Abstract
The Shelterin complex associates with telomeres and plays an essential role in telomere protection and telomerase regulation. In its most abundant form, the complex is composed of six core components: TRF1, TRF2, POT1, TIN2, TPP1 and RAP1. Of these subunits, three can interact directly with either single-stranded (POT1) or double-stranded (TRF1, TRF2) telomeric DNA. In this report, we have developed assays to measure the DNA binding activity of Shelterin complexes in human cell extracts. With these assays, we have characterized the composition and DNA binding specificity of two Shelterin complexes: a 6-member complex that contains all six core components and a second complex that lacks TRF1. Our results show that both of these complexes bind with high affinity (KD = 1.3–1.5 × 10−9 M) and selectively to ds/ss-DNA junctions that carry both a binding site for POT1 (ss-TTAGGGTTAG) and a binding site for the SANT/Myb domain of TRF1 or TRF2 (ds-TTAGGGTTA). This DNA binding specificity suggests the preferential recruitment of these complexes to areas of the telomere where ss- and ds-DNA are in close proximity, such as the 3′-telomeric overhang, telomeric DNA bubbles and the D-loop at the base of T-loops.
INTRODUCTION
Telomeres are essential structures that cap and protect the ends of linear chromosomes. Telomeres hide chromosomal ends from DNA damage sensing mechanisms and the DNA repair machinery (1,2) (http://www.els.net/). When telomeres become dysfunctional, ends are sensed as double-stranded DNA breaks (ds-DNA breaks). This recognition results in the activation of DNA damage checkpoints that trigger senescence or apoptosis as well as attempts by the cells to repair the ends by non-homologous end-joining (NHEJ), a process that creates interchromosomal fusions. The protective function of telomeres is mediated by the activities of telomere-associated protein complexes and by the T-loops (3,4). Telomeres are made of tandem copies of a simple DNA repeat, (TTAGGG)n, which the enzyme telomerase synthesizes (5,6). Most of the telomere is composed of duplex telomeric DNA with the exception of the last 50–500 bases of the G-rich strand, which forms a single-stranded 3′-telomeric overhang (1,2). Evidence suggests that this 3′-overhang is sequestered into a large lariat structure, termed the T-loop (3,7). Formation of this structure is presumed to involve the insertion of the 3′-telomeric overhang into duplex telomeric DNA, its hybridization with the C-rich strand, and dislodgment of the G-rich strand into a displacement loop (D-loop). It has been proposed that T-loops are especially well-adapted to shield the ends of chromosomes from DNA repair and DNA damage-sensing mechanisms (3,7). Several DNA binding proteins interact selectively with telomeric DNA and localize to telomeres. TRF1 and TRF2 (Telomeric Repeat Factors 1 and 2) recognize duplex telomeric DNA whereas POT1 (Protection of Telomere 1) associates with the ss-telomeric DNA present at the 3′-overhang and in the D-loop (8,9). Via protein–protein interactions, these DNA binding factors bring a number of other proteins to telomeres, where they localize to form a variety of complexes (1,10). Among these complexes is the Shelterin/Telosome complex, which plays a key role in telomere capping and telomerase regulation.
The Shelterin/Telosome complex brings the three telomeric DNA binding factors (TRF1, TRF2 and POT1) together in the same large complex (4,10–12). In its most abundant form, the complex is composed of six core components: TRF1, TRF2, POT1, TIN2, TPP1 and RAP1. The scaffolding subunit TIN2 binds simultaneously to TRF1, TRF2 and TPP1, whereas TPP1 connects POT1 with TIN2 (11,13–18). A sixth component, RAP1, is a TRF2-associated factor (19). In addition to this 6-member complex, Shelterin may also be present in the form of sub-complexes that lack either TRF1 or TRF2/RAP1 (10,12,17,20–25). While the importance of these different sub-complexes remains unclear, the particular function of each of the three DNA binding subunits (TRF1, TRF2 and POT1) is well-understood. For example, POT1 serves to hide the 3′-telomeric overhang from telomerase and from DNA damage sensing mechanisms. Hence, the loss of POT1 leads to the activation of the ATR kinase, formation of telomere dysfunction-induced foci (TIFs), and induction of either apoptosis or cell cycle arrest (22,26–28). In cells that lack functional checkpoints, POT1 dysfunction leads to a loss of telomerase regulation that results in longer telomeres (18,29). The same effects, along with the loss of telomeric POT1, have been observed in cells expressing POT1 mutants that fail to interact with TPP1 or after depletion of TRF1, TPP1 or TIN2 (10,13–16,18,22,29–32). These observations are consistent with the idea that POT1 performs its functions as part of the Shelterin complex.
On the other hand, TRF2 serves to block recognition of telomeres as ds-DNA breaks. Loss of TRF2 function leads to the activation of the ATM kinase, formation of TIFs, and induction of either senescence or apoptosis (20,26,33–36). In cells lacking functional checkpoints, telomeres devoid of TRF2 serve as substrates for NHEJ and give rise to interchromosomal fusions (25,37). TRF2 also appears to be important for the formation and maintenance of T-loops. In vitro studies have shown that purified TRF2 can promote the folding of artificial telomeres into T-loop structures, a property not displayed by TRF1 (7,38). Amiard et al. (39) have described properties of TRF2 that may explain its ability to form T-loops. Their results show that DNA is positively supercoiled by the binding of TRF2 but not TRF1. In rotationally restricted duplex DNA molecules, this energy was released by the untwisting of flanking DNA, a process that made the untwisted strands available for hybridization to ss-telomeric DNA probes (39). At the telomeres, this untwisting of the DNA helix at sites flanking TRF2 could promote invasion by the 3′-telomeric overhang. TRF2 also appears to be important for T-loop stability. As recently demonstrated by Fouche et al. (40), the N-terminal basic domain of TRF2 (amino acids 1–44) binds ss-DNA in a sequence-independent manner and can associate with a multitude of DNA topologies, including 3-way and 4-way junctions, model replication forks, chickenfoot structures and ds/ss-junctions. Expression of a TRF2 mutant that lacks this domain results in catastrophic deletions of T-loops through what appears to be XRCC3-mediated homologous recombination (41). Based on these observations, it has been proposed that TRF2 protects telomeres, at least in part, through its ability to promote T-loop formation (3,7). However, what is still unclear is whether these many functions are all being accomplished by TRF2 in the context of the Shelterin complex. One confounding factor is the evidence that TRF2 may be part of other complexes as well. A recent study has found isolated chromatin to contain a vast molar excess of TRF2, TIN2 and RAP1 compared to TRF1, TPP1 and POT1, thereby raising the possibility of a separate TRF2/RAP1/TIN2 complex (24). Moreover, the effects of TRF2 depletion were not recapitulated by the loss of the other Shelterin components TRF1, TPP1 or POT1 (10,18,22,26,29–32). Hence, it may be that telomeres are populated by Shelterin complexes and by other TRF1- and TRF2-containing complexes, each performing their own distinct functions.
Among telomere-associated complexes, Shelterin complexes are unique in that they bring together the DNA binding activities of POT1, TRF1 and TRF2. This arrangement most certainly provides these complexes with their own unique mode of interaction with the telomere, the understanding of which should shed light on how and where these complexes function at the telomere. Current models of the interactions of these complexes with the telomere are extrapolated from the well-characterized DNA binding properties of recombinant TRF1, TRF2 and POT1. Recombinant POT1 binds with high affinity (KD = 9.5 nM) to single-stranded 5′-TTAGGGTTAG-3′ by means of two oligonucleotide/oligosaccharide binding (OB) folds (8,42,43). As shown by X-ray crystallography, the first OB-fold (OB1) interacts with 5′-TTAGGG whereas the second fold (OB2) associates with downstream TTAG-3′ (43). TRF2 and TRF1 share the same basic architecture defined by two conserved regions: a TRFH domain that mediates homodimerization and a carboxy-terminal DNA binding domain of the SANT/Myb family (9). The two factors give rise to homodimers that bind selectively to ds-telomeric DNA (9,44). As determined by SELEX (systematic evolution of ligands by exponential enrichment) and electrophoretic mobility shift assays, telomeric DNA recognition by TRF1 dimers requires two copies of an YTAGGGTTR motif (Y = C/T, R = A/G), which the dimer can recognize without constraints on either spacing or orientation (45). This flexibility has been attributed to the flexible hinge region that separates the dimerization and DNA binding domains. While the interactions of TRF1, TRF2 and POT1 with DNA have been extensively investigated, how these factors combine to dictate the DNA binding properties of Shelterin complexes remains poorly understood. Shelterin complexes are most often depicted as being bound to tracks of ds-telomeric DNA, and in some cases with POT1 interacting simultaneously with the 3′-telomeric overhang. Yet, the modes of interaction of these complexes with the telomere, their DNA binding affinity and specificity are still very poorly defined. Major shortfalls have been the inability to reconstitute these complexes in vitro using purified recombinant proteins and the lack of an assay to detect the activity of the native complexes in cell extracts.
In this article, we describe an electrophoretic mobility shift assay and a pulldown assay which allow detection of the DNA binding activity of Shelterin complexes in human cell extracts. With these assays, we have characterized the composition and DNA binding specificity of Shelterin complexes present in HeLa and HT1080 cell extracts. Our results show that these complexes bind preferentially to DNA fragments that contain a binding site for POT1 and a binding site for the Myb domain of TRF1 or TRF2. This binding specificity would be expected to recruit these complexes to regions of the telomere where ss- and ds-DNA are in close proximity, such as the 3′-telomeric overhang, telomeric DNA bubbles, and the D-loop at the base of T-loops.
MATERIALS AND METHODS
Materials
Oligonucleotides were synthesized by the Eppley Core Facility (University of Nebraska Medical Center, Omaha, NE, USA) or Integrated DNA Technologies Inc. (Coralville, IA, USA). Plasmids pTetFLAGhTRF245-501 (25) and pTethTRF1 (31) were gifts from Dr Titia de Lange (Rockefeller University, New York, NY, USA). Restriction enzyme PpiI was purchased from Fermentas (Hanover, MD, USA), whereas HgaI and MlyI were obtained from New England BioLabs (Beverly, MA, USA). Unless otherwise specified, all other enzymes were either from Invitrogen (Carlsbad, CA, USA) or Promega (Madison, WI, USA). γ-[32P]-ATP was obtained from MP Biologicals (Solon, OH, USA). DMEM, Trypsin–EDTA, Penicillin/Streptomycin and Lipofectamine 2000 were from Invitrogen. Fetal bovine serum, Transferrin and Microcystin-LR were from HyClone (Logan, UT, USA), Collaborative Biomedical Products (Bedford, MA, USA) and Cayman Chemical (Ann Arbor, MI, USA), respectively. Mammalian protease inhibitor cocktail (cat # P8340), 3XFLAG peptide and all other chemicals were from Sigma-Aldrich (St Louis, MO, USA). M-450 magnetic beads coated with a Sheep anti-Mouse IgG antibody were purchased from Dynal Biotech. Inc. (Lake Success, NY, USA).
