Abstract
Mitochondrial DNA replication is performed by a simple machinery, containing the TWINKLE DNA helicase, a single-stranded DNA-binding protein, and the mitochondrial DNA polymerase γ. In addition, mitochondrial RNA polymerase is required for primer formation at the origins of DNA replication. TWINKLE adopts a hexameric ring-shaped structure that must load on the closed circular mtDNA genome. In other systems, a specialized helicase loader often facilitates helicase loading. We here demonstrate that TWINKLE can function without a specialized loader. We also show that the mitochondrial replication machinery can assemble on a closed circular DNA template and efficiently elongate a DNA primer in a manner that closely resembles initiation of mtDNA synthesis in vivo.
INTRODUCTION
During DNA replication, helicases unwind the double-stranded DNA (dsDNA) ahead of the DNA polymerase and thus create the single-stranded DNA (ssDNA) template used to synthesize the complementary DNA strand. TWINKLE is the replicative helicase required for in vivo synthesis of human mitochondrial DNA (mtDNA) (1). On its own, TWINKLE can unwind short stretches of dsDNA in the 5′ to 3′ direction (2). In combination with mtDNA Polymerase γ (POLγ) and the mitochondrial ssDNA-binding protein (mtSSB), TWINKLE forms a processive replication machinery, a replisome, that can synthesize ssDNA molecules of >16 kb (3). Mitochondrial DNA is divided into a heavy (guanine-rich) and a light (cytosine-rich) strand. Heavy-strand DNA synthesis is initiated at the origin of heavy strand mtDNA replication (oriH), located in the main non-coding region of the mtDNA molecule (4). After the replication machinery has completed about two-thirds of heavy-strand DNA synthesis, it passes the origin of light strand mtDNA replication (oriL) and the parental H strand is exposed as a single strand. In its ssDNA conformation, oriL forms a stem-loop structure and the mitochondrial RNA polymerase (POLRMT) can initiate primer synthesis from a poly-dT stretch in the single-stranded loop region (5,6). Primer synthesis proceeds for ~25 bp, after which POLRMT is replaced by POLγ, and light-strand DNA synthesis is initiated (6–8).
POLRMT also generates the primers required for initiation of heavy-strand mtDNA synthesis. The 3′-end of the primer is defined by transcription termination at a conserved sequence element (conserved sequence block II) in the mitochondrial DNA control region (9–12). This site-specific termination event is caused by G-quadruplex structures formed in nascent RNA upon transcription of CSB II (13). Most DNA synthesis events initiated at oriH do not proceed to full circle, but are terminated at the termination-associated sequences (TAS), situated a few hundred base pairs downstream of the initiation site. The newly synthesized DNA strand stays stably hybridized to the parental strand, forming a triple-stranded structure, a displacement loop (D-loop) (4,8). In order to initiate DNA synthesis at oriH, TWINKLE and the other mitochondrial replication factors must be able to load in the D-loop region of the closed mtDNA molecule. How the mitochondrial replisome accomplishes this process and if additional factors are required, have so far not been studied.
Replicative helicases assemble into ring-shaped hexamer structures with a ring diameter of ~12–14 nm (14). Most of them form hexamers even in the absence of DNA, but the presence of a cofactor (NTP or Mg2+) is often needed for oligomerization. The central channel of the hexameric ring has a diameter varying between 2 and 4.5 nm, and accommodates one DNA strand while the second strand is excluded (14–16). Due to their ring-shaped appearance, replicative DNA helicases have to be loaded onto the ssDNA during initiation of replication and in most cases loading involves the action of accessory proteins, denoted helicase loaders (17). Some helicases are present in solution in monomeric form and are assembled around the DNA with the help of helicase loaders, whereas other DNA helicases form preformed hexameric rings that must be opened by a helicase loader to load onto the ssDNA. For instance, in Escherichia coli, the DnaB helicase forms a very stable hexamer that is loaded onto the ssDNA with the help of the DnaC loader in an ATP dependent manner. This event is facilitated by an interaction of the DnaC–DnaB complex with DnaA, the replication initiation protein (18,19). Interestingly, some helicases such as the viral protein SV40 T antigen and T7gp4 can load onto DNA without the aid of a loading factor (14,20,21). Even if a distinct factor is not needed, a separate domain of the helicase itself facilitates the loading process. In addition to its helicase domain, T7gp4 contains a primase domain and it has been suggested that the DNA binding site of the primase domain acts as a helicase loader by making the initial contact with the DNA. DNA binding by the primase domain is followed by a conformational change in the T7gp4 protein, which leads to opening of the ring and entrance of DNA into the central channel, followed by closure of the ring.
TWINKLE forms a stable hexamer or heptamer and unwinds duplex DNA in the 5′ to 3′ direction(22,23). Similar to T7gp4, TWINKLE requires a fork-like structure with both a 5′- and a 3′-single-stranded stretch of DNA to efficiently initiate DNA unwinding (2,24,25). All unwinding experiments performed so far with TWINKLE have been performed with templates containing a free 5′- end. This allows threading of the oligomeric TWINKLE onto the DNA without the need of a change in the protein conformation to let the DNA pass through and bind to the central channel of the protein. TWINKLE is a stable hexameric helicase in solution even in the absence of Mg2+ or NTP or at high ionic force (22). Therefore there must be a precise mechanism that enables TWINKLE to load onto circular DNA in vivo, prior to initiation of mtDNA synthesis (4). In the present study we have investigated the loading of TWINKLE onto a circular ssDNA in vitro. Our results show that TWINKLE is able to load onto circular ssDNA without the help of a loading factor and can support initiation of DNA replication on a closed circular dsDNA substrate in combination with only POLγ.
MATERIALS AND METHODS
Recombinant proteins
For purification of POLγ A, POLγ B and TWINKLE, we infected Sf9 cells with recombinant baculoviruses encoding versions of the individual proteins lacking the N-terminal mitochondrial targeting signal, but with a His6-tags at the C-terminus. A non-tagged mtSSB protein, lacking the N-terminal mitochondrial targeting signal was expressed the same way. POLγ A and B were purified separately as described (26). TWINKLE and mtSSB was purified as in (27). For purification of the T7gp4 protein with a N-terminal His6-tag, we cloned the T7gp4-A gene into the pET-20 b vector (Stratagene) and the protein was expressed in E. coli [BL21(DE3)pLysS]. The bacterial culture was grown in LB medium at 37°C to an A600 of 0.8. Isopropyl β-d-thiogalactopyranoside was added to a final concentration of 1 mM and the cells were cultured for three additional hours at 30°C and harvested by centrifugation. Cells were frozen in liquid nitrogen, thawed in lysis buffer (50 mM NaH2PO4 pH 8.0, 300 mM NaCl, 10 mM imidazole) and incubated on ice for 30 min in the presence of 1 mg/ml lysozyme. The cells were then disrupted by sonication (6 × 20 s) and centrifuged at 10 000g for 30 min. The cleared lysate obtained was mixed with 2 ml of Ni2+-NTA matrix superflow (Qiagen) equilibrated with buffer A (25 mM Tris–HCl pH 8.0, 10% glycerol, protease inhibitors, 10 mM β-mercaptoethanol, 0.4 M NaCl, 10 mM imidazole) and incubated rotating for 60 min at 4°C. The Ni2+-NTA matrix was collected by centrifugation (1500g for 10 min), resuspended in buffer A (10 mM imidazole), poured onto a column and washed with 10 column volumes of buffer A. The protein was eluted with buffer A containing 250 mM imidazole and fractions containing the proteins were combined. T7gp4 was further purified on Heparin Sepharose and Mono Q (GE Healthcare) in a buffer containing 25 mM Tris–HCl pH 8.0, 10% glycerol, 1 mM DTT, 0.5 mM EDTA, protease inhibitors and 0.2 M NaCl. For both columns, the protein was eluted using a 0.2 to 1.2 M NaCl gradient. The purity of T7gp4, estimated by SDS–PAGE with Coomassie blue staining, was >95%.
