Abstract
The synthesis of a caged RNA phosphoramidite building block containing the oxidatively damaged base 5-hydroxycytidine (5-HOrC) has been accomplished. To determine the effect of this highly mutagenic lesion on complementary base recognition and coding properties, this building block was incorporated into a 12-mer oligoribonucleotide for Tm and CD measurements and a 31-mer template strand for primer extension experiments with HIV-, AMV- and MMLV-reverse transcriptase (RT). In UV-melting experiments, we find an unusual biphasic transition with two distinct Tm's when 5-HOrC is paired against a DNA or RNA complement with the base guanine in opposing position. The higher Tm closely matches that of a C-G base pair while the lower is close to that of a C-A mismatch. In single nucleotide extension reactions, we find substantial misincorporation of dAMP and to a lesser extent dTMP, with dAMP almost equaling that of the parent dGMP in the case of HIV-RT. A working hypothesis for the biphasic melting transition does not invoke tautomeric variability of 5-HOrC but rather local structural perturbations of the base pair at low temperature induced by interactions of the 5-HO group with the phosphate backbone. The properties of this RNA damage is discussed in the context of its putative biological function.
INTRODUCTION
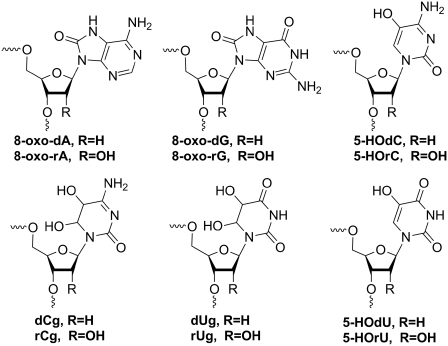
Oxidative damage to DNA bases by reactive oxygen species (ROS) such as hydrogen peroxide, superoxide and hydroxyl radicals are recognized as a major threat to the integrity of the genome. The most abundant base lesions known are 8-oxo-dA, 8-oxo-dG, 5-hydroxy-dU (5-HOdU) and 5-hydroxy-dC (5-HOdC) (Figure 1). Some of them can be highly mutagenic during DNA replication in vivo (1–3). For example, 8-oxo-dG is known to lead to a substantial level of G-T transversions (4). On the other hand 5-HOdU, originating from oxidative deamination of dC via the intermediates 5,6-dihydroxy-5,6-dihydro-2'-deoxycytidine (dCg) and 5,6-dihydroxy-5,6-dihydro-2'-deoxyuridine (dUg), causes high levels of C→T transition mutations (5). While there is some controversy on the level of mutagenicity of the intermediates along the oxidation pathway (5–7), it has been shown in Escherichia coli that 5-HOdC itself also causes C→T transitions, however, at a significantly lower frequency (5). The mutagenicity has been associated with tautomeric variability of the oxidized base (7–9).
Figure 1.
Chemical structures of the main base lesion products arising from reactive oxygen species in DNA and RNA.
Nucleobase damage by ROS, however, is not restricted to DNA but also occurs in RNA. It is conceivable that the base lesions in RNA are the same as that in DNA. Indeed, 8-oxo-rG (10), as well as 8-oxo-rA, 5-hydroxy-U (5-HOrU) and 5-hydroxy rC (5-HOrC) (11), have been isolated from RNA and identified in the past. RNA is even more prone to base oxidation than DNA for the following reasons: (i) the relative abundance of RNA in a cell is higher compared to DNA; (ii) RNA is generally less protected by proteins; and (iii) RNA is not only localized in the nucleus but also in the cytosol where the exposure to ROS, specifically near the mitochondria, is significantly higher (12). While nature has developed a highly efficient DNA repair machinery to protect the genome, the mechanism of which has been intensively studied in the past and is now fairly well understood, the impact on cellular function of base damaged RNA has remained poorly investigated in the past, presumably due to the assumption of its transient nature and its high copy numbers. Moreover, its cellular metabolism (clearance or repair) is virtually unknown.
Oxidatively damaged mRNA is known to cause largely diminished translation efficiency and abnormal protein synthesis, both in vitro and in vivo (13). Given the high oxygen turnover in the brain, the RNA in neural cells is particularly exposed to ROS. It is therefore no surprise that damaged RNA is related to a series of neurodegenerative diseases such as Alzheimer's and Parkinson's disease (14). Particularly in the light of its various functions which are not restricted to the transfer of genetic information from DNA to proteins only (tRNA, rRNA and mRNA), but also include the control of gene expression (non-coding RNAs, miRNAs), a more thorough investigation of the function of base-damaged RNA would be highly welcome.
To understand the properties and the interactions of such damaged bases in more detail, 8-oxo-rA, 8-oxo-rG and 5-HOrU have recently been incorporated into oligoribonucleotides by synthetic means and their base pairing properties with complementary RNA as well as their coding preferences in template–primer extension reactions by reverse transcriptases (RTs) have been studied (15–17). Despite its high mutagenicity the base recognition and coding properties of the 5-HOrC lesion has only scarcely been investigated in DNA in detail (18), and even less in RNA. In early work, poly 5-hydroxycytidylic acid, prepared from 5-HOrCDP by polynucleotide phosphorylase, was reported to show little secondary structure in neutral and acidic solution and no complex formation with poly(G), poly(I) and poly(A) (19).
In an effort to elucidate the biological impact on base recognition, RT and translation of base-damaged RNA, we recently prepared abasic RNA and investigated its chemical stability as well as its trans-lesion synthesis by RTs in a comparative study with DNA (20–22). Here, we report on the synthesis of oligoribonucleotides containing the base lesion product 5-HOrC, on its base recognition properties as determined by UV-melting curves, on its structural properties inferred from CD spectroscopy as well as on its coding preferences in primer/template extension reactions with RTs of three different organisms.