Antibodies and western blot analysis
Western blot analyses were performed as described previously (46). Mouse monoclonal antibodies used in western blots were against the Flag tag (M2, Stratagene, La Jolla, CA, USA) and TPP1 (1D8-1B6, Novus Biologicals). Rabbit polyclonal antibodies were against TRF2 (H-300, Santa Cruz Biotech.), TRF1 (ab1423, Abcam Inc.), RAP1 (A300-306A, Bethyl Lab. Inc.) and Actin (I-19, Santa Cruz Biotech.). Antibodies used for supershifts were the same as above plus antibodies against TIN2 (mouse monoclonal 59B388, Abcam Inc.), POT1 (rabbit polyclonal IMG-5343A, IMGENEX), Vimentin (mouse monoclonal sc-6260, Santa Cruz Biotech.) and c-Fos (mouse monoclonal sc-047, Santa Cruz Biotech). Normal IgG from rabbit and mouse were purchased from Calbiochem and Santa Cruz Biotech, respectively.
siRNA transfections
siGENOME ON-TARGETplus SMARTpool duplexes directed against the human TPP1 (L-014237), TIN2 (L-019951), POT1 (L-004205) and TRF1 (L-010542) mRNAs, as well as the non-targeting SMARTpool (D-001810-10), were from Dharmacon (Lafayette, CO, USA). HT1080-TRF2 cells were transfected with the different SMARTpool using DharmaFECT1 transfection reagent, according to Dharmacon’s instructions. Seventy-two hours following transfection, nuclear extracts were prepared as described below and total RNA was isolated using the TRIzol reagents (Invitrogen) following the manufacturer’s instructions.
Real-time qPCR
Real-time PCR quantification of the GAPDH, TRF2, TRF1, TPP1, TIN2 and POT1 mRNA was performed using a SmartCycler System (Cepheid, Sunnyvale, CA, USA). Reverse Transcriptions were done as described previously using the SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA) (46). The cDNA generated was subjected to SYBR green qPCR on the Cepheid SmartCycler® using software version 2.0 c. Gene expression was assayed using RT2 Real-TimeTM SYBR Green PCR master mix from SA BioSciences (Frederick, MD, USA) according to the manufacturer's protocol using PCR primers for GAPDH (5′-AGGTCGGTGTGAACGGATTTG-3′ and 5′-TGTAGACCATGTAGTTGAGGTCA-3′), TRF2 (5′-AGACTTGGGTGGAAGAGGA-3′ and 5′-TAATCATCACAGCTGTTCGG-3′), TRF1 (5′-TCTGCGGTAACTGAATCCTC-3′ and 5′-GTTACCGGCTGACTCTTTGA-3′), TPP1 (5′-GGGAGGACCAGGAGCAT-3′ and 5′-GGGCCTAGAGAGCTCAGAAT-3′), TIN2 (5′-TTGCCTGGAGACAATATGGT-3′ and 5′-GTCGGCCAGCTAGAGGTT-3′) and POT1 (5′-TGTGGCAAGATCTCTGAAGG-3′ and 5′-TCTGAATGCTGATTGGCTGT-3′). To quantify differences in gene expression, all experiments were designed to include a standard curve of a serially diluted pool made of all cDNAs. For each siRNA SMARTpool transfected, levels of the targeted mRNA and of the TRF2 and GAPDH mRNAs were measured, comparing the targeted cells to those transfected with the non-targeting SMARTpool. For each mRNA, expression in the targeted cells was reported as a fraction of that observed in the control cells transfected with the non-targeting siRNA, with the later arbitrarily set to a value of 1.
Expression vectors
Plasmids pcDNA3.1-Flag-TRF2ΔB and pcDNA3.1-Flag-TRF2 were made from their pCMV1 equivalents, by transfer of their TRF2 cassettes to vector pcDNA3.1(–) (Invitrogen). pCMV1-Flag-TRF2ΔB was made by the transfer of a SacII–BamHI fragment from plasmid pTetFLAGhTRF245-501 (a gift from Titia de Lange, Rockefeller University, NY, USA) (25) to pCMV1. To generate pCMV-Flag-TRF2, plasmid pCMV-Flag-TRF2ΔB was cut with EcoRI (between the Flag tag and TRF2ΔB) and with SacI (within TRF2ΔB) to replace the N-terminus of TRF2ΔB with an EcoRI/SacI-digested PCR product amplified from pCMV-SPORT6-TRF2 (Open Biosystem, BC024890 encoding full-length TRF2) using a T7 primer and primer 5′-CCGGAATTCGGTGGAATGGCGGGAGGAGGCGGGAGT-3′ (underlined EcoRI site in frame with the Flag tag). pcDNA3.1-Flag-TRF2ΔB(R361P) was made by site-directed mutagenesis of pcDNA3.1-Flag-TRF2ΔB with the Quick Change II XL kit (Stratagene) using 5′-GCAGCCCAAGAACAAGCGCATGACAATAAGCCCATTGGTCTTGGAGGAGGACAGCCAGAGTACTGAG-3′ (mutated positions underlined) and its complementary oligo. pcDNA3.1-TRF2ΔB was generated by the insertion into the BamHI/XhoI sites of pcDNA3.1(+) of a BamHI/SalI-digested TRF2ΔB cassette amplified from pcDNA3.1-Flag-TRF2ΔB using primers 5′-CTCGGATCCACCATGGCACGGCTGGAAGAGGCAGTCAATC-3′ and 5′-GGAATTCGTCGACTCAGTTCATGCCAAGTCTTTTCAT-3′ (BamH1 and Sal1 sites underlined). pcDNA3.1-Flag–TIN2 plasmid was a derivative of plasmid pOTB7-TIN2 (Open Biosystem) encoding the longer isoform of TIN2 (NP_001092744). Briefly, TIN2’s open reading frame was PCR amplified with Platinum Pfx DNA polymerase (Invitrogen) using primer Tin2-F (5′-CCGGAATTCATGGCTACGCCCCTGGTGGCG-3′, EcoRI site underlined) and Tin2-R (5′-CGCGGATCCTCACTCCTTTTGCTCTGTGGCAGGCAA-3′). EcoRI-digested product was inserted in EcoRI/PmeI sites of plasmid pcDNA3.1-Flag, in frame with the Flag tag. pcDNA3.1-Flag is a pcDNA3.1(–) vector encoding a Flag epitope located downstream of a T7 promoter and followed by an EcoRI site.
HT1080-TRF2 and HT1080-Vector cells
HT1080 cells were cultured in DMEM supplemented with 10% fetal bovine serum and 100 U/ml penicillin/streptomycin. Subconfluent HT1080 cells were transfected with pcDNA3.1-Flag-TRF2 by transferrin-facilitated lipofection, as described previously (47). Transfected cells were selected with 400 µg/ml G418 and individual colonies isolated by means of cloning rings. Using the M2 anti-Flag antibody, colonies were screened by western blot for the expression of Flag-TRF2. Colonies #1 and 3# were found to be negative and positive, respectively. In the present study, these cells are respectively been used as the HT1080-TRF2 and HT1080-Vector cells.
Transfection of HeLa cells
HeLa cells were cultured in DMEM supplemented with 10% fetal bovine serum and 100 U/ml penicillin/streptomycin. Subconfluent cells were transiently transfected by transferrin-facilitated lipofection, as described previously (47). Cells were transiently transfected with no plasmid (Mock), pcDNA3.1 (empty vector), pcDNA3.1-Flag-TRF2, pcDNA3.1-Flag-TRF2ΔB, pcDNA3.1-Flag-TRF2ΔB(R361P), pcDNA3.1-Flag–TIN2 or with both pcDNA3.1-Flag–TIN2 and pcDNA3.1-TRF2ΔB. Forty-eight hours post-transfection, cells were harvested for preparation of whole cells extracts (WCE) or nuclear extracts (NE).
Preparation of nuclear extracts and whole-cell extracts
Cell extracts were prepared following a modification of a previously published method (12). Forty-eight to seventy-two hours following transfection, cells were washed in cold phosphate-buffered saline (PBS) and resuspended in five packed-cell volumes of hypotonic buffer (10 mM Tris–HCl pH 7.6, 0.5 mM EDTA, 1 mM DTT, 1 μM microcystin-LR, and 1% (v/v) protease inhibitor cocktail). Cells were allowed to swell on ice for 20 min and then lysed in a Dounce homogenizer using an A-type pestle until >90% of cells had released their nucleus. For whole-cell extracts (WCE), an equal volume of high salt buffer (hypotonic buffer containing 1.2 M KCl) was added and the samples were rocked at 4°C for 30 min. After centrifugation at 20 000 g for 30 min (Sorvall SL-50 T rotor at 12 900 rpm), supernatants were harvested and dialyzed at 4°C for 3 h against the storage buffer (20 mM Tris–HCl pH 7.4, 20% glycerol, 0.2 mM EDTA, and 10 mM β-mercaptoethanol). Dialyzed samples were cleared by centrifugation (12 000 g for 10 min), aliquoted, and stored at −80°C. For nuclear extracts (NE), cell lysates obtained after douncing were spun at 2500 rpm for 5 min in an Eppendorf centrifuge to collect nuclei, which were then resuspended in 3 ml of buffer S1 (0.25 M Sucrose, 5 mM Tris–HCl pH 7.4, 1 mM DTT, 10 mM MgCl2) and overlaid on top of a sucrose cushion made of buffer S2 (0.35 M Sucrose, 5 mM Tris–HCl pH 7.4, 1 mM DTT, 0.5 mM MgCl2). After spinning at 3500 rpm for 5 min (Rotor 221), the supernatant was carefully removed and the isolated nuclei resuspended in five pellet volumes of hypotonic buffer (as above). An equal volume of high-salt buffer (as above) was added and the samples were rocked at 4°C for 30 min. Remaining steps were as described for the WCE.
