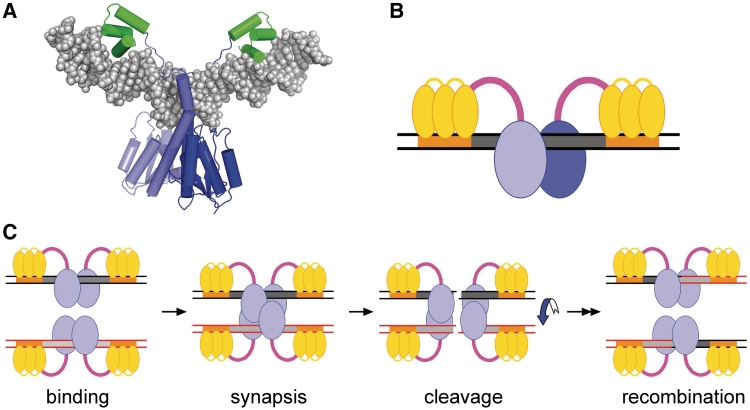
‘Directed evolution’ methods have successfully produced re-targeted recombinase variants (4–8). However, this strategy might not be applicable to targets that differ very substantially from the natural site for the progenitor recombinase (9). An alternative approach, pioneered in this lab, is to direct the recombinase catalytic domain to the chosen target site by linking it to a DNA binding domain which is engineered to recognize an adjacent sequence motif. Initially, we used a catalytic domain derived from the serine recombinase Tn3 resolvase, linked to a zinc-finger DNA binding domain from the mouse transcription factor Zif268 (10) (Figure 1). These zinc-finger recombinases (ZFRs; previously referred to as Z-resolvases) were shown to promote efficient site-specific recombination in Escherichia coli (10). The ‘Z-sites’ recombined by the ZFRs each consisted of a central ‘core’ sequence from binding site I of the Tn3 recombination site res, flanked by 9 bp motifs recognized by the zinc-finger domains (Figure 1B). ZFRs can be modified to target a wide range of different Z-sites, by substituting the zinc-finger domain with modified altered-specificity versions, by mutagenizing the resolvase catalytic domain to alter or reduce its specificity, and by using catalytic domains from other serine recombinases (3,11–13). Tn3 resolvase-derived ZFRs are predicted to bind to Z-sites as dimers (Figure 1B), but might be monomeric in solution (14), and thus could recognize asymmetric Z-sites as on-site heterodimers (12,13). Their mode of target recognition can thus be compared to that of zinc-finger nucleases (15,16).
Figure 1.
ZFR structure and activity. (A) Crystal structure (1GDT) of a γδ resolvase dimer bound to DNA (res site I). The catalytic domains are blue, and the DNA binding domains are green. (B) Cartoon of a ZFR bound to a Z-site. The catalytic domains are blue, the zinc-finger domains are yellow, and the linker peptides between the two domains of each subunit are magenta. The Z-site core sequence is grey, and the motifs bound by the zinc-finger domains are orange. (C) Hypothetical pathway for ZFR-mediated site-specific recombination. Following binding to the Z-sites and synapsis of two sites, the four ZFR subunits each catalyse cleavage of one DNA strand, forming two double-strand breaks with a ZFR covalently linked to each end (the covalent links are not shown). A 180° rotation of one pair of DNA ends and its attached ZFRs relative to the other pair, followed by re-ligation of the broken ends, completes recombination.
Except for a preliminary in vitro experiment (10), all analysis of ZFR activity to date has been in vivo (in E. coli and in human cell lines) (10,12,13,17–19). However, a more detailed understanding of the design, function and mechanism of these enzyme systems is essential if they are to be established as practical tools for manipulation of genomic DNA sequences. Many unanswered questions remain. For example, the design of the linker between the ZFR recombinase and zinc-finger domains (i.e. its length and sequence) may still be far from optimal; the effects of the Z-site length and sequence on activity and specificity are still not well understood; the possible correlation between Z-site core sequence length and the most effective ZFR linker length has not been investigated in any detail; and the steps in the reaction pathway that are affected by ZFR and Z-site design are not known. Furthermore, the known properties of activated serine recombinases suggest that ZFRs may catalyse undesirable side-reactions (such as accumulation of DNA strand breaks) to some extent (20); optimized design of the system components might minimize these problems and thus improve ZFR utility in vivo. These (and other) questions will be best addressed by studying the properties of purified ZFRs in vitro. Here, we report the establishment of an in vitro system for ZFR-mediated recombination, and use it to show how specific reaction steps are affected by ZFR design and Z-site core sequence length.
MATERIALS AND METHODS
Plasmids and in vivo recombination
Plasmids encoding ZFRs, for assays in E. coli or for protein overexpression, were constructed as described previously (10). Recombination substrate plasmids for assays in E. coli and in vitro were constructed as described (10). The Z-site names denote the length of the core sequence between the two 9 bp Zif268-binding motifs (Figure 2B); for example, Z22 has a 22 bp core sequence. In this work, the core sequence is exactly as in Tn3 res site I (21), with the centre of site I at the centre of the Z-site, unless stated otherwise. Plasmid DNA for in vitro assays was purified with a Qiagen midiprep kit. Full sequences of all plasmids are available on request.
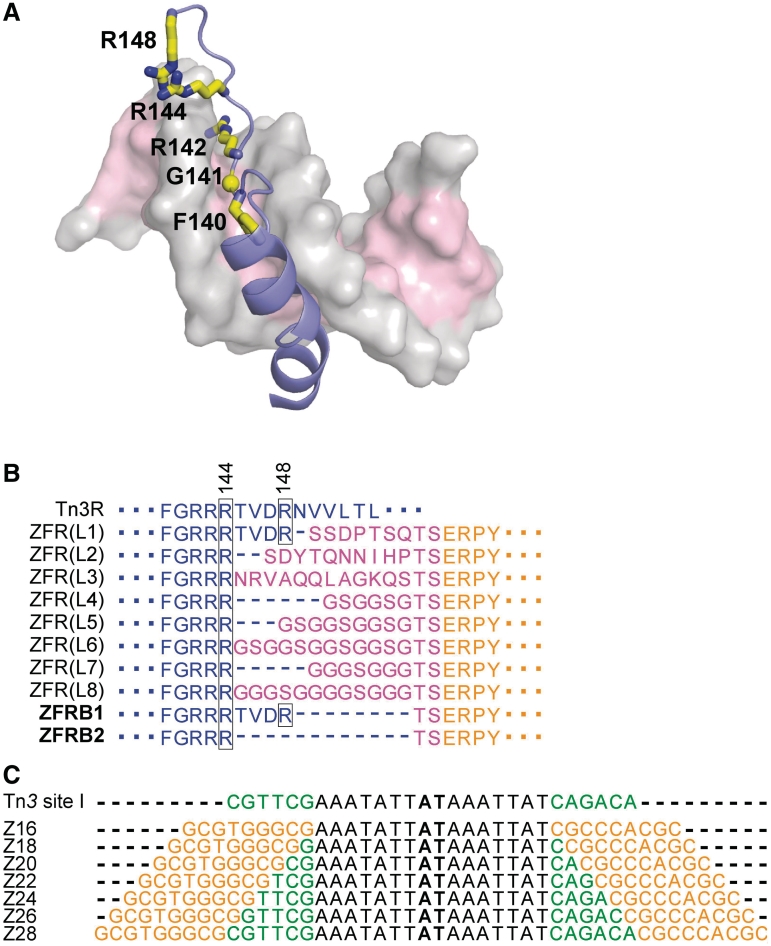
Figure 2.
