Abstract
In vitro-transcribed mRNA has great therapeutic potential to transiently express the encoded protein without the adverse effects of viral and DNA-based constructs. Mammalian cells, however, contain RNA sensors of the innate immune system that must be considered in the generation of therapeutic RNA. Incorporation of modified nucleosides both reduces innate immune activation and increases translation of mRNA, but residual induction of type I interferons (IFNs) and proinflammatory cytokines remains. We identify that contaminants, including double-stranded RNA, in nucleoside-modified in vitro-transcribed RNA are responsible for innate immune activation and their removal by high performance liquid chromatography (HPLC) results in mRNA that does not induce IFNs and inflammatory cytokines and is translated at 10- to 1000-fold greater levels in primary cells. Although unmodified mRNAs were translated significantly better following purification, they still induced high levels of cytokine secretion. HPLC purified nucleoside-modified mRNA is a powerful vector for applications ranging from ex vivo stem cell generation to in vivo gene therapy.
INTRODUCTION
Our understanding of the importance of RNA in biological processes and the therapeutic potential has substantially increased with the discovery of non-coding regulatory RNAs. The use of mRNA has also expanded, including the delivery of mRNA to generate induced pluripotent stem (iPS) cells (1–3) and in vivo administration to express therapeutic proteins (4). The recognition that the immunogenicity of RNA could be reduced by the incorporation of modified nucleosides with a concomitant increase in translation (5), potentially allows efficient expression of intra and extracellular proteins in vivo and ex vivo without activation of innate immune pathways. Unfortunately, modified nucleoside-containing RNA transcribed by phage RNA polymerase transcription still retains a low level of activation of such pathways (3,5–7). The remaining activation of RNA sensors by nucleoside modified RNA could be because the modifications do not completely suppress the RNAs ability to activate sensors or due to contaminants with structures that activate in the presence of nucleoside modification. It is well established that RNA transcribed in vitro by phage polymerase contains multiple contaminants, including short RNAs produced by abortive initiation events (8) and double-stranded (ds)RNAs generated by self-complementary 3′ extension (9), RNA-primed transcription from RNA templates (10) and RNA-dependent RNA polymerase activity (11).
Large quantities of RNA can be easily prepared by in vitro transcription from DNA templates using phage RNA polymerase or solid-phase chemical synthesis. For uses that require further purification, such as NMR (12), crystallography (13) and therapeutic applications (14), a number of techniques have been developed. Preparative denaturing polyacrylamide gel electrophoresis is commonly used to purify in vitro-transcribed RNA, however, this method is suitable only for short RNAs [reviewed in (15)]. Long RNAs can be separated on denaturing agarose gels, but they are not translatable due to covalent modifications introduced by the denaturants glyoxal and formaldehyde (16). Chromatography based on size exclusion can efficiently remove unincorporated nucleoside triphosphates, small abortive transcripts and plasmid template from the desired RNA product under native conditions (17,18), but is limited in its ability to remove contaminants with similar sizes and contaminants complementary to the RNA selected to purify. No technique has been reported for purification and preparative isolation of long in vitro-transcribed mRNA that removes contaminating complementary strands and preserves its translatability.
The development of mRNA to use as a tool to replace intra- and extracellular proteins in vivo and to transdifferentiate, reprogram and differentiate cells ex vivo requires the RNA to have high translatability and no RNA sensor activation. In this report, we identify that contaminants from in vitro-transcribed RNA are a source of innate immune activation and their removal increases RNA translation and eliminates type I interferon and inflammatory cytokine secretion.
MATERIALS AND METHODS
Cells
Human embryonic kidney 293T cells (American Type Culture Collection) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 2 mM l-glutamine (Life Technologies) and 10% fetal calf serum (FCS) (HyClone) (complete medium). Human and murine dendritic cells (DCs) were generated as described (5). Human keratinocytes were obtained from the Skin Disease Research Core (Penn) and grown in MCDB with bovine pituitary extract (140 µg/ml) (Sigma) and 70 µM Ca++ on collagen (0.01 mg/ml) (Invitrogen) coated plates.
mRNA synthesis
mRNAs were transcribed as previously described (5), using linearized plasmids encoding firefly luciferase (pT7TSLuc and pTEVLuc), codon-optimized murine erythropoietin (pTEVmEPO), enhanced green fluorescent protein (pTEVeGFP), Metridia luciferase (pT7TSMetluc) or Renilla luciferase (pT7TSRen and pTEVRen) and T7 RNA polymerase (Megascript, Ambion). All mRNAs were transcribed to contain 30 or 51-nt long poly(A) tails. Additional poly(A) tail was added with yeast poly(A) polymerase (USB) and noted as An. Triphosphate-derivatives of pseudouridine (Ψ) and 5-methylcytidine (m5C) (TriLink) were used to generate modified nucleoside containing RNA. All RNAs were capped using the m7G capping kit with or without 2′-O-methyltransferase (ScriptCap, CellScript) to obtain cap1 or cap0. We did not observed differences in the immunogenicity of cap0- and cap1-containing nucleoside-modified RNAs. All RNAs were analyzed by denaturing or native agarose gel electrophoresis. Pseudouridine-modified mRNAs encoding KLF4, LIN28, cMYC, NANOG, OCT4 and SOX2 were a kind gift of CellScript, Inc.
HPLC purification of RNA
RNA was purified by High performance liquid chromatography (HPLC) (Akta Purifier, GE Healthcare) using a column matrix of alkylated non-porous polystyrene-divinylbenzene copolymer microspheres (2.1 μm) (21 × 100 mm column). Buffer A contained 0.1 M triethylammonium acetate (TEAA), pH = 7.0 and Buffer B contained 0.1 M TEAA, pH = 7.0 and 25% acetonitrile (Transgenomics). Columns were equilibrated with 38% Buffer B, loaded with RNA and run with a single or 2 linear gradients to 55 or 65% Buffer B over 20–30 min at 5 ml/min. RNA analyses were performed with the same column matrix and buffer system using a 7.8 mm × 50 mm column at 1.0 ml/min.
RNA isolation from column fractions
RNA content from desired fractions was concentrated and desalted using Amicon Ultra-15 centrifugal filter units (30K membrane) (Millipore) by successive centrifugation at 4000g for 10 min (4°C) in a Sorvall ST16R centrifuge (Thermo Scientific) and dilution with nuclease free water. The RNA was recovered by overnight precipitation at −20°C in NaOAc (0.3 M, pH 5.5), isopropanol (1 volume) (Fisher) and glycogen (3 µl) (Roche).