Preparation of [32P]-labeled probes
Primers and templates for synthesis of each probe are described in Table 1. Probes 4M-3′, 4M-bl, 4M-5′ and probes listed in Supplementary Figures S2A, S3A and S6A were made in a two-step process (as depicted in Supplementary Figure S1). In the first step, a [32P]-labeled primer was annealed to a single-stranded template and extended with Taq DNA polymerase. In 20 μl of forward reaction buffer (10 mM MgCl2, 5 mM DTT and 70 mM Tris–HCl, pH 7.6), 30 pmol of the appropriate primer were labeled for 15 min at 37°C with 75 μCi of γ-[32P]-ATP (3000 Ci/mmole) and 10 Units of T4 polynucleotide kinase (Invitrogen). Following heat inactivation, this labeling reaction was added to 80 μl of PCR mix (20 mM Tris–HCl, 50 mM KCl, 200 μM of each dNTP, 1.5 mM MgCl2, pH 8.4) containing 10 pmol of the single-stranded DNA template. After 95°C for 3 min, Taq DNA polymerase (5 U) was added and the primer extended for 5 min each at 72°C and then 65°C. Next, the extension product was purified using GENECLEAN Glass Milk (Qbiogene, Irvine, CA, USA) following the manufacturer’s instructions. In the second step, the purified products were digested with outside cutters PpiI, HgaI or MlyI to produce probes ending respectively with a 3′-overhang, 5′-extension, or blunt end. Probes 4M-bl and 4M-5′ shared the same template but were cut with MlyI and HgaI, respectively. After digestion, probes were purified by native gel electrophoresis on an 8% polyacrylamide gel and the eluted probes were desalted on a G50 spin column (GE Healthcare, Piscataway, NJ, USA). Probes D4 and D4-nTel (Figure 4B) were made in the same way, with exception that the extended product did not require enzymatic digestion to produce a 3′-overhang.
Table 1.
Oligonucleotides used for making the longer EMSA probes
 |
Shaded areas are telomeric DNA repeats. Recognition sites for PpiI (single line), HgaI (double line) and MlyI (dotted line) are underlined. Asterisks denote location of the [32P]-phosphate group. Primers are positioned and oriented as they would if annealed to their templates.
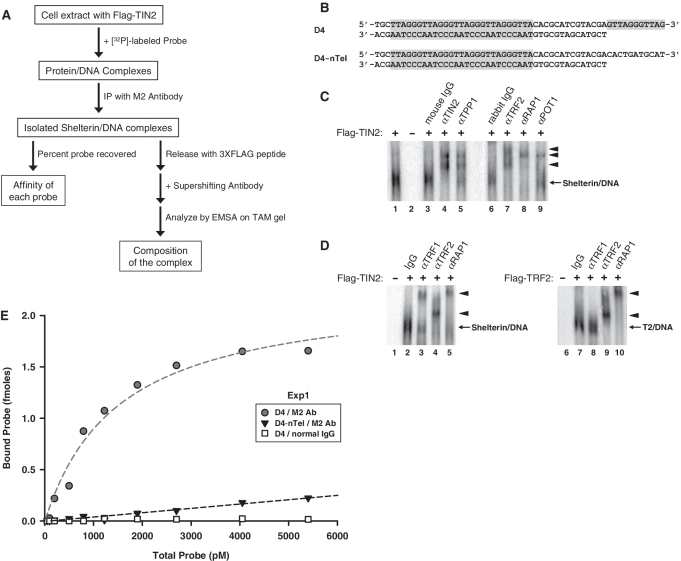
Figure 4.
Composition and DNA binding affinity of the Flag–TIN2 complexes. (A) Experimental plan for characterization of Flag–TIN2 containing Shelterin complexes. Extracts made from HeLa cells transfected with Flag–TIN2 are incubated with the [32P]-labeled probe, after which Flag–TIN2 containing complexes are immunoprecipitated with magnetic beads coated with the anti-Flag M2 antibody. Amount of probe recovered can then be used as a measure of the affinity of each probe for Shelterin complexes (Left arm). Alternatively, the isolated complexes can be released by incubation with an excess of 3XFLAG peptide, after which the eluted complexes are analyzed by native electrophoresis on composite gel containing TAM buffer (Right arm). Exposing the complexes to supershifting antibodies prior to the electrophoresis allows determination of the composition of the complexes. (B) Graphical representations of the probes D4 and D4-nTel. Shaded areas represent telomeric DNA. The probes carry four binding sites (ds-TTAGGGTTA motifs) for the Myb domain of TRF2 followed by a telomeric (D4) or non-telomeric (D4-nTel) 3′-overhang. [32P]-phosphate was at the 5′-end of the bottom strand. (C and D) Subunit composition of the isolated Flag–TIN2 complexes. Extracts of HeLa cells transfected with (+) or without (–) Flag–TIN2 or Flag-TRF2 were incubated with [32P]-probe D4. Protein/DNA complexes containing the Flag-tagged proteins were captured with beads coated with the M2 antibody and released by incubation with an excess of 3XFLAG peptide. After exposure to the indicated antibodies, the eluted complexes were resolved by native electrophoresis in a composite gel containing TAM buffer. Short arrow indicates positions of the supershifted Shelterin/DNA complex. (E) Saturation binding curve of the interaction of Flag–TIN2 complexes with probe D4. Extracts of HeLa cells transfected with Flag–TIN2 were incubated with increasing concentrations of probes D4 or D4-nTel, after which point Flag–TIN2 complexes were recovered with beads coated with either the anti-Flag antibody (M2 Ab) or normal mouse IgG (normal IgG). Scattered plot shows the amount of probe recovered (in fmoles) as a function of the initial probe concentration (pM). Dotted line shows fitting of the data to either a linear curve (Black, D4-nTel) or a one site saturation binding curve (Gray, D4). Nonlinear regression of the D4 data allowed determination of the dissociation constant (Kd = 1.5 ± 0.3 nM) and maximum number of binding complexes (Bmax = 2.3 ± 0.2 fmol/reaction).
Probes listed in Figures 3A and C, 5A and Supplementary Figure S5A were made by the annealing of a [32P]-labeled bottom oligo (Bot) to a 2-fold excess of an unlabeled top oligo (Top). Labeling of the bottom oligos was performed as described above, except that the 20 µl kinase reactions contained 15 pmol of oligo. After heat inactivating the kinase, the top oligo was added (30 pmol) and the two strands were annealed in a PCR machine, performing 95°C for 3 min and the following steps for 1 min each: 72°C, 65°C, 60°C, 55°C, 50°C, 45°C and then 25°C. After annealing, probes were purified by native gel electrophoresis on an 8% polyacrylamide gel and the eluted probes were desalted on a G50 spun column (GE Healthcare).
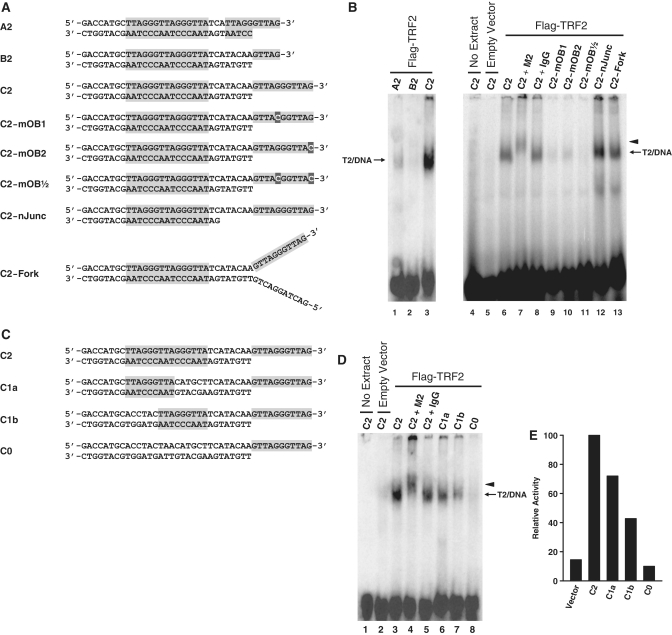
Figure 3.
Characterization of the DNA binding specificity of complex T2. (A and C) Graphical representations of the structures of all probes tested. Probes were designed to harbor different end structures and to contain different number of ds-TTAGGGTTA motifs. Shaded areas represent telomeric DNA. (B) DNA binding by complex T2 requires a functional POT1 binding site. Probes described in A were incubated with no extracts (lane 4) or with nuclear extracts of HT1080-Vector (lane 5) or HT1080-TRF2 cells (lanes 1–3 and 6–13), after which point the protein/DNA complexes were resolved by native electrophoresis in composite gels containing TAM buffer. Five minutes prior to loading, antibodies were added to samples in lanes 7 (anti-Flag M2 antibody) and 8 (Normal mouse IgG). Lanes 1–3 were overexposed to allow detection of the signal of probe A2. Short arrow indicates positions of the supershifted complex T2. (D) DNA binding by complex T2 requires at least one Myb binding site. Probes described in C were incubated with no extracts (lane 1) or with nuclear extracts of HT1080-Vector (lane 2) or HT1080-TRF2 cells (lanes 3–8), after which point the protein/DNA complexes were resolved by native electrophoresis in composite gels containing TAM buffer. Five min prior to loading, antibodies were added to samples in lanes 4 (anti-Flag M2 antibody) and 5 (Normal mouse IgG). Short arrow indicates positions of the supershifted complex T2. (E) Densitometric quantification of the T2/DNA complexes detected in (D).
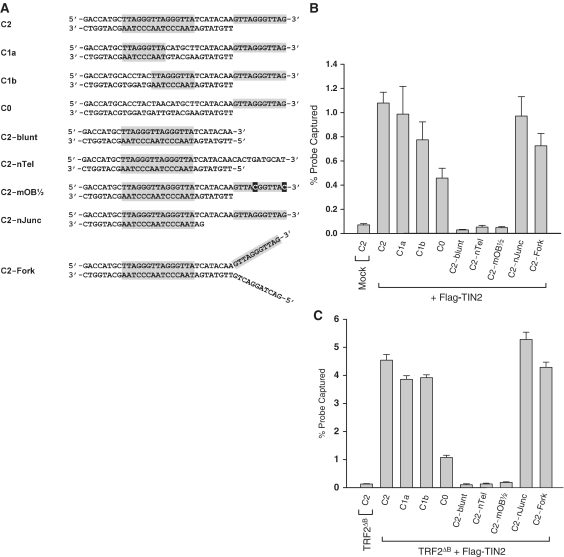
Figure 5.