MATERIALS AND METHODS
Oligonucleotide synthesis and purification
Unmodified DNA and RNA oligonucleotides were from Microsynth (Balgach, Switzerland) and HPLC-purified where necessary. A 12-mer oligoribonucleotide (5′-AUGCUXAGUCGA-3′) and a 31-mer template oligoribonucleotide (5′-UAGUCUGCACXUGCACCAGUCGCUCAGGGAU-3′) were synthesized using building block 6 for X, the natural 2′-TOM protected RNA phosphoramidites and polystyrene (GE Healthcare) or CPG solid supports (GlenResearch). The syntheses were performed on a 1.3 µmole scale on a Pharmacia Gene Assembler Plus DNA synthesizer following standard phosphoramidite protocols with 5-(ethylthio)-1H-tetrazole (0.25 M in CH3CN) as the activator and coupling times of 12 and 6 min for building block 6 and unmodified RNA phosphoramidites, respectively. Oligoribonucleotides were deprotected using 33% NH3/EtOH (3:1) for 16 h at 55°C. Solid supports were then filtered off and washed with water (CPG solid support) or EtOH/CH3CN/H2O 3:1:1 (polystyrene solid support). The solutions were evaporated to dryness at room temperature in a Speedvac evaporator. The resulting pellets were further dried by addition and evaporation of dry ethanol. The pellets were then dissolved in anhydrous DMSO (100 µl) at 65°C and neat triethylamine trihydrofluoride (125 µl) was added. After shaking at 65°C for 90 min (or at 25°C for 24 h for modified oligoribonucleotides), an aqueous solution of NaOAc (3 M, 25 µl) and n-butanol (1 ml) were added and the mixture was chilled on dry ice for 45 min. After centrifugation for 20 min in an Eppendorf centrifuge the liquid was discarded and the pellet washed twice with chilled ethanol (80%, 0.75 ml) and dried under high vacuum at room temperature, yielding the oligonucleotides, in which the nitrobenzyl protecting group on the 5-HOrC unit was still in place (X = 5-nbnOrC). The 31-mer oligoribonucleotide was purified on a preparative 20% denaturing polyacrylamide gel, the product band electroeluted with an Elutrap electroelution system (Schleicher & Schuell) and desalted over NAP-10 columns (GE Healthcare). The 12-mer was HPLC purified on a DNAPac-200 column (4 × 250 mm, Dionex) with a gradient of solvent A (25 mM Tris, pH 8.0) and solvent B (25 mM Tris, 1.25 M NaCl, pH 8.0), 0–50% B in 30 min at room temperature, and finally desalted on a Sep-Pak C18 column (Waters Corp.).
Deprotection of the 2-nitrobenzyl group was achieved by irradiating aliquots of template (300 µl, 50 µM) or 12-mer (300 µl, 200 µM) in H2O for 1 h using a slide projector (25 W tungsten lamp) as described before (20). Wavelengths below 300 nm were filtered off by a glass filter set between the samples and the projector lamp. The fully deprotected oligoribonucleotides (X = 5-HOrC) were then purified by IE-HPLC as described above. Oligonucleotide concentrations were determined using a NanoDrop ND-100 UV/Vis spectrophotometer (NanoDrop Technologies, Inc.). Stock solutions were made from DEPC treated H2O.
UV-melting curves
Solutions were prepared in standard saline buffer (10 mM NaH2PO4, 150 mM NaCl, pH 7.0) with duplex concentrations of 2 µM in 1:1 strand ratio. Thermal melting experiments were carried out on a Varian Cary 100-Bio UV/VIS spectrophotometer (Varian Inc.), equipped with a Peltier element at 260 nm with a heating/cooling rate of 0.5°C/min Tm values were obtained from the maxima of the first derivatives of the melting curves using WinUV software.
Labeling of oligonucleotides
The DNA primer was phosphorylated with [γ-32P]ATP (Hartmann Analytic) and T4 polynucleotide kinase (Fermentas) as follows: DNA primer (30 pmol), [γ-32P]ATP (60 µCi) and T4 PNK (10 U) were incubated in a total volume of 20 µl at 37°C for 30 min in T4 PNK buffer (50 mM Tris–HCl, pH 7.6, 10 mM MgCl2, 5 mM DTT, 0.1 mM spermidine, 0.1 mM EDTA). T4 PNK was inactivated by heating the sample to 90°C for 2 min. The labeled primer was then directly annealed to the RNA template (60 pmol) in the reaction buffer of the respective enzyme (see ‘RT assays’) by heating to 60°C for 5 min and slow cooling to room temperature.
RT assays
RT assays were performed with HIV-1 RT (Worthington Biochemical Corp), AMV RT (Promega Corp.) and MMLV RT (USB Corp./Affymetrix Inc.,) in the following buffers: HIV-1 RT and AMV RT: 50 mM Tris (pH 8.3), 50 mM NaCl, 8 mM MgCl2, 1 mM DTT; M-MLV RT: 50 mM Tris (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM DTT. Final reaction mixtures contained RNA template (100 nM), DNA primer (50 nM) and dNTP (20 µM). After addition of enzyme (0.5–40 U), the mixtures were incubated at 37°C for 1 h. The reactions were quenched with gel loading buffer (98% formamide, 0.05% xylene cyanol (FF), 0.05% bromophenol blue), heated to 90°C for 5 min and applied to a 20% denaturing polyacrylamide gel. Radioactivity was detected and quantified on a Storm 820 phosphorimager with ImageQuant software (GE Healthcare).
