Abstract
Many bacteria and viruses adapt to varying environmental conditions through the acquisition of mosaic genes. A mosaic gene is composed of alternating sequence polymorphisms either belonging to the host original allele or derived from the integrated donor DNA. Often, the integrated sequence contains a selectable genetic marker (e.g. marker allowing for antibiotic resistance). An effective identification of mosaic genes and detection of corresponding partial horizontal gene transfers (HGTs) are among the most important challenges posed by evolutionary biology. We developed a method for detecting partial HGT events and related intragenic recombination giving rise to the formation of mosaic genes. A bootstrap procedure incorporated in our method is used to assess the support of each predicted partial gene transfer. The proposed method can be also applied to confirm or discard complete (i.e. traditional) horizontal gene transfers detected by any HGT inferring method. While working on a full-genome scale, the new method can be used to assess the level of mosaicism in the considered genomes as well as the rates of complete and partial HGT underlying their evolution.
INTRODUCTION
Horizontal gene transfer (HGT) (also called lateral gene transfer) is one of the major mechanisms contributing to microbial genome diversification. HGT is dominant among various groups of genes in prokaryotes (1). The understanding of the key role played by HGT in species evolution has been one of the most fundamental changes in our perception of general aspects of molecular biology in recent years (2,3). HGT can pose several risks to humans, including: cancer triggered by the insertion of transgenic DNA into human cell, antibiotic-resistant genes spreading to pathogenic bacteria, and disease-associated genes spreading and recombining to create new viruses and bacteria (4). Two models of HGT have been considered in the literature (5). First, and the most popular of them, is the traditional model of complete HGT. It assumes that the transferred gene either supplants the orthologous gene of the recipient genome or, when the transferred gene is absent in the recipient genome, is added to it (6). The second model is that of partial gene transfer, implying the formation of ‘mosaic’ genes. A mosaic gene is an allele acquired through transformation or conjugation (e.g. from a different bacterium) and subsequent integration through intragenic recombination into the original host allele (7,8). The term mosaic stems from the pattern of interspersed blocks of sequences having different evolutionary histories but found combined in the resulting allele subsequent to recombination events. The recombined segments can be derived from other strains of the same species or from other more distant bacterial or viral relatives (7,9). When the incoming DNA is significantly different from the host DNA, mosaic genes can express proteins with novel phenotypes (e.g. in the case when the donor DNA derives from a different species or genus). At the time of HGT event, horizontally transferred genes reflect the base composition of the donor genome. However, over time, these sequences ameliorate to reflect the DNA composition of the host genome because the genes affected by HGT are subject to the same mutational processes that influence all genes in the host genome (10).
There is evidence that mosaic genes are constantly generated in populations of transformable organisms, and probably in all genes (11). Mosaic genes have been also observed in non-transformable bacteria but normally at a lower frequency. Zheng et al. (12) reported that mosaic genes account for up to 20% of microbial genomes. For instance, in the naturally competent Neisseria species, mosaic alleles have been observed for many genes, comprising those encoding surface antigens, IgA protease, housekeeping proteins and antibiotic targets (7,10). One of the well-characterized examples of mosaic genes, resulting from partial HGT events, are those that encode the penicillin-resistant binding proteins (PBPs) found in Streptococcus pneumoniae. These high molecular weight proteins are the lethal targets of the B-Lactams of penicillin (10,13). Pneumococci, capable of between-species horizontal transfer, undergo, in all likelihood, even more frequent within-species HGT which contributes to the development of mosaic alleles (7).
While many methods have been proposed to address the issue of the identification and validation of complete HGT events (4,6,14–29), only two methods treat the problem of inferring partial HGT and predicting the origins of mosaic genes (30,31). However, neither of the latter two works discusses the problem of robustness of predicted HGT events or includes a Monte Carlo simulation study which is necessary to test the method's performances in different practical situations.
In this article, we describe a new sliding window-based method for predicting partial HGT events and subsequent intragenic recombination. A sliding window approach has been previously used for detecting recombination (32–36), but none of these studies addresses the problem of inferring partial HGT events. The RDP3 program (36) remains, to date, the most comprehensive tool for characterizing recombination events in DNA-sequence alignments. A method for detecting intragenic recombination, called LikeWind, which is also based on a sliding window procedure and on the inference of a phylogenetic tree for each fixed window position, was described in (33). The main advantages of the method we introduce in this article, over LikeWind and the other existing techniques used to detect recombination, are that our method allows one to detect the sources of transferred sequence fragments and assess the robustness of the obtained solution. A Monte Carlo simulation study was carried out to test the ability of the proposed method to recover correct partial HGTs depending on the number of gene transfers and number of species considered (i.e. tree size). In the ‘Results’ section, the new method is applied to recover partial, and complete, HGT events in the context of the evolution of the genes rbcL [data originally considered in Ref. (37)] and mutU [data originally considered in Ref. (30)].
MATERIALS AND METHODS
A new method for predicting partial horizontal gene transfer events
In this section we describe the main features of the new method for inferring partial HGTs. The main steps of the method intended to provide an optimal scenario of partial transfers of the given gene for the considered group of species, and thus predict putative intragenic recombination events and identify mosaic sequences, are summarized below. The bootstrap validation will be performed for each predicted partial transfer, and only the transfers with significant bootstrap support will be included in the final solution. A sliding window procedure will be carried out to test different fragments of the given multiple sequence alignment (MSA). A method for detecting complete HGTs will be carried out at each step to reconcile the given species tree and partial gene trees inferred from the sequence fragments located within the sliding window (each time its position is fixed).
Preliminary step
Let X be a set of species, MSA be a given multiple sequence alignment of length l, and Si,j be the MSA fragment under examination located between the sites i and j (including both i and j), where 1 ≤ i < j ≤ l. Define the sliding window size w (w = j − i + 1) and the progress step size s. Infer the species phylogenetic tree, denoted T. Usually a morphology-based tree or a molecular tree based on a molecule assumed to be refractory to horizontal gene transfer plays the role of the species tree. For instance, 16S rRNA or 23S rRNA genes may also undergo HGT, but they seem to do it at a relatively low rate (38). The tree T must be rooted with respect to the available evolutionary evidence. If no plausible evidence for rooting T exists, the outgroup or midpoint strategies can be used (6). The tree rooting is necessary because it allows us to take into account the evolutionary time-constraints that should be satisfied when inferring HGTs. These time constraints, which include the same lineage HGTs as well as some criss-crossing transfers, are imposed by the necessity for taxa involved in HGT to be contemporaneous (6,18,20). Fix the sliding window size w and the step size s. In our experiments, the window sizes of l/5, l/4, l/3 and l/2 sites and the sliding window progress step of 10 sites were used.