Characterization of the DNA binding specificity of Flag–TIN2 complexes. (A) Graphical representations of the structures of all tested probes. Probes were designed to harbor different end structures and to contain different number of ds-TTAGGGTTA motifs. Shaded areas represent telomeric DNA. (B) DNA binding specificity of Flag–TIN2 Complexes. Extracts of HeLa cells transfected with no DNA (Mock) or with Flag–TIN2 (+ Flag–TIN2) were incubated with the [32P]-labeled probes described in A, after which Flag-tagged protein/DNA complexes were captured with the M2 antibody and the amount of radioactivity recovered was counted. Results in triplicates show the percent of each probe recovered by the M2 antibody (mean ± standard deviation; n = 3). The percent of probe C2 recovered from the Mock-transfected cell extract defines the value of the background (<0.1%). (C) DNA binding specificity of Flag–TIN2/TRF2ΔB complexes. Extracts of HeLa cells transfected with TRF2ΔB alone (TRF2ΔB) or with TRF2ΔB plus Flag–TIN2 (TRF2ΔB + Flag–TIN2) were incubated with the [32P]-labeled probes described in A, after which Flag-tagged protein/DNA complexes were captured with the M2 antibody and the amount of radioactivity recovered was counted. Results in triplicates show the percent of each probe recovered by the antibody (mean ± standard deviation; n = 3). The percent of probe C2 recovered from the TRF2ΔB alone-transfected cell extract defines the value of the background (<0.2%).
The molarity of each probe was calculated based on the specific activity of the radiolabeled oligo with which each probe was made. To measure the specific activity of this oligo, a 2 µl aliquot of the heat-inactivated kinase reaction (containing either 1.5 or 3 pmol of oligo) was spotted onto a DE81 anion exchange paper (Whatman International, Maidstone, England) and dried for 10 min. After washing away the unincorporated [32P]-ATP three-times for 5 min each in 50 ml of 0.5 M Na2HPO4 (pH 6.8), the remaining radioactivity was measured by scintillation and the results expressed in cpm/pmoles (range was 0.4–1.2 × 106 cpm/pmol). Once purified, EMSA probes were spotted onto a DE81 filter, dried, and counted by scintillation. The molarity of each probe was calculated based on the counts and specific activity of its corresponding primer. Probes were diluted to 60 nM and stored at −20°C.
Electrophoretic mobility shift assay
In a final volume of 20 μl, reactions contained 4 μl of 5X binding buffer (20% glycerol, 5 mM MgCl2, 5 mM DTT, 250 mM NaCl, 50 mM Tris–HCl pH 7.5), 2 μg of sonicated Escherichia coli DNA, and 5 μl of either NE or WCE. After 5 min at room temperature, 5 × 104 cpm of radiolabeled probes were added (probe concentrations ranging from 2 to 4 nM). Reactions were incubated for an additional 25 min, after which point protein/DNA complexes were resolved by native gel electrophoresis. When performing supershift experiments, antibodies were always added for the last 5 min of incubation, prior to the electrophoresis. Depending on the application, electrophoresis took place on either a 4% polyacrylamide gel or a composite gel made of a 0.5% agarose/2.5% polyacrylamide mixture, with the gel containing either TAM (40 mM Tris–acetate, 2 mM MgCl2, pH 8.3) or TBE (45 mM Tris–borate, 2 mM EDTA, pH 8.3) buffer. Gels were run at 180 volts for 2 (TBE) to 3 h (TAM) at 4°C. Gels were subsequently transferred to a DE81 anion exchange chromatography paper (Whatman International), dried, and exposed to a PhosphoImager screen.
Composite gels were made of 0.5% agarose reinforced by the presence of 2.5% polyacrylamide. Gels were made based on modifications of a previously published protocol (48). To make a composite gel in TAM buffer, 0.3 g of Seakem Gold Agarose (Lonza, Rockland, ME, USA) is melted in 30 ml of TAM buffer and kept at 55°C. In TAM buffer, a 5% acrylamide solution (acrylamide:bis-acrylamide ratio of 40:1) is also made and transferred to 55°C. To help keep the agarose and acrylamide solutions and the casting mold at 55°C, we used a hybridization oven set to 55°C. After mixing the two solutions, 60 µl of TEMED and 600 µl of 10% ammonium persulfate are added and the mixture is immediately stirred and poured into a casting mold, pre-warmed at 55°C, with 1.5 mm spacers. Two identical combs of a thickness of 0.75 mm each are inserted together and in registry to create wells with a thickness of 1.5 mm. After inserting the two combs, the gel is allowed to solidify horizontally for 1 h at 4°C. Next, the bottom spacer is removed and the gap filled with 4% agarose in TAM buffer. This plug is needed to support the gel after the plates are mounted vertically. To remove the combs, the middle comb (sandwiched between the outside comb and the back plate) is first removed, leaving the outside comb in place to prevent the wells from collapsing. Then, the second comb is freed by pushing it against the back plate and sliding it out slowly. Next, the gel is mounted vertically onto an electrophoresis apparatus, in our case a Model V16 apparatus by Gibco BRL (Invitrogen). Finally, the tanks are filled with ice-cold TAM buffer, the apparatus is transferred to the cold room, and the wells are cleared of remaining gel using a syringe. After pre-running the gel at 180 volts for 30 min, the EMSA samples are loaded and the gel is allowed to run for an additional 3 h.
Immunoprecipitation of Shelterin/DNA complexes
In a final volume of 50 μl, reactions contained 10 μl of 5 × binding buffer (as described above in the EMSA section), 5 μg of sonicated E. coli DNA, and 12.5 μl of either NE or WCE. After 5 min at room temperature, 1.25 × 105 cpm of radiolabeled probes were added (probe concentrations ranging from 2 to 4 nM). Reactions were incubated for an additional 25 min, after which protein/DNA complexes were captured with magnetic beads coated with the M2 anti-Flag antibody. Ten microliters of M2-coated beads were added and the suspension was rotated at room temperature for an hour. Using a magnet, beads were recovered and washed three times with 500 μl of ice-cold 1× binding buffer containing 0.1% BSA (1× buffer as in the EMSA section). In some experiments, the amount of probe co-precipitated with the captured complexes was counted by scintillation, with the results expressed as the percent of each probe recovered (Mean ± SD, n = 3). In other experiments, the captured complexes were subsequently released by incubation of the beads with 50 μl of 3XFLAG peptide (100 µg/ml in 1× binding buffer). After 10 min at room temperature, the eluted complexes were harvested and subsequently resolved by electrophoresis on composite gels containing TAM buffer. When performing supershift experiments, the eluted complexes were allowed to incubate with the antibodies for 5 min prior to the electrophoresis.
Antibody-coated beads were prepared by mixing 200 μl of M-450 magnetic beads coated with a Sheep anti-Mouse IgG antibody (Dynal Biotech. Inc., Lake Success, NY, USA) with 15 μg of anti-Flag antibody (M2 mouse monoclonal, Stratagene) in 5 ml of PBS containing 0.1% each of NP-40 and BSA. After overnight rotation at room temperature, M2-coated beads were washed three times, after which beads were suspended and stored in 200 μl of the same buffer. Before use, beads were washed three times with ice-cold 1× binding buffer containing 0.1% BSA.
Saturation binding curves
Experiments were performed as described above with the exception that binding reactions were made to contain increasing amounts of [32P]-labeled probes, either D4 or D4-nTel. Probe concentrations were increased in 1.5- to 3-fold increments from 50 pM to 10 nM. Probe concentrations >10 nM could not be tested due to limitations of the methodologies used for probe preparation. Binding reactions (50 μl) were made to contain 12.5 μl of WCEs from Flag–TIN2 transfected HeLa cells. Capture was performed with magnetic beads coated with the anti-Flag M2 antibody or with normal mouse IgG. After washing the beads three times with 500 µl each of ice-cold binding buffer, the amount of radioactivity captured was counted by scintillation. The amount of probe recovered in femto-moles (based on the specific activity of each probe) was plotted as a function of total probe concentration (in pM). The resulting curves were fitted by non-linear regression to a one-site saturation binding curve (Probe recovered = Bmax[probe]/(KD + [probe])). Fitting, performed using SigmaPlot version 11.0, allowed calculation of the dissociation constant (KD) and maximum number of binding complexes (Bmax).
RESULTS
Detection of the DNA binding activity of TRF2-containing Shelterin complex T2
In our initial attempts at detecting the DNA binding activity of Shelterin complexes, radiolabeled telomeric DNA fragments (Figure 1A) were incubated in extracts of HeLa cells previously transfected with TRF2 or TRF2ΔB, a mutant that lacks the N-terminal basic region (Figure 1B–C). This mutant had previously been reported to exhibit increased binding to ds-telomeric DNA (9). Initially, protein/DNA complexes were resolved by electrophoresis in native polyacrylamide gels containing TBE buffer (Tris–Borate–EDTA). Under these electrophoretic conditions, species exhibiting properties of Shelterin complexes were not initially observed (data not shown). But in gels containing TAM buffer (Tris–Acetate–Mg2+), a new complex was detected following transfection of Flag-tagged TRF2ΔB (Figure 1D, top panel, lane 1 versus 2). Unexpectedly, this new complex, designated as T2, associated with probes that possessed a 3′-telomeric overhang (4M-3′, lanes 2) but not with probes carrying either a blunt end (4M-bl, lane 3) or 5′-telomeric extension (4M-5′, lane 4). With magnetic beads coated with an anti-Flag antibody, this complex T2 was captured, eluted with an excess of 3XFLAG peptide, and then resolved onto another TAM gel. As the lower panel of Figure 1D shows, the purified complexes could still distinguish between probes carrying the different end structures. This ability of TRF2ΔB complexes to select ligands based on the overhang was unanticipated, because deletion of the N-terminal basic region should have eliminated the recently described ss-DNA binding domain of TRF2 (40). Accordingly, we then reasoned that TRF2ΔB must have interacted with other cellular components, one of which able to recognize ss-DNA. One possibility was that complex T2 was a Shelterin complex, with its POT1 subunit mediating recognition of the 3′-telomeric overhang.
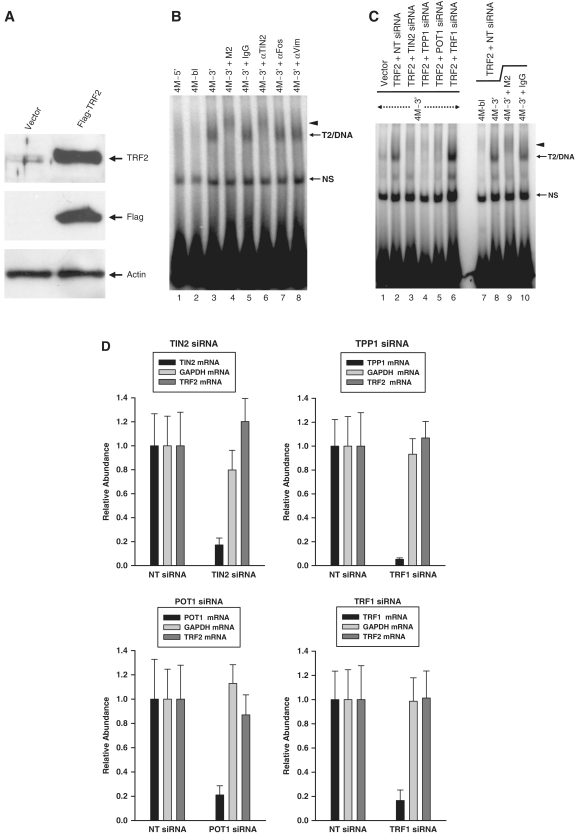
Figure 1.