Dot blot
RNA (200 ng) was blotted onto super charged Nytran, dried, blocked with 5% non-fat dried milk in TBS-T buffer (50 mM Tris–HCl, 150 mM NaCl, 0.05% Tween-20, pH 7.4), and incubated with dsRNA-specific mAb J2 or K1 (English & Scientific Consulting) for 60 min. Membranes were washed six times with TBS-T and reacted with HRP-conjugated donkey anti-mouse Ig (Jackson Immunology), washed six times and detected with ECL Plus Western blot detection reagent (Amersham). Images were captured on a Fujifilm LAS1000 digital imaging system. dsRNA (25 ng) used as a positive control was derived from sense and antisense strands of T7TS UTR sequence (328 bp). Blots were reprobed with 32P-labeled DNA complementary to the 3′-UTR of the RNA to document the presence of RNA.
Complexing of RNA
Lipofectin (Invitrogen) complexing was performed as described previously (5) using 0.8 µl of Lipofectin and 0.1 µg of RNA per well of a 96-well plate. Complexing of RNA to TransIT mRNA (Mirus Bio) was performed according to the manufacturer combining RNA (0.1 µg) with TransIT mRNA (0.3 µl) and boost (0.2 µl) reagents.
Cell transfections
For Lipofectin complexed RNA, medium was removed and 50 µl of complexed RNA was added to 5 x 104 293T or DCs per well. Cells were incubated for 1 h and the Lipofectin-RNA mixture was replaced with 200 µl complete medium. For TransIT complexed RNA, 17 µl of complex was added to cells, 293T, DCs, or 2 × 105 keratinocytes cultured in 183 µl complete medium. Cells were lysed in firefly or Renilla specific lysis reagents (Promega) at 24 h post RNA addition. Aliquots were assayed for enzyme activities using firefly and Renilla luciferase assay systems (Promega) and a LUMAT LB 950 luminometer (Berthold/EG&G; Wallac). Expression of eGFP in DCs was documented using an inverted epifluorescent Nikon microscope mounted with a Nikon D40 digital camera. Murine EPO protein was measured with a specific ELISA assay (R&D Systems).
RNA immunogenicity analyses
DCs (murine or human) (5 × 104 cells/well) in 96-well plates were treated with medium, R-848 (Invivogen), or Lipofectin- or TransIT-complexed RNA or poly(I:C) (Sigma). Supernatant was harvested after 24 h and the levels of IFN-α, IFN-β (PBL InterferonSource), or TNF-α (Biosource International) were measured by ELISA.
Gene array analysis
Human DCs from three donors were generated in 5% FCS. Cells (1 × 106 DCs/well of a 6-well plate) were treated with TransIT-complexed TEVRenA51 RNA with or without modification and with or without purification. Six hours later, RNA was isolated using RNeasy (Qiagen). RNA was amplified with the TargetAmp Nano-g Biotin-aRNA labeling kit (Epicentre) and analyzed on an Illumina Human HT12v4 chip in an Illumina BeadStation 500GX. Raw data was processed by the Bead Studio v.3.0 software. Levels in untreated DC were used as the baseline for the calculation of fold increase.
Northern blot
Samples were processed and analyzed as previously described (6). Probes were derived from plasmids and were specific for the coding regions of human IFN-α13, IFN-β (Open Biosystems), TNF-α, or GAPDH (ATCC).
RESULTS
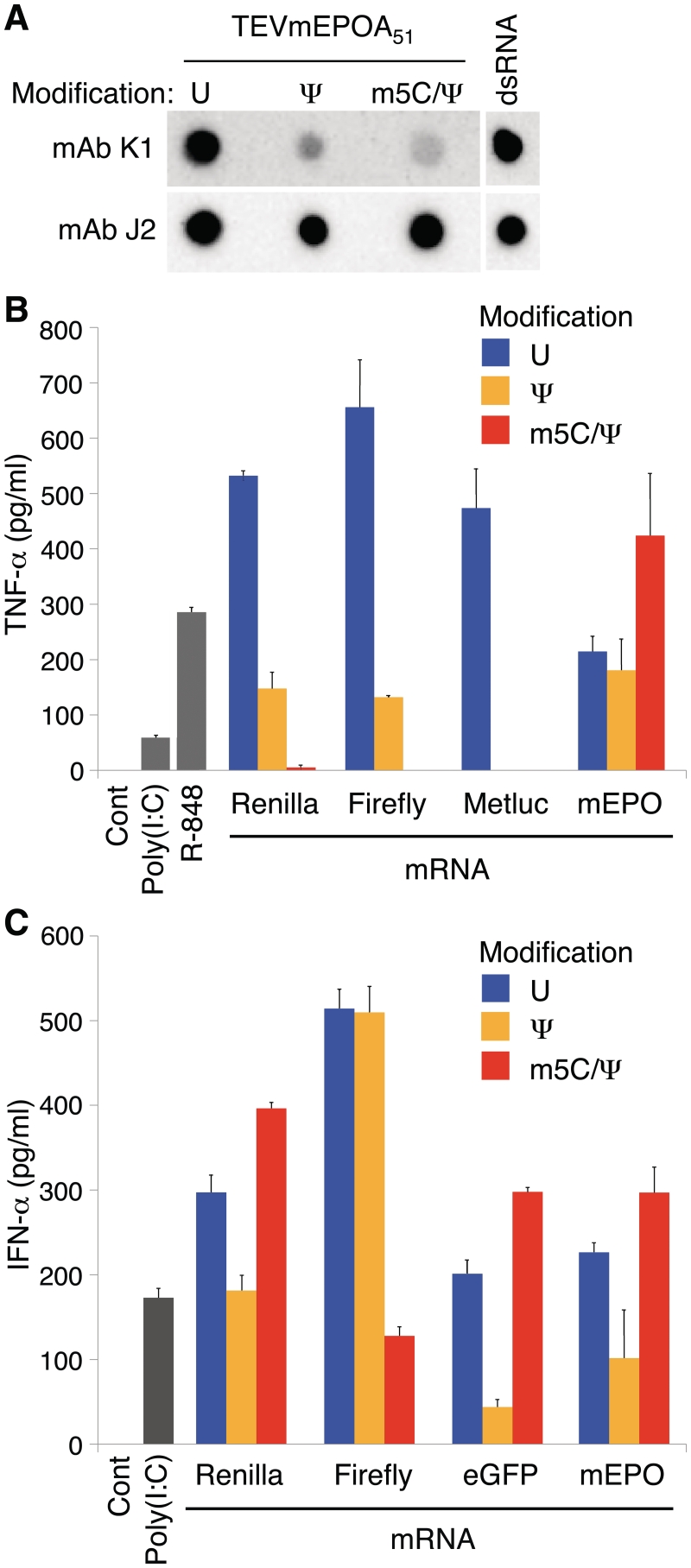
A dot blot assay with J2 and K1 monoclonal antibodies (mAbs) that recognize dsRNA (19) was used to determine whether in vitro-transcribed RNA contains dsRNA. These mAbs recognize continuous double stranded structure of at least 40 bp in length (20), which is not found in any of the coding sequences or UTRs in the mRNAs analyzed in this study. Testing mammalian and reporter protein-encoding in vitro transcripts containing either no nucleoside modifications, pseudouridine- (Ψ), or 5-methylcytidine- (m5C) and Ψ- (m5C/Ψ) nucleoside modifications, we found that all samples contained dsRNA contamination (Figure 1A and data not shown). Recognition of dsRNA by J2 mAb was not affected by the presence of modified nucleosides, while K1 had reduced binding to dsRNA containing Ψ or m5C/Ψ nucleoside modifications.