Variable features of ZFR-mediated recombination. (A) Structure of the ‘arm’ region of resolvase (residues 120–148 are shown) bound to DNA (data are from the crystal structure 1GDT). The protein backbone is shown as cartoon, and the DNA in semi-transparent surface representation. Sidechains of F140, R142, R144 and R148 are shown as sticks, and the α-carbon of G141 is shown as a sphere. (B) Alignment showing the linker sequences of ZFRs tested (and all found to be recombination-proficient) by Akopian et al. (10), and the new ZFRs reported here (names in bold). Resolvase sequence is in blue, linker sequence is in magenta, and Zif268 sequence is in orange. Variants of ZFRB1 are described in the ‘Results’ section. Resolvase residues R144 and R148 are boxed. (C) The recombination sites (Z-sites) used in this study, aligned with site I of Tn3 res (only the top strands are shown). The motifs bound by the Zif268 domains are in orange. The motifs in site I bound by the resolvase C-terminal domain are in green. The central AT dinucleotide of each site is in bold.
Assays for ZFR-mediated recombination in E. coli were essentially as described (13). To determine the extent of recombination, plasmid DNA was isolated. Following growth of transformants on MacConkey agar indicator plates, the cells were recovered from each plate in 1 ml of L-broth. One microlitre of the suspension was used to inoculate 10 ml of L-broth, and this culture was incubated overnight at 37°C with ampicillin and kanamycin selection. Plasmid DNA was prepared using a Qiagen miniprep kit, and analysed by agarose gel electrophoresis.
The method for selection of ZFRs with an enhanced rate of recombination in E. coli was adapted from Burke et al. (22). The coding sequence for ZFRB1 (see ‘Results’ section) was subjected to PCR mutagenesis and then cloned to create a library of mutant expression plasmids. The selection involved a plasmid pStr(Z22), which encodes a strA gene conferring sensitivity to streptomycin flanked by Z22 sites. The Z22 site sequences are as described in Akopian et al. (10). Deletion of strA leads to streptomycin resistance. E. coli cells containing pStr(Z22) were transformed with the library DNA, and expression plasmids were isolated from streptomycin-resistant colonies that appeared at early time points.
ZFR purification
The purification procedure was adapted from that used for Tn3 resolvase mutants, and was essentially as described (20), except that Zn2+ was added to the buffer for refolding the protein after denaturing ion exchange chromatography. A full protocol is in Supplementary Data.
In vitro recombination and cleavage assays
In a typical recombination reaction, 6 µl of ZFR, diluted in a buffer containing 20 mM Tris–HCl (pH 7.5), 1 mM DTT, 1 M NaCl and 50% glycerol to a concentration of ~4 µM, was added to 60 µl of a solution of supercoiled substrate plasmid DNA (25 µg/ml) in a buffer containing 50 mM Tris–HCl (pH 8.2) and 10 mM MgCl2. Note the absence of chelating agents such as EDTA. The mixture was kept at 37°C for 1 h, then the reaction was stopped by heating at 70°C for 10 min. The sample was divided into three 22 µl aliquots which were either untreated, or digested with restriction enzymes, or nicked with DNase I, before separation by agarose gel electrophoresis. Procedures for DNase I nicking and gel electrophoresis were as described (20). The gels were 1.2% w/v agarose except for nicked samples, which were run on 0.7% gels. Excess protein and any recombinase subunits covalently linked to the DNA were destroyed by addition of loading buffer to all samples (20% of the final sample volume), which contained 100 mM Tris–HCl (pH 7.5), 50% v/v glycerol, 0.5% w/v SDS, 1 mg/ml protease K and 0.5 mg/ml bromophenol blue. Assays for ZFR-mediated cleavage followed a similar procedure except that the solution prior to ZFR addition contained plasmid DNA (25 µg/ml), 50 mM Tris–HCl (pH 8.2) and 40% v/v ethylene glycol. Reactions were stopped as above, then SDS + protease K loading buffer was added and the products were separated on 1.2% agarose gels.
Binding/synapsis assays
The procedure was modified from Olorunniji et al. (20). The gel-purified 50 nt oligonucleotide which was to be the Z-site top strand was 32P-phosphorylated at the 5′-end using T4 kinase, and was then annealed with an equimolar amount of unlabelled bottom strand. Diluted ZFR (2.2 µl) was added to 20 µl samples containing 100 nM Z-site DNA, 20 mM Tris–HCl (pH 7.5), 10 µM zinc acetate, 20 µg/ml poly(dI/dC) and 4% w/v Ficoll. The final ZFR concentration was ~800 nM (or as indicated otherwise). The samples were cooled in ice for 10 min, then loaded onto an 8% polyacrylamide gel (30:0.8 acrylamide: bisacrylamide). The gel contained 1× TB buffer (89 mM Tris base, 89 mM boric acid), plus 10% v/v glycerol. The running buffer was 1× TB. Gels were pre-run for 30 min at 200 V (~11 V/cm) and 4°C. Samples were then loaded and the gels were run for a further 4.5 h at 200 V under the same conditions, without buffer recirculation. Gels were dried, and bands were visualized by phosphor-imaging.
Molecular modelling
Models of ZFRs bound to Z-sites with different core sequence lengths were built in PyMol (23) using crystal structures of a γδ resolvase dimer–DNA complex [1GDT; (24)], a synaptic γδ resolvase–DNA intermediate [1ZR4; (25)] and a Zif268 zinc-finger domain–DNA complex [1AAY; (26)]. The required core sequence length was achieved by superimposition of appropriate deoxyribose C-1′ atoms in the region of DNA overlap between the resolvase and Zif268 structures.
RESULTS
Redesigned ZFRs
The first ZFRs (10) utilized the catalytic domain of ‘NM resolvase’, an activated Tn3 resolvase variant with six mutations [R2A E56K G101S D102Y M103I Q105L; (22)]. This domain, from the N-terminus to residue R144 or R148, was followed by a linker of 8–14 amino acids, then the Zif268 DNA binding domain [starting at residue E2, in the domain as used for crystallography (26)] (Figure 2B). The C-terminal cut-off point of the resolvase domain was chosen so as to include all the residues implicated in catalysis and subunit interactions, and most (−R144) or all (−R148) of the ‘arm region’ of resolvase [residues 121–147; (24)]. The arm region interacts with the minor groove near the centre of the recombination site (Figure 2A), and might therefore be important for efficient catalysis. The linkers between the recombinase and zinc-finger domains (Figure 2B) were made quite long and flexible, because it had been thought that conformational plasticity in this part of the protein might be desirable. However, it turned out that the length and sequence of these linkers had surprisingly small effects on ZFR-mediated recombination in E. coli (10), so we speculated that a shorter linker might be tolerated or even advantageous.
We tested two new ZFRs with a short linker (Figure 2B). ZFRB1 consists of NM resolvase sequence from the N-terminus to residue R148, followed by the 2-amino acid linker TS, then the Zif268 DNA binding domain starting at residue E2. ZFRB2 is identical to ZFRB1 except that the NM resolvase sequence is four amino acids shorter, ending at residue R144. The linker sequence TS [also used at the C-terminal ends of the linkers of Akopian et al. (10)] was chosen because it should be quite flexible, consisting of two relatively small hydrophilic residues, and allows a useful SpeI restriction site to be introduced into the DNA sequence.