RESULTS
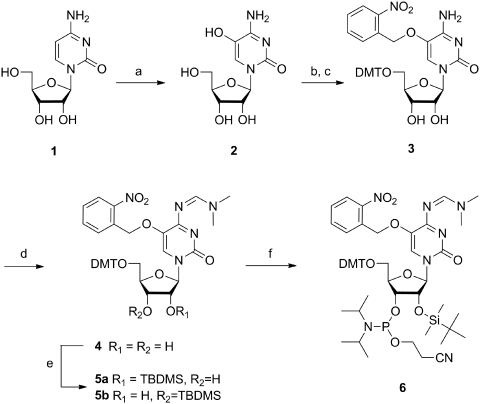
Synthesis of phosphoramidite 6
While the synthesis of a phosphoramidite of 5-HOdC and its incorporation into DNA was already described in the literature (23,24), the phosphoramidite of the RNA building block was not known and had first to be accomplished. Starting from cytidine (1), oxidation at position 5 of the base was achieved in analogy to earlier work with Br2 and H2O (19) giving 5-HOrC (2) (Scheme 1). After standard 5′-O-tritylation, we first attempted to protect the 5-OH group on the base by acylation in analogy to the known DNA building block. While acetylation or benzoylation was in principle successful, both ester functions were rather labile leading to substantial hydrolysis during chromatography. This was also the case for silyl protecting groups such as tert butyldimethylsilyl (TBDMS) or tert butyldiphenylsilyl (TBDPS). We therefore switched to the photochemically cleavable 2-nitrobenzyl group which was smoothly introduced yielding nucleoside 3. This orthogonal protecting group offers the possibility to study oligoribonucleotides in which 5-O is still protected and therefore not ionizable. The 2-nitrobenzyl ether was stable and no specific measures for light protection had to be taken during synthetic manipulations. The N4-amino group of the base was subsequently protected as a formamidine giving nucleoside 4 in excellent yield. The 2′-hydroxy group of the ribose was silylated with TBDMS-Cl in the presence of pyridine and AgNO3, leading to a 2:1 mixture of 2′-O-, and 3′-O-silylated compounds 5a and b that could be separated by column chromatography and the structures unambiguously assigned by 1H,1H-COSY-NMR spectroscopy. Building block 6 for RNA synthesis was finally achieved by standard phosphitylation of 5a. The experimental details and the analytical data of compounds 2–6 are given in the Supplementary Data.
Scheme 1.
(a) 1. Br2, H2O, 0°C; 2. sym-collidine, 40°C, 2 h; 64%; (b) 4,4′-dimethoxytriphenyl chloride, AgNO3, pyridine, 3.5 h, rt; (c) 2-nitrobenzyl bromide, K2CO3, NaI, acetone, 60°C, 3 h, 64% over two steps; (d) N,N-dimethylformamide dimethyl acetal, DMF, 60°C, 1 h, 99%; (e) tert-butyldimethylsilyl chloride, AgNO3, pyridine, THF, 2 h, 50% for 5a; (f) 2-cyanoethyl diisopropylchlorophosphoramidite, N-ethyldiisopropylamine, THF, rt, 16 h, 71%. DMT = 4,4′-dimethoxytriphenyl, TBDMS = tert-butyldimethylsilyl.
Synthesis of oligoribonucleotides
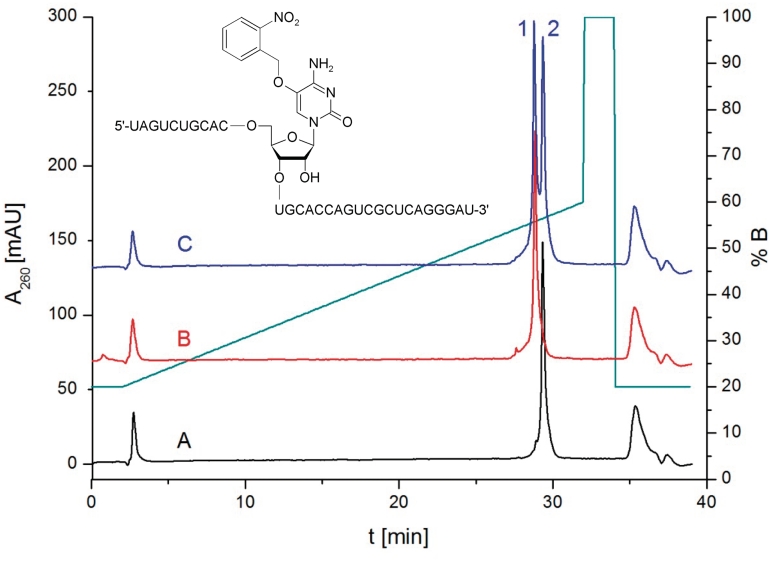
Two sets of oligoribonucleotides containing either a 5-O nitrobenzyl protected (5-nbnOrC) or a free 5-HOrC unit, a 12-mer for Tm and CD measurements and a 31-mer template strand for primer extension reactions were synthesized using building block 6. Coupling yields as determined by trityl assay were >98% at 12 min coupling time. Oligoribonucleotides were first base-, phosphate and 2′-O deprotected yielding the oligoribonucleotides with X = 5-nbnOrC that were purified, and their composition verified by mass spectrometry (Supplementary Table S1). The nitrobenzyl group was then photolytically cleaved by a tungsten slide projector lamp (λ >300 nm) leading to the fully deprotected oligoribonucleotides (X = 5-HOrC). This reaction is clean and goes to completion within 1 h as can be seen from Figure 2 which shows the HPLC traces of the 31-mer template before and after nitrobenzyl deprotection as an example. Successful deprotection was again controlled by mass spectrometry (Supplementary Table S1). The half-live of nitrobenzyl deprotection was determined with a 12-mer oligodeoxyribonucleotide of the same sequence as above, and found to be 11.4 min under the conditions applied (Supplementary Figure S1). Fully deprotected oligoribonucleotides were again purified by HPLC and used as such in the following experiments.
Figure 2.
HPLC traces of 31-mer (5′-UAGUCUGCACXUGCACCAGUCGCUCAGGGAU-3′) before (trace A, X = 5-nbnOrC, Rt = 29.3 min) and after (trace B, X = 5-HOrC, Rt = 28.7 min) nitrobenzyl deprotection; trace C: coinjection of both.
