Abstract
The ability to verify the sequence of a nucleic acid-based therapeutic is an essential step in the drug development process. The challenge associated with sequence identification increases with the length and nuclease resistance of the nucleic acid molecule, the latter being an important attribute of therapeutic oligonucleotides. We describe methods for the sequence determination of Spiegelmers, which are enantiomers of naturally occurring RNA with high resistance to enzymatic degradation. Spiegelmer sequencing is effected by affixing a label or hapten to the 5′-end of the oligonucleotide and chemically degrading the molecule in a controlled fashion to generate fragments that are then resolved and identified using liquid chromatography-mass spectrometry. The Spiegelmer sequence is then derived from these fragments. Examples are shown for two different Spiegelmers (NOX-E36 and NOX-A12), and the specificity of the method is shown using a NOX-E36 mismatch control.
INTRODUCTION
The development of nucleic acid molecules for therapeutic applications requires explicit confirmation of their identity as an integral part of the quality control strategy (1,2). The techniques employed should be specific, determining not only purity and molecular mass, but also nucleobase sequence, nature of sugar moieties and linkages as well as the presence of modifications (2,3). Ideally, the methods used should also be amenable to routine testing.
Many oligonucleotide drugs undergoing clinical trials are modified (4,5), meaning that traditional sequencing techniques utilizing enzymes are often not effective (6,7). Furthermore, approaches using visual readouts such as gel electrophoresis are disfavoured due to artefacts such as band compression caused by nucleic acid structural influences (8–11). Sequence elucidation of oligonucleotides by mass spectrometry (MS) has become the industry standard due to its reproducibility, sensitivity and accuracy. As such, the utilization of fragmentation patterns in MS/MS experiments is increasingly being used, and typically yields data sufficient for sequence confirmation. However, the success of MS/MS sequencing strongly depends on the length and nature of the oligonucleotide, with long RNA molecules (>30 nt) proving particularly challenging to fully sequence at the current state of this method (3,12). As such, the sequencing of long nucleic acid molecules and their modified derivatives remains problematic. In addition, MS/MS experiments usually provide data for sequence confirmation rather than sequence determination (de novo sequencing) where the sequence is not known beforehand.
RNA Spiegelmers (13,14) are structured oligonucleotides of typically 30–60 nt in length that possess an l-configuration, i.e. the mirror (German translation = Spiegel) image of naturally occurring RNA. These molecules are designed to bind to target molecules with high affinity and specificity (15). Due to their l-configuration, Spiegelmers have an unprecedented stability in biological media as nucleases and other enzymes do not recognise or metabolise them. Due to the lack of enzymatic recognition, several of the standard sequencing techniques available for elucidating the primary structure of unmodified d-RNA are not applicable to Spiegelmers. For instance, Sanger sequencing or variations thereof cannot be used as the polymerases employed do not recognize l-configured nucleic acid molecules. Enzymatic stability also prevents sequence analysis through nuclease digestion. In addition, Spiegelmers are generally of a length that exceeds the capability of current sequence confirmation MS/MS techniques.
Spiegelmer drugs that are currently in clinical development are conjugated at one terminus to a 40-kDa branched polyethylene glycol (PEG) moiety in order to enhance the pharmacokinetic profile of the molecule. As both the polydispersity of large PEG molecules (>20 kDa) and their propensity to form adducts with cations significantly complicate their corresponding mass spectra, the required sequence information is derived from the intermediate prePEG precursor. Such a strategy is valid since the primary sequence of the Spiegelmer is not altered during the PEGylation and subsequent processing steps. The intermediate, prepared through standard solid support synthesis, deprotection and purification protocols feature a handle, such as a primary amino modification, upon which the PEG moiety can be attached (Table 1).
Table 1.
List of Spiegelmers and the corresponding sequences
| Name | Sequence |
|---|---|
| NOX-E36 Int. | NH2-(CH2)6-OP(O)(OH)-GCA CGU CCC UCA CCG GUG CAA GUG AAG CCG UGG CUC UGC G |
| NOX-E36 Int. mutant | NH2-(CH2)6-OP(O)(OH)-GCA CGU CCC CUA CCG GUG CAA GUG AAG CCG UGG UCC UGC G |
| NOX-A12 Int. | NH2-(CH2)6-OP(O)(OH)O-GCG UGG UGU GAU CUA GAU GUA UUG GCU GAU CCU AGU CAG GUA CGC |
Nucleotides are depicted in bold.
Underlined sequence sections represent the CU switches of ‘NOX-E36 int. mutant’ as compared to the original sequence: ‘NOX-E36 Int.’
We describe herein methods for identifying the sequence of mirror-image RNA oligonucleotides. The l-RNA oligonucleotide is labelled at the 5′-end courtesy of the pre-installed 5′-amino functionality, and subjected to a controlled random fragmentation process leading to fragments that can be resolved and identified through liquid chromatography (LC) and electrospray ionization–mass spectrometry (ESI–MS). The resulting sequencing ladder enables Spiegelmer identity elucidation through the mass difference between sequential fragments. Due to the high specificity and robustness of the method, it is easy to detect point mutations or other modifications to the sequence or perform de novo sequencing on the RNA oligonucleotide.