NTPase activities
NTPase activities were determined by colorimetry using the ‘malachite green phosphate assay kit’ (BioAssay Systems). In this assay, the inorganic phosphate liberated during nucleotide hydrolysis forms a colored product with malachite green. The formation of the colored product was measured on a spectrophotometer at 620 nm. Before the reaction was performed, TWINKLE was dialyzed against a buffer containing 25 mM Tris–HCl pH 7.5, 10% glycerol, 0.5 mM EDTA, 1 mM dithiothreitol and 400 mM NaCl for 4 h at 4°C. The NTP hydrolysis reaction was performed in a 20 µl reaction mixture containing 20 mM Tris–HCl pH 7.5, 4.5 mM MgCl2, 1 mM dithiothreitol, 0.5 mM of the indicated nucleotide and 200 fmol of TWINKLE hexamer in the presence or absence of 188 fmol M13mp18 ssDNA. The final NaCl concentration was 50 mM. The reactions were incubated at 25°C or 42°C for 45 min. The reaction mixture was then diluted 4-fold in water and terminated by the addition of 20 µl of Malachite Green Reagent. After 20 min of incubation at room temperature for color development, the absorbance at 620 nm was measured on a spectrophotometer. The quantity of phosphate released was determined using a standard curve generated with free phosphate according to the instructions of the manufacturer.
Helicase assay
DNA substrates used for the different helicase assays were prepared by annealing the following oligonucleotides (60 nt) to M13mp18 ssDNA (7249 nt): 5′-tailed template: ACA TGA TAA GAT ACA TGG ATG AGT TTG GAC AAA CCA CAA CGT AAA ACG ACG GCC AGT GCC; and 3′-tailed template: GTA AAA CGA CGG CCA GTG CCC AAC ACC AAA CAG GTT TGA GTA GGT ACA TAG AAT AGT ACA. The DNA substrates formed contain a 20-bp double-stranded region and a 40-nt single-stranded tail (Figure 2A). The reaction mixture (15 µl) contained 20 mM Tris–HCl pH 7.5, 5 mMMgCl2, 4 mM DTT, 100 µg/ml BSA, 3 mM NTP, 5 fmol of DNA substrate, and the amount of TWINKLE indicated in the figure legends. Individual reactions were incubated for 45 min at the temperature indicated in the figure legends and stopped by the addition of 3 µl of stop solution [90 mM EDTA (pH 8.0), 6% SDS, 30% glycerol, 0.025% bromophenol and 0.025% xylene cyanol]. The reaction products were separated by electrophoresis through a 10% non-denaturing polyacrylamide gel. The gel was dried onto DE81 (Whatman) and autoradiographed overnight at −80°C with an intensifying screen.
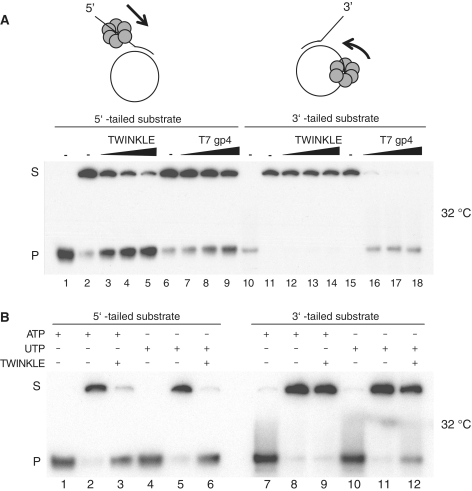
Figure 2.
TWINKLE can load onto a circular ssDNA and unwind duplex DNA. (A) Helicase activity of TWINKLE and T7gp4. The DNA substrates used in this study are composed of a 20-bp duplex region and a 40-nt single-stranded overhang (5′ or 3′). The helicase activity is measured as the ability of the protein to separate the annealed 60-nt radiolabeled oligonucleotide from the circular M13ssDNA. The reactions were performed at 32°C as described in ‘Materials and Methods’ section using the 5′-tailed substrate (lanes 1–9) or the 3′-tailed substrate (lanes 10–18) in the presence of 3 mM ATP and increasing amounts of TWINKLE (150, 300 and 600 fmol) or 3 mM dTTP and increasing amounts of T7gp4 (250, 500 and 1000 fmol). Lanes 1 and 10, 5′-tailed and 3′-tailed substrates heated to 100°C before loading; lanes 2 and 6, untreated 5′-tailed DNA substrate; lanes 11 and 15, untreated 3′-tailed DNA substrate; S, double-stranded substrate; P, single-stranded product. (B) Loading of TWINKLE on ssDNA in presence of UTP. Helicase activity of TWINKLE (300 fmol) was measured at 32°C using the 5′-tailed substrate (lanes 1–6) or the 3′-tailed substrate (lanes 7–12) in presence of 3 mM ATP or 3 mM UTP. Lanes 1, 4, 7 and 10, DNA substrates heated to 100°C before loading.
DNA replication assay
The bubble template used for the DNA replication assay was produced as described (28), but with some modifications. ssDNA produced from pBluescript II SK+ and pBluescript II SK− was isolated according to the manufacturer’s protocol (Stratagene). Single-stranded SK− DNA was linearized by annealing an oligonucleotide (5′-TTC GAT ATC AAG CTT ATC GAT ACC-3′) and then cleaving the partial duplex with HindIII. Linear SK− ssDNA and circular SK+ ssDNA were then mixed in a 1:1.25 ratio and annealed over night at 37°C. The material was run on a 1% agarose gel and the annealed product was purified with a gel extraction kit (Qiagen). After T4 DNA ligase treatment, the ligated DNA (bubble template) was gel purified as above. The bubble template was hybridized to an oligonucleotide (5′-GGCGAA CGT GGC GAG AAA GGAAGG G-3′), which was complementary to the ssDNA-bubble, in a 1:1 molar ratio. The mixture was incubated at 65°C and then left to cool to 20°C.