Transfected Flag-TRF2ΔB forms a complex that binds selectively to telomeric DNA fragments that carry a 3′-overhang. (A) Graphical representation of probes 4M-3′, 4M-bl and 4M-5′. Shaded areas represent telomeric DNA. The probes carried four binding sites for the Myb domain of TRF2 (ds-TTAGGGTTA motifs numbered 1–4) followed by different end structures: 3′-telomeric overhang (4M-3′), blunt (4M-bl) or 5′-telomeric overhang (4M-5′). [32P]-phosphate was at the 5′-end of the top strand. (B) TRF2 constructs used in transient transfection. Full-length TRF2 contains a basic domain that binds ss-DNA independently of sequence (Basic), a TRF homology region (TRFH/Dimerization) that serves as dimerization interface, a domain of interaction with TIN2 (aa 350–367), and a Myb domain that binds to ds-TTAGGGTTA (Myb). Interaction with TIN2 allows the recruitment of TRF2 in Shelterin complexes. In these complexes, TPP1 and TIN2 act scaffolds linking TRF2 to the ss-DNA binding protein POT1, which binds to 5′-TTAGGGTTAG-3′. TRF2ΔB lacks amino acids 2-44 containing the sequence-independent ss-DNA binding domain of TRF2. TRF2ΔB (R361P) contains an additional mutation that blocks the association of TRF2ΔB with TIN2. All constructs were tagged at the N-terminus with the Flag epitope. (C) Western blot analysis of HeLa cells transiently transfected with the different TRF2ΔB constructs. Whole cell extracts prepared for EMSA (50 µg) were probed with the anti-Flag M2 antibody. (D) Transfected TRF2ΔB is in a complex that binds selectively to telomeric DNA fragments carrying a 3′-overhang. Top panel: WCEs from mock-transfected (lane 1) and TRF2ΔB-transfected HeLa cells (lanes 2–4) were incubated with the indicated [32P]-probes (4M-3′, 4M-blunt, 4M-5′) and the protein/DNA complexes that formed were resolved by electrophoresis in TAM buffer. T2, TRF2-containing complex T2. NS, non-specific band. Bottom panel: after incubation of the extracts with the indicated probes, protein/DNA complexes containing Flag-TRF2ΔB were captured using magnetic beads coated with an anti-Flag antibody, were eluted with an excess of 3XFLAG peptide, and were subsequently resolved by electrophoresis in TAM buffer. (E) Effects of R361P mutation on formation of complex T2. HeLa cells were mock-transfected or transfected with Flag-TRF2ΔB or Flag-TRF2ΔB(R361P). WCEs were prepared and incubated with probe 4M-3’, after which point protein/DNA complexes were resolved by electrophoresis in a native polyacrylamide gel containing TAM buffer. Complex T2 failed to be detected in extracts of HeLa cells transfected with the R361P mutant.
In Shelterin complexes, TIN2 is a central component that couples TRF1 and TRF2 with the ss-DNA binding activity of the TPP1/POT1 dimer. To determine if TIN2 was an essential component of complex T2, experiments were repeated with TRF2ΔB(R361P), a TRF2ΔB protein carrying a mutation in its TIN2-interacting domain (49) (Figure 1B; aa 350–367). In TAM gels, complex T2 was detected following transfection of Flag-TRF2ΔB but not after Flag-TRF2ΔB(R361P) (Figure 1C, E), thereby suggesting that the interaction with TIN2 is required for formation of the complex. Using an anti-TIN2 antibody, we then asked if TIN2 could be detected as a component of the complex. For these experiments, an HT1080 cell line expressing a Flag-tagged full-length TRF2 was made (HT1080-TRF2 cells). In these cells, total TRF2 was six times higher than in the control HT1080-Vector cells (Figure 2A). Incubating probe 4M-3′ with extracts of HT1080-TRF2 cells led to the detection of complex T2, as observed by EMSA in TAM gels (Figure 2B; lanes 3–8). Again, the complex failed to associate with probes lacking a 3′-telomeric overhang, such as 4M-5′ (lane 1) or 4M-bl (lane 2). Most significantly, complex T2 was supershifted by the TIN2 antibody (lane 6) as well as the Flag antibody (lane 4). Yet, neither the normal mouse IgG (lane 5) nor two other unrelated antibodies (lanes 7 and 8) could supershift the complex. These results along with the effects of the R361P mutation identified TIN2 as an important component of complex T2.
Figure 2.
Complex T2 formed by Flag-tagged TRF2 is a member of the Shelterin family. (A) Western blot analysis of HT1080-TRF2 and HT1080-Vector cells. WCEs prepared for EMSA (50 µg each) were analyzed by western blotting to detected total TRF2 (anti-TRF2 antibody) and the stably transfected Flag-TRF2 (anti-Flag M2 antibody). Membranes were re-probed with an antibody against β-actin. (B) Complex T2 formed by Flag-TRF2 contains the Shelterin scaffolding component TIN2. Extracts of HT1080-TRF2 cells were incubated with the indicated probes, after which point protein/DNA complexes were resolved by native electrophoresis in a composite gel containing TAM buffer. Five minutes prior to loading, the indicated antibodies were added to samples in lanes 4–8 (1 µg each of anti-Flag M2 antibody, normal mouse IgG, or antibodies against TIN2, c-Fos, or Vimentin). Short arrow indicates positions of the supershifted complex T2. NS, non-specific band. (C) Loss of Shelterin components prevents formation of complex T2. HT1080-TRF2 cells were transiently transfected with siRNA smartpools against TPP1, TIN2, POT1 or TRF1 or with non-targeting (NT) siRNA. Nuclear extracts prepared from HT1080-Vector cells (Vector) or from HT1080-TRF2 cells (TRF2) transfected with the different siRNA were incubated with probe 4M-3′, after which protein/DNA complexes were separated by native electrophoresis in composite gel containing TAM buffer. As expected, complex T2 was more abundant in cells expressing Flag-TRF2 (lane 1 versus 2), was selective for telomeric DNA fragments that carried a 3′-overhang (lane 7 versus 8), and was supershifted by the anti-Flag antibody (lane 9 versus 10). Complex T2 was reduced in HT1080-TRF2 cells transfected with siRNA smartpools directed against TIN2, TPP1 or POT1 (lanes 3–5 versus lane 2). Short arrow indicates positions of the supershifted complex T2. NS, non-specific band. (D) Relative abundance of the TPP1, TIN2, POT1 and TRF1 mRNA in the siRNA-transfected cells. For each transfection, abundance of the targeted mRNA was measured by real-time PCR, comparing cells transfected with the targeting and non-targeting (NT) siRNA. For each mRNA, abundance in cells treated with the non-targeting siRNA was set to 1. To show that targeting was specific, TRF2 and GAPDH mRNAs were also measured, both of which found to be unaffected by the different siRNA treatments.
In Shelterin complexes, TRF2 and POT1 are connected by a protein bridge made of the TIN2 and TPP1 proteins (11,13–18). If T2 is a Shelterin complex, then the knockdown of TIN2, TPP1 or POT1 should block formation of the complex. Our results shown in Figure 2C are in agreement with this prediction. In HT1080-TRF2 cells transfected with the non-targeting siRNA, complex T2 was detected (lane 2). The complex was supershifted by the Flag antibody (lane 9) and interacted with probe 4M-3′ (lanes 8–10) but not with probe 4M-bl lacking a 3′-overhang (lane 7). In Figure 2C, formation of this complex was blocked by the separate knockdown of TIN2 (lane 3), TPP1 (lane 4) or POT1 (lane 5). In these Shelterin-depleted samples, the activity of complex T2 was decreased to a level comparable (POT1-depleted) or even lower (TIN2- and TPP1-depleted) than in the HT1080-Vector cells (lane 1). In contrast, knocking down TRF1 did not reduce the activity of complex T2, thereby suggesting that TRF1 is either absent from the complex or not required for DNA binding (lane 6). To confirm the effectiveness of the siRNA treatments, we employed real-time PCR. As shown in Figure 2D, the siRNA targeting TIN2, TPP1, POT1 and TRF1 were found to have reduced the levels of their target mRNAs by 5.8-, 19.1-, 4.7- and 6.0-fold, respectively. Yet, none of these siRNAs affected the levels of either the TRF2 or GAPDH mRNA. These results indicate that T2 is a Shelterin complex and show that formation of the complex requires the presence of TIN2, TPP1 and POT1, but not TRF1.
DNA binding specificity of complex T2
Next, we defined the DNA binding specificity of complex T2 using HT1080-TRF2 extracts as a source of complexes. In Figure 3A–B, we show that with probes carrying two Myb sites, extending the size of the overhang to 11 bases produces a more than 10-fold increase in affinity (Figure 3B, probe C2 versus A2 and B2). Extending the overhang also made the interaction with complex T2 stable enough to allow detection in TBE gels (data not shown). This increase in binding/stability was consistent with POT1 mediating the recognition of the 3′-overhang. Maximum binding to POT1 requires a 5′-TTAGGGTTAG-3′ motif, which POT1 recognizes by means of its two OB-folds, OB1 and OB2 (8,42,43). OB1 recognizes 5′-TTAGGG and accounts for most of the interactions whereas OB2 binds to the terminal TTAG-3′ (43). Consistent with a role for POT1, point mutations designed to reduce recognition of the overhang by OB1 (C2-mOB1, lane 9), OB2 (C2-mOB2, lane 10) or both domains of POT1 (C2-mOB1/2, lane 11) significantly reduced the affinity of these probes to complex T2. Next, we determined that binding did not require the immediate proximity of the junction between ds- and ss-DNA (ds/ss-junction). In previous probes, this junction was right next to the POT1 site (ss-TTAGGGTTAG-3′). In probe C2-nJunc, we repositioned the POT1 site at the end of a longer overhang, six bases away from the ds/ss-junction. This modification did not reduce binding but rather had the opposite effect (C2-nJunc, lane 12), perhaps by making the overhang more readily available to POT1. After that, we extended the bottom strand to create probe C2-Fork, a forked DNA structure that carried the POT1 site. Consistent with the known ability of POT1 to associate with telomeric DNA bubbles (50), complex T2 exhibited strong binding to this probe as well (C2-Fork, lane 13). Taken together, these results show that the presence of a functional POT1 site is required for the detection of the interaction of complex T2 with DNA. Finally, we also performed cross-linking experiments with a radiolabeled telomeric DNA junction that carried a photoactivatable BrdU base in its 3′-telomeric overhang (Supplementary Figure S2A–B). After UV cross-linking, immunoprecipitation of Flag-TRF2ΔB complexes and SDS–PAGE electrophoresis, a cross-linked protein of the same size as POT1 (74 kDa) was detected (Supplementary Figure S2C). Cross-linking of this protein was not observed in either the mock-transfected cells or cells transfected with the Flag-TRF2ΔB(R361P) mutant. This result supports the hypothesis that POT1 is the subunit of complex T2 that mediates the recognition of the 3′-telomeric overhang.