Figure 1.
In vitro-transcribed RNA is immunogenic and contains dsRNA contaminants. (A) 200 ng of in vitro transcripts encoding mEPO and containing the indicated modified nucleosides were blotted and analyzed with K1 and J2 dsRNA-specific mAbs. The dsRNA positive control contained a 328 bp long dsRNA (25 ng). (B) DCs were treated with Lipofectin-complexed Renilla luciferase (T7TSRenA30), firefly and Metridia luciferases (T7TSLucA30, T7TSMetlucA30), and mEPO (TEVmEPOA51) mRNAs. TNF-α levels were measured in the supernatants at 24 h. (C) DCs were treated with TransIT-complexed in vitro transcripts encoding Renilla and firefly luciferases (T7TSRenA30, T7TSLucA30), eGFP (TEVeGFPA51) and mEPO (TEVmEPOA51). IFN-α levels were measured in the supernatants at 24 h. Error bars are standard error of the mean. Data shown is from one experiment that is representative of 3–6.
Others and we have previously demonstrated that incorporation of modified nucleotides into RNA reduced its ability to activate RNA sensors including Toll-like receptor (TLR)3, TLR7 and TLR8 (21), retinoic acid inducible gene I (RIG-I) (22) and RNA-dependent protein kinase (PKR) (6,23). Monocyte-derived DCs that express these and all other known RNA sensors (24) were used to measure residual immune activation present in modified nucleoside-containing RNA. A series of RNAs with coding sequences for mammalian and reporter proteins flanked by different 5′- and 3′-UTRs were analyzed. The RNAs were cell-delivered following complexing with Lipofectin, a cationic lipid, or TransIT, a membrane active polymer and lipid mixture. RNA complexed with Lipofectin induced high levels of TNF-α and moderate levels of IFN-α, while RNA complexed with TransIT induced low levels of TNF-α and high levels of IFN-α (data not shown) with some donor-dependent variation. Typically, Lipofectin-complexed RNA with Ψ or m5C/Ψ modifications induced less TNF-α. (Figure 1B), while TransIT-complexed RNA with or without nucleoside modification induced variable, sequence-dependent effects on IFN-α secretion (Figure 1C). These data suggest that the presence of dsRNA and potentially other contaminants in in vitro-transcribed RNA could be responsible for innate immune activation.
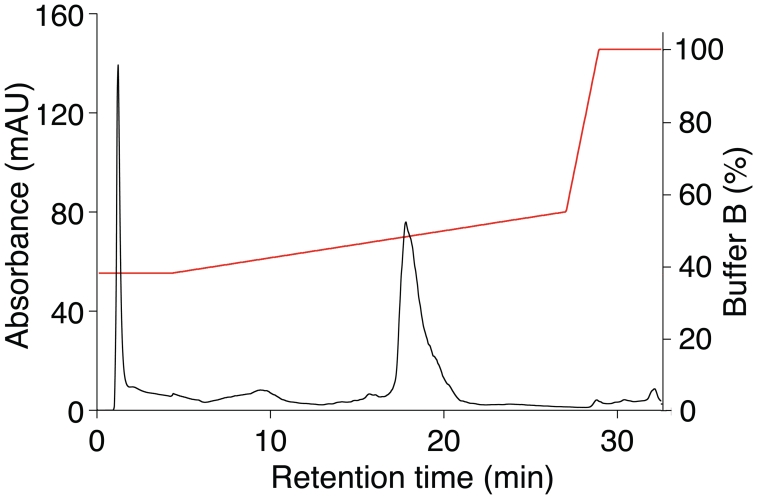
Multiple HPLC bead matrix compositions and buffer systems were screened and alkylated non-porous polystyrene-divinylbenzene copolymer matrix and triethylammonium acetate buffer with an acetonitrile gradient was identified as a system capable of removing dsRNA and other contaminants from in vitro-transcribed RNA. The HPLC chromatogram of Ψ-modified mRNA encoding enhanced green fluorescent protein (eGFP) demonstrated a major peak (Figure 2), which was collected and identified as the expected RNA product using agarose gel electrophoresis. Additional UV-absorbing products with shorter and longer retention times relative to the main RNA product could also be observed. Reanalysis of the purified RNA by HPLC demonstrated a single peak with the same retention time. RNAs with or without nucleoside modification encoding different sequences yielded similar patterns with varying relative heights for the preceding and succeeding peaks.
Figure 2.
HPLC purification of RNA identifies contaminants eluting before and after the expected product. Chromatogram of Ψ-modified TEVeGFPAn mRNA. RNA was applied to the HPLC column and eluted using a linear gradient of Buffer B (0.1 M TEAA, pH 7.0, 25% acetonitrile) in Buffer A (0.1 M TEAA, pH 7.0). The gradient spanned 38–55% Buffer B over 22 min (red line). Absorbance at 260 nm was analyzed (black line), which demonstrated the expected sized RNA as well as smaller and larger RNA species. Data shown are from one experiment that is representative of over 200.