The DNA replication reaction mixture (15 µl) contained 20 mM Tris–HCl pH 7.5, 7 mM MgCl2, 5 mM DTT, 100 µg/ml BSA, 4 mM UTP, 100 µM dATP, 100 µM dTTP, 100 µM dGTP, 1 µM dCTP, 2 µCi [α-32P] dCTP, 70/220 fmol POLγ A/B and 10 fmol of bubble template. The incubation temperatures as well as the amount of TWINKLE and mtSSB are indicated in the figure legends. Reactions were terminated at the indicated times by addition of 200 µl of stop buffer (10 mM Tris–HCl pH 8, 200 mM NaCl, 1 mM EDTA, 0.1 mg/ml glycogen). The samples were treated with 200 µg/ml of Proteinase K and incubated at 42°C for 1 h. After ethanol precipitation the pellets were dissolved in 10 µl H2O and 10 µl gel-loading buffer (98% formamide, 10 mM EDTA pH 8, 0.025% xylene cyanol FF, 0.025% bromophenol blue), heated at 95°C for 3 min and separated on a 6% denaturing polyacrylamide gel in 1× TBE.
Electrophoresis mobility shift assay
The ssDNA-binding affinity of TWINKLE was assayed by an electrophoresis mobility shift assay (EMSA) using three different probes: a ssDNA linear substrate (5′-GAT37CAT ACCCCTATGAGGGGTATGT38AT-3′); a dsDNA linear substrate (5′-GAT37 CAT ACCCCTATGAGGGGTATG T38AT-3′ annealed to 5′-ATA38 CAT ACC CCT CAT AGG GGT ATG A37TC-3′); and a closed circular ssDNA substrate (5′-GAG GGG TAT GT80 CAT ACC CCT AT-3′). The three probes were labeled in the 5′ end with [γ-32P] ATP using the T4 polynucleotide kinase. The closed circular substrate was constructed by treating the oligonucleotide with CircLigaseTMssDNA ligase (Epicentre). Unligated oligonucleotides were removed by addition of Exonuclease I to the sample, followed by incubation for 1 h at 37°C. The closed circular oligonucleotides were separated on a 10% denaturating polyacrylamide gel and purified by extraction (29). Binding reactions were carried out in 15 µl volumes containing 10 fmol of the indicated DNA template, 20 mM Tris–HCl pH 7.5, 5 mM MgCl2, 4 mM DTT, 100 µg/ml BSA, 10% glycerol and 3 mM UTP (UTP was omitted from the reaction when indicated in the figure legend). Proteins were added as indicated in the figure legends and reactions were incubated at 4°C or 42°C for 10 min. The final NaCl concentration was 50 mM. When indicated TWINKLE was cross-linked at 4°C for 10 min (1 µl of 10% glutaraldehyde was added to the reaction mixture). The samples were loaded directly on a 6% native polyacrylamide gel in 0.5× TBE or, when indicated, 3 µl of stop buffer [90 mM EDTA (pH 8.0), 6% SDS, 30% glycerol, 0.25% bromophenol blue, 0.25% xylene cyanol) was added and the samples were heated at 95°C for 5 min prior to loading.
RESULTS
TWINKLE can load on closed circular ssDNA
TWINKLE is a stable hexamer/heptamer in solution (22,23). We wanted to investigate if the TWINKLE oligomer could load on a circular ssDNA template, i.e. a DNA template without free ends. To this end, we engineered a 100-nt long ssDNA substrate that adopted a circular conformation through annealing of a short complementary region (9 bp, Supplementary Figure S1A). The template was radioactively labeled using polynucleotide kinase and subsequently ligated to create a circular substrate. The substrate was treated with exonuclease I to remove any contaminating linear DNA molecules and subsequently purified by denaturing polyacrylamide gel electrophoresis (Supplementary Figure S1B). We incubated this DNA template with TWINKLE for 10 min at the indicated temperatures and monitored binding by separation on a non-denaturing polyacrylamide gel (Figure 1A, upper panel). As controls in the binding reactions, we used linear dsDNA (100 bp, Figure 1A, middle panel) and ssDNA (100 nt, Figure 1A, lower panel) molecules, which contained free 5′-ends on which TWINKLE could be threaded. We found that TWINKLE could bind the linear substrates at both 4°C and 42°C. Interestingly, under the same conditions, TWINKLE could also bind to the closed circular ssDNA template (Figure 1A, upper panel). We concluded that TWINKLE did not need a free 5′-end in order to load on ssDNA. The TWINKLE helicase preparations used for these experiments did not contain any apparent nuclease activities (data not shown).
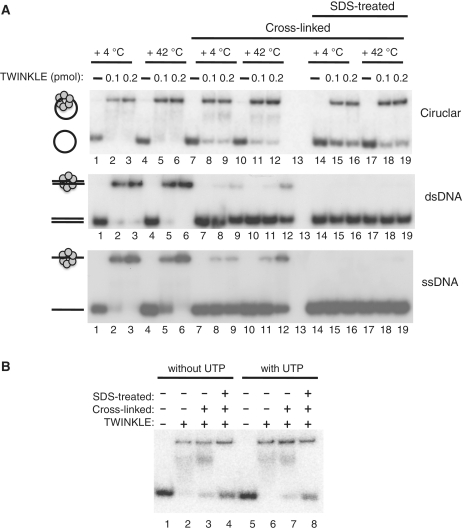
Figure 1.
TWINKLE can efficiently load onto circular DNA substrates. TWINKLE interactions with circular ssDNA, linear ssDNA and linear dsDNA. (A) TWINKLE–DNA interactions were monitored by EMSA using 32P-labeled circular ssDNA (100 nt, upper panel), linear dsDNA (100 bp, middle panel) or linear ssDNA (100 nt, lower panel) as described in ‘Materials and Methods’ section. Incubation temperatures and the amount of TWINKLE (0.1 or 0.2 pmol) are indicated at the top of the figure. Lanes 7–12 and 14–19, were cross-linked with glutaraldehyde and lanes 14–19; were further treated with SDS and heated at +95°C for 5 min. (B) The binding of TWINKLE to circular ssDNA is nucleotide independent. Binding reactions between TWINKLE and closed circular ssDNA in the absence (lanes 1–4) or presence of UTP (lanes 5–8) were performed as described in ‘Materials and Methods’ section. Lanes 1 and 5, DNA substrate alone; lanes 2–4 and 6–8, were incubated with 200 fmol TWINKLE at +4°C; lanes 3–4 and 7–8, were cross-linked with glutaraldehyde; and lanes 4 and 8 were further treated with SDS and heated at +95°C for 5 min.