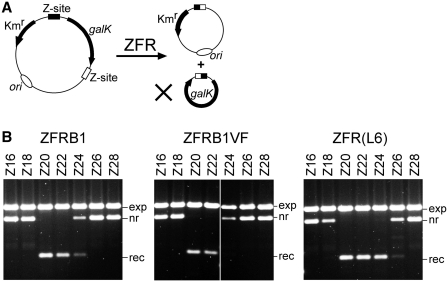
The new ZFRs were tested first in E. coli. The substrate plasmids each have two identical Z-sites flanking a galK marker gene. The Z-site core sequence length was varied from 16 to 28 bp in 2 bp increments, by putting sequence from the centre of Tn3 res site I between the Zif268 binding motifs (Figure 2C). Thus, the longest site tested, Z28, has a complete copy of site I (28 bp) at its centre. Strains containing substrate plasmids were transformed with a ZFR expression plasmid, and transformants were selected on MacConkey agar indicator plates. Deletion of the galK gene by ZFR-mediated recombination (Figure 3A) results in pale-coloured colonies, whereas galK+ colonies are red. The colony colour gives a visual indication of recombination activity, but to estimate activity more accurately we recovered the plasmid DNA (see ‘Materials and Methods’ section). ZFRB2 was completely inactive on all substrates, but ZFRB1 was recombination-proficient; pGal(Z20) and pGal(Z22) were completely resolved, pGal(Z24) was partially resolved, and the other plasmids were not resolved at all (Figure 3B).
Figure 3.
ZFR-mediated recombination at Z-sites in E. coli. (A) The assay used to observe ZFR-mediated recombination in E. coli. Deletion of a galK marker gene causes a reduction in plasmid size and a change in colony colour on MacConkey indicator plates from red to pale. (B) Gels showing plasmid DNA recovered from E. coli cells after ZFR expression. Each substrate plasmid contained two identical Z-sites as cartooned in Figure 3A. The Z-site sequences are shown in Figure 2C. The ZFRs are described in the ‘Results’ section and Figure 2B. The annotation of the bands is as follows: exp, ZFR expression plasmid; nr, non-recombinant (substrate) plasmid; rec, recombinant plasmid (galK segment deleted).
For the in vitro studies described below, we purified the following proteins:
ZFRB1 (see above).
ZFRB1SA, which is identical to ZFRB1 except for a mutation of the active site serine residue (S10) to alanine which renders the protein catalytically inactive.
ZFRB1WT, which is identical to ZFRB1 except that the recombinase domain is that of wild-type Tn3 resolvase, not the activated resolvase mutant NM. This protein has no recombination activity on Z-sites in E. coli (data not shown).
ZFRB1VF, identical to ZFRB1 except for a mutation V107F in the resolvase domain. This mutant was selected for fast recombination activity in an E. coli assay (see ‘Materials and Methods’ section). When tested on the pGalK substrates, ZFRB1VF activities were similar to those of ZFRB1 (Figure 3B).
ZFR(L6). This protein has the highest activity in E. coli of the variants created by Akopian et al. (10). It has a long (14 amino acids) linker between the resolvase and Zif268 domains (Figure 2B). In E. coli, ZFR(L6) had recombination activity on pGalK substrates with a wider range of core sequence lengths than ZFRB1 (Figure 3B).
We also carried out comparative experiments with NM resolvase, purified as described previously (20).
In vitro recombination by ZFRs
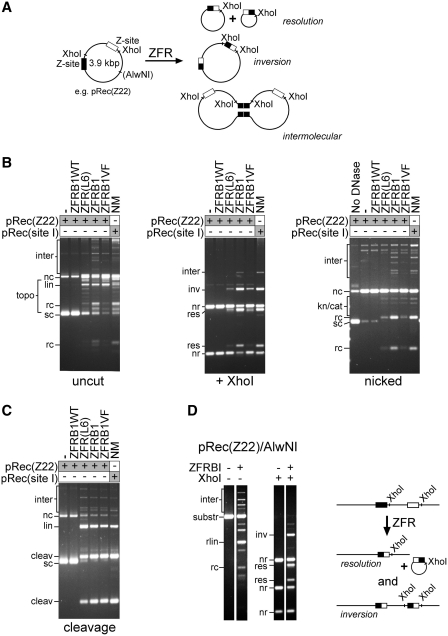
First, we tested the activity of each ZFR on a supercoiled plasmid pRec(Z22), which contains two copies of Z22 (the Z-site that was recombined most efficiently in E. coli) (Figure 4A). Preliminary titration experiments were carried out with each ZFR to assess the dependence of activity on protein concentration (Supplementary Figure S1). pRec(Z22) was then incubated with an optimal concentration of each ZFR (~1 µM) at 37°C for 1 h. Each sample was divided into three aliquots; one was not treated further, one was digested with XhoI, and one was treated with DNase I to nick covalently closed DNA. The aliquots were then loaded onto agarose gels and subjected to electrophoresis (Figure 4B). For comparison, a similar substrate pRec(site I) containing two copies of Tn3 res site I was treated with NM resolvase under the same conditions.
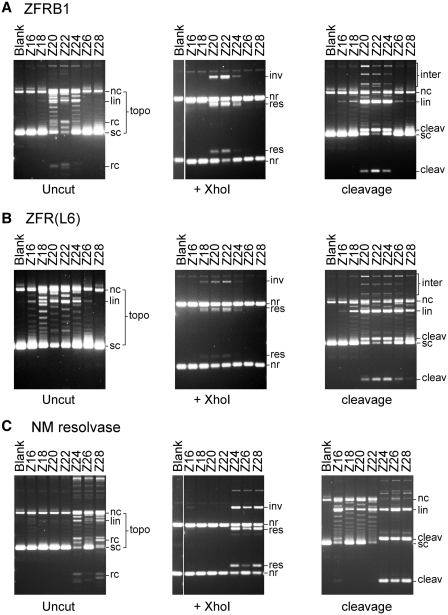
Figure 4.
ZFR-mediated recombination in vitro. (A) In vitro recombination of a plasmid substrate. In parts B and C, the plasmid used (pRec(Z22)) contained two Z22 sites. In this diagram, the resolution and inversion recombinant products are shown, along with one example of a product of intermolecular recombination. Sites for restriction enzymes used in the analysis are marked. (B) Reaction products after treatment of pRec(Z22) with ZFRs were visualized by agarose gel electrophoresis either without further treatment (left panel), or after digestion with XhoI (centre panel), or after nicking with DNase I to remove supercoiling (right panel). The ZFR used is indicated above each lane. The right-hand lane of each panel shows the products from pRec(site I), which has two copies of Tn3 res site I instead of Z-sites, after treatment with the activated Tn3 resolvase mutant NM. The bands are annotated as follows: sc, supercoiled circle; nc, nicked circle; lin, linearized plasmid; rc, recombinant circle; topo, this zone contains several bands including topoisomers of substrates and products, and linearized recombinant DNA; inter, this zone contains the large products of intermolecular recombination; nr, non-recombinant; res, recombinant (resolution); inv, recombinant (inversion); kn/cat, this zone contains bands assigned as nicked knotted and catenated reaction products. Other, faint bands in the restriction digests derived from cleavage products are not labelled. (C) Reactions of pRec(Z22) with ZFRs in 40% ethylene glycol buffer, which inhibits re-ligation of cleaved intermediates. The annotation is as in part B, with the addition of: cleav, product of double-strand cleavage at both recombination sites. Cleavage at only one site gives a full-length linear product (labelled lin). All samples were treated with protease K (see ‘Materials and Methods’ section), so the ZFR subunits covalently linked to the cleaved DNA ends have been destroyed. (D) ZFRB1-mediated recombination of linearized pRec(Z22). The annotation of bands is as in (B and C). The expected products of intramolecular recombination are illustrated in the scheme to the right of the gel image. The bands are labelled as in (B), with the additional label rlin, linear ‘resolution’ product.