MATERIALS AND METHODS
Synthesis of amino-modified Spiegelmers
The Spiegelmers were produced by solid-phase synthesis with an Äkta oligopilot 100 synthesizer (GE Healthcare, Freiburg, Germany) using 2′-TBDMS RNA phosphoramidite chemistry (16). l-rA(N-Bz)-, l-rC(Ac)-, l-rG(N-ibu)- and l-rU- phosphoramidites were purchased from ChemGenes (Wilmington, MA, USA). The 5′-amino-modifier was purchased from American International Chemicals Inc. (Framingham, MA, USA). Synthesis of the Spiegelmers was started on l-ribo nucleoside modified CPG with a pore size of 1000 Å (Link Technology, Glasgow, UK). For coupling (15 min per cycle), 0.3 M benzylthiotetrazole (American International Chemicals Inc., Framingham, MA, USA) in acetonitrile and 3.5 equivalents of the respective 0.2 M phosphoramidite solution in acetonitrile were used. A capping–oxidation–capping cycle was used. Further standard solvents and reagents for oligonucleotide synthesis were purchased from Biosolve (Valkenswaard, The Netherlands). The Spiegelmers were synthesized DMT-ON; after deprotection, the crude oligonucleotide was purified via preparative reversed-phase high-performance liquid chromatography (RP-HPLC) using Source 15RPC medium (GE Healthcare) (17). The 5′-DMT-group was removed with 80% acetic acid (90 min at room temperature, RT). Subsequently, aqueous 2 M NaOAc was added and the Spiegelmer was desalted by tangential-flow filtration using a 5-kDa molecular weight cut-off (MWCO) regenerated cellulose membrane (Millipore, Bedford, MA, USA). Table 1 lists the all sequences that were synthesized.
Labelling of amino-modified Spiegelmers
Labelling with cleavable biotin affinity tag
Ten milligrams (250 ODs) of amino-modified Spiegelmer were placed in a reaction tube and dissolved in 260 µl Theorell and Stenhagen's Universal buffer pH 8.5 (33 mM sodium citrate, 33 mM sodium phosphate, 57 mM sodium borate, pH 8.5). To this were added 200 µl N,N-dimethylformamide. The solution was vortexed and spun down, whereupon 2.2 mg biotin disulphide N-hydroxy-succinimide ester (Sigma, Taufkirchen, Germany) pre-dissolved in 50 µl DMF was added. The solution was incubated at RT for 60 minutes, whereupon an aliquot was taken and analysed by anion-exchange HPLC [Dionex DNA-Pac 200 column, Buffer A: 100 mM Tris (Applichem, Darmstadt, Germany); 10%ACN in H2O. Buffer B: 100 mM Tris; 1 M NaCl and 25 mM NaClO4 (Sigma-Aldrich, Taufkirchen, Germany); 10%ACN in H2O; both buffers were adjusted to pH 8.5 with aqueous conc. HCl (Merck, Darmstadt, Germany); gradient 10–30% B in 6 min then 30–70% B in 35 min, temperature of column 80°C] which determined that the reaction was complete. The crude reaction mixture was desalted using a NAP-25 column (GE Healthcare, Freiburg, Germany) and lyophilized.
Labelling with fluorescein dye
Two-hundred and twelve ODs of aminomodified NOX-E36 (NOX-E36 Int., Table 1) were placed in a reaction tube and dissolved in 250 µl H2O. To this were added 3 mg fluorescein-5-isothiocyanate (FITC Isomer I) (Sigma-Aldrich) pre-dissolved in 250 µl N,N-dimethylformamide. The solution was vortexed and spun down, whereupon 6 mg sodium bicarbonate (Merck) was added. The solution was incubated at RT for 6 h, whereupon the crude mixture was desalted by size-exclusion chromatography using an NAP-25 column and lyophilized. The lyophilisate was redissolved in water and purified via preparative RP-HPLC using a Source 15RPC medium (GE Healthcare) and was desalted via size-exclusion chromatography using NAP-25 columns.
Controlled fragmentation of Spiegelmers
Biotin-labelled Spiegelmers
To 20 µl biotin-labelled Spiegelmer (0.5 OD/µl) was added 30 µl sterilized water and 2.5 µl 0.5 M K2CO3 (Merck) at RT. The solution was vortexed and then incubated on an Eppendorf Thermomixer Comfort machine (Eppendorf, Hamburg, Germany) at 70°C at 1350 r.p.m. for 12.5 min (NOX-E36 derivatives), or 20 min (NOX-A12) whereupon it was frozen in liquid nitrogen and allowed to thaw out. Then 4 µl 1 M AcOH was added (≈pH 7) to quench the reaction and the solution was vortexed and spun down.
FITC-labelled NOX-E36
To 13.5 µl (1 OD) of FITC-labelled NOX-E36 was added 1.5 µl 0.5 M K2CO3 at RT. The solution was vortexed and then incubated on an Eppendorf Thermomixer Comfort machine (Eppendorf) at 70°C at 1350 r.p.m. for 5 min whereupon it was frozen in liquid nitrogen and allowed to thaw out. Then 2.5 µl 1 M AcOH was added to quench the reaction (final pH ≈ 7) and the solution vortexed, spun down and then analysed by liquid chromatography-mass spectrometry (LCMS).