Step k
Fix the position of the sliding window in the interval [i, j], where i = 1 + s(k − 1) and j = i + w − 1; k also corresponds to the window rank (Figure 1). If i + w − 1 > l and i + w − 1 − l ≥ w/2, then j = l, otherwise stop the algorithm here (i.e. short window sizes usually lead to trees with low bootstrap support and hence to doubtful HGTs). Infer a partial gene tree T ′ characterizing the evolution of the MSA fragment located within the interval [i, j]. In this study, the PhyML method (39) was used to reconstruct partial gene trees. Apply an existing HGT detection method to infer an optimal scenario of partial HGTs associated with the interval [i, j]. Here we used the HGT-Detection method described in Ref. (6) in the context of complete HGT, but any other HGT inferring method can be carried out instead. The HGT-Detection method was shown to be faster and, in most instances, as accurate as the popular LatTrans (20) and RIATA–HGT (4) methods. Here, the bipartition dissimilarity measure introduced in (6) was used as an optimization criterion. It takes into account the degree of similarity between the topologically closest subtrees in two phylogenetic trees and can be viewed as a refinement of the popular Robinson and Foulds topological distance (40).
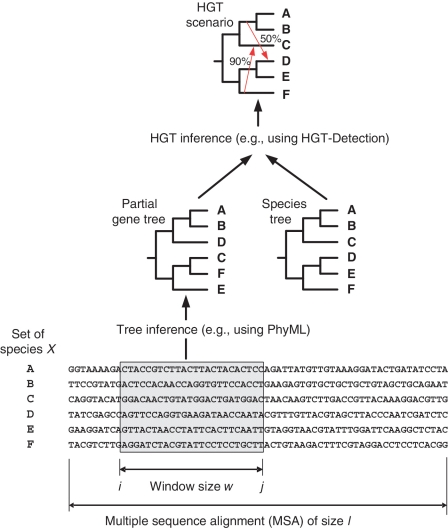
Figure 1.
Partial gene tree is inferred using the sequences located within the sliding window of size w. The PhyML method (39) was used to reconstruct partial gene trees. The HGT-Detection method (6) was then applied to infer complete HGTs.
In addition, the following procedure for assessing the reliability of obtained partial transfers (i.e. HGT bootstrap support), which takes into account the uncertainty of partial gene trees as well as the number of occurrences of the selected transfers in all minimum-cost HGT scenarios found for the given pair of species and partial gene trees, was carried out. In the bootstrap procedure, only the sequence data used to build the partial gene tree T ′, inferred from the sequences located within the sliding window, were pseudo-replicated. The species tree T was fixed and thus taken as an a priori assumption of the method. We first executed our program with the exhaustive search option providing the list of all minimum-cost HGT scenarios. This option consists of verifying at each step of the algorithm all possible HGTs that satisfy the evolutionary constraints. Once the list of all possible minimum-cost HGT scenarios for the trees T and T ′ was established, the HGT bootstrap score of each individual partial transfer was computed. Formulas 1 and 2 were used to compute the bootstrap score HGT_BS of the partial transfer t:
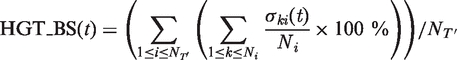 |
(1) |
and
 |
(2) |
where NT’ is the number of partial gene trees (i.e. number of HGT bootstrap replicates) generated from pseudo-replicated sequences and Ni is the number of minimum-cost scenarios obtained when carrying out the algorithm with the species tree T and partial gene tree Ti′. Among the obtained partial HGTs, only the transfers with significant bootstrap scores were retained. Obviously, a short window size produced partial gene trees with much greater variability, and hence lower bootstrap supports for HGT histories.
Final step
Establish a list of predicted partial HGT events. Identify the overlapping intervals giving rise to the identical partial transfers (i.e. the same donor and recipient and the same direction). Re-execute the HGT detection method for all overlapping intervals (considering their total length in each case) that support the identical partial HGTs. If such partial HGTs are found again for the sequence fragment located within the overlapped intervals, assess their bootstrap support and, depending of the obtained support, include them in the final solution or discard them. If a window located in the middle of a larger interval does not suggest the transfer that is indicated (with a certain significant bootstrap support) on the interval's ends, the entire, larger, interval is tested for the presence of significant HGTs.
The time complexity of the proposed method is as follows:
| (3) |
where w is the size of the sliding window, s is the sliding window progress step, C(Phylo_Inf) is the time complexity of the tree inferring method used to infer phylogenies from sequence fragments located within the sliding window, C(HGT_Inf) is the time complexity of the complete HGT detection method used to infer HGTs for the given species tree and partial species trees inferred from sequence fragments located within the sliding window, r is the number of replicates in bootstrapping.
Given that the time complexity of PhyML (39) is O(pnw), where p represents the number of refinement steps being performed, and the time complexity of HGT-Detection (6) is O(τ×n4), the exact time complexity of our implementation is as follows:
| (4) |
where n is the number of species, and τ is the average number of transfers found for a sequence fragment located within the sliding window of size w.
For instance, the running time of the algorithm for the numerical example considered in the Results section and involving an MSA of 30 mutU DNA sequences of length 384 sites, three different window sizes: 100, 150 and 200 sites, the advancement step of 10 sites and 100 replicates in HGT and PhyML bootstrapping, was 4 min and 33 s when executed on a PC computer equipped with the Intel Core i7-2635QM (2.0 GHz) processor and 4 Gb of RAM.
RESULTS
Simulation study
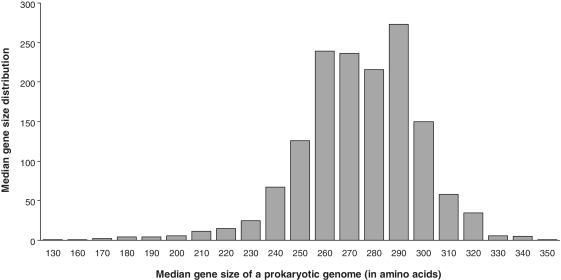
A Monte Carlo simulation study was conducted to test the ability of the new method to recover correct partial HGTs. We examined how the proposed method performs depending on the number of observed species and number of generated partial transfers. First of all, we calculated the distribution of median gene sizes of prokaryotic genomes (Figure 2) considering 1494 complete microbial genomes available in the GenBank database in April 2011 (for more details, see: http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi). The ‘average median gene size’ (i.e. the average here was taken over all calculated median gene sizes; Figure 2) of a microbial genome was 268 amino acids (the standard deviation was equal to 24 and the average size of a prokaryotic gene was 315 amino acids). The determined average median gene size of a prokaryotic genome was then used as a benchmark for MSA length in our simulations. Mention that the size of a transferred DNA fragment varies from organism to organism, and can be, in some situations, larger than a single gene [e.g. it is in the range 5–10 kb for some pathogenic bacteria; (41)]. Such longer transferred fragments can be detected using the new method by treating the complete genomes of involved organisms on the gene-by-gene basis. The most significant among the obtained partial transfers can then be merged to form longer sequence segments affected by HGT. Furthermore, any existing method for the identification of complete HGTs (e.g. LatTrans, RIATA–HGT, HGT-Detection) can be used to confirm or discard complete HGTs detected by our method.
Figure 2.
Distribution of median gene sizes of prokaryotic genomes computed on the basis of 1494 complete microbial genomes available in April 2011 in the GenBank database [(46), for more details, see: http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi]. The height of each column corresponding to the graduation mark k of the abscissa axis represents the number of genomes whose median gene sizes are located in the interval [k-5; k + 4]. For instance, the column corresponding to the mark 300 of the abscissa axis accounts for the genomes whose median gene sizes comprise between 295 and 304 amino acids.