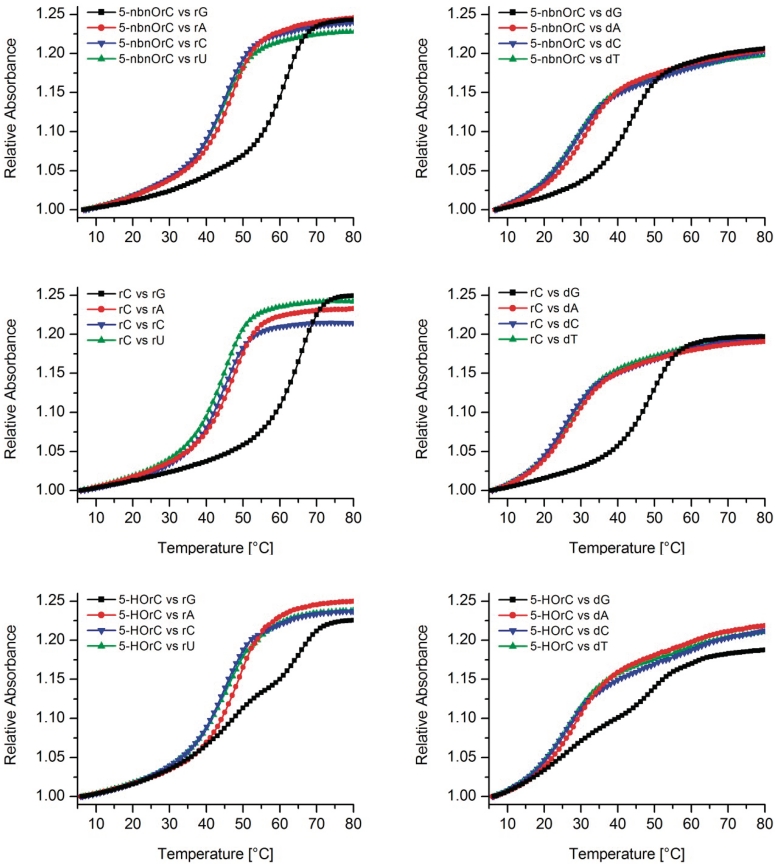
UV-melting curves
Melting curves of the 12-mers were measured at 260 nm with complementary RNA and DNA and compared to those of the unmodified duplexes (X = rC, Figure 3). The corresponding Tm data are summarized in Table 1. Melting of the 12-mer with and without nitrobenzyl protecting group in the absence of complementary strands gave no sigmoidal transition, thus excluding self-structure formation of the single strand (data not shown). From the Tm data it becomes clear that the 5-nitrobenzyl protected 12-mer (X = 5-nbnOrC) shows strong destabilization when paired against rA, rC or rU (Tm = 44.9–47.1°C) but only a slight destabilization compared to the unmodified duplex (X = rC) when paired against rG (ΔTm 5-nbnOrC versus rC = −3.9°C). The same trend albeit with generally lower Tm's is also observed if the complement is DNA. Thus 5-nbnOrC behaves by and large as cytosine in nucleobase recognition.
Figure 3.
Tm curves for duplexes of 5′-AUGCUXAGUCGA-3′ with complementary RNA (left column) and complementary DNA (right column) displaying all four natural nucleobases opposite X; top row: X = 5-nbnOrC; middle row: X = rC and bottom row: X = 5-HOrC; duplex conc.: 2 µM in 10 mM NaH2PO4, 150 mM NaCl, pH 7.0.
Table 1.
Tm data (°C, 260 nm) of duplexes of 5′-AUGCUXAGUCGA-3′ with 5′-UCGACUYAGCAU-3′ (RNA) or 5′-TCGACTYAGCAT-3′ (DNA) as complement; duplex concentration: 2 µM in 10 mM NaH2PO4, 150 mM NaCl, pH 7.0
| X | RNA complement (Y) |
DNA complement (Y) |
||||||
|---|---|---|---|---|---|---|---|---|
| rG | rA | rC | rU | dG | dA | dC | dT | |
| rC | 65.7 | 47.8 | 45.6 | 46.0 | 49.9 | 28.4 | 26.7 | 27.6 |
| 5-nbnOrC | 61.8 | 47.1 | 44.8 | 44.9 | 43.6 | 31.3 | 28.1 | 28.3 |
| 5-HOrC, Tm1 Tm2 | 65.2 47.7 | 47.4 | 44.0 | 44.6 | 48.6 25.9 | 27.2 | 25.4 | 25.4 |
| rU | 54.6 | 59.2 | – | – | 32.0 | 42.3 | – | – |
| 5-HOrUa | 54.2 | 58.8 | – | – | 32.5 | 42.6 | – | – |
aSynthesized in analogy to known methods (15).
The situation is similar in the case of the fully deprotected 12-mer (X = 5-HOrC) when paired against rA, rC or rU, but significantly different when paired against rG. In this case two transitions were observed in which Tm1 (47.7°C) closely matches that of a complementary rC-rA base mismatch whereas Tm2 (65.2°C) is reminiscent of that of a unmodified duplex with a matched rC-rG base pair. The equivalent behavior was found again with DNA as complement. Also here, two transitions were observed only with dG as the complementary base. This biphasic melting behavior could be reproduced with different samples over multiple heating and cooling cycles with some variations on the relative hyperchromicities of the two transitions (Supplementary Figures S4–S5).
To exclude deamination and thus 5-HOrU formation to occur under Tm-conditions we incubated in a control experiment the free nucleoside 2 in Tm buffer for 24 h at 80°C and compared the 1H and 13C-NMR spectra with those of 5-HOrU from an authentic sample. In the carbon NMR we could identify small signals arising from 5-HOrU, however, at an abundance of <5% (Supplementary Figures S2 and S3) suggesting that 5-HOrC to 5-HOrU conversion during Tm measurements is negligible. We also measured melting curves with duplexes containing an authentic 5-HOrU unit paired against G or A (X = 5-HOrU, Table 1), the Tms of which are consistently different from that of the 5-HOrC containing duplexes. Based on these results we rule out partial hydrolysis of 5-HOrC to be the cause for the biphasic melting behavior.
To investigate whether the two transitions are associated with different ionization states of the 5-OH group we measured Tm's with complementary rG and dG at pH 5.5, 7.0 and 8.5 (Supplementary Figures S4 and S5). In all cases, we found two transitions. The relative intensities of the hyperchromicities of these two transitions between continuous rounds of heating and cooling were lower at pH 5.5 as compared to pH 7.0 or 8.5. The two Tm's were always of the same magnitude as described in Table 1, irrespective of the pH. We thus conclude that there are no fundamental pH-dependent differences in this range, perhaps with the exception that at pH 5.5 the relative differences between the hyperchromicities of the two transitions between consecutive cooling/heating cycles are smaller as compared to the measurements at higher pH.