To define the ds-DNA requirements for complex T2 binding, probes that contained variable numbers of Myb sites (ds-TTAGGGTTA) were tested. Starting with probes carrying an overhang of 5 bases only, HT1080-TRF2 extracts were subjected to EMSA in TAM gels. No difference in complex T2 affinity was noted for probes carrying 4 Myb sites or just 2 sites (Supplementary Figure S3A–B). Yet, removing one or the other Myb site from a probe that contained two of these sites abrogated complex T2 binding, thereby indicating that two such sites were required for binding (Supplementary Figure S3A–B). However, much different results were obtained with probes that carried a full POT1 site. Starting with probe C2 containing two Myb sites (Figure 3C), we made derivatives carrying one site (probe C1a), the other site (probe C1b), or no Myb site (probe C0). In TAM gels, a complex T2 was detected in HT1080-TRF2 cell extracts incubated with probe C2 (Figure 3D, lanes 3–5) but not probe C0 (lane 8). Probe C1a (lane 6) exhibited less than a 2-fold decrease in binding compared to the C2 probe (Figure 3E). Probes C1b, whose single Myb site was located closer to the 3′-overhang, had a much lower affinity for complex T2 (lane 7, Figure 3E). These results show that when a fully functional POT1 site is present, a single appropriately positioned Myb site is sufficient to mediate binding to complex T2.
Composition and DNA binding specificity of Flag–TIN2 complexes
The apparent low affinity of complex T2 for telomeric DNA molecules lacking a POT1 site (Figure 3B, C2-mOB½) was unexpected. So we reasoned that the EMSA conditions may have been too stringent to allow binding mediated by TRF2 alone. To address this possibility, we wanted to measure DNA binding under conditions that did not require electrophoretic migration in a specific buffer. Extracts of Flag–TIN2 transfected HeLa cells were incubated with the radiolabeled probes and the Shelterin/DNA complexes that formed were captured using the anti-Flag antibody (Figure 4A). The amount of radioactive DNA recovered was then used as a measure of the affinity of the Flag–TIN2 complexes for each probe (Figure 4A, left arm) (Results of Figures 4E, 5B, 5C). As a first step towards using this assay, EMSA was employed to characterize the composition of the immunoprecipitated complexes (Figure 4A, right arm). After incubation of probe D4 (Figure 4B) with an extract of Flag–TIN2 transfected HeLa cells, Shelterin/DNA complexes were immunoprecipitated, eluted with an excess of 3XFLAG peptide, and analyzed by EMSA. Prior to loading the gel, the eluted complexes were incubated with antibodies against each of the Shelterin subunits. As Figure 4C shows, a radiolabeled complex was recovered by the antibody from an extract of Flag–TIN2 transfected cells (lane 1) but not from the Mock-transfected cells (lane 2). Most importantly, the captured complex was supershifted by antibodies against TIN2 (lane 4), TPP1 (lane 5), TRF2 (lane 7), RAP1 (lane 8) and POT1 (lane 9) but not by the normal IgG (lanes 3 and 6). These results confirm that the captured complexes were members of the Shelterin family. Shelterin complexes have been proposed to exist in several forms depending on the presence of TRF1 alone, TRF2 alone or both factors (10,12,17,20–25). Figure 4D shows that the isolated complexes were supershifted by both the TRF1 and TRF2 antibodies (Left panel; lanes 3 and 4). The antibodies against TRF2 (lane 4) and RAP1 (lane 5) supershifted almost all of the purified complexes, whereas the anti-TRF1 antibody supershifted only half of the complexes (lane 3). These results indicate that more than half of these complexes contained both TRF1 and TRF2 as part of the same complex. In contrast, a T2 complex recovered from HT1080-TRF2 cells by immunoprecipitation of the Flag-TRF2 protein was supershifted by antibodies against TRF2 (lane 9) and RAP1 (lane 10) but not by the anti-TRF1 antibody (lane 8), thereby confirming the absence of TRF1 in this complex (Figure 4D, right panel).
Next, we performed saturation binding experiments to quantify the difference in affinity conferred by the POT1 site, and measured this difference in the context of probes containing an excess of Myb binding sites. To achieve this, we compared probes D4 and D4-nTel, both of which containing 4 Myb sites (Figure 4B). Extracts of Flag–TIN2 transfected HeLa cells were incubated with increasing concentrations of each probe (from 50 to 5400 pM), after which the protein/DNA complexes were immunoprecipitated with the anti-Flag M2 antibody. In two separate experiments, plotting the amount of probe recovered as a function of the total probe concentration revealed a saturable number of binding sites for probe D4 (Figure 4E and data not shown). Non-linear regression to a one-site saturation binding curve allowed for the calculation of the dissociation constant (KD). The apparent KD of the Flag–TIN2 complexes for probe D4 was calculated to be in the low nanomolar range (Exp 1: 1.5 nM; Exp 2: 1.3 nM). With probe D4-nTel, detectable binding above the background required much higher concentrations of probe, all above the KD value of probe D4 (Figure 4E, data not shown). Probe D4-nTel binding increased linearly up to the highest concentration of probe tested (5400 pM) with no evidence of saturation. These results show that if Shelterin complexes bind to probe D4-nTel, they do so with a KD at least one order of magnitude higher than with probe D4. These results reiterate the strong preference of Shelterin complexes for telomeric DNA molecules that carry a functional POT1 site.
Next, we reexamined the requirements for Shelterin DNA binding using the Flag–TIN2 pulldown assay. In triplicates, each of the probe listed in Figure 5A were co-precipitated with Flag–TIN2 present in HeLa cell extracts and the amount of radiolabeled probe recovered was counted (Figure 5B). Overall, the results almost perfectly reflected those obtained by EMSA for complex T2 (Figure 3A–D). Probe C2 containing a POT1 site and two Myb sites was recovered efficiently from the cells transfected with Flag–TIN2 (C2 in Flag–TIN2) but not from the mock-transfected cells (C2 in Mock). The same efficient recovery was observed for probes C1a and C1b, both of which carrying a POT1 site but lacking one or the other Myb site. Under these immunoprecipitation conditions, even probe C0 containing no discernible Myb site exhibited a detectable, albeit reduced, binding activity. But again, probes that did not possess a functional POT1 site (C2-blunt, C2-nTel or C2-mOB½) showed no evidence of binding. Nonetheless, efficient recovery was again observed for probes C2-nJunc and C2-Fork, both of which containing a functional POT1 site. In Figure 5C, we repeated these experiments with cells co-transfected with TRF2ΔB as a mean of maximizing the DNA binding activity of TRF2. Again, probe C2 was efficiently recovered only when Flag–TIN2 was present (TRF2ΔB + Flag–TIN2 extracts) and not from extracts lacking the tagged protein (TRF2ΔB extracts). Co-transfection of TRF2ΔB with Flag–TIN2 increased probe recovery by 4- to 5-fold (Figure 5C) compared to cells transfected with Flag–TIN2 alone (Figure 5B). But overall, the DNA binding specificity of the complexes were largely unaffected by the presence of TRF2ΔB. As Figure 5C shows, optimal binding required a functional POT1 site and at least one of the Myb sites (as in probes C2, C1a, C1b, C2-Junk and C2-Fork). These results reiterate the strong preference of Shelterin complexes for ds/ss-telomeric DNA junctions that carry a binding site each for POT1 and the Myb domain of TRF1 or TRF2.
DISCUSSION
With radiolabeled probes containing ds/ss-telomeric junctions, we were able to detect the DNA binding activity of Shelterin complexes in cell extracts. Using these and other probes, we have defined the composition and DNA binding specificity of these complexes. Our results describe the properties of two Shelterin sub-complexes that differ in the presence or absence of TRF1, and both of these complexes were determined to have a strong preference for DNA fragments that contain both ss- and ds-telomeric DNA. More specifically, high affinity binding was found to require a binding site each for POT1 (ss-TTAGGGTTAG) and the Myb domain of either TRF1 or TRF2 (ds-TTAGGGTTA). This DNA binding specificity would be expected to recruit these complexes to areas of the telomere where ss- and ds-telomeric DNA are in close proximity, such as the 3′-overhang, DNA bubbles and the D-loop at the base of T-loops.
In extracts of cells transfected with Flag-TRF2, a Shelterin complex was detected by mean of its binding to a radiolabeled telomeric DNA junction. This complex was supershifted with antibodies against TIN2, TRF2 and RAP1 but not by an anti-TRF1 antibody, which we know can shift TRF1-containing complexes (Figures 2B and 4D). Moreover, the activity of the complex was abrogated by the knockdown of TIN2, TPP1 and POT1 but was unaffected by the loss of TRF1 expression (Figure 2C). These results are consistent with the interpretation that this complex contains the TRF2, RAP1, TIN2, TPP1 and POT1 proteins. Hence, this complex could be described as a Shelterin sub-complex that contains TRF2 but lacks TRF1. A tentative model for the structure of this T2 complex, based on the current literature, is presented in Figure 6A. The model proposes a 5-member complex that contains a dimer of TRF2, its associated RAP1 subunits, and one molecule each of TIN2, TPP1 and POT1.
Figure 6.