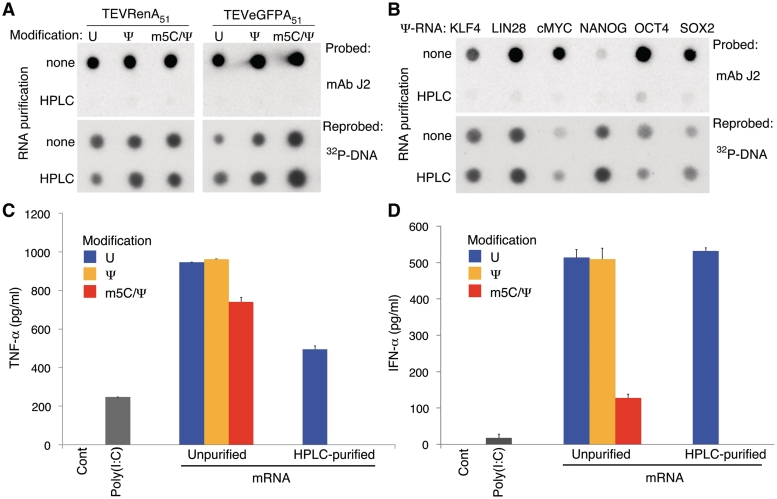
HPLC purification of both unmodified and nucleoside-modified RNA reduced staining by dsRNA-specific mAb to baseline levels (Figure 3A). Analysis of Ψ-modified RNA encoding clinically relevant proteins demonstrated that the amounts of dsRNA contamination in the in vitro transcripts were dependent on the sequence, but HPLC could successfully remove the contaminants from all of them (Figure 3B). Next, the HPLC-purified RNAs were tested on human DCs. No TNF-α or type-I interferons (IFN-α and β) were induced following transfection of HPLC-purified Ψ- or m5C/Ψ-modified RNAs that were complexed with Lipofectin or TransIT, respectively (Figure 3C and D and data not shown). Similarly, no cytokine induction could be detected when HPLC-purified modified nucleoside-containing RNAs were transfected into murine DCs. HPLC purification similarly ablated IFN-α secretion from DCs transfected with the clinically relevant Ψ-nucleoside modified mRNAs complexed to TransIT used in Figure 3B. However, HPLC-purified RNA without nucleoside modification remained potent inducers of TNF-α and IFN-α (Figure 3C and D).
Figure 3.
HPLC purification of in vitro-transcribed nucleoside modified mRNA removes dsRNA contaminants and eliminates immunogenicity. (A) 200 ng of RNA encoding the indicated protein and containing the indicated modified nucleosides with or without HPLC purification were blotted and analyzed with the J2 dsRNA-specific mAb. (B) 200 ng of RNA encoding the indicated protein and containing Ψ-modifications with or without HPLC purification were blotted and analyzed with the J2 dsRNA-specific mAb. Blots were reprobed with a 32P-labeled probe for the 3′-UTR of the RNAs to control for amount of RNA analyzed. (C) DCs were treated with TEVRenA51 RNA containing the indicated nucleoside modifications with or without HPLC purification and complexed to Lipofectin. TNF-α levels were measured in the supernatants at 24 h. Differences in the effect of nucleoside modification on immunogenicity of Renilla-encoding mRNA compared to Figure 1B is likely due to donor variation and differences in UTRs of the RNAs. (D) DCs were treated with TEVLucA51 RNA containing the indicated nucleoside modifications with or without HPLC purification and complexed to TransIT. IFN-α levels were measured in the supernatants at 24 h. Error bars are standard error of the mean. Data shown is from one experiment that is representative of 3 or more.
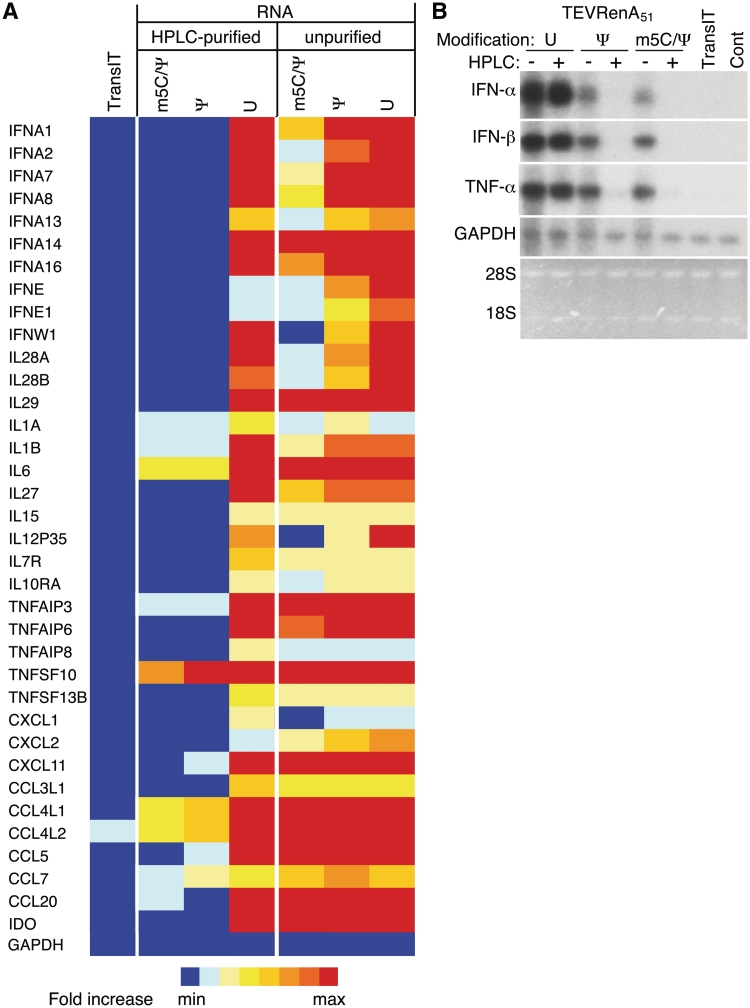
The impact of HPLC purification of unmodified, Ψ- and m5C/Ψ-modified RNA on gene expression in human DCs was analyzed using gene arrays. Total cellular RNA isolated from DCs from three different donors 6 h after cells were transfected with TransIT-complexed RNA, were analyzed on an Illumina Human HT12v4 chip. RNA modified with Ψ or m5C/Ψ nucleosides induced less expression of type I interferons, interleukins, tumor necrosis factor (TNF) family members, chemokines and markers associated with DC activation, while HPLC purification of Ψ- and m5C/Ψ-modified RNA further reduced induction of these genes to the levels observed in cells treated only with TransIT (Figure 4A). The same sets of total RNA from DCs that were tested on the gene arrays were also analyzed for levels of IFN-α, IFN-β and TNF-α mRNA by northern blot. Lower levels of IFN-α, IFN-β and TNF-α mRNAs were detectable in DCs treated with nucleoside modified as compared to unmodified RNA. More importantly, none of these cytokine mRNAs were detectable when DCs were transfected with HPLC-purified RNAs containing Ψ or m5C/Ψ modification. However, HPLC purified RNA without nucleoside modification remained a potent inducer of IFN-α, IFN-β and TNF-α mRNAs (Figure 4B).
Figure 4.
HPLC purification of in vitro-transcribed nucleoside-modified mRNA eliminates activation of genes associated with RNA sensor activation. (A) Heatmap representing changes in expression of genes activated by RNA sensors were derived from microarray analyses of DCs treated for 6 hr with TransIT alone or transit-complexed TEVRenA51 RNA with the indicated modifications with or without HPLC purification. RNA from medium-treated cells was used as the baseline for comparison. (B) Northern blot of RNA from DCs treated with medium or TransIT alone or TransIT-complexed TEVRenA51 RNA with the indicated modifications with or without HPLC purification and probed for IFN-α, IFN-β, TNF-α and GAPDH mRNAs.