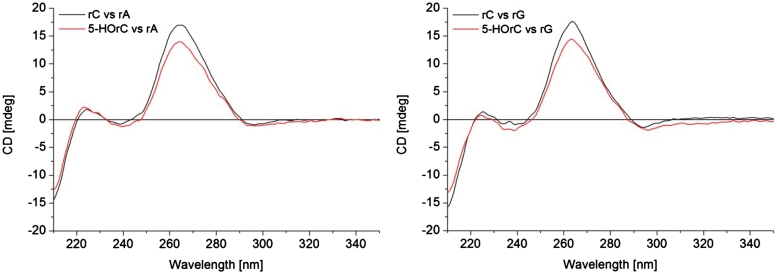
A CD-spectroscopic investigation was performed to determine whether pairing of 5-HOrC against rA or rG is associated with structural differences in comparison to the unmodified duplexes with a rC-rA mismatch or a rC-rG match (Figure 4). All CD spectra are very similar and suggest classical A-duplex conformation. The only difference is a slightly decreased positive cotton effect at 263 nm in both duplexes containing the 5-HOrC residue which is most likely due to differences in the UV absorption spectra between rC and 5-HOrC (see Supplementary Data). Thus the CD spectra suggest no major structural perturbations imposed by the 5-HOrC unit, irrespective of whether the opposing base is A or G.
Figure 4.
CD spectra of 12-mer duplexes: left: 5′-AUGCUXAGUCGA-3′ and 5′-UCGACUAAGCAU-3′ with X = rC or 5-HOrC; right: 5′-AUGCUXAGUCGA-3′ and 5′-UCGACUGAGCAU-3′ with X = rC or 5-HOrC. Duplex concentration: 2 µM (1:1 strand ratio) in 10 mM NaH2PO4, 150 mM NaCl, pH 7.0; T = 25°C.
Primer template extension experiments
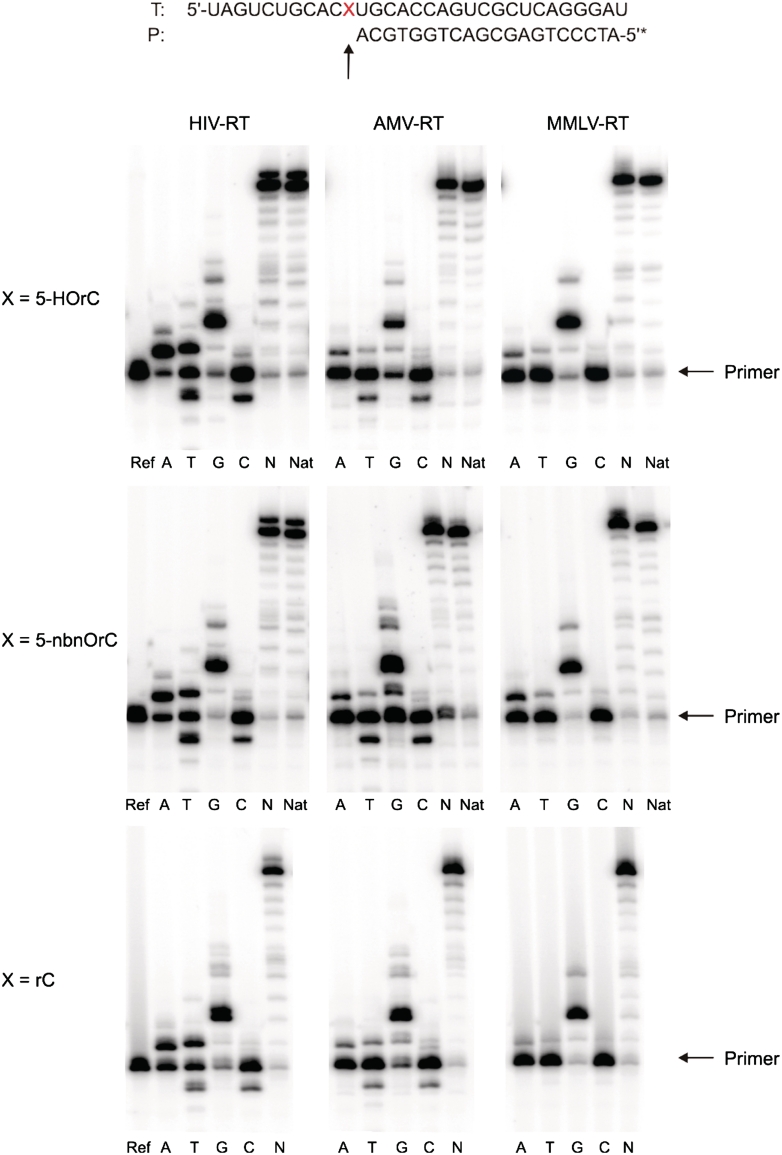
To elucidate the readout of the 5-HOrC lesion by RTs, we performed standing start primer extension reactions on the 31-mer template with a corresponding 5′-32P-radiolabeled DNA primer, dNTPs and three different RTs that vary in their transcription fidelity (HIV-RT, AMV-RT and MMLV-RT, Figure 5). Quantitative data of dNTP incorporation opposite the lesion are given in the Supplementary Table S2.
Figure 5.
Autoradiograms of standing start primer extension reactions for the reverse transcriptases indicated. Reactions were performed at 37°C for 1 h. Enzyme concentrations: HIV-RT, 0.5 U; AMV-RT, 8 U; MMLV-RT, 40 U. Ref: primer without enzyme; A, T, G, C: reactions in presence of the according dNTP; N: reactions in presence of all four dNTPs; Nat: unmodified RNA template (X = U) and all four dNTPs.
As expected all three RTs readily insert dGMP opposite the 5-HOrC lesion with high efficiency, and all enzymes proceed to full-length extension in the presence of all 4 dNTPs (Figure 5, top). But all three enzymes also insert a dAMP residue to a substantial amount opposite the lesion. In the case of HIV-RT, this proceeds about as efficiently as dGMP incorporation (63% for dAMP versus 64% for dGMP) while for the other two enzymes this process is ~2- to 3-fold less efficient. Interestingly, HIV-RT inserts also dTMP with about half of the efficiency of dGMP while AMV- and MMLV-RT incorporate only traces of dTMP (~10%) relative to dGMP. The remaining pyrimidine nucleotide, dCMP, is hardly incorporated by any of the three enzymes. Thus the order of preferential incorporation is G ~ A>T>>C for HIV-RT and G>A>T>>C for AMV-RT and MMLV-RT.