Isolation of 5′–fragments generated from controlled fragmentation
Binding of biotinylated fragments to Neutravidin beads
Neutravidin Agarose beads were treated as follows: 150 µl of Neutravidin bead slurry (Pierce, Milwaukee, MI, USA) was put in a 500 -µl reaction tube. The beads were spun down and the supernatant was carefully removed. Whereupon 300 µl 1 M Tris HCl pH 8.0 (Ambion, Huntindon, UK) was added. The slurry was vortexed, spun down and the supernatant was carefully removed. The beads were then washed 2 × 300 µl in the same manner with sterile H2O. A biotin-labelled Spiegelmer that had undergone the controlled fragmentation and quench was then added to the beads and the resulting slurry was mixed vigorously (1350 r.p.m.) at 10°C for 2 h.
Washing of bound fragments of NOX-E36 derivatives
The beads with bound fragments of NOX-E36 derivatives were isolated through filtration using a spin microfuge tube (Ultrafree-MC GV, 0.22 µm; Millipore, Schwalbach, Germany) and washed with 2 × 300 µl sterile H2O.
Washing of bound fragments of NOX-A12
The beads with bound fragments of NOX-A12 were spun down and the supernatant was removed. A 1 × 300 µl 8 M urea was added and the mixture was vortexed and spun down. The supernatant was carefully removed and the beads were washed a further four times with sterilized water (200 µl).
Cleavage of the biotinylated fragments from the Neutravidin beads
The disulphide linker of the bound biotin-labelled fragments was cleaved using a 0.05 M sodium phosphate buffer (pH 8.5), 100 µl with 5 µl 1 M DTT solution. This was vigorously mixed at 25°C for 2 h on an Eppendorf Thermomixer Comfort machine. The slurry was filtered using a spin microfuge tube (Ultrafree-MC GV, 0.22 µm; Millipore), and the beads were washed with a further 50 µl sterile water. A UV measurement was taken to determine the optical density units at 260 nm, and 0.25 ODs of this solution was analysed by LCMS.
LCMS analysis of fragments for Spiegelmer identification
Analysis of Spiegelmer fragments generated from the protocols above were analysed using an LCMS system (Agilent Technologies, Waldbronn, Germany) comprised of a 1200-series rapid resolution pump, autosampler with sample cooler (set to 6°C), column oven set to 65°C equipped with a six-port valve, Diode Array Detector (DAD) and a 6520 Quadrupole-Time Of Flight (Q-TOF) mass spectrometer equipped with electrospray ionization source. Data were acquired in the negative ion mode between m/z 50 and 3200. The gas temperature was 350°C, drying gas flow 12 l/min., nebulizer pressure 60 PSI, capillary voltage 4000 V, and fragmentation voltage was set to 350 V. For separation, an Acquity BEH C18 Column (1.7 µm, 130 Å pore size, 2.1 × 30 mm; Waters, Eschborn, Germany) operated at a flow rate of 0.2 ml/min. has been used.
The sample was analysed by injecting and trapping the Spiegelmer fragments onto the LC column while non-volatile salts were eluted to waste via the DAD and the six-port valve. After this desalting process (0.5 min), the effluent was delivered to the ESI-MS by switching the six-port valve. A gradient was run from 0–20% B in 0.5 to 22 min, 20–30% B in 40 min. Buffer A consisted of 1% methanol and 100 mM hexafluoroisopropanol (Biosolve), 10 µM ethylenediaminetetraacetic acid (EDTA) (NH4+ form) and 10 mM Triethylamine (Sigma-Aldrich) in water. Buffer B consisted of 10 mM Triethylamine, 100 mM hexafluoroisopropanol, 10 µM EDTA (NH4+ form) and 50% methanol in water. For fragments that originated from biotinylated Spiegelmer, mass spectra from the total ion chromatogram (TIC) were derived for each peak and then deconvoluted. For fragments that originated from FITC-NOX-E36, the mass spectra from the TIC were derived for each peak observed in the UV chromatogram extracted at 495 nm, and then deconvoluted.
Deconvolution for smaller fragments was performed using resolved isotope deconvolution, whereby the monoisotopic mass has been reported up to a molecular weight where the monoisotopic mass could be clearly identified. The exact Spiegelmer fragment monoisotopic mass could be identified up to 5–8 kDa, depending on signal intensity. For larger Spiegelmer fragments, average mass deconvolution was applied (mass step 1.0, varying deconvolution range and varying m/z range depending on Spiegelmer fragment envelope).
RESULTS AND DISCUSSION
Initial attempts to confirm sequence identity of Spiegelmers focused on using state-of-the-art MS/MS techniques on the amino-modified precursor of exemplary Spiegelmers. However, the data obtained did not cover the whole sequence, leaving a gap of 10–15 nt in the middle unidentified (data not shown). Recent advancements in MS/MS techniques have enabled longer RNA molecules to be sequenced (12).