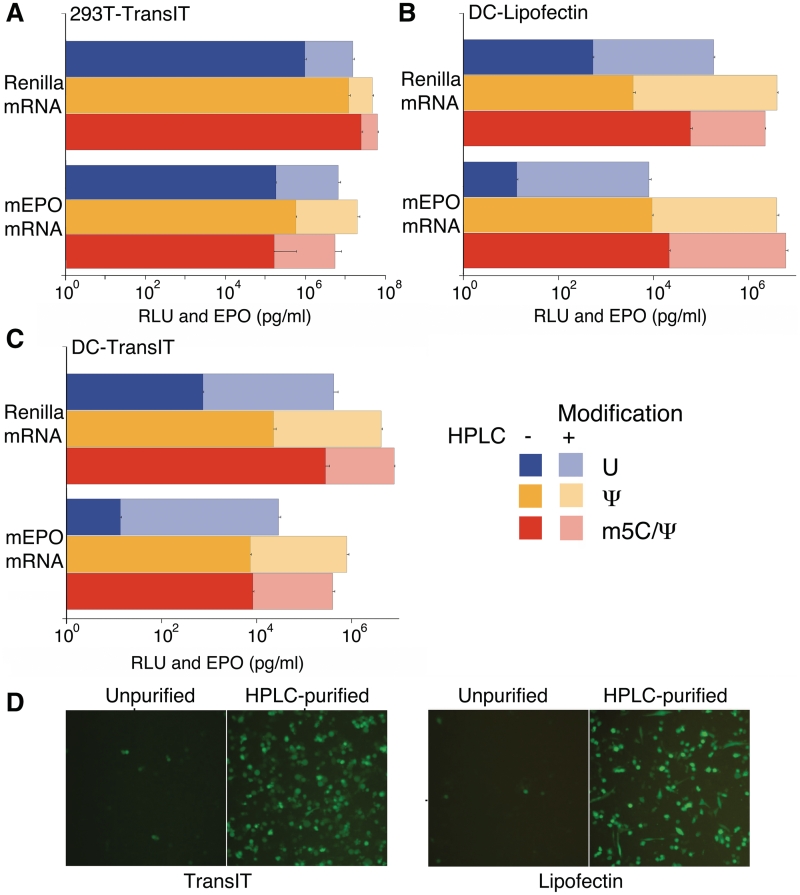
To determine whether HPLC purification affected translatability of in vitro transcripts, a series of mRNAs were tested following cell delivery. HPLC-purified Renilla and mouse erythropoietin (mEPO) mRNAs were translated at 2- to 20-fold higher levels compared to unpurified RNA when delivered to 293 T cells by TransIT (Figure 5A). In primary human DCs, the translational enhancement was more robust, resulting in up to a 1000-fold increase when the same sets of unpurified and HPLC-purified mRNAs were transfected with Lipofectin (Figure 5B) or TransIT (Figure 5C). Similar increases in translation were observed for other mRNAs after HPLC purification, including mRNAs encoding firefly luciferase, human EPO, macaque EPO and Metridia luciferase, and other cell types, including mouse embryonic fibroblasts and human primary keratinocytes. Translation levels were much higher with Ψ- and m5C/Ψ-modified eGFP mRNA when the mRNA was HPLC-purified prior to transfection of human DCs (Figure 5D and data not shown).
Figure 5.
HPLC purification of in vitro transcribed mRNA enhances translation. 293T (A) and human DCs (B and C) were transfected with TransIT- (A and C) or Lipofectin- (B) complexed TEVRenA51 or TEVmEPOA51 mRNA with the indicated modifications with or without HPLC purification and analyzed for Renilla luciferase activity or levels of supernatant-associated mEPO protein at 24 h. (D) Human DCs were transfected with Ψ-modified TEVeGFPAn mRNA with or without HPLC purification (0.1 µg/well) complexed with Lipofectin or TransIT and analyzed 24 h later. Error bars are standard error of the mean. Data shown is from one experiment that is representative of three or more.
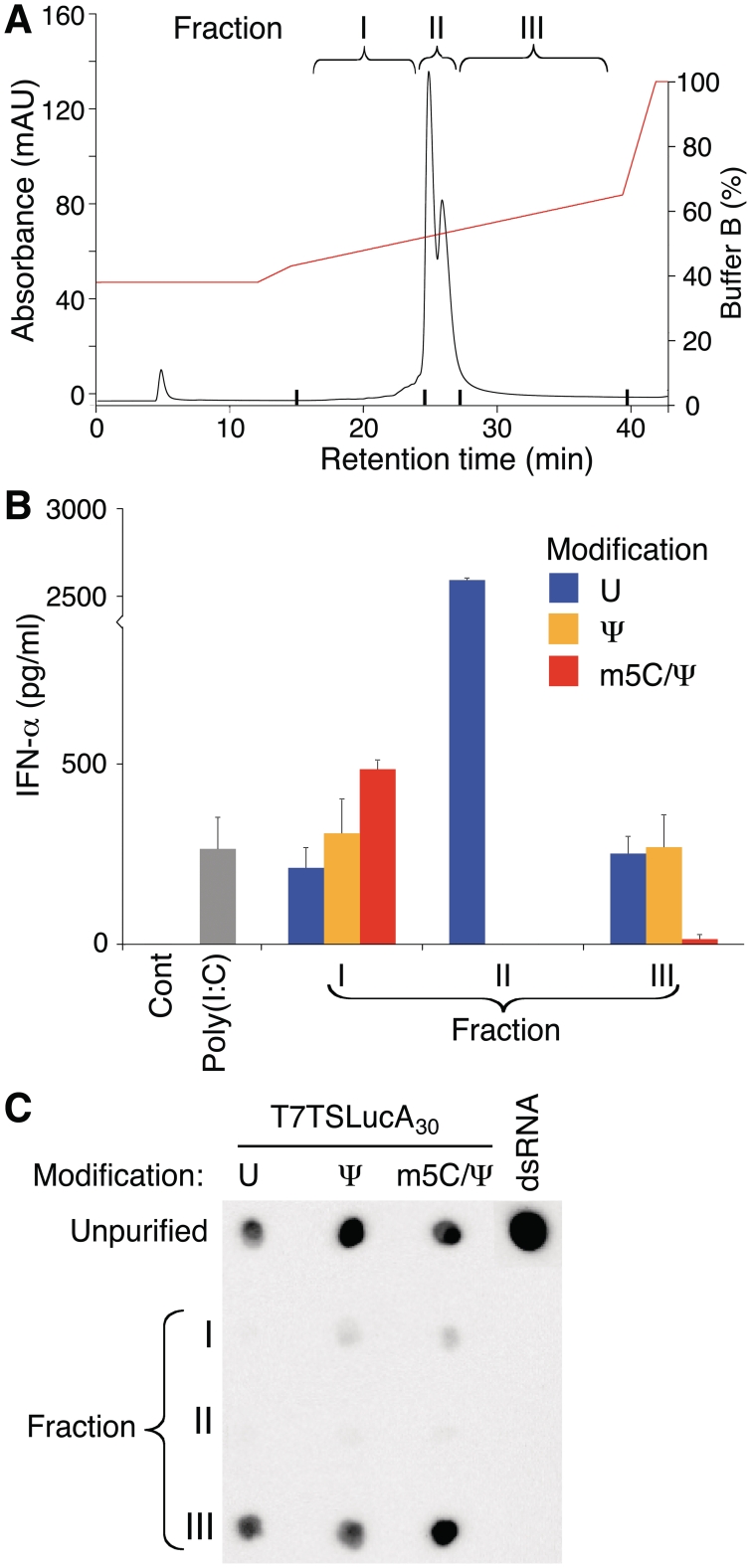
To characterize the contaminants being removed by HPLC purification, three fractions corresponding to RNAs eluting from the column prior to the major transcription product (fraction I), the full-length transcription product (fraction II), and RNAs eluting after the expected transcription product (fraction III) were collected (Figure 6A). Nucleoside-modified RNA occasionally demonstrated a second smaller peak overlying the large peak and isolation and purification of both peaks demonstrated similar RNA lengths on denaturing and non-denaturing agarose gel electrophoresis and RNAs with similar levels of translation and immunogenicity. RNA purified from the three fractions was analyzed for immunogenicity. RNA in fractions I and III induced IFN-α secretion from transfected DCs, while the purified full-length RNA (fraction II) was not immunogenic when it contained Ψ- or m5C/Ψ-nucleoside modifications (Figure 6B). Fraction I RNA had low levels of staining with the J2 mAb, while fraction III RNA had high levels of staining similar to the unpurified RNA (Figure 6C).