The simulation protocol included four main steps described below. First, random binary species trees with 8, 16, 32 and 64 leaves were generated using the procedure described by Kuhner and Felsenstein (42). The branch lengths of the species trees were generated using an exponential distribution. Following the approach of Guindon and Gascuel (43), we added some noise to the branches of the species phylogenies in order to provide a deviation from the molecular clock hypothesis. The trees yielded by this procedure had depth of O[log (n)], where n is the number of species (i.e. number of leaves in a binary phylogenetic tree).
Second, we carried out the SeqGen program (44) to generate random multiple sequence alignments of protein sequences along the branches of the species trees constructed at the first step. The SeqGen program was used with the JTT model of proteins substitution (45). Protein sequences with 268 amino acids (i.e. average median gene size of a prokaryotic genome) were generated.
Third, having the sequences corresponding to the nodes of each species tree T, we, in turn, generated gene trees with the same number of leaves by performing random SPR (Subtree Prune and Regraft) moves of its subtrees. A model satisfying all plausible evolutionary constraints was implemented to generate random HGTs. For each species tree, 1–4 random SPR moves were performed and different gene trees T′, encompassing 1–4 partial HGT events, were created. For each gene tree, the sequence fragments involved in the transfer(s) were identified and the corresponding sequence(s) in the subtree(s) affected by HGT were regenerated using SeqGen. Two different sizes of transferred fragments, 89 and 134 amino acids, corresponding respectively to one-third and one-half of the total gene length, were tested in our simulations. The tests conducted with two different transferred fragment sizes were carried out separately. When more than one HGT was generated, the sequence fragments affected by HGT could overlap. Thus, the obtained MSAs, each MSA included the sequences corresponding to the leaves of a gene tree, comprised blocks of amino acids affected by HGT.
Fourth, we carried out the new method for each generated species tree and the associated set of MSAs affected by partial HGT(s). The size of the sliding window was set to 100 sites; 100 replicates of each partial gene tree T′ were generated to assess the bootstrap support of its branches, first, and the support of the obtained partial transfers, second. Partial gene trees whose average bootstrap support was <60% were ruled out from the analysis. Among the obtained HGTs only the transfers with bootstrap scores of 90% and higher were considered as significant and retained in the final solution.
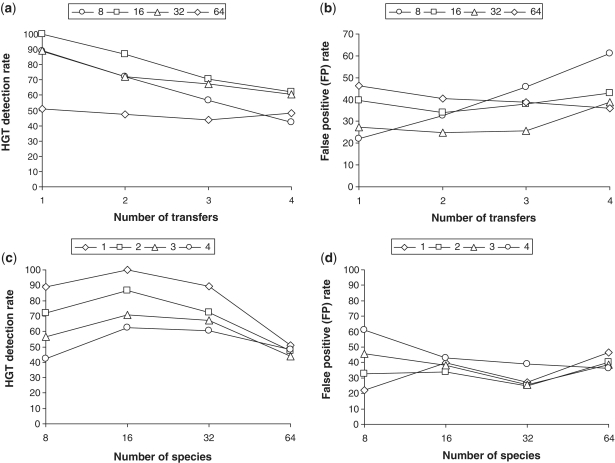
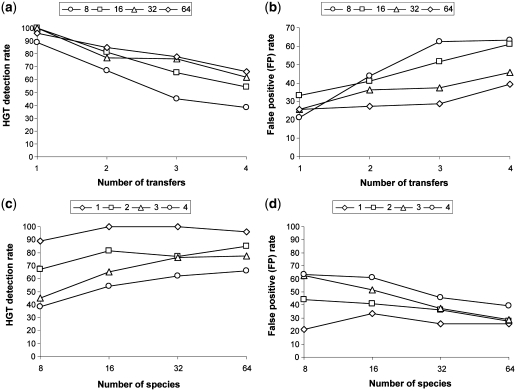
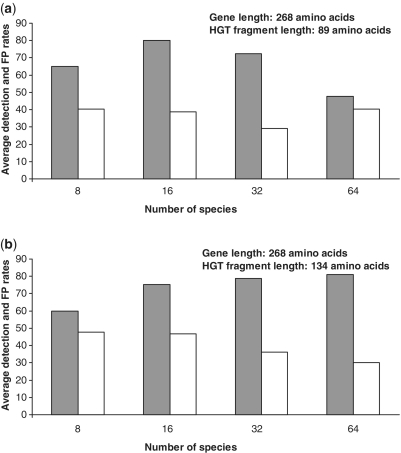
Finally, we estimated the detection rate (i.e. true positives) and the false positive rate depending on the number of species and generated transfers. The obtained average performances of the new method are illustrated in Figures 3 and 4. For each set of parameters (tree size; number of generated HGTs), 100 replicated data sets were generated. On the other hand, Figure 5 highlights the difference between the average detection rate and average false positive rate with respect to the number of species. Figures 3 and 5a show that the best detection rates for the transferred fragments of size 89 amino acids were obtained for the 16-species trees. The results vary from 100% for one transfer to 69% for four transfers, giving a 79.9% partial HGT recovery on average. The best average false positive rate of 29.2% was obtained for the 32-species trees (Figure 5a). For the transferred fragments of size 134 amino acids, the best results were obtained for the 64-species trees (Figure 4). The average partial HGT detection rate for this size of trees was 81.1% and average false positive rate was 30.2% (Figure 5b). The average here was computed from the results obtained for 1–4 generated HGTs. Similar trends can be observed for the other tree sizes. According to our additional tests, these results can be improved by adjusting the simulation parameters with respect to the nature of the studied sequences.
Figure 3.
Average HGT detection rate depending on the number of transfers (a), and number of species (c). Average false positive (FP) rate depending on the number of transfers (b), and number of species (d). Each reported value represents the average result obtained for random trees with 8, 16, 32 and 64 leaves (cases a and b), and 1–4 HGTs (cases c and d); 100 replicates were generated for each parameter combination. The presented results were obtained with the sequences of length 268 amino acids (i.e. median prokaryotic genome gene size) and HGT fragments of 89 amino acids (i.e. one-third of the total gene length).
Figure 4.
Average HGT detection rate depending on the number of transfers (a), and number of species (c). Average false positive (FP) rate depending on the number of transfers (b), and number of species (d). Each reported value represents the average result obtained for random trees with 8, 16, 32 and 64 leaves (cases a and b), and 1–4 HGTs (cases c and d); 100 replicates were generated for each parameter combination. The presented results were obtained with the sequences of length 268 amino acids (i.e. median prokaryotic genome gene size) and HGT fragments of 134 amino acids (i.e. one-half of the total gene length).
Figure 5.
Average, over 1–4 generated transfers, HGT detection rate (grey columns) and false positive rate (white columns) depending on the number of species, obtained for the sequences of length 268 amino acids (i.e. median prokaryotic genome gene size) and HGT fragments of: (a) 89 amino acids (i.e. one-third of the total gene length) and (b) 134 amino acids (i.e. one-half of the total gene length).