It was therefore deemed necessary to break the Spiegelmer molecule into fragments, but the ability to connect these fragments to provide a sequencing ladder is required to provide the complete primary structure. Although it has been shown that l-oligonucleotides have a limited lability towards Snake Venom Phosphodiesterase (18), sequential degradation, as reported for d-RNA (19) with this enzyme, would require very long incubation times, thus making this route unfeasible. However, Spiegelmers are susceptible to chemical degradation to the same extent as the corresponding d-nucleic acid material. Chemical sequencing of RNA has been previously reported by the use of nucleobase specific chemical reactions that can be used to effect strand scission at specific sites along the nucleotide backbone (20). However, this approach was disfavoured due to the many reactions required, the lack of absolute nucleobase specificity and the potential for artefactual results by virtue of the nucleobase modification and cleavage steps.
Chemical cleavage of RNA can be effected with acidic or basic solutions. This indiscriminate cleavage creates a mix of fragments representing random strand scission 3′ to every nucleotide, resulting in the formation of 5′, 3′ and internal fragments. In order to extract a meaningful pattern that would yield a sequencing ladder, it occurred to us that the pre-existence of the 5′-amino functionality on the non-PEGylated Spiegelmer intermediate could be exploited by attaching an affinity tag (ligand) hapten to it. Upon careful fragmentation of the Spiegelmer, labelled species could be subsequently isolated e.g. through immobilization. Non-labelled species, in this case 3′- and internal fragments, could be washed away, and the bound fragments then released from the support providing a sequencing ladder of 5′-fragments that could be analysed further with LC coupled to MS. The LC-MS approach utilizing reversed-phase ion-pairing chromatography facilitates the identification of the individual fragments by attributing corroborating properties to the fragments e.g. the tendency for longer oligonucleotide fragments to have a longer column retention time than shorter oligonucleotide fragments, and the separation of the fragments would promote their isolated analysis by ESI-MS, thus minimizing ion-suppression phenomena due to co-eluting compounds.
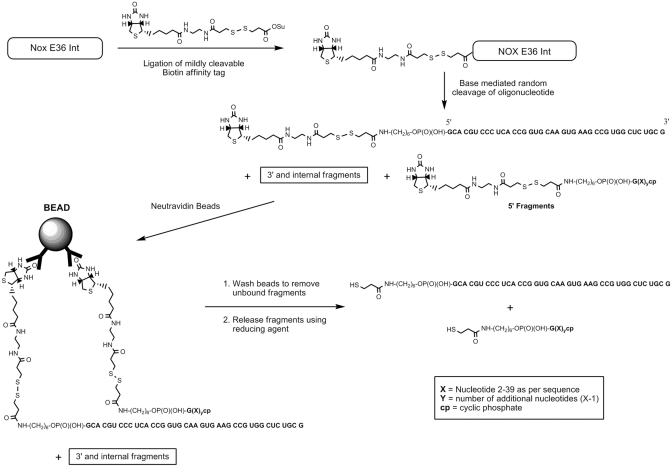
As proof of principle, 5′-aminomodified NOX-E36 intermediate (NOX-E36 Int.) was labelled with (2-[Biotinamido]ethylamido)-3,3′-dithiodipropionic acid N-hydroxysuccinimide ester (see Figure 1 for schematic) in the universal buffer of Theorell & Stenhagen adjusted to pH 8.5. This biotin label derivative possesses an internal disulphide bond that can be cleaved under mild conditions. After desalting the crude mixture using a size-exclusion column, the material was chemically cleaved in a random fashion with 25 mM potassium carbonate at 70°C. The cleavage time was carefully optimized so as not to drive the cleavage to completion, and to ensure that sufficient amounts of fragments are generated. Optimal cleavage times lengthened with an increase in Spiegelmer concentration and purity, and varied according to Spiegelmer sequence and salt content. Upon reaching the desired incubation time, solutions were quenched by snap cooling with liquid nitrogen and then the basic solution was neutralised with aqueous acetic acid. This was found to be a milder, more reliable quenching method than simply adding the quenching solution to the hot solution.
Figure 1.
Schematic of proof of principle evaluation with NOX-E36-Int. 5′–fragments in this schematic have been encompassed using the formula ‘R’-NH-(CH2)6-OP(O)(OH)-G(X)ycp where ‘R’ is either the structurally drawn cleavable biotin affinity tag or cleaved fragment. Letters in bold and underlined, respectively, represent the nucleic acid sequence, X indicates the identity of the particular nucleotide (A, C, G, U) read from the sequence 5′- to 3′- and y represents how many additional nucleotides are in the fragment over the first fragment. For example, for fragment 2: y = 1 (one extra nucleotide). The extra nucleotide(s) X over the first fragment is one (C) therefore the fragment, fragment 2 is ‘R’-NH-(CH2)6-OP(O)(OH)-GCcp. The annotation cp in subscript denotes 2′3′ cyclic phosphate. The l-RNA sequence of the Spiegelmer is in bold letters. FLP = Full Length (undegraded) Product.