To determine how an alkyl residue at 5-O in the base, rendering it non-ionizable, influences the coding properties we performed the same experiments on the template with X = 5-nbnOrC (Figure 5, center). Again, full-length extension of the primer in the presence of all dNTPs occurs readily with all three enzymes. The order and efficiency of incorporation of the single dNTPs follows closely that observed for the 5-HOrC containing template in the case of AMV- and MMLV-RT. With HIV-RT, however, the extent of misincorporation of dA is reduced compared to 5-HOrC.
The same experiment with a template containing a natural rC unit as a control (Figure 5, bottom) also reveals significant dAMP and dTMP misincorporation with HIV-RT. However, the extent of misincorporation of dA is significantly reduced compared to 5-HOrC (49% for dAMP versus 77% for dGMP). Insertion of dT proceeds for all bases investigated to about the same extent. Again, MMLV- and AMV-RT display higher fidelity with almost exclusive incorporation of dGMP and only traces of dAMP and dTMP.
These experiments clearly show that there is substantial miscoding propensity for dA and to a lesser extent also for dT during cDNA synthesis opposite 5-HOrC by all three RTs investigated. For HIV-RT, the ratio of incorporation of dA versus dG opposite to 5-HOrC is approximately 0.98 whereas for rC it is 0.63. That of dT is about equal. Interestingly, the presence of a substituent at O-5 of the base, as in 5-nbnOrC, shows only slightly enhanced misincorporation of dAMP compared to rC, suggesting that either the ionization state or the proton itself in 5-HOrC influences the coding preferences in a subtle way. Our results are also in agreement with earlier work on primer template extension in DNA with the Klenow fragment of DNA polymerase I, where in one sequence context also increased misincorporation of dA opposite 5-HOdC was observed.
Our results reflect similar behavior to standing start primer template extension reactions on a DNA template by Klenow fragment (Kf) DNA polymerase, where also substantial misincorporation of dAMP was observed opposite 5-HOdC and to a much lesser extent opposite a natural dC residue (18).
DISCUSSION
In this article, we describe for the first time the incorporation of the RNA lesion product 5-HOrC into oligoribonucleotides by RNA phosphoramidite chemistry. The synthetic problem that had to be solved was the choice of an appropriate protecting group for the 5-OH group. Standard acyl groups such as benzoyl used for the corresponding 2′-deoxynucleoside building block (23,24), or silyl groups proved too labile in our hands. This is likely due to the rather high acidity of this phenolic hydroxyl group [pKa 7.37 in the case of the corresponding 2′-deoxynucleoside (9)], which renders it a good leaving group and the ester an activated ester. The choice of the caged benzyl group used in this study thus fulfills two purposes: (i) it confers chemical stability during building block and oligoribonucleotide synthesis and (ii) it is orthogonal in its deprotection conditions to the other RNA protecting groups which allows also the isolation of the 5-O-substituted oligonucleotides. This can be of interest for further studies where protonation state dependent tautomerism (7), or the redox chemistry of this base in the context of oligoribonucleotides is of interest (25).
Clearly the most striking result of this work is the biphasic melting behavior of duplexes with a 5-HOrC-rG or a 5-HOrC-dG base pair, giving rise to two Tm's where the higher one is close to that of a matched rC-rG or rC-dG base pair, respectively, while the lower one is close to that of a mismatched 5-HOrC-rA or 5-HOrC-dA pair, respectively. The absence of this effect in duplexes where 5-HOrC is replaced by 5-nbnOrC clearly demonstrates that either the ionization state of this group or the presence or absence of a proton in this position significantly influences its base recognition properties.
Several hypotheses can be formulated to explain this biphasic melting behavior. One of these invokes a chemical deamination from 5-HOrC to 5-HOrU under the conditions of Tm-analysis. This can be disproved based on the fact that the Tm profiles of duplexes containing authentic 5-HOrU in place of 5-HOrC are significantly different from each other (Table 1). We also determined the stability of nucleoside 2 at neutral pH and 80°C over 24 h and found <5% 5-HOrC to 5-HOrU conversion by NMR. Taken all together, deamination cannot be the cause of the biphasic melting behavior. On another note the results on the stability of 2 are in agreement with earlier experiments on the oxidation of poly d(C-G) in which it was shown that deamination occurs exclusively on the level of the dihydroxylated dCg (Figure 1) but not on 5-HOdC (6). Thus, most of the mutagenicity (C→T transitions) seems to be due to the intermediate dCg and must correlate with the rate of hydrolysis of the amino group relative to the rate of dehydration to 5-HOrC of the dCg intermediate.
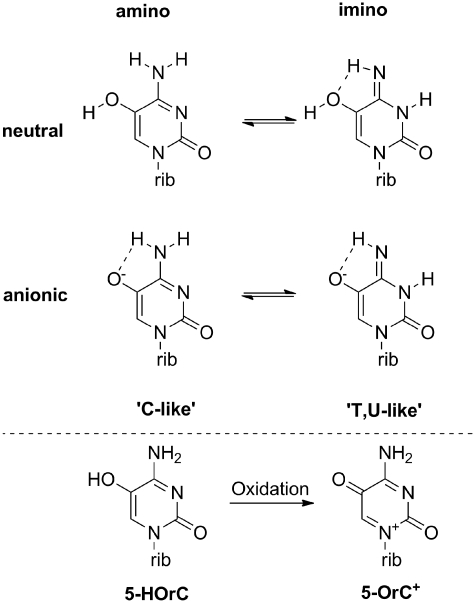
An obvious and tempting hypothesis arises from invoking changes in the tautomeric equilibrium of 5-HOrC, either depending on the protonation state of the 5-OH group or on temperature. It has been shown before that 5-HOdC can besides the amino form also adopt a minor imino tautomeric form (Figure 6, top). While the amino form emulates the hydrogen bonding pattern of C the imino form emulates that of T(U). Theoretical calculations in the gas phase favor the amino over the keto tautomer in the neutral form (8). UV resonance Raman spectroscopy of 5-HOdC shows enhanced presence of the imino tautomer by ~100-fold compared to dC at physiological pH and temperature, which results in a frequency of occurrence of this tautomer of 0.3–0.7%. The tendency toward the imino form slightly increases with temperature and pH (7). An alternative 15N-NMR investigations shows no substantial difference in tautomeric form between the neutral and anionic form of 5-HOdC, with the amino form being the only detectable form (9). The sensitivity of this method, however, might not be high enough to detect alternative tautomeric forms in the below 1% range. Although the tendency of increased imino tautomeric form at higher pH and higher temperature fits the trend of our observations it seems unlikely that its extent under any conditions applied would be detectable in a UV-melting curve.