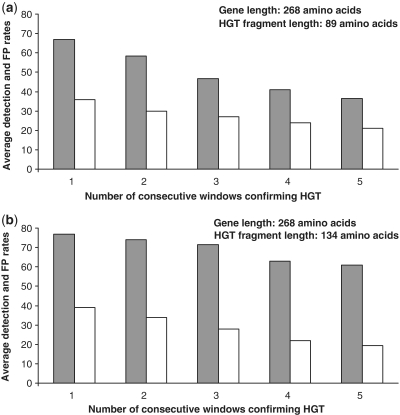
Mention that high false positive rate obtained for the small trees (i.e. with 8 and 16 leaves) was mainly due to the fact that multiple minimum-cost HGT scenarios (i.e. solutions including the same minimum number of transfers) often exist in the case when small phylogenies are affected by several (e.g. 3 or 4) transfers (6, 20). For instance, Figure OA6 (e) in (6) shows that in case of complete gene transfers, we have only up to 40% of chances to obtain the same (correct) HGT scenario for 10-species trees and up to 47% for 20-species trees (the results in Figure OA6 are shown for the HGT-Detection and LatTrans algorithms). In order to lower the false positive rate that is higher for smaller trees (Figure 5), we conducted an additional simulation. Note that the results presented in Figures 3–5 correspond to the strategy in which any transfer with bootstrap scores of 90% and higher found for ‘at least one fixed window position’ was considered as significant. Such a strategy allows for a high hit detection rate but is also capable of generating some false positives transfers. We also considered algorithmic strategies where an HGT was recognized as significant if and only if it was found for ‘at least 2, 3, 4 or 5 consecutive fixed window positions’. Such consecutive windows were overlapping each other because the window progress step of 10 sites was used in our simulations. Figure 6 illustrates the evolution of the average HGT detection rate (grey columns) and average false positive rate (white columns) depending on the number of consecutive windows for all of which the same transfer was detected. The averages here were taken over the results obtained for all considered trees sizes (8, 16, 32 and 64-species trees) and 1, 2, 3 and 4 generated HGTs; 100 trees were generated and tested for each combination of these parameters. The results presented in Figure 6 suggest that the strategy considering several consecutive windows can be effective for decreasing the false positive rate, especially in the case of longer transferred fragments (i.e. 134 amino acids). Certainly, the improvement in the false positives rate was obtained at the expense of the detection rate. For the transferred fragments of 89 amino acids, the false positive rate decreased from 37.6% to 21.3%, while for the 134 amino acids fragments, it decreased from 39.4% to 19.1%, for one and five consecutive windows, respectively. The largest difference between the average false positive and false negative rates was obtained with one window, for the transferred fragments of 89 amino acids (31.3%), and with three consecutive windows for the fragments of 134 amino acids (44.0%). The lowest average false positive rate of 5.3% was obtained, while considering five consecutive windows, for 64-species trees and 134 amino acids fragments. This means that for longer transferred sequences and larger trees one should look for a result confirmation over a few consecutive window positions in order to validate the obtained transfers. The option allowing for validating the obtained HGTs for a series of consecutive window positions was included in our software available at: http://www.trex.uqam.ca.
Figure 6.
Average HGT detection rate (grey columns) and false positive rate (white columns), computed over 1–4 generated transfers and trees with 8, 16, 32 and 64 leaves, depending on the number of consecutive windows confirming the same HGT, obtained for the sequences of length 268 amino acids (i.e. median prokaryotic genome gene size) and HGT fragments of: (a) 89 amino acids (i.e. one-third of the total gene length) and (b) 134 amino acids (i.e. one-half of the total gene length).
The presented simulation results suggest that the new method can be useful for detecting partial transfers, and thus for identifying mosaic genes, especially when large trees and long sequence fragments affected by HGT are considered. With smaller transferred sequence fragments (i.e. one third of the total gene length), the best HGT detection rates were found for the trees with 16 and 32 leaves, whereas with larger transferred fragments (i.e. one-half of the total gene length), the best results were obtained for 64-leaf trees. While, on average, the HGT detection rates obtained for partial HGTs were slightly lower than those obtained by the LatTrans (20) and HGT-Detection algorithms for complete gene transfers [see Figure OA6 in Ref. (6)], we should notice that the problem of detecting partial HGTs is much more complex than the problem of inferring complete gene transfers. This complexity is due, first, to high similarity of short sequence fragments located in the sequence blocks affected by HGT and, second, to a possible overlap of the latter blocks which can disguise real gene transfers.
Detecting partial transfers of the gene rbcL
First, we applied the new method to analyze the Proteobacteria, Cyanobacteria and plastid data originally examined by Delwiche and Palmer (37). The latter authors discussed the hypothesis of HGT of the rubisco genes versus the hypothesis of ancient gene duplication followed by partial gene loss. Delwiche and Palmer (37) inferred a maximum parsimony phylogeny of the gene rbcL (large subunit of rubisco) for 48 organisms, including 42 taxa for Form I and 6 taxa for Form II of rubisco. They pointed out that the classification based on the gene rbcL contained numerous conflicts compared to the classification based on 16S ribosomal RNA and other evidence. The aligned rbcL amino acid sequences comprising 532 bp considered by Delwiche and Palmer and reanalyzed in this study can be found at: http://www.life.umd.edu/labs/delwiche.
To perform the analysis, we retained 42 of 48 organisms from the original study: all the taxa of Form I of rbcL were examined, whereas the 6 taxa of Form II, used by Delwiche and Palmer (37) to root the gene tree, were discarded. For the species Chromatium and Hydrogenovibrio two different copies of the rubisco gene, denoted, respectively, Chromatium A and L, and Hydrogenovibrio L1 and L2, were considered in the original study. Thus, in this example, the gene phylogeny comprised 42 organisms, while the species phylogeny only 40. It is worth noting that the new method was adapted to the case when the species and gene trees have different number of leaves. The ML tree of the gene rbcL inferred using the PhyML method (39) is shown in Figure 7. This tree is very similar to the maximum parsimony gene tree obtained by Delwiche and Palmer (see Figure 2 in Ref. 37). The organisms Pseudomonas and endosymbiont of Alviniconcha, denoted as uncertain in Figure 2 of Delwiche and Palmer (37), were later classified as β-proteobacteria.
Figure 7.
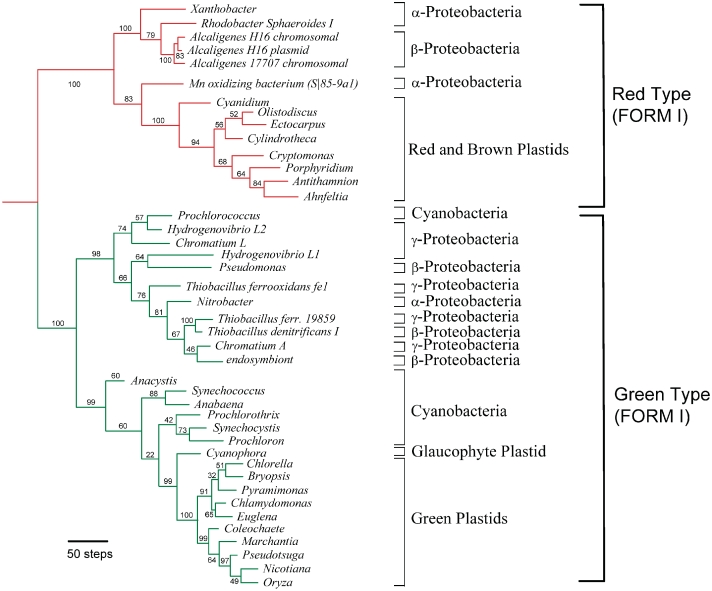
ML tree of the gene rbcL for 42 bacteria and plastids inferred from the rubisco amino acid sequences with 532 bases using the PhyML method (39). Taxa classification based on 16S rRNA and other evidence is indicated to the right. Numbers above the branches are their bootstrap scores calculated using 100 replicates.