We next examined if DNA was located in the center of the TWINKLE hexameric ring. To this end we used glutaraldehyde, which preferentially will create protein–protein cross-links. If DNA is located in the central channel, a cross-linked TWINKLE hexameric ring will remain locked onto a circular DNA molecule also after treatment with sodium dodecyl sulfate (SDS), whereas it will fall of a DNA substrate with free ends. In the absence of SDS treatment, cross-linked TWINKLE remained bound to both the linear and circular substrates (Figure 1A). The amount of shifted linear DNA was however lower after treatment with the cross-linking agent (middle and lower panels) compared to the circular template (upper panel). This finding suggests that cross-linked TWINKLE can dissociate from the linear substrate by falling off one of the ends, but once dissociated, the cross-linked oligomer will have difficulties in reloading onto the DNA substrates. Furthermore, when the cross-linked samples were treated with SDS and heated at 95°C for 5 min, we could only observe binding of TWINKLE to the circular template. Glutaraldehyde-treatment of TWINKLE prior to incubation with the template prevented binding to circular ssDNA (data not shown). These results thus demonstrate that the TWINKLE hexamer is locked onto the circular template, supporting the notion that ssDNA is located in the central channel of the TWINKLE ring (Figure 1A, lanes 14–19). SDS-treatment completely abolished binding to the linear template, suggesting that there are no apparent cross-links between protein and DNA under the conditions used for these experiments.
We also performed a competition experiment in the absence of a cross-linking agent, to compare the stability of TWINKLE binding to linear and circular ssDNA templates. As a non-radioactive competitor we used 100-fold molar excess of the linear ssDNA substrate. We observed no apparent disassembly of TWINKLE from the circular ssDNA (60 min incubation at 25°C), whereas TWINKLE binding to linear ssDNA was lost already after 1 min incubation (data not shown). The high stability of the TWINKLE hexamer on circular ssDNA is in good agreement with the processive nature of TWINKLE observed at the mtDNA replication fork in the presence of POLγ and mtSSB (3).
Finally, we investigated if a nucleotide cofactor could influence TWINKLE binding to the circular template. Our experiments demonstrated that TWINKLE binding to ssDNA is not stimulated in the presence of a nucleotide cofactor (Figure 1B). From our experiments, we could thus conclude that TWINKLE could bind and encircle ssDNA independently of nucleotides and additional loading factors. Our findings are partially in conflict with a previous report from our laboratory, which demonstrated that ATP or a non-hydrolyzable ATP analog (ATPγS) could stimulate TWINKLE binding to ssDNA (22). The molecular explanation for the discrepancy between our current findings and previous reports is still unclear, but it is possible that older purification methods generated a somewhat unstable TWINKLE protein conformation, which could be activated/stabilized by addition of a nucleotide cofactor (22,27). Others have also demonstrated that TWINKLE can efficiently interact with linear ssDNA in the absence of a nucleotide cofactor (23).
TWINKLE can initiate unwinding on a closed circular ssDNA molecule
We next investigated if TWINKLE could initiate DNA unwinding on a circular DNA substrate. We constructed two different substrates, by annealing radioactively labeled 60-nt long oligonucleotides to a circular single-stranded M13 DNA molecule. One substrate contained a duplex region of 20-bp and a 40-nt 5′-single-stranded overhang (Figure 2A, lanes 1–9). On the 5′-tailed substrate, the TWINKLE ring can be threaded on the free ssDNA tail and as would be expected, the substrate was efficiently unwound, resulting in the accumulation of a displaced labeled oligonucleotide (Figure 2A, lanes 3–5). For comparison, we used the T7gp4 protein, which was also able to unwind the 5′-tailed substrate (Figure 2A, lanes 7–9). The helicase activity of T7gp4 was measured in the presence of dTTP instead of ATP, since the T7gp4 protein preferentially utilizes dTTP to unwind duplex DNA in vitro (24,30,31).
We next used a substrate with a duplex region of 20-bp and a 40-nt 3′-single-stranded overhang (Figure 2A, lanes 10–18). TWINKLE and T7gp4 are both 5′ to 3′ helicases and to unwind the 3′-tailed substrate they would need to load onto the closed circular DNA strand. The 3′-tailed substrate is therefore more similar to the in vivo situation, where TWINKLE is expected to load onto a closed circular mtDNA molecule. T7gp4 was perfectly able to unwind the 3′-tailed substrate (Figure 2A, lanes 16–18), which corroborates previous studies showing that T7gp4 could load onto circular DNA without help of any accessory factors (24). In contrast, TWINKLE did not display strand displacement activity on the 3′-tailed substrate in the presence of ATP as a source of energy (Figure 2A, lanes 12–14). Since we had previously demonstrated that UTP was a better energy source than ATP for the TWINKLE helicase activity (2), we also performed the assay in the presence of UTP (Figure 2B). Interestingly, we now found that TWINKLE was able to unwind the 3′-tailed substrate, albeit with lower efficiency than that observed for the 5′-tailed substrate (Figure 2B, compare lanes 6 and 12). The results thus suggest that TWINKLE can be loaded on a circular ssDNA and perform displacement activity, but with very low efficiency, under the conditions used.
Temperature dependency of TWINKLE-helicase activity on circular DNA
The observation that TWINKLE can load on a circular ssDNA and perform unwinding in presence of UTP but not ATP (Figure 2B) might suggest that TWINKLE needs more energy to perform this task. We therefore monitored DNA unwinding as a function of increasing temperatures, ranging from 20°C to 45°C (Figure 3A). TWINKLE could effectively unwind the 5′-tailed DNA substrate even at low temperatures and already at 20°C, >50% of the template was unwound (Figure 3A, lane 3). Nearly no strand displacement activity could be measured at the low temperatures on the 3′-tailed DNA substrate (Figure 3A, lanes 11 and 12). However, at higher temperature, TWINKLE was able to unwind the duplex DNA (Figure 3A, lanes 15 and 16) with almost the same efficiency as that observed for the 5′-tailed DNA substrate. At 37°C, we observed 57% unwinding of the 3′-tailed DNA substrate compared to 90% unwinding for the 5′-tailed template.
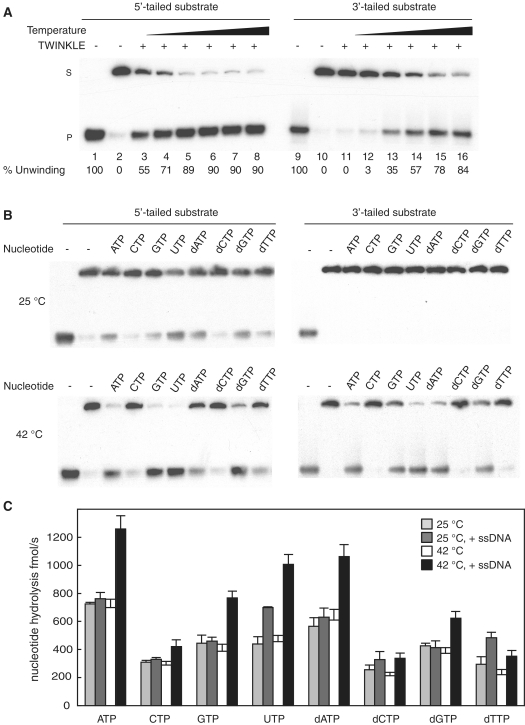
Figure 3.