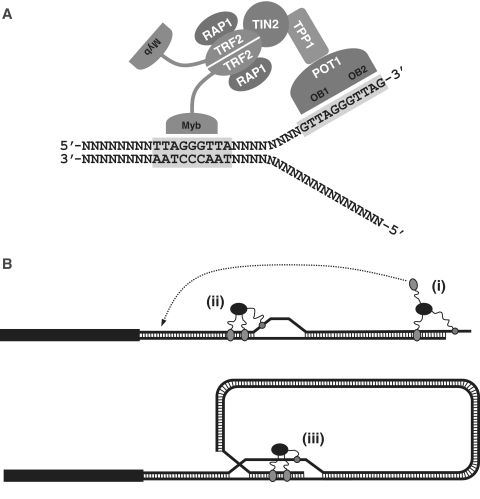
Modeling the interactions of Shelterin complexes with ds- and ss-telomeric DNA. (A) Simultaneous interactions of Shelterin complexes with both ds- and ss-telomeric DNA. Binding of these multi-subunit complexes to DNA requires a binding site for POT1 (5′-TTAGGGTTAG-3′) and at least one binding site for the Myb domains of either TRF1 or TRF2 (ds-TTAGGGTTA motif). Once bound to telomeric DNA, other Myb domains present in the complexes could further stabilize the protein/DNA complex by establishing additional contacts with ds-telomeric DNA. B) Implications of this model on the role(s) played at telomeres by the Shelterin complexes. The DNA binding specificity of Shelterin complexes would be expected to recruit these complexes to regions of telomeres where ss- and ds-telomeric DNA are present in close proximity, including the 3′-telomeric overhang (i), telomeric DNA bubbles (ii), and the D-loop at the base of T-loops (iii). At these locations, Shelterin complexes would be ideally positioned to control the accessibility of the 3′-telomeric overhang (i) to telomerase and T-loop forming machinery. Unoccupied Myb domains in complexes bound to the 3′-telomeric overhang could potentially be available to mediate long-range interactions with upstream ds-telomeric DNA (dotted arrow). These long-range interactions could help initiate the formation of T-loops. Finally, through their simultaneous interactions with both ds- and ss-telomeric DNA, Shelterin complexes could help stabilize pre-existing D-loops and T-loops (iii) as well as telomeric DNA bubbles (ii). Stabilization of these bubbles could promote T-loop formation if the stabilized bubbles are persisting long enough to have a chance to collide with the 3′-telomeric overhang.
An unexpected finding was the more than 10-fold higher level of complex T2 activity in cells transfected with TRF2 (Figures 1D, E, 2C, 3B, D and Supplementary Figure S3B). We did not anticipate that transfection of one subunit alone, TRF2, would suffice to increase the activity of the complex if the other subunits are required. A recent report by Takai et al. (24) provides a plausible explanation to these observations. If in total cell extracts TRF2, RAP1 and TIN2 are in vast molar excess compared to TPP1 and POT1, this apparently is not true of the soluble protein fraction not bound to chromatin. As it turns out, TRF1 and TRF2 are almost exclusively associated with the chromatin, most likely due to their direct binding to DNA, with virtually none detected in the soluble fraction (24). As a consequence, this soluble fraction is rich in RAP1, TIN2, TPP1 and POT1 but lacks TRF1 and TRF2. Under these conditions, transfection of an exogenous TRF2 could potentially saturate all DNA binding sites, causing the excess TRF2 to accumulate in the soluble fraction where the factor could interact with RAP1, TIN2, TPP1 and POT1 to form T2 complexes. If correct, this model would predict that the transfection of TRF1, like TRF2, should lead to dramatic increases in Shelterin DNA binding activity, in this case by the accumulation of TRF1/TIN2/TPP1/POT1 sub-complexes. In agreement with this prediction, large increases in Shelterin DNA binding activity were also observed following TRF1 transfection but not after the transfection of RAP1 or TPP1 (Supplementary Figure S4). Evidence for the occurrence of separate sub-complexes containing TRF1/TIN2/TPP1/POT1 or TRF2/RAP1/TIN2/TPP1/POT1 have also been observed after deletion or inhibition of TRF2 or TRF1 (10,12,17,20–25). However, an unresolved question is whether these sub-complexes would also be present in cells expressing physiological levels of TRF1 and TRF2. Additional studies will be needed to address this question, but when we compared the DNA binding specificity of complex T2 with that of the larger 6-member complex, no significant differences were observed (Figures 3A–D versus 5A–C).
Our second approach to investigate the binding properties of Shelterin complexes was to quantify the amounts of radiolabeled DNA probes co-precipitated with Flag–TIN2 containing complexes (Figure 4A). TIN2 is a central component of Shelterin complexes but the factor could also be part of other undescribed DNA binding complexes. Performing saturation binding curves with telomeric DNA fragments with and without a POT1 site determined that Flag–TIN2 complexes possess both a low-affinity and a high-affinity DNA binding activity (Figure 4E). The high-affinity activity required the presence of a POT1 site and displayed apparent Kd values (1.3 to 1.5 nM) that were lower than those previously reported for POT1, TRF1 or TRF2 alone (39,43). Characterization of this high-affinity activity by EMSA indicated that it was attributable to Shelterin complexes (Figure 4C–D). After co-precipitation with [32P]-labeled ds/ss-telomeric junction, the high-affinity complexes were released by incubation with the Flag peptide. Once analyzed by EMSA, the eluted complexes were supershifted by antibodies against TRF1 and TRF2 as well as antibodies for TIN2, RAP1, TPP1 and POT1. Antibodies against TIN2, TRF2 and RAP1 supershifted all of the purified complexes, whereas those against TRF1, TPP1 and POT1 supershifted only a fraction of the complexes. Could this result suggest that some of the complexes are missing TRF1 or the TPP1/POT1 heterodimer? Not necessarily, as some of the antibodies may have lacked the required affinity to produce complete supershifts. This must have been the case of the TPP1 and POT1 antibodies, since binding by the purified complexes was still very much dependent on the presence of a binding site for POT1. In the pulldown assays, for instance, probe recovery was most dramatically affected by the loss of the 3′-telomeric overhang—whether deleted, converted to non-telomeric DNA, or point mutated to block recognition by POT1 (Figure 5A–C). Most significantly, loss of the POT1 site eliminated binding by most, if not all, of the immunoprecipitated complexes. These results are consistent with the interpretation that these complexes are made of a TRF2/RAP1/TIN2 core complex in association with the TPP1/POT1 heterodimer, with at least half of these complexes also containing TRF1. In the present literature, Shelterin complexes are often depicted as containing a dimer each of TRF2 and TRF1, both interacting simultaneously with TIN2 to produce a 6-member complex. In cells transfected with Flag–TIN2, at least half of the Shelterin complexes co-precipitated with Flag–TIN2 had properties consistent with those of the 6-member complex (Figure 4D).
Using the EMSA and pulldown assays, we have characterized the DNA binding specificity of complex T2 and of the Flag–TIN2 complexes, respectively. In both assays, the presence of a binding site for POT1 was determined to be an absolute requirement for high-affinity binding. A complete loss of binding affinity resulted from the loss of this site, whether deleted, converted to non-telomeric DNA, or point mutated to block recognition by POT1 (Figures 3A–B and 5A–C). In contrast, the distance between this site and the ds/ss-junction could be changed with little effects on binding. In 80% of native telomeres, this junction is positioned 2 bases upstream of the first POT1 (Supplementary Figure S5A) (51). In most of our probes, this spacing was only of one nucleotide and in probe C2-nJunc, the spacing was of seven bases (Figure 3A). Both probes were recognized with high affinity by the Shelterin complexes (Figures 3B, 5B–C). Similar results were obtained in experiments that varied spacing from 1 to 2, 4 or 7 bases (Supplementary Figure S5A–B). Finally, we also have examined the requirements for the presence of Myb sites. Not as stringent as for the essential POT1 site, these requirements were found to vary depending on the assay. Under the conditions of the pulldown assay, a single Myb site was necessary and sufficient for high-affinity binding (Figure 5A–C). Binding affinity was only marginally higher when two sites were present, whereas the loss of all Myb sites reduced binding significantly. Nonetheless, probe C0 containing no discernible Myb sites still displayed a low but detectable level of activity. This residual activity might represent binding mediated by POT1 alone or alternatively, might be due to the occurrence in probe C0 of a cryptic Myb site. Under the conditions of the EMSA assay, a single Myb site was also determined to be necessary and sufficient for binding, but the location of this site was critical (Figure 3C–D). Binding was significantly lower when the single Myb site was closest to the 3′-telomeric overhang, possibly due to steric hindrance caused by the proximity of the Myb and POT1 sites. At native telomeric junctions, where the complex would always find an appropriately positioned Myb site, a single Myb domain would therefore be expected to suffice in providing stable binding. Taken together, data from the two assays show that Shelterin complexes bind preferentially to ds/ss-DNA junctions that contain a POT1 binding site (ss-TTAGGGTTAG) and at least one site for the Myb domain of TRF1 or TRF2 (ds-TTAGGGTTA). If many of these complexes contain a multitude of Myb domains, then clearly, occupancy of all of these domains is not required for high-affinity binding to DNA. Once these complexes are bound to a telomeric junction, then the other Myb domains could potentially be available to interact with distant sites, and, in the process, promote the looping of the telomere.
For the recruitment of Shelterin complexes, the telomere could be depicted as offering both lower- and higher-affinity binding sites. Internal tracks of telomeric repeats, which likely serve as anchor for a variety of complexes, would offer an excess of the lower affinity sites. At these sites, Shelterin complexes would likely compete for binding with a number of other TRF1/TRF2-containing complexes. Based on our findings, telomeres would also offer a limited number of higher-affinity sites. At these sites, Shelterin complexes would be expected to be more stably bound, thus spending more time on DNA before falling off. It should be noted that more stable pools of telomeric complexes have been reported previously for TRF2. In these photo-bleaching experiments, two distinct pools of TRF2 were noted at the telomeres (52). One fraction exchanged rapidly (residence time of ~44 s) but a second smaller fraction was far more stable with dynamics similar to POT1 (residence time of ~11 min). It may be that this second fraction was part of Shelterin complexes that were bound to higher affinity sites. How many of these higher affinity sites are likely to be present at the telomere? The ds/ss-junction next to the 3′-telomeric overhang would certainly create one such site (Figure 6B; i). At this location, a Shelterin complex would be optimally positioned to perform many of its key functions. There, the complex could control the availability of the 3′-telomeric overhang to telomerase, T-loop forming machinery, and DNA damage sensing mechanisms (RPA/ATR, ATM). And since we show that junction binding requires no more than one Myb domain, then the other such domains in an already bound complex could potentially be available to interact with other upstream sites and, in the process, loop the telomere to initiate T-loop formation (Figure 6B; dotted arrow). High-affinity binding sites might also be present at other telomeric substructures if they contain ds- and ss-telomeric DNA in close proximity to each other. In follow-up experiments, we have examined the flexibility with which high-affinity binding sites are recognized by Shelterin complexes. Starting with a telomeric DNA junction carrying a sub-optimal POT1 site, we varied the orientation and distance separating the POT1 site and two Myb sites (Supplementary Figure S6A). Surprisingly, binding took place with little constraints on either distance or orientation (Supplementary Figure S6B), a capability likely made possible by the long and flexible hinge region of TRF1 and TRF2. With this flexibility, other high-affinity sites could potentially be recognized at the base of T-loops, within the displacement loop (Figure 6B; iii). This D-loop consists of ss- and ds-telomeric DNA fibers that run parallel to each other. Interacting simultaneously with the two fibers, Shelterin complexes could lock the D-loop in its current conformation, and in so doing, stabilize the T-loop. In Figures 3B, 5B and C, we show that Shelterin complexes can interact with forked DNA junctions, which are structures that may exist as part of telomeric DNA bubbles. Bubbles that form in duplex telomeric DNA as a result of DNA breathing or biochemical activities (TRF2-induced supercoiling, DNA helicases) could therefore provide other high affinity sites (Figure 6B; ii). By stabilizing these bubbles, Shelterin complexes could promote T-loop formation if the bubbles are stabilized long enough to collide with the 3′-telomeric overhang.