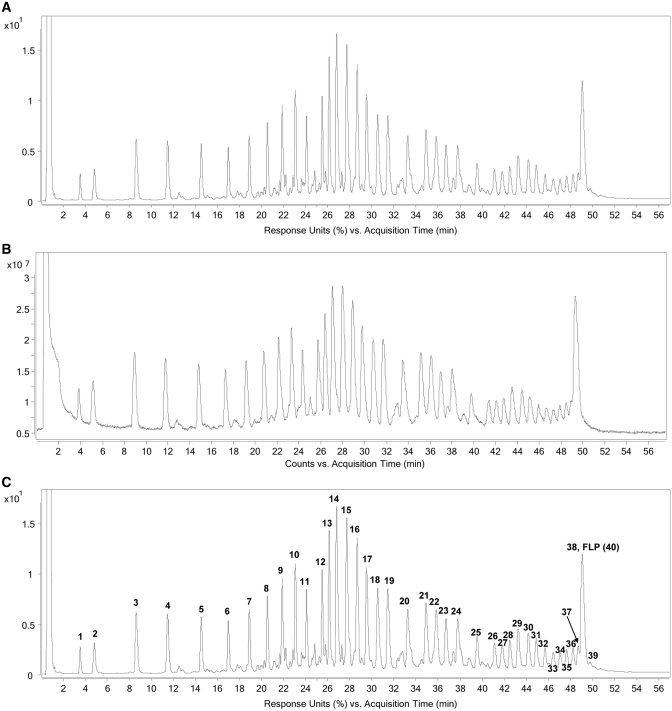
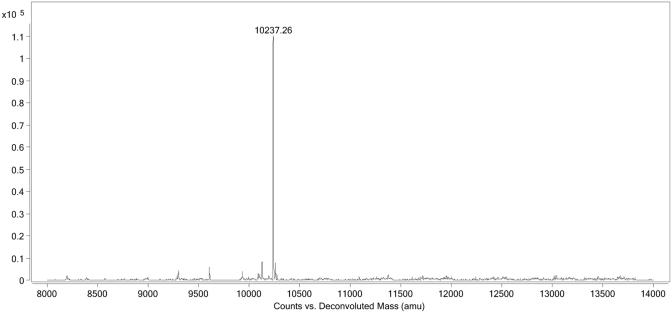
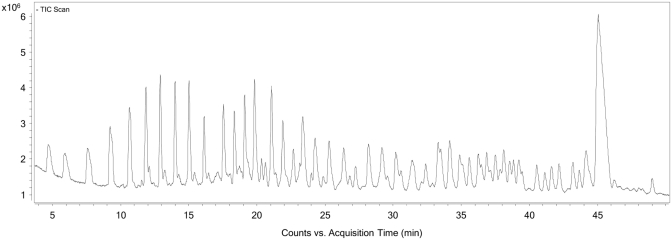
Subsequent to the fragmentation and quench, which produces 5′-fragments, 3′-fragments, and random internal fragments of NOX-E36 Int., the resulting solution was then incubated with Neutravidin beads whereby the 5′-(biotinylated) fragments and remaining intact biotinylated NOX-E36 Int. (i.e. full-length product) are selectively immobilized on the beads by virtue of the hapten biotin. The unbound fragments, i.e. 3′-fragments and random internal fragments that do not possess the affinity tag, are then washed away. Release of the immobilized fragments by cleaving the internal disulphide bond of the label's linker under mild reducing conditions (50 mM DTT in 50 mM sodium phosphate buffer) furnished the corresponding thiol derivatives of the fragments (Figure 1). The liberated fragments were then analysed by LC-MS using a Q-TOF electrospray ionization mass spectrometer. The UV and total ion chromatogram (TIC) both show discrete peaks that correspond to derivatives of all labelled species: 5′-fragments and undegraded NOX-E36 Int. (Figure 2). The exact or average mass(es) observed in the discrete peaks were obtained through deconvolution of the derived mass spectra pertaining to each discrete peak (see Figure 3 for an exemplary spectrum). Where possible, monoisotopic resolved masses were obtained, otherwise the corresponding average molecular mass values were acquired.
Figure 2.
This figure shows the UV (260 nm) (A) and total ion chromatogram (TIC) (B) obtained from the LC-MS experiment after biotin labelled NOX-E36 Int. was subjected to the sequencing protocol as described. The former after sequence confirmation is annotated (C), to show the position/retention time of the fragments.
Figure 3.
Deconvoluted molecular weight of a fragment (mass peak value = 10 237.26 Da), in this case Fragment 31.
As previously mentioned, the LC-ESI-MS approach facilitates fragment identity confirmation/elucidation. It is of interest to note that unlike other sequencing methods (3,21,22), all fragments are retained on the column and all information is available in one LC-MS measurement. This significantly improves method robustness and convenience. Some fragments co-elute with the full-length product (FLP). Due to the phenomenon of ion suppression, this can impact on the sensitivity or accuracy of the data acquisition of low-abundant species. However, this can be overcome by controlling the fragmentation time so that the FLP:N-1 or N-2 ratio is optimal. Interestingly, in this LC method, the penultimate fragment of NOX-E36 elutes later than the FLP. This can be readily explained by taking into account the relative phosphate charges (and hence ion pairs with the buffer) and the number of nucleobases. For fragment 39, the number of phosphates is equal to the FLP but it possesses one nucleobase less (in this case a guanosine).