Figure 6.
Analysis of RNA contaminants removed by HPLC purification. (A) One hundred microgram of Ψ-modified T7TSLucA30 RNA was applied to the HPLC column and 3 fractions were collected, all RNAs eluting before the main transcription product (I), the expected RNA (II), and all RNAs eluting after the main transcription product (III). The gradient began at 38% Buffer B and increased to 43% Buffer B over 2.5 min and then spanned 43–65% Buffer B over 22 min. Unmodified and m5C/Ψ-modified T7TSLucA30 RNA had similar fractions obtained. (B) The RNAs from each fraction were complexed to TransIT and added to DCs and IFN-α in the supernatant was measured 24 h later. Error bars are standard error of the mean. (C) 200 ng of RNA from the 3 fractions and the starting unpurified RNA were blotted and analyzed with the J2 dsRNA-specific mAb.
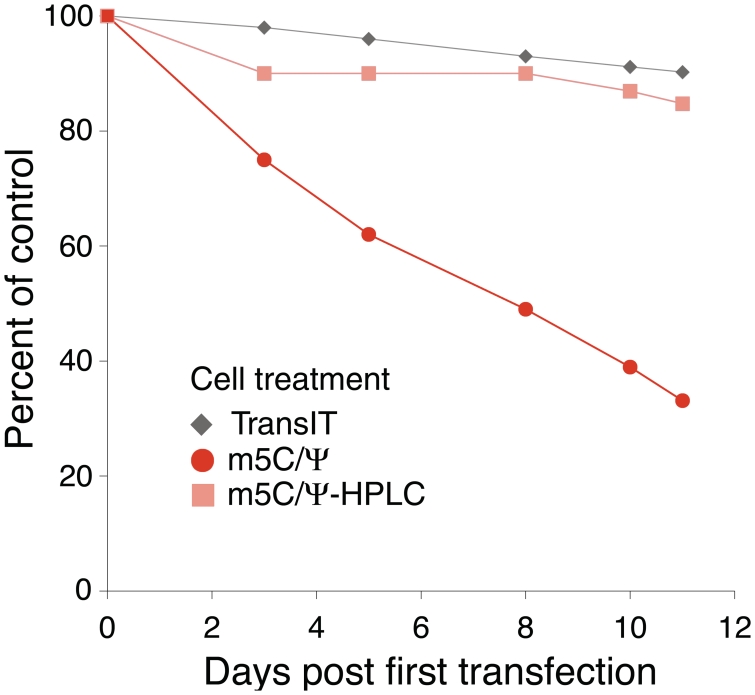
Primary human keratinocytes and murine fibroblasts treated once with unpurified, unmodified RNA delivered by TransIT complexing demonstrated detachment from the collagen-coated plastic base as evidence of cell death. A second delivery of unmodified RNA 24 h later resulted in the termination of the culture. Substantially less toxicity was observed when the RNA contained Ψ- or m5C/Ψ-nucleoside modifications, but repeated daily delivery of m5C/Ψ-nucleoside modified mRNA to keratinocytes reduced the final cell number by 75% on day 11 (Figure 7). HPLC purification of the RNA greatly reduced toxicity in treated keratinocytes where unmodified mRNA caused minimal cell rounding and death. Daily treatment of keratinocytes for 10 days with HPLC-purified m5C/Ψ-modified mRNA complexed to TransIT showed no signs of cell toxicity and the rate of proliferation was similar to that obtained with TransIT alone treated cells (Figure 7).
Figure 7.
Daily transfection with HPLC-purified m5C/Ψ-modified mRNA does not reduce cell proliferation. Primary keratinocytes were transfected daily with TransIT alone or m5C/Ψ-modified RNA-encoding Renilla luciferase with or without HPLC purification complexed with TransIT. Every 2–3 days, cultures were split and equal numbers of cells for each condition were plated. Total cell numbers for each condition were divided by the total cell number in untreated cells to calculate the percent of control proliferation.