The corresponding species tree (Figure 8, undirected branches) was reconstructed and rooted using the appropriate information from the NCBI taxonomic browser (46). Since in this study we were mostly interested in identifying the transfers between different groups of organisms, we deliberately kept intact in the species tree, with respect to the topology of the gene tree, the positions of the organisms belonging to the same group. For instance, the topologies of the clades of Green plastids, Cyanobacteria, and Red and Brown plastids were identical in the gene and species phylogenies shown in Figures 7 and 8, respectively. A number of important topological conflicts between the species and gene trees can be observed. For example, there exists a large clade in the gene tree with bootstrap support of 98% (Figure 6), including one α-proteobacterium, three β-proteobacteria, six γ-proteobacteria and one cyanobacterium. Such topological conflicts can be explained either by frequent HGT events (partial or complete) or by ancient gene duplication followed by gene losses [these two hypotheses are not mutually exclusive; see reference (37) for more details]. Below, we consider only the HGT hypothesis to explain topological incongruence between the species and gene phylogenies.
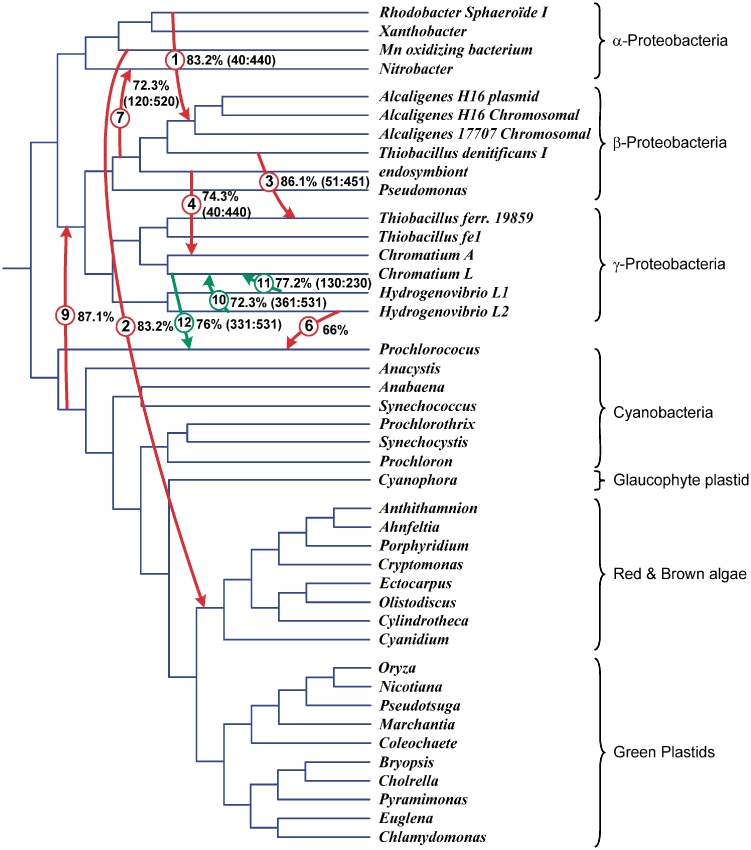
Figure 8.
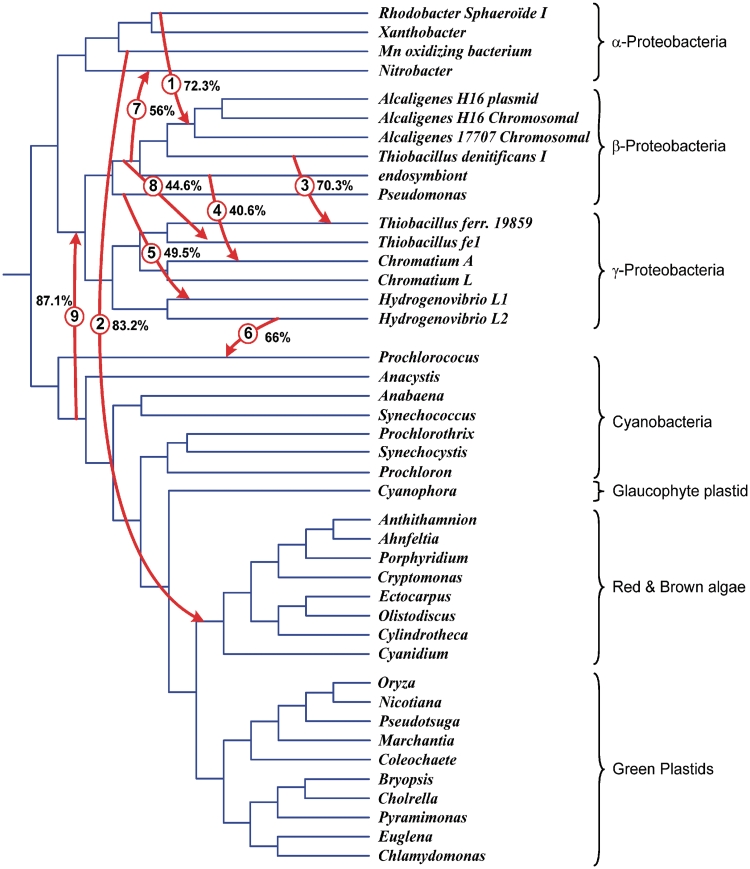
Species tree for the 42 bacteria and plastid organisms from Figure 7 with 9 HGT branches (denoted by arrows) representing complete horizontal transfers of the gene rbcL. This scenario was a unique shortest complete HGT scenario found for the given pair of species and gene trees. Bootstrap support of complete HGT events is indicated.
First, we carried out the HGT-Detection method for predicting complete HGTs (6); the bipartition dissimilarity criterion was used for optimization. The minimum-cost transfer scenario with nine HGTs necessary to reconcile the species and gene phylogenies is shown in Figure 8 (HGTs are depicted by numbered arrows). The optimality of this solution was confirmed by the LatTrans algorithm (20) based on the exhaustive search. The bootstrap support of the obtained complete HGTs was also computed.
Second, we carried out the new method for predicting partial HGTs. We used the sliding windows of the size 200, 300 and 400 sites with the progress step of 10 sites. Partial trees corresponding to the subsequences located within the sliding window were inferred using the PhyML method (39) with the JTT model of proteins substitution (45). For the windows smaller than 200 sites, the average bootstrap score of the branches of partial trees was often smaller than 50% because of the strong similarity between the examined amino acid sequences. The HGT-Detection method (6) with the bipartition dissimilarity option was then performed to infer partial HGTs for each position of the sliding window. As a final result, we retained 10 partial transfers illustrated in Figure 9 (all partial transfers with bootstrap scores lower than 60% were discarded). Some of these transfers were indeed complete transfers.
Figure 9.