In general, the masses observed are those of the 2′,3′-cyclic phosphates. As expected, fragment mass increases with an increase in retention time. The exception to this rule is the presence of low abundance of fragments whereby the cyclic phosphate has been hydrolysed. The hydrolysed 2′ (3′) phosphate derivatives, which further confirm the identity of the fragments generated, typically elute later than the parent 2′,3′-cyclic phosphate and often have a retention time greater than the subsequent fragments’ cyclic phosphate derivative. This is presumably due to the added charge/ion pair generated by the hydrolysis.
The masses of the fragments generated can be compared to the calculated masses of the predicted 5′–fragments of the thiol-derived NOX-E36 intermediate to confirm the sequence (Table 2). Alternatively, by virtue of the sequencing ladder generated, Spiegelmer identity can be derived without prior knowledge of the sequence due to the differences between the fragments generated (sequence ladder). In this scenario, the first fragment of the nucleic acid molecule, representing the terminal 5′-nucleotide, can be predicted and verified, and the incremental differences of the subsequent fragments reveal their identity. A target mass list that can be used for de novo sequencing of Spiegelmers using the aforementioned biotin method is depicted in Table 3. When employing this de novo method, the incremental difference between fragments is derived using the calculated mass of the (elucidated) previous fragment. This prevents accumulations of experimental errors (e.g. −0.2-Da error followed by a +0.2-Da error) that may exceed the threshold for unambiguous discrimination between a ribo-C and ribo-U nucleoside, whose masses differ by just 1 Da. Employing either data processing method readily confirmed the sequence of NOX-E36 within the required accuracy range.
Table 2.
Sequence confirmation of NOX-E36 Int
| Fragmenta/RT (min) | Structureb | Observed (Da) | Calculated (Da) |
|---|---|---|---|
| 1/3.57 | R-Gcp | 612.12 | 612.12 |
| 2/4.85 | R-GCcp | 917.16 | 917.16 |
| 3/8.65 | R-GCAcp | 1246.21 | 1246.21 |
| 4/11.49 | R-GCACcp | 1551.25 | 1551.25 |
| 5/14.53 | R-GCACGcp | 1896.30 | 1896.30 |
| 6/16.99 | R-GCACGUcp | 2202.32 | 2202.32 |
| 7/18.91 | R-GCACGUCcp | 2507.36 | 2507.37 |
| 8/20.53 | R-GCACGUCCcp | 2812.40 | 2812.41 |
| 9/21.88 | R-GCACGUCCCcp | 3117.44 | 3117.45 |
| 10/23.07 | R-GCACGUCCCUcp | 3423.46 | 3423.47 |
| 11/24.08 | R-GCACGUCCCUCcp | 3728.51 | 3728.52 |
| 12/25.50 | R-GCACGUCCCUCAcp | 4057.55 | 4057.57 |
| 13/26.13 | R-GCACGUCCCUCACcp | 4362.60 | 4362.61 |
| 14/26.81 | R-GCACGUCCCUCACCcp | 4667.64 | 4667.65 |
| 15/27.73 | R-GCACGUCCCUCACCGcp | 5012.68 | 5012.70 |
| 16/28.67 | R-GCACGUCCCUCACCGGcp | 5357.72 | 5357.75 |
| 17/29.52 | R-GCACGUCCCUCACCGGUcp | 5663.75 | 5663.77 |
| 18/30.53 | R-GCACGUCCCUCACCGGUGcp | 6008.81 | 6008.82 |
| 19/31.45 | R-GCACGUCCCUCACCGGUGCcp | 6313.85 | 6313.86 |
| 20/33.25 | R-GCACGUCCCUCACCGGUGCAcp | 6642.90 | 6642.91 |
| 21/34.91 | R-GCACGUCCCUCACCGGUGCAAcp | 6971.96 | 6971.96 |
| 22/35.84 | R-GCACGUCCCUCACCGGUGCAAGcp | 7317.01 | 7317.01 |
| 23/36.72 | R-GCACGUCCCUCACCGGUGCAAGUcp | 7623.05 | 7623.04 |
| 24/37.79 | R-GCACGUCCCUCACCGGUGCAAGUGcp | 7968.13 | 7968.08 |
| 25/39.53 | R-GCACGUCCCUCACCGGUGCAAGUGAcp | 8297.15 | 8297.14 |
| 26/41.13 | R-GCACGUCCCUCACCGGUGCAAGUGAAcp | 8626.19 | 8626.19 |
| 27/41.81 | R-GCACGUCCCUCACCGGUGCAAGUGAAGcp | 8971.29 | 8971.24 |
| 28/42.51 | R-GCACGUCCCUCACCGGUGCAAGUGAAGCcp | 9276.31 | 9276.28 |
| 29/43.26 | R-GCACGUCCCUCACCGGUGCAAGUGAAGCCcp | 9581.24 | 9581.32 |
| 30*/44.18 | R-GCACGUCCCUCACCGGUGCAAGUGAAGCCGcp | 9931.04 | 9931.02 |
| 31*/44.89 | R-GCACGUCCCUCACCGGUGCAAGUGAAGCCGUcp | 10237.26 | 10237.19 |
| 32*/45.69 | R-GCACGUCCCUCACCGGUGCAAGUGAAGCCGUGcp | 10582.54 | 10582.40 |
| 33*/46.43 | R-GCACGUCCCUCACCGGUGCAAGUGAAGCCGUGGcp | 10927.70 | 10927.60 |
| 34*/47.05 | R-GCACGUCCCUCACCGGUGCAAGUGAAGCCGUGGCcp | 11232.86 | 11232.78 |
| 35*/47.65 | R-GCACGUCCCUCACCGGUGCAAGUGAAGCCGUGGCUcp | 11538.97 | 11538.95 |
| 36*/48.23 | R-GCACGUCCCUCACCGGUGCAAGUGAAGCCGUGGCUCcp | 11844.12 | 11844.13 |
| 37*/48.71 | R-GCACGUCCCUCACCGGUGCAAGUGAAGCCGUGGCUCUcp | 12150.36 | 12150.29 |
| 38*/49.06 | R-GCACGUCCCUCACCGGUGCAAGUGAAGCCGUGGCUCUGcp | 12495.72 | 12495.50 |
| 39*/49.79 | R-GCACGUCCCUCACCGGUGCAAGUGAAGCCGUGGCUCUGCcp | 12800.76 | 12800.68 |
| FLP*/49.06 | R-GCACGUCCCUCACCGGUGCAAGUGAAGCCGUGGCUCUGCG | 13083.85 | 13083.92 |
This table shows the retention time, structure and deconvoluted observable masses (exact or average) obtained from the TIC along with the corresponding calculated values of the expected fragments.