ZFRB1 recombined pRec(Z22) efficiently, giving a very similar product distribution to the NM resolvase reaction on pRec(site I) (Figure 4B). The expected resolution and inversion products were observed, along with smaller amounts of intermolecular recombination products. The resolution products were predominantly free circles, though a small amount of topologically complex species could be seen on nicking with DNase I. The predominance of topologically simple products may be a consequence of multiple rounds of recombination (20). In the ‘uncut’ samples, each recombinant circle product appeared as several bands. These bands are covalently closed topoisomers along with some nicked circles, and therefore both strands of the recombinant Z-site DNA have been re-ligated in a large fraction of the products. The topoisomers were converted to nicked circles on treatment with DNase I. There was also a noticeable amount of linearized substrate and linearized recombinant-length DNA in the uncut samples (Figure 4B), which might derive from incompletely ligated reaction intermediates.
Surprisingly, ZFRB1VF, which had been isolated as a rate-enhanced mutant in vivo, gave significantly lower amounts of recombinant products than its parent ZFRB1. There were substantial amounts of nicked substrate DNA in the uncut sample, suggesting ZFR-mediated abortive cleavage of only one DNA strand. ZFR(L6) was also much less proficient for recombination than ZFRB1, and gave greater amounts of nicked substrate. Both ZFRB1VF and ZFR(L6) gave some linear products, as did ZFRB1 (see above). ZFRB1WT, whose catalytic domain is that of wild-type Tn3 resolvase, was completely inactive in this assay, showing that activating mutations in the catalytic domain are essential for ZFR recombination proficiency.
In a buffer containing a high concentration of ethylene glycol and no Mg2+, re-ligation of strands by resolvase is suppressed, and resolvase-linked products of cleavage at the recombination sites accumulate (27,20). We tested the ZFRs under these conditions (Figure 4C). ZFRB1, ZFRB1VF and ZFR(L6) all cleaved pRec(Z22) very efficiently; nearly all the substrate DNA was cleaved at either one or both Z-sites. ZFRB1VF had slightly higher cleavage activity than ZFRB1 (i.e. more products cleaved at both Z-sites), ZFR(L6) had lower activity and ZFRB1WT was completely inactive (as expected; see above). There was no evidence for any off-site ZFR activity, which could have been revealed by the presence of extra linear DNA bands in the cleavage assays. Likewise, the ZFRs had no recombination or cleavage activity on pRec(site I) (data not shown), even though the two copies of res site I in this plasmid are the natural target for the resolvase domains of the ZFRs.
The foregoing assays used a supercoiled plasmid substrate. In many potential applications of ZFRs (for example, in eukaryotic cells), the substrate DNA would not be supercoiled, so it was important to demonstrate that non-supercoiled DNA is also a substrate in vitro. Linear substrate DNA was prepared by restriction enzyme digestion of pRec(Z22). The purified linear DNA was then treated with ZFRB1. After 1 h, much of the substrate DNA had reacted to give products of resolution, inversion and intermolecular recombination (Figure 4D). The DNA segment excised in the resolution reaction was present as a relaxed circle.
Effects of Z-site core sequence length
Our experiments in E. coli (Figure 3; see also Ref. 10) revealed that ZFR activity depended on the Z-site core sequence length. We therefore tested the effect of core sequence length in vitro. The in vitro results were entirely consistent with the in vivo observations. Only pRec(Z20), pRec(Z22) and pRec(Z24) were recombined by ZFRB1, and of these pRec(Z22) was recombined most efficiently (Figure 5A). Similarly, only pRec(Z20), pRec(Z22) and pRec(Z24) were cleaved efficiently by ZFRB1 in ethylene glycol buffer, though traces of linearized full-length plasmid DNA were observed with the other substrates. ZFR(L6), which has a longer interdomain linker than ZFRB1 (Figure 2B) had a broader spectrum of activity (as was also seen in vivo); pRec(Z18), pRec(Z20), pRec(Z22) and pRec(Z24) were all recombined (Figure 5B), and in ethylene glycol buffer, ZFR(L6) promoted some DNA cleavage of all the substrates.
Figure 5.
ZFR-mediated recombination at Z-sites with different core sequence lengths. (A) Reactions catalysed by ZFRB1 on a set of pRec substrate plasmids, each containing two identical Z-sites as indicated above the lanes. For economy of space only one untreated sample [‘blank’; pRec(Z16) substrate plasmid] is shown in each panel. The layout of the figure and annotation of bands are as in Figure 4. (B) Reactions catalysed by ZFR(L6) on the same set of plasmids. (C) Reactions catalysed by NM resolvase on the same set of plasmids.
As a control, all seven of the pRec(Z) substrates were treated with NM resolvase, which should not recognize the Zif268-binding motifs at each end of the Z-sites. pRec(Z24), pRec(Z26), and pRec(Z28), whose Z-sites include most or all of the 28 bp Tn3 res site I, supported NM resolvase-mediated recombination (Figure 5C). pRec(Z24) is a better substrate than pRec(Z26), probably because some basepairs in the Zif268-binding motifs of Z24 accidentally match site I (Figure 2C). Similar results were obtained in in vivo assays (data not shown). More surprisingly, NM resolvase in ethylene glycol buffer promoted some double-strand cleavage and topoisomerization of all the pRecZ substrates, even those with short Z-site core sequences. Bands of the sizes predicted for cleavage at both Z-sites [seen, for example, in the pRec(Z16) products] show that at least some of the DNA cleavage is Z-site-specific. NM resolvase thus retains a residual capacity to recognize and cleave its natural target DNA sequence even when the specific motifs recognized by its C-terminal helix-turn-helix have been replaced by zinc-finger binding motifs.
Binding and synapsis of Z-sites by ZFRs
To ascertain the basis of the relationship between Z-site length and ZFR-mediated recombination efficiency, we need to know the stage in the reaction pathway that is affected. We therefore analysed binding and synapsis of Z-sites by ZFRs.