TWINKLE unwinding of a closed circular template is stimulated at higher temperatures. (A) The helicase activity of TWINKLE (300 fmol) was monitored using the 5′-tailed substrate (lanes 1–8) or the 3′-tailed substrate (lanes 9–16) in the presence of 3 mM UTP and at increasing temperatures (20°C, 25°C, 32°C, 37°C, 41°C and 45°C). After 45 min of incubation at the indicated temperature, the reaction products were separated on a 12.5% non-denaturating polyacrylamide gel. Lanes 1 and 9, DNA substrates heated at 100°C before loading; lanes 2 and 10, DNA substrate incubated at 45°C in the absence of TWINKLE; S, double-stranded substrate; P, single-stranded product. (B) Helicase assays were performed using the 5′-tailed or the 3′-tailed substrates in the presence of 200 fmol of TWINKLE and 3 mM of the indicated nucleotide at 25°C (upper panel) or at 42°C (lower panel). (C) Hydrolysis of different nucleotides by TWINKLE were measured as described in ‘Materials and Methods’ section. The reactions were performed in the presence of 200 fmol of TWINKLE, 0.5 mM of the indicated nucleotide, and in the absence (light gray and white blocks) or presence (dark gray and black bars) of M13ssDNA (188 fmol) at 25°C (light and dark gray bars) or at 42°C (white and black bars).
Nucleotide dependency of the unwinding activity of TWINKLE
We next investigated the effects of different nucleotide cofactors on TWINKLE unwinding (Figure 3B). With the exception of CTP and dCTP, TWINKLE could use all nucleotides tested as an energy source to unwind the 5′-tailed substrate at 25°C. The highest levels of unwinding were observed in the presence of ATP, UTP, GTP and dATP (Figure 3B, upper left panel). The overall enzyme activity was higher at 42°C, but the nucleotide dependence was the same as that observed at 25°C (Figure 3B, lower left panel). Interestingly, we observed no unwinding on the 3′-tailed substrate when the reaction was conducted at 25°C (Figure 3B, upper right panel), regardless of the nucleotide used. In contrast, at 42°C (or at 37°C, data not shown), TWINKLE was able to unwind the 3′-tailed template with a similar nucleotide preference as the one observed with the 5′-tailed substrate.
Nucleotide hydrolysis of TWINKLE
Our results so far demonstrated that TWINKLE is less effective on the 3′-tailed than on the 5′-tailed substrate, but the observed difference was apparently not due to ineffective loading of TWINKLE on the circular template (Figure 1). Instead, the difference could be due to a problem with translocation on ssDNA, since TWINKLE may need to translocate extensive regions of ssDNA before it reaches duplex DNA and can initiate unwinding on the 3′-tailed substrate, whereas it only needs to translocate a short 40-nt long stretch on the 5′-tailed substrate. In agreement with this notion, ssDNA has previously been shown to be a relatively poor stimulator of the TWINKLE ATPase activity, compared to that observed with the related T7gp4 protein (32). This low stimulatory effect could be explained by inefficient translocation of TWINKLE on an ssDNA template. To compare the ability of different nucleotides to stimulate the translocation of TWINKLE, we therefore measured ssDNA dependent hydrolysis in the presence of different nucleotides (Figure 3C). At 25°C and in absence of DNA (Figure 3C, light gray bars), the nucleotide hydrolysis activity of TWINKLE is very low in the presence of CTP, dCTP and dTTP, slightly higher in presence of GTP, UTP and dGTP and maximal in presence of ATP or dATP. In the absence DNA (Figure 3C, white bars), the nucleotide hydrolysis rates at 42°C are roughly similar to those measured at 25°C, demonstrating that higher temperature does not have any major impact on TWINKLE nucleotide hydrolysis in isolation. We next added ssDNA to monitor stimulation of the NTPase activity. Interestingly, addition of ssDNA did not stimulate the NTPase activity further at 25°C, except in the presence of UTP (Figure 3C, dark gray bars). Furthermore, even if ssDNA has a general stimulatory effect on the NTPase activity at 42°C, the stimulation is the strongest for ATP, GTP, UTP and dATP and less pronounced for dGTP. The NTP requirements observed in these data were in nice agreement with those observed for DNA unwinding on the 3′-tailed substrate (Figure 3B). This close correlation supports the notion that poor translocation explains why TWINKLE unwinds the 3′-tailed substrate with lower efficiency compared to the 5′-tailed substrate.
TWINKLE in combination with POLγ can support DNA replication on a circular dsDNA template
Given our findings, it should be possible to initiate DNA synthesis using only mtDNA replication factors and a closed template that mimicked the in vivo situation. To address this possibility, we designed a ‘bubble template’, a double-stranded circular DNA template containing a non-annealed region of 457 nt (a ‘bubble’). To initiate DNA synthesis, the replication machinery requires a primer and we therefore annealed a 25-nt long oligonucleotide in the ‘bubble’ region (Figure 4A). POLγ can bind to the annealed primer and can synthesize DNA until it reaches the double-stranded region, leading to the formation of a 129-nt product (indicated in Figure 4B and C). POLγ displays a delicate balance between its DNA polymerase and 3′ to 5′ exonuclease activities. In our reactions we used dNTP concentrations >1 µM, which favors net polymerization and prevents the exonuclease activity (26,33). Addition of TWINKLE to the reaction, allowed for DNA unwinding and for continued DNA synthesis into the dsDNA region of the bubble template. In the presence of TWINKLE we observed the formation of long DNA products (Figure 4B, lanes 2–4). We could thus conclude that TWINKLE can be loaded on a structure that is similar to the D-loop structure in vivo and support DNA synthesis. In the experiments we used UTP as the source of energy, but ATP and GTP could also support initiation of DNA replication on the bubble DNA template (Supplementary Figure S1C).
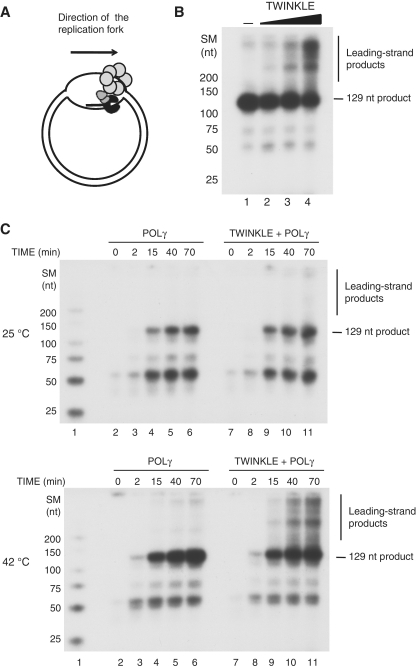
Figure 4.
TWINKLE in combination with POLγ can support DNA replication on a circular dsDNA template. (A) The bubble template was prepared as described in ‘Materials and Methods’ section. The tentative localization of TWINKLE (light gray hexamer) and POLγ (dark gray and black heterotrimer) are indicated. The bubble region is symmetric and may contain one annealed primer on each strand, allowing initiation of DNA synthesis in opposite directions. (B) DNA replication assays were performed using the bubble template (10 fmol), POLγ (70 fmol), and increasing amounts of TWINKLE (0, 50, 200 and 750 fmol) for 60 min at 42°C. (C) DNA replication assays were performed as in (B) but in presence or absence of 750 fmol TWINKLE. The reactions were incubated for 0, 2, 15, 40 or 70 min at either 25°C (upper panel) or 42°C (lower panel). SM, size marker.