aFragment number (in bold) corresponds to fragment size according to the sequence.
bStructure and sequence of fragments: R = HS-(CH2)2C(O)-NH(CH2)6-OP(O)(OH)-, cp in subscript annotation denotes 2′3′–cyclic phosphate, l-RNA sequence of Spiegelmer is in bold letters; FLP = Full Length (undegraded) Product.
All masses observed are exact isotopic masses unless indicated (*) whereby the observed mass is the corresponding average molecular mass.
Calculated masses are switched to average molecular weight accordingly.
Table 3.
Target Mass list for de novo sequencing using the immobilization method exemplified in Figure 1
| Nucleoside | Fragment 1 |
Δ mass (Fa+1 – Fa) |
Δ mass (FLP – Fp) |
|||
|---|---|---|---|---|---|---|
| Exact | Average | Exact | Average | Exact | Average | |
| A | 596.12 | 596.49 | 329.05 | 329.21 | 267.10 | 267.24 |
| C | 572.11 | 572.46 | 305.04 | 305.18 | 243.09 | 243.22 |
| G | 612.12 | 612.49 | 345.05 | 345.21 | 283.09 | 283.24 |
| U | 573.09 | 573.45 | 306.03 | 306.17 | 244.07 | 244.20 |
All values are in daltons.
Fragment 1 is the smallest 5′–fragment, masses displayed are those of the cyclic phosphate derivative.
‘Δ mass (Fa+1 – Fa)’ denotes the mass difference between sequential 2′3′–cyclic phosphate containing fragments i.e. where ‘a+1’ is not the Full Length (undegraded) Product (FLP).
‘Δ mass (FLP – Fp)’ denotes the mass difference attributable to the 3′–terminal nucleoside.
To demonstrate the selectivity of the method, and the ability to sequence de novo, an amino-modified NOX-E36 derivative that has a UC and a CU switch in the NOX-E36 sequence (underlined, Table 1), NOX-E36 Int. mutant, was synthesized. Due to the 1-Da difference, the cytosine/uridine switch is the most challenging to detect and therefore such a switch was placed at both ends of the molecule. This derivative was processed in an identical manner to NOX-E36 Int as previously described. Figure 4 shows the corresponding TIC displaying the fragments. Masses were obtained from all fragments, and using the ‘Target Mass list’ (Table 3), Fragment 1 and hence the first nucleotide was readily identified within the allowed error margin. As depicted in Table 4, the calculated value of this now elucidated fragment is used as a reference for the next fragment. Hence, the identity of the second nucleotide is obtained by subtracting the observed mass of Fragment 2 from the calculated mass of the previous elucidated fragment, Fragment 1. In an iterative process, once the identity of the second nucleotide has been elucidated, the theoretical mass of this fragment is calculated and used for the identification of the third fragment. This process continues up to the last nucleotide, whereby the mass difference here amounts to the value of the nucleoside itself due to a lack of 3′-cyclic phosphate (see Table 3). As can be seen in Table 4, following a de-novo sequencing protocol, both switches were readily identified.
Figure 4.
TIC obtained from NOX-E36 Int. mutant (Table 1) after sequencing protocol (from labelling to analysis) has been carried out.
Table 4.