Figure 6.
Top: relevant tautomeric forms of 5-HOrC; bottom: the two known redox states of 5-HOrC.
It is known that 5-HOrC is a redox active nucleoside which can be oxidized with agents such as Fe3+ to 5-oxo-cytidine (5-OrC+, Figure 6, bottom) (25). Therefore the question arises whether this oxidized form of 5-HOrC is associated with the unusual melting behavior. We rule out this hypothesis primarily based on the reason that the conditions during nbnOrC deprotection and subsequent manipulations for melting curve measurements are not oxidizing, and that the hydrogen bonding pattern of 5-OrC+ is expected to be tautomerically stable and identical to that of a natural rC unit which would not explain the biphasic transition.
We also questioned whether temperature-dependent changes in the syn/anti ratios (syn at low and anti at high temperatures) of the glycosidic torsion angle in 5-HOrC could be a reason for the biphasic melting transition. This cannot be ruled out completely but seems less likely. Given that the hydroxy group is on C-5 and not C-6, it is difficult to envisage a structural or stereoelectronic force that would drive the base in syn conformation which is generally the less preferred conformation for pyrimidine nucleosides. In addition, the related base 5-nbnOrC where the bulky aryl group could lead to steric interactions with the phosphodiester backbone in the anti conformation, required for pairing with G, behaves as rC and does not show biphasic melting behavior. Also, the ab initio calculations of 5-HOdC did not give any indications on a predominance of the syn-conformer, neither in the neutral nor in the ionized form (9).
Yet another, and probably more plausible, explanation could be that the ionization state of the 5-OH group influences indirectly the glycosidic torsion angle of the nucleoside when incorporated into a DNA or RNA backbone. The presence of a 5′-phosphate unit is known to increase the pKa of 5-HOrC by 1.1 units to 8.5, relative to the free nucleoside. This increase has been associated with electrostatic repulsion of the base oxyanion with the negatively charged 5′-phosphate unit (9). On the other hand, ab initio calculations on 5-HOdCMP have shown that in the neutral form the 5-OH group can form a strong H-bond with a 5′-phosphate oxygen. It may therefore well be that this hydrogen bond drives the glycosidic torsion angle in a geometry which is unfavorable for correct base pair formation with G within an oligoribonucleotide duplex. Upon thermal denaturation of this H-bond during heating the glycosidic bond could relax, thus enabling efficient G-base recognition. Such a process could be a reason for the observed double transition in the melting curves. The local structural reorganizations are likely to be too small to be detectable by CD spectroscopy, explaining the almost identical CD spectra of duplexes containing either a 5-HOrC-rG or a rC-rG base pair.
We currently favor this last hypothesis but we are aware that there may exist alternative explanations yet to be discovered. For this, however, more biophysical data including high resolution structural data on 5-HOrC containing duplexes is needed. We note that besides tautomeric variability such structural effects could also determine the differences in the coding properties of 5-HOrC units during RT.
Given the base recognition and coding bias of 5-HOrC, as described here, the question arises on the consequences on transcription and translation. In forms of life where RNA is the genetic material, the expected higher C-T transition rate may have direct consequences on genomic stability and evolution. In ribosomal translation of mRNA such a lesion may influence the protein expression profile. It has been shown previously in in vitro translation experiments of oxidatively damaged mRNA coding for luciferin or GFP that abortion or alteration of protein synthesis occurs, however, without detailed knowledge of the underlying biochemical mechanism (13,26). Clearly, codon–anticodon misrecognition might lead to misincorporation of amino acids into a growing peptide chain which accidentally might change its structure and/or function much in the same way as a genetically encoded protein mutation.
CONCLUSIONS
In this work, we describe the synthesis of RNA containing the oxidized base 5-hydroxycytidine. The synthesis uses a phosphoramidite building block containing a photochemically cleavable protecting group at O-5 of the base. In UV-melting curves of RNA duplexes, we find an unusual biphasic melting behavior when a 5-HOrC unit is paired against rG or dG. We exclude chemical instability and tautomeric variability of 5-HOrC to be responsible for this but currently favor a structurally based mismatch to rG induced by a phosphate oxygen/5-OH hydrogen bond, perturbing the glycosidic bond angle of this base in the duplex. In RT primer template extension experiments, we find substantial misincorporation of dAMP opposite the lesion. The non-negligible propensity of base misrecognition clearly points to consequences during ribosomal translation if such a lesion were to occur on a mRNA. We are currently investigating this with oxidatively damaged synthetic mRNAs in an in vitro translation system.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: University of Bern and Swiss National Science Foundation (grant. No. 200020-130373).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Alessandro Calabretta for providing the Tm data of the duplex containing 5-HOrU (Table 1).