Species tree for the 42 bacteria and plastids from Figure 7 with 10 HGT branches (denoted by arrows) representing partial horizontal transfers of the gene rbcL. Partial HGTs having their analogs among complete HGTs received the same numbers as in Figure 8; HGTs absent in Figure 8 are numbered 10–12. Bootstrap support of partial HGT events and affected intervals of the original MSA are indicated. For complete HGTs 2, 6 and 9 (affecting the whole MSA) the interval is not indicated.
Thus, the proposed technique for inferring partial HGTs allowed us to refine the results of a method predicting complete transfers. Some of the detected complete HGTs were confirmed (i.e. HGTs 2, 6 and 9), some of them were discarded (i.e. HGTs 5 and 8 with low bootstrap support), and some of them were reclassified as partial transfers (i.e. HGTs 1, 3, 4 and 7). Moreover, the three new (partial) HGTs were found (i.e. HGTs 10, 11 and 12). For instance, the rbcL gene of Chromatium L is composed of the sequence polymorphisms stemming from Hydrogenovibrio L1 (on the interval 130:230) and L2 (on the interval 361:531) as well as from the original sequence (on the intervals 1:129 and 231:360). Obviously, the bootstrap scores of partial transfers, found for a part of the MSA, were higher than the corresponding bootstrap scores of complete transfers, found for the whole MSA.
The transfers shown in Figures 8 and 9 include one of the main HGTs predicted by Delwiche and Palmer [see Figure 4 in Ref. (37) and the following discussion]: Between α-proteobacteria and Red and Brown algae (complete HGT 2 with bootstrap support of 83.2%). The exact transfer between Cyanobacteria and the ancestor of β- and γ-proteobacteria (complete HGT 9 with 87.1% bootstrap support) was not predicted by Delwiche and Palmer (37), but the latter study discussed the possibility of a close ancient transfer between Cyanobacteria and the ancestor of γ-proteobacteria. The obtained partial HGT scenario does not include, however, any HGT from γ-proteobacteria to α- and β-proteobacteria hypothesized by Delwiche and Palmer (37). To resolve multiple topological conflicts between the species and gene phylogenies, our scenario relies on HGTs from β-proteobacteria to α- and γ-proteobacteria, and from α- to β-proteobacteria.
Detecting partial transfers of the gene mutU
Second, we examined the evolution of the bacterial mismatch repair (MMR) gene mutU of Escherichia coli originally discussed by Denamur et al. (30). Denamur et al. explored the hypothesis that MMR deficiency emerging in nature has left some ‘imprint’ in the bacterial genomes and showed that individual functional MMR genes, when compared to housekeeping genes, exhibit high sequence mosaicism derived from different phylogenetic lineages. The E. coli MMR genes, mutS, mutL, mutH and mutU (uvrD), and two control genes (mutT and recD), were partially sequenced from 30 natural isolates in order to test the transfer hypothesis. Denamur et al. (30) compared the obtained gene phylogenies to the whole genome reference tree and found numerous topological conflicts that ranged from single (for mutT) to multiple (for mutS). To test whether these topological conflicts were due to HGT or tree reconstruction artefacts, the latter authors applied the incongruence length difference (ILD) method (47) and concluded that the MMR gene trees, when compared to the whole genome tree, exhibit significant incongruence due, most likely, to horizontal gene transfer. Supplementary Figure S1 reports the hypothetical partial horizontal transfers of the gene mutU within the E. coli evolutionary tree found in Ref. (30). Because of the highest level of mosaicism within MMR genes, the strain ECOR 37 does not have a clear phylogenetic position within the E. coli stain phylogeny (see Supplementary Figure S1, where this strain is not included in the set of tree leaves).
The new method was applied on the MSA of the gene mutU MMR, using three different window sizes: 100, 150 and 200 sites and the advancement step of 10 sites. The total length of the mutU MSA was 384 nt. The aligned sequences of the gene mutU that we examined can be found at: http://www.info2.uqam.ca/~makarenkov_v/mutU.txt. To build the mutU tree, we used the HKY85 (48) substitution model and the default settings of PhyML. Because of the strong similarity between the DNA sequences, multiple unresolved partial gene trees were found. All partial gene trees whose average bootstrap score was under 50% were ruled out from the analysis (i.e. not treated by the HGT detection method).
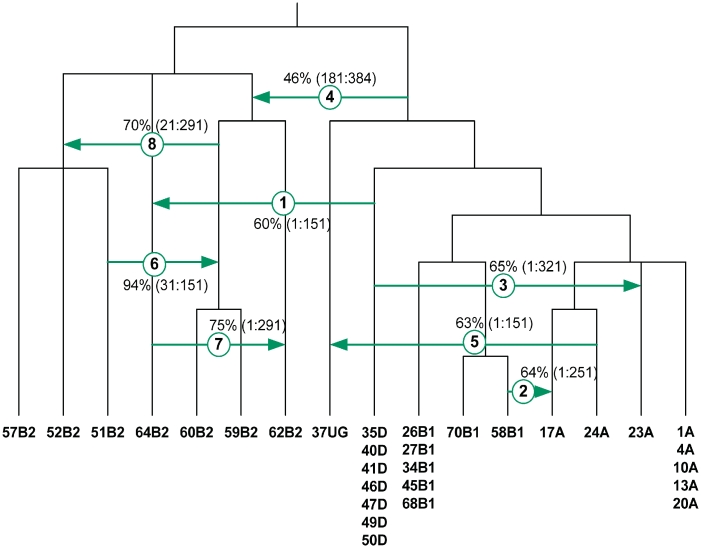
Figure 10 presents the eight most significant transfers inferred by the new method (the transfers whose bootstrap support was greater than 40% are represented). For each transfer, its direction, involved species, bootstrap support and associated interval of the original MSA are depicted.
Figure 10.
Hypothetical partial transfers of the gene mutU predicted by the new method. Partial HGTs are denoted by arrows. Bootstrap support of partial HGT events and affected intervals of the original MSA are indicated.
For instance, HGTs 1, 3 and 4, with bootstrap support of 60%, 65% and 46%, respectively, correspond to three similar transfers found by Denamur et al. (30), Supplementary Figure S2; in the latter study, an exact analogue of HGT 4 was not determined, but a very close transfer was found. HGT 2 detected by the new method was also identified by Denamur et al. (30), but it goes in the opposite direction in that study. It is worth noting that all eight transfers found by Denamur et al. (30) were also predicted by the new method, but four of them are not represented in Figure 10 as a consequence of their low bootstrap support. We also found four new partial gene transfers (HGTs 5, 6, 7 and 8) with high bootstrap scores (63%, 94%, 75% and 70%, respectively). Mention that the solution found by the HGT-Detection method for inferring complete transfers (6) included only HGTs 2 and 3 from Figure 10. The other transfers found by HGT-Detection were different from those represented in Figure 10 and usually had a low bootstrap support.