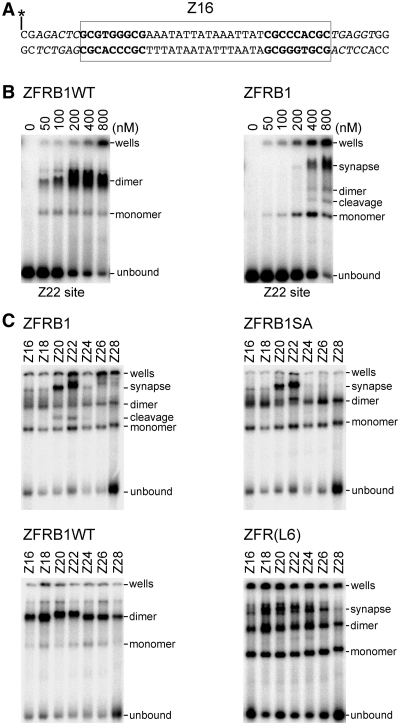
Wild-type Tn3 resolvase solution monomers bind cooperatively to a DNA fragment containing site I of res to form a dimer complex. Traces of a monomer–DNA complex can also be detected. The activated mutant NM resolvase can also form monomer and dimer complexes with site I, but two dimer–site I complexes can then interact to form a synapse containing a resolvase tetramer. These complexes can be observed by non-denaturing gel electrophoresis (14,20,28). Structures of resolvase–site I complexes show pronounced bending of the site I DNA (24,25,29,30). Our ZFRs have resolvase catalytic domains, so they were predicted to make analogous complexes with Z-sites. Radiolabelled 50 bp substrates, each containing a centrally positioned Z-site, were prepared by annealing oligonucleotides (Figure 6A). When ZFRB1WT (wild-type Tn3 resolvase catalytic domain) was added to a Z22 substrate, gel electrophoresis revealed lower mobility bands corresponding to binding of a ZFR monomer or a dimer (Figure 6B), as predicted. When ZFRB1 (activated NM resolvase catalytic domain) was assayed similarly, an additional strong band with the low mobility expected for a synaptic complex was observed (Figure 6B). When a longer, unlabelled Z22 oligonucleotide was included in the reaction mixture, this band was shifted to an even lower mobility species, confirming that it contains two DNA molecules (data not shown).
Figure 6.
Binding and synapsis of Z-sites by ZFRs. (A) The substrates used for the bandshift experiments. All the substrates are exactly 50 bp, with a centrally positioned Z-site. The diagram shows the Z16 site (boxed), with the motifs recognized by the Zif268 domains in bold. The longer Z-sites are similar except that the core sequence is longer (Figure 2C), and one or more of the immediately flanking basepairs at each end of the site (in italics) is deleted. The asterisk indicates the position of the 5′-32P label. (B) Binding of the Z22 site by varying concentrations of ZFRB1WT (left panel) and ZFRB1 (right panel). Estimated ZFR concentrations are indicated above each lane. The band marked ‘cleavage’ is a labelled half-site cleavage product with a ZFR monomer covalently attached via the active site serine residue; see text for details. (C) Binding of Z-sites (as indicated above the lanes) by ZFRB1 (top left), ZFRB1SA (top right), ZFRB1WT (bottom left) and ZFR(L6) (bottom right).
We then compared ZFR binding and synapsis of the Z-sites with different core sequence lengths. A single optimal concentration (~800 nM) of each ZFR was used (Figure 6C). ZFRB1WT bound to all the Z-sites to form monomer and dimer complexes, although the longest site (Z28) was not bound as efficiently as the others. The sites that recombine most efficiently (Z20, Z22) formed lower mobility dimer complexes than the other sites, consistent with increased bending of the Z-site DNA. When the active recombinase ZFRB1 was used, synaptic complexes were observed. The Z-sites that gave the most retarded dimer complexes with ZFRB1WT (Z20, Z22) also gave the strongest and lowest mobility synapse bands. In some lanes, there were multiple bands with the low mobility associated with a synaptic complex. We therefore propose that the synapses may exist in different conformational states. Additional faint higher mobility bands in the Z20, Z22 and Z24 lanes are interpreted as products of double-strand cleavage of the Z-site. When the catalytically defective mutant ZFRB1SA was used instead of ZFRB1, the pattern of complexes was similar, but the presumed cleavage product bands were absent.
Binding and synapsis by ZFR(L6) were also assayed, to determine whether the long linker peptide between the recombinase and zinc-finger domains in this protein (Figure 2B) affected the pattern of complex formation. ZFR(L6) converted less of the DNA to synaptic complexes than ZFRB1 (Figure 6C), but the range of Z-site lengths that supported significant ZFR(L6)-mediated synapsis was wider than for ZFRB1. These observations parallel the recombination and cleavage assays (Figures 4B and 5B), supporting the hypothesis that the level of ZFR catalytic activity reflects the extent of synapse formation.
DISCUSSION
Design of ZFRs
It was shown previously that residues 1–144 of resolvase are sufficient for ZFR activity (10). However, ZFRB2 which has a very short (two residues) linker between residue R144 and the zinc-finger domain is inactive. In contrast, residues 1–148 of resolvase can be connected to the zinc-finger domain by the same two-amino acid linker to give a very efficient recombinase (ZFRB1). We propose (see below) that the linker of ZFRB2 is not long enough to allow the two domains to interact properly with any Z-site. The extra four amino acids in ZFRB1 might contribute to its high recombination activity by extending the Tn3 resolvase domain to 148 residues, as well as by lengthening the protein. Analogous active ZFRs with a 5-amino acid linker between Tn3 resolvase residues 1–144 and a zinc-finger domain have been described (12,17–19).
An E. coli-based assay was used to select ZFRB1 mutants with rate-enhanced activity (deletion of a marker gene). However, subsequent in vitro analysis of one of these mutants (ZFRB1VF) revealed undesirable side-reactions (see below).
ZFR recombination and cleavage in vitro
If ZFRs are to become practical tools for programmed DNA rearrangements at chosen genomic target sites, it is important that we fully understand their activities and mechanisms of action. With an in vitro system, we can study the basic properties of ZFRs in much greater detail than is possible in vivo.
Tn3 resolvase-derived ZFRs can catalyse efficient, site-specific reciprocal recombination in vitro. The DNA ends are properly re-ligated; double-strand break intermediates do not accumulate to a high level except when induced by special reaction conditions. The activity of the most efficient ZFR tested here (ZFRB1) is strikingly similar to that of the activated Tn3 resolvase variant (NM) from which its catalytic domain is derived. As we had predicted, activating mutations in the ZFR catalytic domain are essential; ZFRB1WT is identical to ZFRB1 except that it has the wild-type Tn3 resolvase catalytic domain, and it has no recombination activity.
ZFR(L6), first characterized by Akopian et al. (10), which has a longer interdomain linker (Figure 2B), was less efficient in vitro than ZFRB1, but it was also less sensitive to the length of the Z-site core sequence (as discussed in more detail below). We also tested a ZFRB1 mutant, ZFRB1VF, which was selected for its enhanced rate of recombination in E. coli. The single additional mutation V107F in ZFRB1VF is at a position where activating mutations have been observed previously (22). ZFRB1VF gave low levels of recombinant products in vitro and caused accumulation of substrate plasmid DNA containing single- or double-strand breaks. This increased propensity to leave strand breaks at the Z-sites might be undesirable in applications of ZFRs, as it could lead to genetic damage.
The ZFRs are highly specific for recombination at Z-sites; there was no evidence for off-site activity at ‘pseudosites’, which would have produced extra bands on the gels shown in Figure 4. We estimate that we could have observed cleavage of <5% of the DNA at any pseudosite. ZFRs thus appear to have higher site specificity than the parent NM resolvase, which does display some off-site activity (Figure 4; Ref. 20). All the active ZFRs promoted very efficient cleavage of a substrate with Z22 sites under reaction conditions where re-ligation of DNA ends is inhibited (Figure 4C). This suggests that the lower recombination efficiency of ZFRB1VF and ZFR(L6) is because of a defect in steps after cleavage.