In the present report, we have developed assays with which to measure the DNA binding activity of Shelterin complexes in human cell extracts. Our results indicate that these complexes bind preferentially to DNA fragments that contain a binding site each for POT1 and the Myb domain of TRF1 or TRF2. This binding specificity is predicted to recruit these complexes to areas of the telomere where ss- and ds-DNA are in close proximity, such as the 3′-telomeric overhang, telomeric DNA bubbles and the D-loop at the base of T-loops. At these locations, Shelterin complexes would be ideally positioned to control the access of telomerase to telomeres as well as promote the formation and stability of T-loops.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Health, the research effort (grants P50CA127297, P30CA036727); fellowship (T32CA009476 to A.S.F); Susan G. Komen Breast Cancer Foundation (BCTR0202153). Funding for open access charge: National Institutes of Health (P50CA127297).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank Titia de Lange (Rockefeller University, New-York, NY, USA) for the pTetFLAGhTRF245-501 and pTethTRF1 plasmids. We also wish to thank Ying Yan for helpful discussions and comments. We also wish to acknowledge the UNMC Eppley Cancer Center Molecular Biology Core and the University of Nebraska Medical Center DNA Sequencing Core Facility.
REFERENCES
- 1.de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–540. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- 2.Ouellette MM, Choi KH. Encyclopedia of Life Sciences. Chichester, UK: John Wiley & Sons Ltd; 2007. [Google Scholar]
- 3.de Lange T. T-loops and the origin of telomeres. Nat. Rev. Mol. Cell Biol. 2004;5:323–329. doi: 10.1038/nrm1359. [DOI] [PubMed] [Google Scholar]
- 4.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 5.Collins K. The biogenesis and regulation of telomerase holoenzymes. Nat. Rev. Mol. Cell Biol. 2006;7:484–494. doi: 10.1038/nrm1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ouellette MM, Wright WE, Shay JW. Targeting telomerase-expressing cancer cells. J. Cell, Mol. Med. 2011;15:1433–1442. doi: 10.1111/j.1582-4934.2011.01279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 8.Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 9.Broccoli D, Smogorzewska A, Chong L, de Lange T. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 1997;17:231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- 10.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 11.O'Connor MS, Safari A, Xin H, Liu D, Songyang Z. A critical role for TPP1 and TIN2 interaction in high-order telomeric complex assembly. Proc. Natl Acad. Sci. USA. 2006;103:11874–11879. doi: 10.1073/pnas.0605303103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu D, O'Connor MS, Qin J, Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J. Biol. Chem. 2004;279:51338–51342. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- 13.Houghtaling BR, Cuttonaro L, Chang W, Smith S. A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr. Biol. 2004;14:1621–1631. doi: 10.1016/j.cub.2004.08.052. [DOI] [PubMed] [Google Scholar]
- 14.Kim SH, Beausejour C, Davalos AR, Kaminker P, Heo SJ, Campisi J. TIN2 mediates functions of TRF2 at human telomeres. J. Biol. Chem. 2004;279:43799–43804. doi: 10.1074/jbc.M408650200. [DOI] [PubMed] [Google Scholar]
- 15.Kim SH, Kaminker P, Campisi J. TIN2, a new regulator of telomere length in human cells. Nat. Genet. 1999;23:405–412. doi: 10.1038/70508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu D, Safari A, O'Connor MS, Chan DW, Laegeler A, Qin J, Songyang Z. PTOP interacts with POT1 and regulates its localization to telomeres. Nat. Cell Biol. 2004;6:673–680. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
- 17.Ye JZ, Donigian JR, van Overbeek M, Loayza D, Luo Y, Krutchinsky AN, Chait BT, de Lange T. TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J. Biol. Chem. 2004;279:47264–47271. doi: 10.1074/jbc.M409047200. [DOI] [PubMed] [Google Scholar]
- 18.Ye JZ, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, de Lange T. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004;18:1649–1654. doi: 10.1101/gad.1215404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li B, Oestreich S, de Lange T. Identification of human Rap1: implications for telomere evolution. Cell. 2000;101:471–483. doi: 10.1016/s0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- 20.Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 21.Celli GB, Denchi EL, de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat. Cell Biol. 2006;8:885–890. doi: 10.1038/ncb1444. [DOI] [PubMed] [Google Scholar]
- 22.Hockemeyer D, Palm W, Else T, Daniels JP, Takai KK, Ye JZ, Keegan CE, de Lange T, Hammer GD. Telomere protection by mammalian Pot1 requires interaction with Tpp1. Nat. Struct. Mol. Biol. 2007;14:754–761. doi: 10.1038/nsmb1270. [DOI] [PubMed] [Google Scholar]
- 23.Loayza D, De Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;423:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 24.Takai KK, Hooper S, Blackwood S, Gandhi R, de Lange T. In vivo stoichiometry of shelterin components. J. Biol. Chem. 2010;285:1457–1467. doi: 10.1074/jbc.M109.038026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 26.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 27.Veldman T, Etheridge KT, Counter CM. Loss of hPot1 function leads to telomere instability and a cut-like phenotype. Curr. Biol. 2004;14:2264–2270. doi: 10.1016/j.cub.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 28.Yang Q, Zheng YL, Harris CC. POT1 and TRF2 cooperate to maintain telomeric integrity. Mol. Cell. Biol. 2005;25:1070–1080. doi: 10.1128/MCB.25.3.1070-1080.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loayza D, De Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;424:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 30.Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, Schnapp G, de Lange T. Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 2000;20:1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 32.Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O'Connor MS, Songyang Z. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 33.d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 34.Karlseder J, Broccoli D, Dai Y, Hardy S, de Lange T. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 1999;283:1321–1325. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- 35.Smogorzewska A, De Lange T. Different telomere damage signaling pathways in human and mouse cells. EMBO J. 2002;21:4338–4348. doi: 10.1093/emboj/cdf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr. Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 37.Konishi A, de Lange T. Cell cycle control of telomere protection and NHEJ revealed by a ts mutation in the DNA-binding domain of TRF2. Genes Dev. 2008;22:1221–1230. doi: 10.1101/gad.1634008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stansel RM, de Lange T, Griffith JD. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 2001;20:5532–5540. doi: 10.1093/emboj/20.19.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amiard S, Doudeau M, Pinte S, Poulet A, Lenain C, Faivre-Moskalenko C, Angelov D, Hug N, Vindigni A, Bouvet P, et al. A topological mechanism for TRF2-enhanced strand invasion. Nat. Struct. Mol. Biol. 2007;14:147–154. doi: 10.1038/nsmb1192. [DOI] [PubMed] [Google Scholar]
- 40.Fouche N, Cesare AJ, Willcox S, Ozgur S, Compton SA, Griffith JD. The basic domain of TRF2 directs binding to DNA junctions irrespective of the presence of TTAGGG repeats. J. Biol. Chem. 2006;281:37486–37495. doi: 10.1074/jbc.M608778200. [DOI] [PubMed] [Google Scholar]
- 41.Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 42.Loayza D, Parsons H, Donigian J, Hoke K, de Lange T. DNA binding features of human POT1: a nonamer 5′-TAGGGTTAG-3′ minimal binding site, sequence specificity, and internal binding to multimeric sites. J. Biol. Chem. 2004;279:13241–13248. doi: 10.1074/jbc.M312309200. [DOI] [PubMed] [Google Scholar]
- 43.Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 44.Fairall L, Chapman L, Moss H, de Lange T, Rhodes D. Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2. Mol. Cell. 2001;8:351–361. doi: 10.1016/s1097-2765(01)00321-5. [DOI] [PubMed] [Google Scholar]
- 45.Bianchi A, Stansel RM, Fairall L, Griffith JD, Rhodes D, de Lange T. TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J. 1999;18:5735–5744. doi: 10.1093/emboj/18.20.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee KM, Nguyen C, Ulrich AB, Pour PM, Ouellette MM. Immortalization with telomerase of the Nestin-positive cells of the human pancreas. Biochem. Biophys. Res. Commun. 2003;301:1038–1044. doi: 10.1016/s0006-291x(03)00086-x. [DOI] [PubMed] [Google Scholar]
- 47.Joshee N, Bastola DR, Cheng PW. Transferrin-facilitated lipofection gene delivery strategy: characterization of the transfection complexes and intracellular trafficking. Hum. Gene Ther. 2002;13:1991–2004. doi: 10.1089/10430340260355392. [DOI] [PubMed] [Google Scholar]
- 48.Suh MH, Ye P, Datta AB, Zhang M, Fu J. An agarose-acrylamide composite native gel system suitable for separating ultra-large protein complexes. Anal. Biochem. 2005;343:166–175. doi: 10.1016/j.ab.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 49.Chen Y, Yang Y, van Overbeek M, Donigian JR, Baciu P, de Lange T, Lei M. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science. 2008;319:1092–1096. doi: 10.1126/science.1151804. [DOI] [PubMed] [Google Scholar]
- 50.Wei C, Price CM. Cell cycle localization, dimerization, and binding domain architecture of the telomere protein cPot1. Mol. Cell. Biol. 2004;24:2091–2102. doi: 10.1128/MCB.24.5.2091-2102.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sfeir AJ, Shay JW, Wright WE. Fine-tuning the chromosome ends: the last base of human telomeres. Cell Cycle. 2005;4:1467–1470. doi: 10.4161/cc.4.11.2161. [DOI] [PubMed] [Google Scholar]
- 52.Mattern KA, Swiggers SJ, Nigg AL, Lowenberg B, Houtsmuller AB, Zijlmans JM. Dynamics of protein binding to telomeres in living cells: implications for telomere structure and function. Mol. Cell. Biol. 2004;24:5587–5594. doi: 10.1128/MCB.24.12.5587-5594.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.