DISCUSSION
Modified nucleoside-containing mRNA has previously been used for the induction of iPS cells from fibroblasts with very high efficiency (3). The authors determined that m5C/Ψ-nucleoside modified mRNA yielded the least amount of RNA sensor activation and the highest level of translation, but needed to add the B18R protein, a vaccinia virus decoy receptor for type I interferon (25), for optimal iPS cell generation. We previously reported that modified nucleoside-containing mRNA was efficiently delivered to primary dividing and non-dividing cells and produced high levels of encoded protein in an easily controlled manner (5). In addition, the RNA had reduced innate immune sensor activation. The residual amount of activation of modified nucleoside-containing mRNA depended on the sequence. In this report, we identify that m5C/Ψ-nucleoside-modified RNA often has the least ability to induce RNA sensor activation, but even with these modifications, certain RNA sequences induce high levels of cytokine production (Figure 1B and C). HPLC purification removes dsRNA and other contaminants from in vitro-transcribed RNAs containing Ψ or m5C/Ψ nucleosides, yielding RNA with the highest levels of translation, up to 1000-fold more than non-HPLC purified RNA with no release of type I IFNs or TNF-α and no significant induction of genes associated with RNA sensor activation. The purification procedure can be easily scaled to produce large amounts of RNA necessary for therapeutic applications and can be completed quickly and efficiently.
The data suggests that different types of immunogenic contaminants are present in in vitro-transcribed mRNA. A series of RNAs that eluted before the major transcription product, suggesting a smaller size, induced high levels of IFN-α, but had minimal staining with dsRNA-specific mAbs. A second series of RNAs that eluted after the main transcription product were immunogenic when the RNA was unmodified or contained Ψ-modifications and had levels of dsRNA staining similar to the unpurified RNA (Figure 6). Long RNA (2 kb) with an added shorter length (276 nt) of complimentary RNA eluted at high concentrations of acetonitrile, i.e. had a longer retention time, suggesting that ds structures delay elution from the column matrix (K.K., H.M. and D.W., preliminary data). Three of the known mechanisms that produce contaminants during in vitro transcription, self-complementary 3′ extension (9), RNA-primed transcription from RNA templates (10) and RNA-dependent RNA polymerase activity (11) can result in dsRNA of various lengths. If these contaminants contain the main transcription product, they would likely elute after the main transcription product. Interestingly, fraction III RNA from the purification of m5C/Ψ-modified RNA stained with dsRNA mAb, but induced little IFN-α (Figure 6). dsRNA containing nucleoside modifications, including Ψ, has reduced ability to activate PKR (23), but whether m5C/Ψ-nucleoside modifications in dsRNA block its ability to induce type I IFNs through RNA sensors is unknown. The nature of the contaminants that elute prior to the main product is unknown. They have minimal staining with dsRNA specific mAb, but these mAbs require extended lengths of dsRNA for binding (20). We cannot rule out that shorter segments of dsRNA are present in the RNA that elutes prior to the main transcription product.
HPLC purification of mRNAs enhanced their translation up to a 1000-fold in primary cells. The level of enhancement was greatest when the mRNA was unmodified, decreased when Ψ was incorporated and decreased further when m5C and Ψ were present (Figure 5A–C). These differences in translational enhancement of the mRNAs are likely due to the RNA sensors PKR and oligoadenylate synthetase (OAS) that directly decrease translation with activation. We previously demonstrated that incorporation of m5C and Ψ into long mRNA reduced its ability to activate PKR (6). Similarly, we recently demonstrated that RNA with Ψ-modifications in the absence of dsRNA contaminants induced less activation of OAS compared to unmodified mRNA (26). It is also possible that RNA samples with nucleoside modifications contain a smaller amount of short dsRNA contaminants as the dsRNA-specific mAbs do not recognize dsRNA shorter than 40 bp. An additional mechanism for the inhibition of translation by mRNA containing dsRNA contaminants could involve RNA interference. dsRNA greater than 27 bp in length is a substrate for Dicer (27), whose action can lead to a specific suppression of translation through the RNAi pathway (28). dsRNA contaminants homologous to the desired mRNA would lead to a specific suppression of translation in addition to the non-specific suppression through RNA sensors of the innate immune system.
The observation that the complexing agent used with identical RNAs alters the type of cytokines released from DCs could be caused by an effect of the complexing agent on the interaction between the RNA and endosomal RNA sensors, the location of RNA after cytoplasmic entrance, or the amount of time the RNA remains in the endosome after endocytosis. RNA sensing TLRs require acidification of endosomes to signal (29). A complexing agent that allows endocytosed RNA to exit the endosome and enter the cytoplasm before acidification and TLR signaling would result in reduced TNF-α secretion, as was observed with TransIT-complexed mRNA. In a comparison of cationic liposomes to linear polyethyleneimine (PEI) (a cationic polymer) delivery of CpG containing DNA, it was found that PEI-DNA induced less TNF-α, which was associated with a faster exit from endosomes, while cationic lipid delivery resulted in DNA remaining in vesicular structures for extended periods of time and high levels of TNF-α (30). We similarly observed that cationic liposome complexed RNA induced higher levels of TNF-α compared to the cationic polymer with lipid TransIT. Another possibility for the discrepant cytokine response could be due to the sizes of the complexed mRNA particles. A recent report by Retting et al. (31) demonstrated that nanometric particles induced IFN-α, while larger, micrometric particles induced TNF-α even when the particles were made with the same RNA and complexing agent.
Current gene therapy methods in clinical trials use viral vectors that can activate innate immune sensors (32) and integrate into host cell DNA (33) with potentially serious complications. Although the use of mRNA for such therapies seems a safer alternative, activation of RNA sensors that alter cellular activities and impair the intended function of the delivered mRNA (3) limit its usefulness. With the sequencing of the human genome and the increasing ease and ability to sequence the genome of individuals, new approaches are needed for the treatment of inherited genetic disorders and therapies that require in vivo expression of extracellular or intracellular proteins. In addition, advances in regenerative medicine therapies where transient expression of transcription factors ex vivo transdifferentiate, reprogram and differentiate cells are leading to new approaches for organ and tissue restoration with the hope of treating many diseases including diabetes, spinal cord injury, neurologic syndromes and organ repair. As we report here, purification of modified nucleoside-containing mRNA avoids RNA sensor activation and increases the translational levels of the encoded protein. These mRNAs can be delivered repeatedly without toxicity or the need to add inhibitors of innate immune activation and can be used for both ex vivo and in vivo therapies.
FUNDING
Funding for open access charge: National Institutes of Health (grant number HL87688 to K.K., AI050484, DE019059 to D.W.).
Conflict of interest statement. Drs Weissman and Kariko are named on a patent for modified RNA submitted by the University of Pennsylvania.
ACKNOWLEDGEMENTS
We thank Dr Louise Showe and the Wistar Genomics Core Facility for performing the gene array analyses, Houping Ni for technical assistance and the Skin Disease Research Core at Penn for primary keratinocytes.