Most of our assays involved the use of negatively supercoiled DNA substrates. However, a linear DNA molecule was also recombined efficiently (Figure 4D), showing that ZFR recombination does not require supercoiling. Further experiments will be necessary to determine any more subtle effects of DNA topology, which might be important for specific applications of ZFRs.
Dependence of ZFR activity on Z-site core sequence length
Our results (Figures 3 and 5) dramatically confirm the surprisingly specific dependence of ZFR recombination and cleavage activity on the length of the Z-site core sequence, first noted by Akopian et al. (10). All the ZFR variants that we have tested recombine Z22 sites most efficiently, in E. coli and in vitro. Sites with 21 and 23 bp core sequences (Z21, Z23) were recombined slightly less efficiently than Z22 sites in E. coli (data not shown). The preference for the Z22 site is due to its length, not any specific sequence motif, because the same length was preferred when a set of Z-sites with substantially different core sequences was tested [(10), and data not shown].
When we began this work, we had expected to find that ZFRs with short inter-domain linkers would prefer to act on Z-sites with short core sequences, and that as the core sequence length was increased, longer linkers would be preferred that can stretch between the zinc-finger domains at the ends of the Z-site and the catalytic domains at the centre (Figure 1B). In vivo analysis failed to reveal any such correlation. However, in vitro, we do see significant effects of the linker length. The ZFR(L6) linker is 8 amino acids longer than that of ZFRB1 (Figure 2B). ZFRB1 recombination and cleavage activity was restricted to the Z20, Z22 and Z24 substrates (Figure 5A), whereas ZFR(L6) also recombined pRec(Z18), and gave cleavage products with all substrates except pRec(Z16) (Figure 5B). We suggest that the linker length is the main cause of this difference between ZFRB1 and ZFR(L6) activity (although we cannot exclude the possibility that the amino acid sequence of the linker is a significant factor). ZFRs with long linkers might therefore be useful to target sites where the motifs to be recognized by the zinc-finger domains are not spaced exactly 22 bp apart, but they might also have reduced specificity.
Our results reveal that effects of core sequence length are apparent even at the early reaction steps of binding and synapsis (Figure 6). The Z-sites that are recombined most efficiently (Z20, Z22) also give the strongest, lowest mobility synapse bands and the lowest mobility dimer complex bands; the low electrophoretic mobility suggesting increased DNA bending. In contrast, the longest Z-site (Z28) was bound rather poorly by the ZFRs.
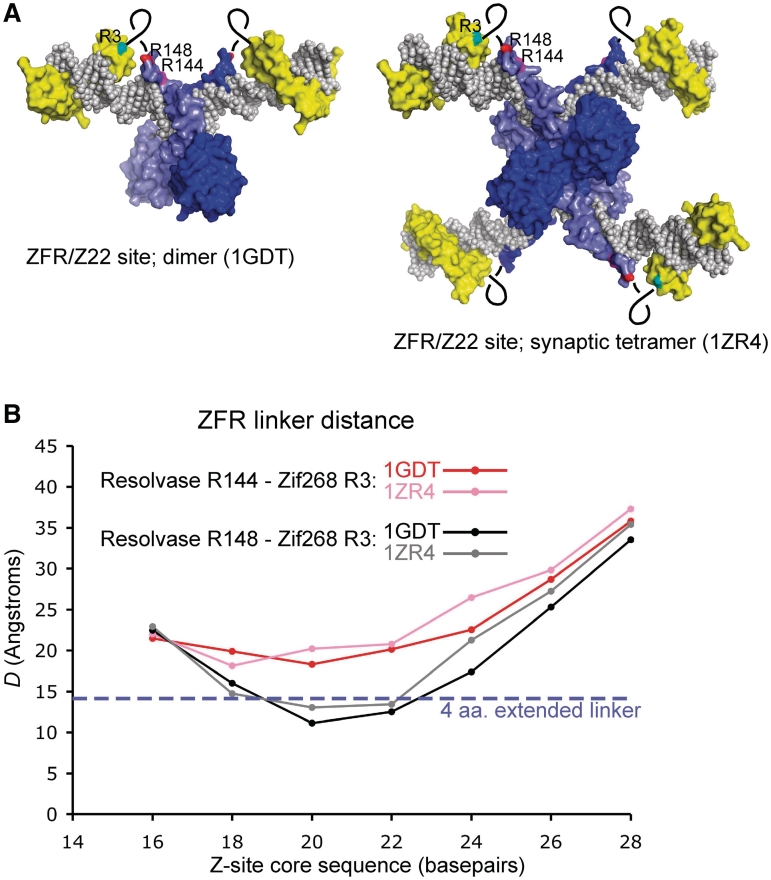
To gain structural insight into these effects, complexes of ZFRs with all the Z-sites (Z16–Z28) were modelled using available crystal structures of a γδ resolvase dimer–DNA complex, a synaptic γδ resolvase–DNA intermediate and a Zif268 zinc-finger domain–DNA complex (see ‘Materials and Methods’ section for details). No steric clashes within the two ZFR subunits bound to any single Z-site were predicted. Likewise, no clashes between any of the four subunits in a synapse of two Z-sites were predicted (Figure 7A; and data not shown). The straight-line distance D between the α-carbon atoms of amino acids that would be at either end of the ZFR linker (R144 or R148 of the resolvase domain and R3 of the Zif268 domain) was measured from the models (Figure 7B). The feasibility of positioning the two domains of any particular ZFR in the modelled arrangements can be estimated from D, assuming a maximum extended linker peptide length of ~3.6 Å per amino acid. In ZFRB1 the extended length of the linker between the α-carbon atoms of resolvase R148 and Zif268 R3 (R-T-S-E-R) would be ~14.4 Å. The models predict that this linker would be long enough to reach between the two domains when ZFRB1 binds to Z20 or Z22 (the sites that recombine most efficiently), but not when it binds to the other Z-sites (Figure 7B). We therefore speculate that the ZFRB1 linker is too short for optimal productive binding to the Z-sites other than Z20 or Z22. Parts of the resolvase and/or zinc-finger domains might have to flex and/or alter their secondary structure from that seen in the crystals in order to allow the protein to span the shorter and longer Z-sites, and this might reduce proficiency for synapsis and recombination. On the longest sites, ZFR subunits bound at each end via their zinc finger domains might be too distant for a strong dimer interaction of the catalytic domains; hence binding is weaker.
Figure 7.
Modelling the interactions of ZFRs with Z-sites. (A) Molecular models of a ZFR bound to the Z22 site. The left panel shows a model of a ZFR dimer bound to Z22. The model is built from the resolvase–DNA structure 1GDT and two copies of the Zif268 domain–DNA structure 1AAY. The right panel shows a ZFR synaptic tetramer, built from four copies of 1AAY and the resolvase synaptic complex structure 1ZR4. The resolvase domains are blue, the Zif268 domains are yellow, and the DNA is grey. The positions of resolvase residues R144 and R148, and Zif268 domain residue R3, are indicated by the magenta, red and cyan blotches respectively. The black loops indicate the approximate locations of the ZFR linker peptide. (B) The distance D between the α-carbon atoms of resolvase residue R148 (or R144) and Zif268 domain residue R3 predicted from the molecular models, plotted against the Z-site core sequence length. Note that E2, the first residue of the Zif268 domain in the ZFRs (Figure 2B), could not be used for the measurements as it was not resolved in the 1AAY crystal structure. The plotted D values are averages of the values from the two crystallographically non-identical ZFRs in each of the 1GDT-based models, or from the four non-identical ZFRs in each of the 1ZR4-based models.