REFERENCES
- 1.De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–185. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- 2.Darwanto A, Ngo L, Sowers LC. Chapter 6 Pyrimidine Damage and Repair. In: James CF, editor. Advances in Molecular Toxicology. Vol. 2. Elsevier; 2008. pp. 153–182. [Google Scholar]
- 3.Marnett LJ. Oxyradicals and DNA damage. Carcinogenesis. 2000;21:361–370. doi: 10.1093/carcin/21.3.361. [DOI] [PubMed] [Google Scholar]
- 4.Wang D, Kreutzer DA, Essigmann JM. Mutagenicity and repair of oxidative DNA damage: insights from studies using defined lesions. Mutat. Res. 1998;400:99–115. doi: 10.1016/s0027-5107(98)00066-9. [DOI] [PubMed] [Google Scholar]
- 5.Kreutzer DA, Essigmann JM. Oxidized, deaminated cytosines are a source of C → T transitions in vivo. Proc. Natl Acad. Sci. USA. 1998;95:3578–3582. doi: 10.1073/pnas.95.7.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tremblay S, Wagner JR. Dehydration, deamination and enzymatic repair of cytosine glycols from oxidized poly(dG-dC) and poly(dI-dC) Nucleic Acids Res. 2008;36:284–293. doi: 10.1093/nar/gkm1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suen W, Spiro TG, Sowers LC, Fresco JR. Identification by UV resonance Raman spectroscopy of an imino tautomer of 5-hydroxy-2′-deoxycytidine, a powerful base analog transition mutagen with a much higher unfavored tautomer frequency than that of the natural residue 2′-deoxycytidine. Proc. Natl Acad. Sci. USA. 1999;96:4500–4505. doi: 10.1073/pnas.96.8.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cysewski P. Molecular perspective review of biochemical role of nucleobases modified by oxidative stress. Comput. Meth. Sci. Technol. 2010;16:51–72. [Google Scholar]
- 9.La Francois CJ, Jang YH, Cagin T, Goddard WA, Sowers LC. Conformation and proton configuration of pyrimidine deoxynucleoside oxidation damage products in water. Chem. Res. Toxicol. 2000;13:462–470. doi: 10.1021/tx990209u. [DOI] [PubMed] [Google Scholar]
- 10.Schneider JE, Phillips JR, Pye Q, Maidt ML, Price S, Floyd RA. Methylene blue and rose bengal photoinactivation of RNA bacteriophages: comparative studies of 8-oxoguanine formation in isolated RNA. Archives Biochem. Biophys. 1993;301:91–97. doi: 10.1006/abbi.1993.1119. [DOI] [PubMed] [Google Scholar]
- 11.Yanagawa H, Ogawa Y, Ueno M. Redox ribonucleosides. Isolation and characterization of 5-hydroxyuridine, 8-hydroxyguanosine, and 8-hydroxyadenosine from Torula yeast RNA. J. Biol. Chem. 1992;267:13320–13326. [PubMed] [Google Scholar]
- 12.Li Z, Wu J, DeLeo CJ. RNA damage and surveillance under oxidative stress. IUBMB Life. 2006;58:581–588. doi: 10.1080/15216540600946456. [DOI] [PubMed] [Google Scholar]
- 13.Shan X, Chang Y, Lin CL. Messenger RNA oxidation is an early event preceding cell death and causes reduced protein expression. FASEB J. 2007;21:2753–2764. doi: 10.1096/fj.07-8200com. [DOI] [PubMed] [Google Scholar]
- 14.Nunomura A, Honda K, Takeda A, Hirai K, Zhu X, Smith MA, Perry G. Oxidative damage to RNA in neurodegenerative diseases. J. Biomed. Biotech. 2006;2006:82323. doi: 10.1155/JBB/2006/82323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui S, Kim Y-H, Jin C-H, Kim SK, Rhee M-h, Kwon O-S, Moon BJ. Synthesis and base pairing properties of DNA–RNA heteroduplex containing 5-hydroxyuridine. BMB Reports. 2009;42:373–379. doi: 10.5483/bmbrep.2009.42.6.373. [DOI] [PubMed] [Google Scholar]
- 16.Kim SK, Kim JY, Baek AK, Moon BJ. Base pairing properties of 8-oxo-7,8-dihydroadenosine in cDNA synthesis by reverse transcriptases. Bioorg. Mred. Chem. Lett. 2002;12:1977–1980. doi: 10.1016/s0960-894x(02)00297-4. [DOI] [PubMed] [Google Scholar]
- 17.Kim SK, Lee SH, Kwon O-S, Moon BJ. DNA.RNA heteroduplex containing 8-oxo-7,8-dihydroguanosine: Base pairing, structures, and thermodynamic stability. J. Biochem. Mol. Biol. 2004;37:657–662. doi: 10.5483/bmbrep.2004.37.6.657. [DOI] [PubMed] [Google Scholar]
- 18.Purmal AA, Kow YW, Wallace SS. Major oxidative products of cytosine, 5-hydroxycytosine and 5-hydroxyuracil, exhibit sequence context-dependent mispairing in vitro. Nucleic Acids Res. 1994;22:72–78. doi: 10.1093/nar/22.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eaton MA, Hutchinson DW. Poly(5-hydroxycytidylic acid) Biochim. Biophys. Acta. 1973;319:281–287. doi: 10.1016/0005-2787(73)90167-6. [DOI] [PubMed] [Google Scholar]
- 20.Küpfer PA, Leumann CJ. The chemical stability of abasic RNA compared to abasic DNA. Nucleic Acids Res. 2007;35:58–68. doi: 10.1093/nar/gkl948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Küpfer PA, Leumann CJ. Photochemically induced RNA and DNA abasic sites. Nucleosides, Nucleotides Nucleic Acids. 2007;26:1177–1180. doi: 10.1080/15257770701527711. [DOI] [PubMed] [Google Scholar]
- 22.Küpfer PA, Crey-Desbiolles C, Leumann CJ. Trans-lesion synthesis and RNaseH activity by reverse transcriptases on a true abasic RNA template. Nucleic Acids Res. 2007;35:6846–6853. doi: 10.1093/nar/gkm767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morningstar ML, Kreutzer DA, Essigmann JM. Synthesis of oligonucleotides containing two putatively mutagenic DNA lesions: 5-hydroxy-2'-deoxyuridine and 5-hydroxy-2'-deoxycytidine. Chem. Res.Toxicol. 1997;10:1345–1350. doi: 10.1021/tx970052a. [DOI] [PubMed] [Google Scholar]
- 24.Romieu A, Gasparutto D, Molko D, Cadet J. A convenient synthesis of 5-Hydroxy-2′-deoxycytidine phosphoramidite and its incorporation into oligonucleotides. Tetrahedron Lett. 1997;38:7531–7534. [Google Scholar]
- 25.Yanagawa H, Ogawa Y, Ueno M, Sasaki K, Sato T. A novel minimum ribozyme with oxidoreduction activity. Biochemistry. 1990;29:10585–10589. doi: 10.1021/bi00499a002. [DOI] [PubMed] [Google Scholar]
- 26.Shan X, Lin C-lG. Quantification of oxidized RNAs in Alzheimer's disease. Neurobiol. Aging. 2006;27:657–662. doi: 10.1016/j.neurobiolaging.2005.03.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.