DISCUSSION
We described a new method for predicting partial HGT events followed by intragenic recombination and thus for identifying the origins of mosaic genes. To the best of our knowledge, this relevant problem has not been properly addressed in the literature [for instance, the two existing partial HGT detection methods, (30) and (31), do not include any validation of the obtained gene transfers or Monte Carlo simulations]. The proposed method is based on a sliding window procedure that progressively analyzes the fragments of the given sequence alignment. The size of the sliding window should be adjusted with respect to the existing information about the genes and species under study. The use of smaller sizes of the sliding window allows one to detect smaller partial transfers with a better accuracy (i.e. HGTs affecting shorter intervals of the given multiple sequence alignment), but this also increases the running time of the method. For each fixed window position, a corresponding partial tree is inferred and a scenario of partial HGT events is determined by reconciling the obtained partial gene tree and the given species tree. A bootstrap procedure, allowing one to assess the bootstrap support of partial HGTs by taking into account the uncertainty of partial gene trees, was also developed. Another advantage of the presented method over the existing sliding window techniques used to detect recombination (33–37) is that it also allows for detecting the source (i.e. from which donor species the transferred fragments arrived) of the transferred sequences. The described method was included in the T-REX package (49) available at: http://www.trex.uqam.ca.
Both examples considered in the ‘Results’ section suggest that the new method can be also useful for confirming or discarding complete HGTs inferred by any existing HGT detection method. Our study of the evolution of the gene rbcL for 40 species of Proteobacteria, Cyanobacteria and plastids (37) and that of the mismatch repair (MMR) gene mutU for 30 E. coli strains (30) showed that most of the predicted gene transfers (i.e. six out of eight for each data set) may have been, in fact, partial HGTs. The conducted simulations showed that with smaller transferred sequence fragments, the best HGT detection rates were found for the trees having 16 and 32 leaves, whereas with larger transferred fragments the best results were obtained for 64-leaf trees. The following general trend can be formulated when analyzing the results presented in Figures 3–5: longer transferred sequence fragments and larger trees provide a much better HGT recovery and a smaller number of false positives. The problem occurring when considering short sequence fragments is that partial phylogenies inferred from them usually have low bootstrap support, and consequently provide a low confidence level of detected HGTs. The simulation results also suggest that in case of longer transferred sequences and larger trees one should look for a result confirmation over a few consecutive window positions in order to validate the obtained transfers.
The results of crosses with either the same donor or the same recipient show that recombination frequency decreases exponentially with increasing sequence divergence (50). Thus, the recombination success is strongly dependent on percent of nucleotide identity, which implies that recombination breakpoints occur only in the most conserved parts of a gene. This feature can be integrated into the described method by considering a more comprehensive statistical model taking into account the sequence divergence parameter. On the other hand, information about the obtained partial HGTs and their bootstrap scores can be incorporated in an extended evolutionary model that takes into account horizontal gene transfer, ancient gene duplication and gene loss (e.g. topological incongruence giving rise to predicted partial and/or complete transfers with low bootstrap support may be due to ancient gene duplication followed by partial gene loss). The determined bounds of transferred fragments can be examined in more details by comparing the corresponding 3D conformations. The discussed method can be also applied on a full-genome scale to estimate the proportion of mosaic genes in each studied genome as well as the rates of partial and complete HGTs between involved species. Several relevant statistics regarding the position and functionality of genetic fragments affected by horizontal gene transfer along with the rates of intraspecies (i.e. HGT between strains of the same species) and interspecies (i.e. HGT between distinct species) transfers can be estimated using the discussed technique. An alternative approach that can be also envisaged would be based on a Hidden Markov Model applied along the given MSA with the hidden state representing the HGT history of each considered sequence fragment. As any method of phylogenetic analysis, the presented algorithm for detecting partial gene transfers is subject to some artifacts. The main of them are long-branch attraction, unequal evolutionary rates and situations when possible HGT events almost coincide with speciation events. In the future, it will be important to investigate the impact of these artifacts on the identification of mosaic genes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Natural Sciences and Engineering Research Council of Canada (NSERC); Nature and Technologies Research Funds of Quebec (FQRNT).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Hervé Philippe and two anonymous reviewers for their helpful comments.
REFERENCES
- 1.Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2129. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 2.Koonin EV. Horizontal gene transfer: the path to maturity. Mol. Microbiol. 2003;50:725–727. doi: 10.1046/j.1365-2958.2003.03808.x. [DOI] [PubMed] [Google Scholar]
- 3.Doolittle WF, Boucher Y, Nesbø CL, Douady CJ, Anderson JO, Roger AJ. How big is the iceberg of which organellar genes in nuclear genomes are but the tip? Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2003;358:39–57. doi: 10.1098/rstb.2002.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakhleh L, Ruths D, Wang LS. RIATA-HGT: a fast and accurate heuristic for reconstructing horizontal gene transfer. In: Wang L, editor. Lecture Notes in Computer Science. Kunming, China: Springer; 2005. pp. 84–93. [Google Scholar]
- 5.Makarenkov V, Kevorkov D, Legendre P. Phylogenetic network reconstruction approaches. Bioinformatics. 2006;6:61–97. [Google Scholar]
- 6.Boc A, Philippe H, Makarenkov V. Inferring and validating horizontal gene transfer events using bipartition dissimilarity. Syst. Biol. 2010;59:195–211. doi: 10.1093/sysbio/syp103. [DOI] [PubMed] [Google Scholar]
- 7.Hollingshead SK, Becker R, Briles DE. Diversity of PspA: mosaic genes and evidence for past recombination in Streptococcus pneumoniae. Infect. Immun. 2000;68:5889–5900. doi: 10.1128/iai.68.10.5889-5900.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhaxybayeva O, Lapierre P, Gogarten JP. Genome mosaicism and organismal lineages. Trends Genet. 2004;20:254–260. doi: 10.1016/j.tig.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 9.Gogarten JP, Doolittle WF, Lawrence JG. Prokaryotic evolution in light of gene transfer. Mol. Biol. Evol. 2002;19:2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- 10.Lawrence JG, Ochman H. Amelioration of bacterial genomes: rates of change and exchange. J. Mol. Evol. 1997;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- 11.Maiden M. Horizontal genetic exchange, evolution, and spread of antibiotic resistance in bacteria. Clin. Infect. Dis. 1998;27:12–20. doi: 10.1086/514917. [DOI] [PubMed] [Google Scholar]