REFERENCES
- 1.Angel M, Yanik MF. Innate immune suppression enables frequent transfection with RNA encoding reprogramming proteins. PLoS One. 2010;5:e11756. doi: 10.1371/journal.pone.0011756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yakubov E, Rechavi G, Rozenblatt S, Givol D. Reprogramming of human fibroblasts to pluripotent stem cells using mRNA of four transcription factors. Biochem. Biophys. Res. Commun. 2010;394:189–193. doi: 10.1016/j.bbrc.2010.02.150. [DOI] [PubMed] [Google Scholar]
- 3.Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kormann MS, Hasenpusch G, Aneja MK, Nica G, Flemmer AW, Herber-Jonat S, Huppmann M, Mays LE, Illenyi M, Schams A, et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 2011;29:154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- 5.Kariko K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson BR, Muramatsu H, Nallagatla SR, Bevilacqua PC, Sansing LH, Weissman D, Kariko K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38:5884–5892. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kariko K, Weissman D. Naturally occurring nucleoside modifications suppress the immunostimulatory activity of RNA: implication for therapeutic RNA development. Curr. Opin. Drug Discov. Devel. 2007;10:523–532. [PubMed] [Google Scholar]
- 8.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Triana-Alonso FJ, Dabrowski M, Wadzack J, Nierhaus KH. Self-coded 3′-extension of run-off transcripts produces aberrant products during in vitro transcription with T7 RNA polymerase. J. Biol. Chem. 1995;270:6298–6307. doi: 10.1074/jbc.270.11.6298. [DOI] [PubMed] [Google Scholar]
- 10.Nacheva GA, Berzal-Herranz A. Preventing nondesired RNA-primed RNA extension catalyzed by T7 RNA polymerase. Eur. J. Biochem. 2003;270:1458–1465. doi: 10.1046/j.1432-1033.2003.03510.x. [DOI] [PubMed] [Google Scholar]
- 11.Arnaud-Barbe N, Cheynet-Sauvion V, Oriol G, Mandrand B, Mallet F. Transcription of RNA templates by T7 RNA polymerase. Nucleic Acids Res. 1998;26:3550–3554. doi: 10.1093/nar/26.15.3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thakur CS, Sama JN, Jackson ME, Chen B, Dayie TK. Selective 13C labeling of nucleotides for large RNA NMR spectroscopy using an E. coli strain disabled in the TCA cycle. J. Biomol. NMR. 2010;48:179–192. doi: 10.1007/s10858-010-9454-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pallan PS, Egli M. Selenium modification of nucleic acids: preparation of oligonucleotides with incorporated 2′-SeMe-uridine for crystallographic phasing of nucleic acid structures. Nat. Protoc. 2007;2:647–651. doi: 10.1038/nprot.2007.75. [DOI] [PubMed] [Google Scholar]
- 14.Fotin-Mleczek M, Duchardt KM, Lorenz C, Pfeiffer R, Ojkic-Zrna S, Probst J, Kallen KJ. Messenger RNA-based vaccines with dual activity induce balanced TLR-7 dependent adaptive immune responses and provide antitumor activity. J. Immunother. 2011;34:1–15. doi: 10.1097/CJI.0b013e3181f7dbe8. [DOI] [PubMed] [Google Scholar]
- 15.Summer H, Gramer R, Droge P. Denaturing urea polyacrylamide gel electrophoresis (Urea PAGE) J. Vis. Exp. 2009;29:1485. doi: 10.3791/1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masuda N, Ohnishi T, Kawamoto S, Monden M, Okubo K. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res. 1999;27:4436–4443. doi: 10.1093/nar/27.22.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKenna SA, Kim I, Puglisi EV, Lindhout DA, Aitken CE, Marshall RA, Puglisi JD. Purification and characterization of transcribed RNAs using gel filtration chromatography. Nat. Protoc. 2007;2:3270–3277. doi: 10.1038/nprot.2007.480. [DOI] [PubMed] [Google Scholar]
- 18.Kim I, McKenna SA, Puglisi EV, Puglisi JD. Rapid purification of RNAs using fast performance liquid chromatography (FPLC) RNA. 2007;13:289–294. doi: 10.1261/rna.342607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schonborn J, Oberstrass J, Breyel E, Tittgen J, Schumacher J, Lukacs N. Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res. 1991;19:2993–3000. doi: 10.1093/nar/19.11.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonin M, Oberstrass J, Lukacs N, Ewert K, Oesterschulze E, Kassing R, Nellen W. Determination of preferential binding sites for anti-dsRNA antibodies on double-stranded RNA by scanning force microscopy. RNA. 2000;6:563–570. doi: 10.1017/s1355838200992318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kariko K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 22.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 23.Nallagatla SR, Bevilacqua PC. Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. RNA. 2008;14:1201–1213. doi: 10.1261/rna.1007408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kramer M, Schulte BM, Eleveld-Trancikova D, van Hout-Kuijer M, Toonen LW, Tel J, de Vries IJ, van Kuppeveld FJ, Jansen BJ, Adema GJ. Cross-talk between human dendritic cell subsets influences expression of RNA sensors and inhibits picornavirus infection. J. Innate Immun. 2010;2:360–370. doi: 10.1159/000300568. [DOI] [PubMed] [Google Scholar]
- 25.Symons JA, Alcami A, Smith GL. Vaccinia virus encodes a soluble type I interferon receptor of novel structure and broad species specificity. Cell. 1995;81:551–560. doi: 10.1016/0092-8674(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 26.Anderson BR, Muramatsu H, Jha BK, Silverman RH, Weissman D, Kariko K. Nucleoside modifications in RNA limit activation of 2′-5′ oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr586. (in press) PMID: 21813458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 28.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 29.Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, Iwasaki A, Flavell RA. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl Acad. Sci. USA. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saito Y, Higuchi Y, Kawakami S, Yamashita F, Hashida M. Immunostimulatory characteristics induced by linear polyethyleneimine-plasmid DNA complexes in cultured macrophages. Hum. Gene Ther. 2009;20:137–145. doi: 10.1089/hum.2008.013. [DOI] [PubMed] [Google Scholar]
- 31.Rettig L, Haen SP, Bittermann AG, von Boehmer L, Curioni A, Kramer SD, Knuth A, Pascolo S. Particle size and activation threshold: a new dimension of danger signaling. Blood. 2010;115:4533–4541. doi: 10.1182/blood-2009-11-247817. [DOI] [PubMed] [Google Scholar]
- 32.Raper SE, Chirmule N, Lee FS, Wivel NA, Bagg A, Gao GP, Wilson JM, Batshaw ML. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Mol. Genet. Metab. 2003;80:148–158. doi: 10.1016/j.ymgme.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 33.Hacein-Bey-Abina S, Hauer J, Lim A, Picard C, Wang GP, Berry CC, Martinache C, Rieux-Laucat F, Latour S, Belohradsky BH, et al. Efficacy of gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 2003;363:355–364. doi: 10.1056/NEJMoa1000164. [DOI] [PMC free article] [PubMed] [Google Scholar]