To further verify that TWINKLE could support initiation of DNA synthesis on a circular substrate, we performed time course experiments at 25°C and 42°C.
At 25°C, POLγ was able to utilize the primer and synthesize DNA until it reached the double-stranded region (Figure 4C, upper panel lanes 2–6). Addition of TWINKLE did not have any significant effect on the DNA synthesis reaction (Figure 4C, upper panel lanes 7–11). In contrast, when the temperature is increased to 42°C (and to 37°C, data not shown), TWINKLE supports the DNA synthesis by POLγ and allows the polymerase to utilize dsDNA (Figure 4C, lower panel lanes 9–11). We conclude from these experiments that TWINKLE is unable to efficiently translocate on a DNA template at lower temperatures even in the presence of POLγ.
mtSSB does not prevent loading of TWINKLE on ssDNA
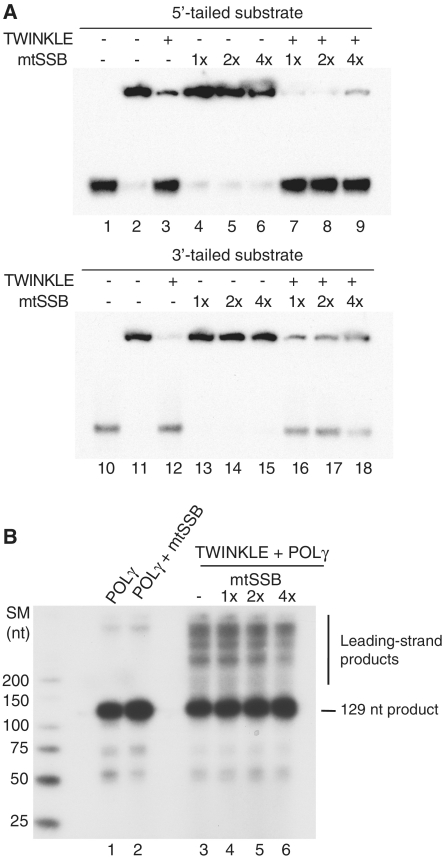
In vivo, the long stretches of ssDNA formed during mtDNA replication are stabilized and protected by the mitochondrial ssDNA binding protein (mtSSB). In some other systems e.g. bacteriophage T4 and HSV 1, specialized enzymes (gene 59 and UL8 respectively) are required to load the replicative helicase on SSB coated ssDNA (34,35). We therefore investigated if ‘coating’ by mtSSB could prevent TWINKLE from loading and initiating unwinding on a ssDNA template. The tailed DNA substrates (Figure 2A) were therefore pre-incubated with mtSSB, before TWINKLE was added to the reactions. We found that TWINKLE could efficiently load and unwind the duplex DNA even when ssDNA was fully coated by mtSSB (Figure 5A, 1×). Only at very high mtSSB concentration (4-fold excess, 4×), a slight inhibition of the unwinding activity could be noticed (Figure 5A, lanes 9–18).
Figure 5.
mtSSB does not prevent the loading of TWINKLE onto ssDNA. (A) Helicase assays were performed as described in ‘Materials and Methods’ section. Reaction mixtures contained 5 fmol of 5′-tailed or 3′-tailed DNA substrates (Figure 2A), 200 fmol of TWINKLE, and increasing amounts of mtSSB (2.5, 5 and 10 pmol of the mtSSB tetramer). The saturation levels of mtSSB relative the DNA substrates are indicated and based on the assumption that one mtSSB tetramer covers 60 nt (44). The DNA substrates were first incubated with mtSSB at room temperature for 10 min. TWINKLE was then added to the reactions and the helicase assay was performed at 42°C for 45 min in the presence of 3 mM UTP. (B) DNA replication assays were performed as described in ‘Materials and Methods’ section. The reaction mixture contained 10 fmol of the bubble template (Figure 4A), 70 fmol of POLγ, 750 fmol of TWINKLE, and increasing amounts of mtSSB (150, 300 and 600 fmol of mtSSB tetramer). The saturation levels of mtSSB relative the DNA substrates are indicated and the reactions were incubated at 37°C for 1 h.
We also monitored if mtSSB could affect initiation of DNA synthesis on the bubble template. As observed in Figure 5B, mtSSB did not prevent loading of TWINKLE and initiation of DNA synthesis on the bubble template, even if a slight reduction in DNA synthesis levels could be observed at very high mtSSB concentrations (Figure 5B, lane 6). We could therefore conclude that TWINKLE can support DNA synthesis on an mtSSB coated DNA substrate.
DISCUSSION
In mitochondria, transcription and initiation of DNA replication are linked. The mitochondrial RNA polymerase (POLRMT) is not only required for transcription but also generates the RNA primers used to initiate DNA synthesis at oriH and oriL (4,6,12,36). Leading-strand DNA synthesis initiated at oriH is primed by an RNA primer of ~100-nt formed by transcription from the light-strand promoter (LSP) (12,37,38). The majority of the DNA synthesis events initiated at oriH is terminated ~600-bp downstream the origin and the products stay stably hybridized to the parental strand, forming a triple-stranded structure, denoted the displacement loop (D-loop) (4). Even if our laboratories have reconstituted both mitochondrial transcription and mtDNA replication in vitro, we have so far not been able to couple these two events and obtain transcription-dependent initiation of DNA replication at oriH in vitro. One possible explanation for this observation could be the need for a helicase loading factor that actively loads TWINKLE on to the closed mtDNA template. In the current manuscript, we demonstrate that this is not the case, since similar to T7gp4, TWINKLE can load onto DNA by itself (14,39). TWINKLE can efficiently place itself onto circular ssDNA even in absence ofNTPs as a source of energy. Since TWINKLE is a closed circular hexamer/heptamer in solution there must be a transient parting of one of the subunit interfaces to insert DNA in to the central channel (22,23). We also demonstrate that TWINKLE efficiently can load and translocate on mtSSB-coated substrates. This finding is in contrasts to other systems, e.g. bacteriophage T4 and Herpes simplex virus type I, in which SSB coated substrates have been shown to inhibit the helicase activity. In these systems but not in the bacteriophage T7 system, a specific protein is required to assemble the helicase onto SSB-coated ssDNA (34,35). It should be emphasized that even if our findings conclusively demonstrate that TWINKLE can function in the absence of a specialized helicase-loading factor, we cannot exclude the possibility that other factors influence, e.g. stimulate helicase loading in vivo.