- 12.Zheng Y, Roberts RJ, Kasif S. Segmentally variable genes: a new perspective on adaptation. PLoS Biol. 2004;2:452–464. doi: 10.1371/journal.pbio.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Claverys JP, Prudhomme M, Mortier-Barrière I, Martin B. Adaptation to the environment: Streptococcus pneumoniae, a paradigm for recombination-mediated genetic plasticity? Mol. Microbiol. 2000;35:251–259. doi: 10.1046/j.1365-2958.2000.01718.x. [DOI] [PubMed] [Google Scholar]
- 14.Hein J. A heuristic method to reconstructing the evolution of sequences subject to recombination using parsimony. J. Mol. Evol. 1993;36:396–405. [Google Scholar]
- 15.von Haeseler A, Churchill GA. Network models for sequence evolution. J. Mol. Evol. 1993;37:77–85. doi: 10.1007/BF00170465. [DOI] [PubMed] [Google Scholar]
- 16.Page RDM. Maps between trees and cladistic analysis of historical associations among genes, organism and areas. Syst. Biol. 1994;43:58–77. [Google Scholar]
- 17.Mirkin BG, Muchnik I, Smith TF. A biologically consistent model for comparing molecular phylogenies. J. Comput. Biol. 1995;2:493–507. doi: 10.1089/cmb.1995.2.493. [DOI] [PubMed] [Google Scholar]
- 18.Maddison WP. Gene trees in species trees. Syst. Biol. 1997;46:523–536. [Google Scholar]
- 19.Charleston MA. Jungle: a new solution to the host/parasite phylogeny reconciliation problem. Math. Biosci. 1998;149:191–223. doi: 10.1016/s0025-5564(97)10012-8. [DOI] [PubMed] [Google Scholar]
- 20.Hallett M, Lagergren J. Efficient algorithms for lateral gene transfer problems. In: El-Mabrouk N, Lengauer T, Sankoff D, editors. Proceedings of the Fifth Annual International Conference on Research in Computational Biology. New York: ACM Press; 2001. pp. 149–156. [Google Scholar]
- 21.Boc A, Makarenkov V. New efficient algorithm for detection of horizontal gene transfer events. In: Benson G, Page R, editors. Algorithms in Bioinformatics. Budapest, Hungary: Springer; 2003. pp. 190–201. [Google Scholar]
- 22.MacLeod D, Charlebois RL, Doolittle F, Bapteste E. Deduction of probable events of lateral gene transfer through comparison of phylogenetic trees by recursive consolidation and rearrangement. BMC Evol. Biol. 2005;5:27. doi: 10.1186/1471-2148-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsirigos A, Rigoutsos I. A new computational method for the detection of horizontal gene transfer events. Nucleic Acids Res. 2005;33:922–933. doi: 10.1093/nar/gki187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsirigos A, Rigoutsos I. A sensitive, support-vector-machine method for the detection of horizontal gene transfers in viral, archaeal and bacterial genomes. Nucleic Acids Res. 2005;33:3699–3707. doi: 10.1093/nar/gki660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beiko RG, Hamilton N. Phylogenetic identification of lateral genetic transfer events. BMC Evol. Biol. 2006;6:15. doi: 10.1186/1471-2148-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin G, Nakhleh L, Snir S, Tuller T. Efficient parsimony-based methods for phylogenetic network reconstruction. Bioinformatics. 2006;23:123–128. doi: 10.1093/bioinformatics/btl313. [DOI] [PubMed] [Google Scholar]
- 27.Jin G, Nakhleh L, Snir S, Tuller T. Inferring phylogenetic networks by the maximum parsimony criterion. Mol. Biol. Evol. 2007;241:324–337. doi: 10.1093/molbev/msl163. [DOI] [PubMed] [Google Scholar]
- 28.Linz S, Radtke A, von Haeseler A. A maximum likelihood framework to measure horizontal gene transfer. Mol. Biol. Evol. 2007;24:1312–1319. doi: 10.1093/molbev/msm052. [DOI] [PubMed] [Google Scholar]
- 29.Than C, Nakhleh L. In Proceedings of the 6th Asia Pacific Bioinformatics Conference. Kyoto, Japan: 2008. SPR-based tree reconciliation: non-binary trees and multiple solutions; pp. 251–260. [Google Scholar]
- 30.Denamur E, Lecointre G, Darlu P, Tenaillon O, Acquaviva C, Sayada C, Sunjevaric I, Rothstein R, Elion J, Taddei F, et al. Evolutionary implications of the frequent horizontal transfer of mismatch repair genes. Cell. 2000;103:711–721. doi: 10.1016/s0092-8674(00)00175-6. [DOI] [PubMed] [Google Scholar]
- 31.Makarenkov V, Boc A, Delwiche CF, Diallo AB, Philippe H. New efficient algorithm for modeling partial and complete gene transfer scenarios. In: Batagelj V, Bock HH, Ferligoj A, Ziberna A, editors. Data Science and Classification. Springer; 2006. pp. 341–349. [Google Scholar]
- 32. Ray,S.C. (1998) SimPlot for Windows (version 1.6). Berlin, Germany, Baltimore, Md.
- 33.Archibald JM, Roger AJ. Gene duplication and gene conversion shape the evolution of archaeal chaperonins. J. Mol. Biol. 2002;316:1041–1050. doi: 10.1006/jmbi.2002.5409. [DOI] [PubMed] [Google Scholar]
- 34.Paraskevis D, Deforche K, Lemey K, Magiorkinis, Hatzakis A, Vandamme AM. SlidingBayes: exploring recombination using a sliding window approach based on Bayesian phylogenetic inference. Bioinformatics. 2005;21:1274–1275. doi: 10.1093/bioinformatics/bti139. [DOI] [PubMed] [Google Scholar]
- 35.Lee WH, Sung WK. RB-finder: an improved distance-based sliding window method to detect recombination breakpoints. J. Comput. Biol. 2008;15:881–898. doi: 10.1089/cmb.2007.0154. [DOI] [PubMed] [Google Scholar]
- 36.Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delwiche CF, Palmer JD. Rampant horizontal transfer and duplication of Rubisco genes in Eubacteria and Plastids. Mol. Biol. Evol. 1996;13:873–882. doi: 10.1093/oxfordjournals.molbev.a025647. [DOI] [PubMed] [Google Scholar]
- 38.Acinas SG, Marcelino LA, Klepac-Ceraj V, Polz MF. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 2004;38:2629–2635. doi: 10.1128/JB.186.9.2629-2635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guindon S, Gascuel O. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 40.Robinson DR, Foulds LR. Comparison of phylogenetic trees. Math. Biosciences. 1981;53:131–147. [Google Scholar]
- 41.Smith JM, Feil EJ, Smith NH. Population structure and evolutionary dynamics of pathogenic bacteria. Bioessays. 2000;22:1115–1122. doi: 10.1002/1521-1878(200012)22:12<1115::AID-BIES9>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 42.Kuhner M, Felsenstein J. A simulation comparison of phylogeny algorithms under equal and unequal evolutionary rates. Mol. Biol. Evol. 1994;11:459–468. doi: 10.1093/oxfordjournals.molbev.a040126. [DOI] [PubMed] [Google Scholar]
- 43.Guindon S, Gascuel O. Efficient biased estimation of evolutionary distances when substitution rates vary across sites. Mol. Biol. Evol. 2002;19:534–543. doi: 10.1093/oxfordjournals.molbev.a004109. [DOI] [PubMed] [Google Scholar]
- 44.Rambaut A, Grassly NC. Seq-Gen: an application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Comput. Appl. Biosci. 1997;13:235–238. doi: 10.1093/bioinformatics/13.3.235. [DOI] [PubMed] [Google Scholar]
- 45.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 46.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2009;37:D26–D31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Farris JS, Kallersjo M, Kluge AG, Bult C. Testing significance of incongruence. Cladistics. 1994;10:3, 315–319. [Google Scholar]
- 48.Hasegawa M, Hirohisa K, Taka-aki Y. Dating the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 49.Makarenkov V. T-Rex: reconstructing and visualizing phylogenetic trees and reticulation networks. Bioinformatics. 2001;17:664–668. doi: 10.1093/bioinformatics/17.7.664. [DOI] [PubMed] [Google Scholar]
- 50.Vulic M, Dionisio F, Taddei F, Radman M. Molecular keys to speciation: DNA polymorphism and the control of genetic exchange in enterobacteria. Proc. Natl Acad. Sci. USA. 1997;94:9763–9767. doi: 10.1073/pnas.94.18.9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.