The structural models predict that the ZFRB2 linker between resolvase R144 and Zif268 R3 (also R-T-S-E-R; ~14.4 Å) is not long enough to allow proper interaction with any of the Z-sites (Figure 7B), in accord with our results showing that ZFRB2 is inactive in E. coli. The corresponding (R144-R3) linker length for ZFR(L6) is 16 amino acids, which when fully extended would be ~57 Å long. This is more than D even for the longest site (Z28; D ~35 Å). Actually, the linker would need to be much longer than 35 Å for proper binding to Z28, because the N-terminus of the Zif268 domain and the C-terminus of the resolvase domain are on opposite sides of the double helix, so the linker would need to wrap around the DNA. Inspection of the Z28-ZFR models suggests that the ZFR(L6) linker should be long enough, yet binding, synapsis, cleavage and recombination of the Z28 site are defective. At present, we can only speculate on the reasons for this defective interaction. The effective maximum length of the ZFR(L6) linker might be less than the fully extended length, due to secondary structure; or, a negative entropy change upon stretching out the linker might disfavour binding to the long Z-sites.
In summary, we can trace the dependence of recombination efficiency on Z-site core sequence length back to effects on binding of the ZFR to a single Z-site, prior to synapsis or any catalytic steps. However, we still cannot give a complete explanation of why a core sequence of exactly 22 bp between the zinc-finger binding motifs is preferred by all the ZFRs we have tested, regardless of ZFR linker length. The structural models predict that binding of very long Z-sites may be abnormal or inefficient, thus reducing recombination efficiency. But why do the shorter Z-sites (Z16, Z18) bind ZFR efficiently, yet fail to synapse and recombine? One speculation proposes that synapsis and catalysis depend on proper interaction of the resolvase arm region with the minor groove of the DNA, as shown in Figure 2A. In the shorter Z-sites, the all-AT DNA sequence contacted by the arm region is adjacent to the all-GC motif recognized by the first finger of the Zif268 domain. It might be unfavourable to abut the DNA bound by the arm region, with its unusually narrow minor groove, to the DNA bound by the Zif268 domain, which also has an unusual structure with a rather wide minor groove (26). An answer to this question will require further experiments.
Use of ZFRs in biotechnology
We aim to gain a detailed understanding of the properties and design of ZFRs in order to allow them to become useful tools for the promotion of specific genetic rearrangements (3). In this section, we outline some of the main obstacles in the way towards widespread applicability of ZFRs, which might be overcome by approaches involving further in vitro analysis.
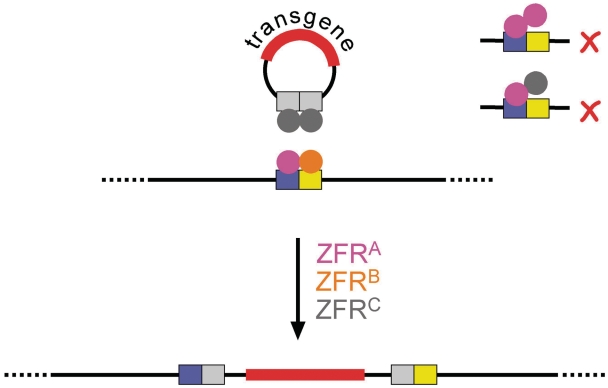
A typical application might be to promote integration of a transgene at a specific site in the genome of an organism. In most cases, it would be necessary to use two ZFRs with different DNA binding specificities to target a natural sequence, one to bind at each end of the ‘Z-site’. Another recombinase with a compatible catalytic domain, which recognizes a site on the transgenic DNA, might also be needed (Figure 8). To promote such a reaction effectively, several issues will need to be addressed. First, the ZFR catalytic domains must promote recombination at the core sequence of the chosen target ‘Z-site’, and this sequence might be very different from that of Tn3 res site I. Towards this end, modifications of the catalytic domain that broaden or alter specificity for the Z-site core sequence have been reported recently (12,13,19), but it is not clear yet if these specificity changes are accompanied by undesirable effects on catalysis (for example, incomplete re-ligation of broken DNA strands, as we found with the ZFRB1VF variant). Second, strategies will be needed to favour ZFR-mediated integration over the reverse reaction, deletion. ZFRs can promote integration in vivo (18,31) and equivalent intermolecular reactions in vitro (see for example Figure 4B). However, the Z-sites flanking the integrated DNA can recombine again, so recombination will tend towards an equilibrium of integration and deletion products. Deletion is thermodynamically favoured because the two separate deletion product DNA molecules have higher entropy than the single integration product molecule. No advances on this problem have been reported to date. A third imperative will be to minimize inappropriate subunit interactions when two or more different ZFRs are present, as in the scenario shown in Figure 8. If all the ZFR catalytic domains are identical, a single ZFR subunit bound correctly at one end of a target site might dimerize with a subunit whose zinc-finger domain is not specific for the other end of the site. Although these undesirable interactions are reversible, they might nevertheless inhibit recombination, reduce the effective ZFR concentration, and increase the likelihood of off-site reactions. Analogous problems arise in applications of zinc-finger nucleases (ZFNs), and protein modifications to block the formation of inappropriate ZFN dimers have been described (32–35); similar approaches should be applicable to ZFRs. Fourth, off-target activity must be limited to a very low level if ZFRs are to be used in demanding applications such as gene therapy, where unintended genetic damage could have very serious consequences. The natural biochemistry of resolvase and other possible progenitor serine recombinases, requiring on-site dimerization and dimer–dimer synapsis prior to catalysis (20), suggests that ZFRs might have the potential for very high target specificity, perhaps more so than ZFNs (36). However, no critical experimental evidence on this issue is available as yet. The level of off-target activity will almost certainly be affected by aspects of ZFR design such as the origin of the catalytic domain, the activating mutations in the catalytic domain, and the characteristics of the inter-domain linker peptide, and these effects could be quantified by in vitro analysis.
Figure 8.
ZFR-mediated site-specific transgene integration. Cartoon showing hypothetical ZFR-mediated integration of a transgenic construct into a genomic target site. The genomic site is recognized by a heterodimer of the magenta and orange ZFR subunits, whereas the site on the transgene DNA is recognized by a homodimer of the grey ZFR. The diagram on the right illustrates how, in this scenario, inappropriate on-site dimerization could lead to unproductive complexes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Wellcome Trust (studentships 078766 to J.-G.S., 075617 to M.M.P., 026339 to A.A., 069113 to A.H.); Biotechnology and Biosciences Research Council (BBSRC) (BB/E022200/1 to A.McP., F.J.O.; BB/F021593/1 to A.McP., D.W.). Funding for open access charge: BBSRC grant (BB/F021593/1).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are very grateful to Chris Proudfoot, Martin Boocock, Mary Burke and Sean Colloms, for advice and practical help; and to Sean Colloms and Martin Boocock for critical reading of the article.