As also demonstrated here, TWINKLE binds in a very stable manner to a circular ssDNA molecule. This finding suggests that once the hexameric ring has closed around the ssDNA in the central channel, TWINKLE becomes processive and stays on the DNA until it reaches a free DNA end or DNA replication is terminated. Given the physical properties of the protein, it would be of interest to investigate the behavior of TWINKLE when the replisome encounters a roadblock, such as DNA damage. Does TWINKLE stall until other enzymes take care of the damage and then restarts unwinding? Or are there specialized proteins required for the regulated disassembly of TWINKLE at a stalled replication forks or after DNA replication has been completed? These questions may also be of relevance for our understanding of normal mtDNA replication and D-loop formation. The mechanisms that govern replication termination and the fate of the replication machinery at the end of the D-loop are still not understood in mammalian cells. In sea urchin, mtDBP protein has been suggested to terminate DNA synthesis at a specific site in the genome and stimulate D loop formation (40). MtDBP displays a contrahelicase activity, which can inhibit the activity of the replicative helicase and thus block replication fork progression (41). In mammalian cells, a related mechanism may exist, since short conserved DNA elements, denoted TAS are located at the 3′ end of the nascent D-loop H strand (42). In addition, an unidentified 48-kDa protein has been shown bind to the TAS sequence in bovine mitochondria (43). The fate of TWINKLE and other components of the mitochondrial replisome at the mammalian TAS sequence remain unclear. Does TWINKLE encounter a contrahelicase related to mtDBP and does this lead to dissociation of TWINKLE from the template, or does the mitochondrial replisome remain bound at the TAS sequence, waiting for a signal that leads to continued DNA synthesis? In the current report, we have demonstrated that it is possible to initiate DNA synthesis on a closed circular template with an artificial D-loop structure. With this bubble template, we can now introduce specific mtDNA elements and investigate their effects of mtDNA replication. We can thus investigate if the TAS sequence itself promotes site-specific termination of DNA synthesis or if an additional TAS-binding protein is required. Using our reconstituted in vitro system and the bubble template, we can directly search for protein factors required for TAS-dependent termination of DNA replication in mitochondrial extracts.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swedish Research Council (grant numbers 2010-3545 to M.F., and 2009-4848 to C.G.); Swedish Cancer Society (grant numbers 2007/1216 and 2007/1240); the Swedish Strategic Foundation (FFL-3); European Research Council (starting grant no. 261248 and advanced grant no. 268897). Funding for open access charge: Swedish Research Council (grant numbers 2010-3545).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Spelbrink JN, Li FY, Tiranti V, Nikali K, Yuan QP, Tariq M, Wanrooij S, Garrido N, Comi G, Morandi L, et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat. Genet. 2001;28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 2.Korhonen JA, Gaspari M, Falkenberg M. TWINKLE Has 5′ -> 3′ DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J. Biol. Chem. 2003;278:48627–48632. doi: 10.1074/jbc.M306981200. [DOI] [PubMed] [Google Scholar]
- 3.Korhonen JA, Pham XH, Pellegrini M, Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 2004;23:2423–2429. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 5.Berk AJ, Clayton DA. Mechanism of mitochondrial DNA replication in mouse L-cells: asynchronous replication of strands, segregation of circular daughter molecules, aspects of topology and turnover of an initiation sequence. J. Mol. Biol. 1974;86:801–824. doi: 10.1016/0022-2836(74)90355-6. [DOI] [PubMed] [Google Scholar]
- 6.Fuste JM, Wanrooij S, Jemt E, Granycome CE, Cluett TJ, Shi Y, Atanassova N, Holt IJ, Gustafsson CM, Falkenberg M. Mitochondrial RNA polymerase is needed for activation of the origin of light-strand DNA replication. Mol. Cell. 2010;37:67–78. doi: 10.1016/j.molcel.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 7.Wong TW, Clayton DA. In vitro replication of human mitochondrial DNA: accurate initiation at the origin of light-strand synthesis. Cell. 1985;42:951–958. doi: 10.1016/0092-8674(85)90291-0. [DOI] [PubMed] [Google Scholar]
- 8.Clayton DA. Replication and transcription of vertebrate mitochondrial DNA. Annu. Rev. Cell Biol. 1991;7:453–478. doi: 10.1146/annurev.cb.07.110191.002321. [DOI] [PubMed] [Google Scholar]
- 9.Gillum AM, Clayton DA. Mechanism of mitochondrial DNA replication in mouse L-cells: RNA priming during the initiation of heavy-strand synthesis. J. Mol. Biol. 1979;135:353–368. doi: 10.1016/0022-2836(79)90441-8. [DOI] [PubMed] [Google Scholar]
- 10.Cantatore P, Attardi G. Mapping of nascent light and heavy strand transcripts on the physical map of HeLa cell mitochondrial DNA. Nucleic Acids Res. 1980;8:2605–2625. doi: 10.1093/nar/8.12.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montoya J, Christianson T, Levens D, Rabinowitz M, Attardi G. Identification of initiation sites for heavy-strand and light-strand transcription in human mitochondrial DNA. Proc. Natl Acad. Sci. USA. 1982;79:7195–7199. doi: 10.1073/pnas.79.23.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pham XH, Farge G, Shi Y, Gaspari M, Gustafsson CM, Falkenberg M. Conserved sequence box II directs transcription termination and primer formation in mitochondria. J. Biol. Chem. 2006;281:24647–24652. doi: 10.1074/jbc.M602429200. [DOI] [PubMed] [Google Scholar]
- 13.Wanrooij PH, Uhler JP, Simonsson T, Falkenberg M, Gustafsson CM. G-quadruplex structures in RNA stimulate mitochondrial transcription termination and primer formation. Proc. Natl Acad. Sci. USA. 2010;107:16072–16077. doi: 10.1073/pnas.1006026107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Picha KM, Ahnert P, Patel SS. DNA binding in the central channel of bacteriophage T7 helicase-primase is a multistep process. Nucleotide hydrolysis is not required. Biochemistry. 2000;39:6401–6409. doi: 10.1021/bi992857i. [DOI] [PubMed] [Google Scholar]
- 15.Egelman EH, Yu X, Wild R, Hingorani MM, Patel SS. Bacteriophage T7 helicase/primase proteins form rings around single-stranded DNA that suggest a general structure for hexameric helicases. Proc. Natl Acad. Sci. USA. 1995;92:3869–3873. doi: 10.1073/pnas.92.9.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morris PD, Raney KD. DNA helicases displace streptavidin from biotin-labeled oligonucleotides. Biochemistry. 1999;38:5164–5171. doi: 10.1021/bi9822269. [DOI] [PubMed] [Google Scholar]
- 17.Davey MJ, O’Donnell M. Replicative helicase loaders: ring breakers and ring makers. Curr. Biol. 2003;13:R594–R596. doi: 10.1016/s0960-9822(03)00523-2. [DOI] [PubMed] [Google Scholar]
- 18.Bujalowski W. Expanding the physiological role of the hexameric DnaB helicase. Trends Biochem. Sci. 2003;28:116–118. doi: 10.1016/S0968-0004(03)00034-3. [DOI] [PubMed] [Google Scholar]
- 19.Mott ML, Erzberger JP, Coons MM, Berger JM. Structural synergy and molecular crosstalk between bacterial helicase loaders and replication initiators. Cell. 2008;135:623–634. doi: 10.1016/j.cell.2008.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fanning E, Knippers R. Structure and function of simian virus 40 large tumor antigen. Annu. Rev. Biochem. 1992;61:55–85. doi: 10.1146/annurev.bi.61.070192.000415. [DOI] [PubMed] [Google Scholar]
- 21.Ahnert P, Picha KM, Patel SS. A ring-opening mechanism for DNA binding in the central channel of the T7 helicase-primase protein. EMBO J. 2000;19:3418–3427. doi: 10.1093/emboj/19.13.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farge G, Holmlund T, Khvorostova J, Rofougaran R, Hofer A, Falkenberg M. The N-terminal domain of TWINKLE contributes to single-stranded DNA binding and DNA helicase activities. Nucleic Acids Res. 2008;36:393–403. doi: 10.1093/nar/gkm1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ziebarth TD, Gonzalez-Soltero R, Makowska-Grzyska MM, Nunez-Ramirez R, Carazo JM, Kaguni LS. Dynamic effects of cofactors and DNA on the oligomeric state of human mitochondrial DNA helicase. J. Biol. Chem. 2010;285:14639–14647. doi: 10.1074/jbc.M109.099663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel SS, Picha KM. Structure and function of hexameric helicases. Annu. Rev. Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 25.Matson SW, Tabor S, Richardson CC. The gene 4 protein of bacteriophage T7. Characterization of helicase activity. J. Biol. Chem. 1983;258:14017–14024. [PubMed] [Google Scholar]
- 26.Farge G, Pham XH, Holmlund T, Khorostov I, Falkenberg M. The accessory subunit B of DNA polymerase gamma is required for mitochondrial replisome function. Nucleic Acids Res. 2007;35:902–911. doi: 10.1093/nar/gkl1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korhonen JA, Pande V, Holmlund T, Farge G, Pham XH, Nilsson L, Falkenberg M. Structure-function defects of the TWINKLE linker region in progressive external ophthalmoplegia. J. Mol. Biol. 2008;377:691–705. doi: 10.1016/j.jmb.2008.01.035. [DOI] [PubMed] [Google Scholar]
- 28.Harrington C, Perrino FW. Initiation of RNA-primed DNA synthesis in vitro by DNA polymerase alpha-primase. Nucleic Acids Res. 1995;23:1003–1009. doi: 10.1093/nar/23.6.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Matson SW, Richardson CC. DNA-dependent nucleoside 5′-triphosphatase activity of the gene 4 protein of bacteriophage T7. J. Biol. Chem. 1983;258:14009–14016. [PubMed] [Google Scholar]
- 31.Satapathy AK, Crampton DJ, Beauchamp BB, Richardson CC. Promiscuous usage of nucleotides by the DNA helicase of bacteriophage T7: determinants of nucleotide specificity. J. Biol. Chem. 2009;284:14286–14295. doi: 10.1074/jbc.M900557200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holmlund T, Farge G, Pande V, Korhonen J, Nilsson L, Falkenberg M. Structure-function defects of the twinkle amino-terminal region in progressive external ophthalmoplegia. Biochim. Biophys. Acta. 2009;1792:132–139. doi: 10.1016/j.bbadis.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 33.Atanassova N, Fuste JM, Wanrooij S, Macao B, Goffart S, Backstrom S, Farge G, Khvorostov I, Larsson NG, Spelbrink JN, et al. Sequence-specific stalling of DNA polymerase gamma and the effects of mutations causing progressive ophthalmoplegia. Hum. Mol. Genet. 2011;20:1212–1223. doi: 10.1093/hmg/ddq565. [DOI] [PubMed] [Google Scholar]
- 34.Cha TA, Alberts BM. Effects of the bacteriophage T4 gene 41 and gene 32 proteins on RNA primer synthesis: coupling of leading- and lagging-strand DNA synthesis at a replication fork. Biochemistry. 1990;29:1791–1798. doi: 10.1021/bi00459a018. [DOI] [PubMed] [Google Scholar]
- 35.Falkenberg M, Bushnell DA, Elias P, Lehman IR. The UL8 subunit of the heterotrimeric herpes simplex virus type 1 helicase-primase is required for the unwinding of single strand DNA-binding protein (ICP8)-coated DNA substrates. J. Biol. Chem. 1997;272:22766–22770. doi: 10.1074/jbc.272.36.22766. [DOI] [PubMed] [Google Scholar]
- 36.Wanrooij S, Fuste JM, Farge G, Shi Y, Gustafsson CM, Falkenberg M. Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro. Proc. Natl Acad. Sci. USA. 2008;105:11122–11127. doi: 10.1073/pnas.0805399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee DY, Clayton DA. RNase mitochondrial RNA processing correctly cleaves a novel R loop at the mitochondrial DNA leading-strand origin of replication. Genes Dev. 1997;11:582–592. doi: 10.1101/gad.11.5.582. [DOI] [PubMed] [Google Scholar]
- 38.Kang D, Miyako K, Kai Y, Irie T, Takeshige K. In vivo determination of replication origins of human mitochondrial DNA by ligation-mediated polymerase chain reaction. J. Biol. Chem. 1997;272:15275–15279. doi: 10.1074/jbc.272.24.15275. [DOI] [PubMed] [Google Scholar]
- 39.Yu X, Hingorani MM, Patel SS, Egelman EH. DNA is bound within the central hole to one or two of the six subunits of the T7 DNA helicase. Nat. Struct. Biol. 1996;3:740–743. doi: 10.1038/nsb0996-740. [DOI] [PubMed] [Google Scholar]
- 40.Fernandez-Silva P, Polosa PL, Roberti M, Di Ponzio B, Gadaleta MN, Montoya J, Cantatore P. Sea urchin mtDBP is a two-faced transcription termination factor with a biased polarity depending on the RNA polymerase. Nucleic Acids Res. 2001;29:4736–4743. doi: 10.1093/nar/29.22.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polosa PL, Deceglie S, Roberti M, Gadaleta MN, Cantatore P. Contrahelicase activity of the mitochondrial transcription termination factor mtDBP. Nucleic Acids Res. 2005;33:3812–3820. doi: 10.1093/nar/gki693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doda JN, Wright CT, Clayton DA. Elongation of displacement-loop strands in human and mouse mitochondrial DNA is arrested near specific template sequences. Proc. Natl Acad. Sci. USA. 1981;78:6116–6120. doi: 10.1073/pnas.78.10.6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Madsen CS, Ghivizzani SC, Hauswirth WW. Protein binding to a single termination-associated sequence in the mitochondrial DNA D-loop region. Mol. Cell. Biol. 1993;13:2162–2171. doi: 10.1128/mcb.13.4.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang C, Curth U, Urbanke C, Kang C. Crystal structure of human mitochondrial single-stranded DNA binding protein at 2.4A resolution. Nat. Struct. Biol. 1997;4:153–157. doi: 10.1038/nsb0297-153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.