REFERENCES
- 1.Grindley NDF, Whiteson KL, Rice PA. Mechanism of site-specific recombination. Ann. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 2.Wirth D, Gama-Norton L, Riemer P, Sandhu U, Schucht R, Hauser H. Road to precision: recombinase-based targeting technologies for genome engineering. Curr. Opin. Biotechnol. 2007;18:411–419. doi: 10.1016/j.copbio.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 3.Akopian A, Stark WM. Site-specific recombinases as instruments for genomic surgery. Adv. Genet. 2005;55:1–23. doi: 10.1016/S0065-2660(05)55001-6. [DOI] [PubMed] [Google Scholar]
- 4.Buchholz F, Stewart AF. Alteration of Cre recombinase site specificity by substrate-linked protein evolution. Nat. Biotechnol. 2001;19:1047–1052. doi: 10.1038/nbt1101-1047. [DOI] [PubMed] [Google Scholar]
- 5.Sclimenti CR, Thyagarajan B, Calos MP. Directed evolution of a recombinase for improved genomic integration at a native human sequence. Nucleic Acids Res. 2001;29:5044–5051. doi: 10.1093/nar/29.24.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santoro SW, Shultz PG. Directed evolution of the site specificity of Cre recombinase. Proc. Natl Acad. Sci. USA. 2002;99:4185–4190. doi: 10.1073/pnas.022039799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolusani S, Ma C, Paek A, Konieczka JH, Jayaram M, Voziyanov Y. Evolution of variants of yeast site-specific recombinase Flp that utilize native genomic sequences as recombination target sites. Nucleic Acids Res. 2006;34:5259–5269. doi: 10.1093/nar/gkl548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarkar I, Hauber I, Hauber J, Buchholz F. HIV-1 Proviral excision using an evolved recombinase. Science. 2007;316:1912–1915. doi: 10.1126/science.1141453. [DOI] [PubMed] [Google Scholar]
- 9.Surendranath V, Chusainow J, Hauber J, Buchholz F, Habermann BH. SeLOX - a locus of recombination site search tool for the detection and directed evolution of site-specific recombination systems. Nucleic Acids Res. 2010;38:W293–W298. doi: 10.1093/nar/gkq523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akopian A, He J, Boocock MR, Stark WM. Chimeric recombinases with designed DNA sequence recognition. Proc. Natl Acad. Sci. USA. 2003;100:8688–8691. doi: 10.1073/pnas.1533177100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu. Rev. Biochem. 2009;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- 12.Gaj T, Mercer AC, Gersbach CA, Gordley RM, Barbas CF., III Structure-guided reprogramming of serine recombinase DNA sequence specificity. Proc. Natl Acad. Sci. USA. 2011;108:498–503. doi: 10.1073/pnas.1014214108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Proudfoot CM, McPherson AL, Kolb AF, Stark WM. Zinc finger recombinases with adaptable DNA sequence specificity. PLoS ONE. 2011;6:e19537. doi: 10.1371/journal.pone.0019537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nöllmann M, Byron O, Stark WM. Behavior of Tn3 resolvase in solution and its interaction with res. Biophys. J. 2005;89:1920–1931. doi: 10.1529/biophysj.104.058164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat. Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 16.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 17.Gordley RM, Smith JD, Gräslund T, Barbas CF., III Evolution of programmable zinc finger-recombinases with activity in human cells. J. Mol. Biol. 2007;367:802–813. doi: 10.1016/j.jmb.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 18.Gordley RM, Gersbach CA, Barbas CF., III Synthesis of programmable integrases. Proc. Natl Acad. Sci. USA. 2009;106:5053–5058. doi: 10.1073/pnas.0812502106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gersbach CA, Gaj T, Gordley RM, Barbas CF., III Directed evolution of recombinase specificity by split gene reassembly. Nucleic Acids Res. 2010;38:4198–4206. doi: 10.1093/nar/gkq125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olorunniji FJ, He J, Wenwieser SVCT, Boocock MR, Stark WM. Synapsis and catalysis by activated Tn3 resolvase mutants. Nucleic Acids Res. 2008;36:7181–7191. doi: 10.1093/nar/gkn885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grindley NDF, Lauth MR, Wells RG, Wityk RJ, Salvo JJ, Reed RR. Transposon-mediated site-specific recombination: identification of three binding sites for resolvase at the res sites of γδ and Tn3. Cell. 1982;30:19–27. doi: 10.1016/0092-8674(82)90007-1. [DOI] [PubMed] [Google Scholar]
- 22.Burke ME, Arnold PH, He J, Wenwieser SVCT, Rowland SJ, Boocock MR, Stark WM. Activating mutations of Tn3 resolvase marking interfaces important in recombination catalysis and its regulation. Mol. Microbiol. 2004;51:937–948. doi: 10.1046/j.1365-2958.2003.03831.x. [DOI] [PubMed] [Google Scholar]
- 23.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- 24.Yang W, Steitz TA. Crystal structure of the site-specific recombinase γδ resolvase complexed with a 34 bp cleavage site. Cell. 1995;82:193–208. doi: 10.1016/0092-8674(95)90307-0. [DOI] [PubMed] [Google Scholar]
- 25.Li W, Kamtekar S, Xiong Y, Sarkis G, Grindley NDF, Steitz TA. Structure of a synaptic γδ resolvase tetramer covalently linked to two cleaved DNAs. Science. 2005;309:1210–1215. doi: 10.1126/science.1112064. [DOI] [PubMed] [Google Scholar]
- 26.Elrod-Erickson M, Rould MA, Nekludova L, Pabo CO. Zif268 protein-DNA complex refined at 1.6 Å: a model system for understanding zinc finger-DNA interactions. Structure. 1996;4:1171–1180. doi: 10.1016/s0969-2126(96)00125-6. [DOI] [PubMed] [Google Scholar]
- 27.Boocock MR, Zhu X, Grindley NDF. Catalytic residues of γδ resolvase act in cis. EMBO J. 1995;14:5129–5140. doi: 10.1002/j.1460-2075.1995.tb00195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bednarz AL, Boocock MR, Sherratt DJ. Determinants of correct res site alignment in site-specific recombination by Tn3 resolvase. Genes Dev. 1990;4:2366–2375. doi: 10.1101/gad.4.12b.2366. [DOI] [PubMed] [Google Scholar]
- 29.Nöllmann M, He J, Byron O, Stark WM. Solution structure of the Tn3 resolvase-crossover site synaptic complex. Mol. Cell. 2004;16:127–137. doi: 10.1016/j.molcel.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 30.Kamtekar S, Ho RS, Cocco MJ, Li W, Wenwieser SVCT, Boocock MR, Grindley NDF, Steitz TA. Implications of structures of synaptic tetramers of γδ resolvase for the mechanism of recombination. Proc. Natl Acad. Sci. USA. 2006;103:10642–10647. doi: 10.1073/pnas.0604062103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gersbach CA, Gaj T, Gordley RM, Mercer AC, Barbas CF., III Targeted plasmid integration into the human genome by an engineered zinc finger recombinase. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr421. doi:10.1093/nar/gkr421 [Epub ahead of print, 7 June 2011] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 33.Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat. Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 34.Söllu C, Pars K, Cornu TI, Thibodeau-Beganny S, Maeder ML, Joung JK, Heilbronn R, Cathomen T. Autonomous zinc-finger nuclease pairs for targeted chromosomal deletion. Nucleic Acids Res. 2010;38:8269–8276. doi: 10.1093/nar/gkq720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, Miller JC, Urnov FD, Gregory PD, Holmes MC. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat. Methods. 2011;8:74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- 36.Halford SE, Catto LE, Pernstich C, Rusling DA, Sanders KL. The reaction mechanism of FokI excludes the possibility of targeting zinc finger nucleases to unique DNA sites. Biochem. Soc. Trans. 2011;39:584–588. doi: 10.1042/BST0390584. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.