De novo sequencing of ‘NOX-E36 Int. mutant’ (Table 1) readily reveals both CU mutations compared to the parent NOX-E36 Int. Spiegelmer
| Fragment # | RT in TICa (min) | Type of Observable mass | Deconvoluted Observed mass (Da) | ΔMassb (Da) | ΔMass identified asc | Calculated massd |
Absolute errore (Da) | |
|---|---|---|---|---|---|---|---|---|
| Exact (Da) | Average (Da) | |||||||
| 1 | 3.87 | Exact | 612.12 | 612.12 | G | 612.12 | – | 0.00 |
| 2 | 5.17 | Exact | 917.16 | 305.04 | C | 917.16 | – | 0.00 |
| 3 | 9.02 | Exact | 1246.21 | 329.05 | A | 1246.21 | – | 0.00 |
| 4 | 11.84 | Exact | 1551.25 | 305.04 | C | 1551.25 | – | 0.00 |
| 5 | 14.84 | Exact | 1896.30 | 345.05 | G | 1896.30 | – | 0.00 |
| 6 | 17.25 | Exact | 2202.32 | 306.02 | U | 2202.32 | – | 0.00 |
| 7 | 19.14 | Exact | 2507.36 | 305.05 | C | 2507.37 | – | 0.01 |
| 8 | 20.73 | Exact | 2812.40 | 305.03 | C | 2812.41 | – | –0.01 |
| 9 | 22.08 | Exact | 3117.44 | 305.03 | C | 3117.45 | – | –0.01 |
| 10 | 23.21 | Exact | 3422.48 | 305.03 | C | 3422.49 | – | –0.01 |
| 11 | 24.26 | Exact | 3728.51 | 306.02 | U | 3728.52 | – | −0.01 |
| 12 | 25.65 | Exact | 4057.56 | 329.04 | A | 4057.57 | – | –0.01 |
| 13 | 26.29 | Exact | 4362.60 | 305.03 | C | 4362.61 | – | –0.01 |
| 14 | 26.98 | Exact | 4667.64 | 305.03 | C | 4667.65 | – | –0.01 |
| 15 | 27.87 | Exact | 5012.68 | 345.03 | G | 5012.70 | – | –0.02 |
| 16 | 28.76 | Exact | 5357.74 | 345.04 | G | 5357.75 | – | –0.01 |
| 17 | 29.61 | Exact | 5663.75 | 306.00 | U | 5663.77 | – | –0.02 |
| 18 | 30.59 | Exact | 6008.80 | 345.03 | G | 6008.82 | – | –0.02 |
| 19 | 31.47 | Exact | 6313.86 | 305.04 | C | 6313.86 | – | 0.00 |
| 20 | 33.23 | Exact | 6642.92 | 329.06 | A | 6642.91 | – | +0.01 |
| 21 | 34.84 | Exact | 6971.96 | 329.05 | A | 6971.96 | – | 0.00 |
| 22 | 35.73 | Exact | 7316.99 | 345.03 | G | 7317.01 | – | –0.02 |
| 23 | 36.58 | Exact | 7623.02 | 306.01 | U | 7623.04 | – | –0.02 |
| 24 | 37.62 | Exact | 7968.07 | 345.03 | G | 7968.08 | – | –0.01 |
| 25 | 39.32 | Exact | 8297.13 | 329.05 | A | 8297.14 | – | –0.01 |
| 26 | 40.88 | Exact | 8626.23 | 329.09 | A | 8626.19 | – | +0.04 |
| 27 | 41.56 | Exact | 8971.30 | 345.11 | G | 8971.24 | – | +0.06 |
| 28 | 42.22 | Exact | 9276.28 | 305.04 | C | 9276.28 | – | 0.00 |
| 29 | 42.95 | Exact | 9581.26 | 304.98 | C | 9581.32 | – | –0.06 |
| 30 | 43.84 | Exact | 9926.35 | 345.03 | G | 9926.37 | 9931.02 | –0.02 |
| 31 | 44.54 | Average | 10237.26 | 306.24 | U | – | 10237.19 | +0.07 |
| 32 | 45.31 | Average | 10582.53 | 345.34 | G | – | 10582.40 | +0.13 |
| 33 | 46.09 | Average | 10927.69 | 345.29 | G | – | 10927.60 | +0.09 |
| 34 | 46.66 | Average | 11233.75 | 306.15 | U | – | 11233.77 | –0.02 |
| 35 | 47.21 | Average | 11538.95 | 305.18 | C | – | 11538.95 | 0.00 |
| 36 | 47.76 | Average | 11844.25 | 305.30 | C | – | 11844.13 | +0.12 |
| 37 | 48.28 | Average | 12150.34 | 306.21 | U | – | 12150.29 | +0.05 |
| 38 | 48.58* | Average | 12495.64 | 345.35 | G | – | 12495.50 | +0.14 |
| 39 | 49.26** | Average | 12800.80 | 305.30 | C | – | 12800.68 | +0.12 |
| FLP | 48.58 | Average | 13084.04 | 283.36 | G | – | 13083.92 | +0.12 |
aRetention time values are those taken from the TIC.
bΔ mass is calculated by subtracting the calculated mass value of the previous fragment from the observed value.
cSee Table 3 for cipher.
dCalculated mass values are obtained by calculating the theoretical mass of the confirmed sequence.
eAbsolute error is calculated by subtracting the target Δ mass value (Table 3) from the Δ mass value obtained.
*Fragment 38 co-elutes with the full-length (undegraded) released product (FLP). **Fragment 39 has a retention time greater than the FLP, which in this case is HS-(CH2)2C(O)-NH(CH2)6-OP(O)(OH)-GCACGUCCCCUACCGGUGCAAGUGAAGCCGUGGUCCUGCG.
The C/U switches compared to ‘NOX-E36 Int.’ (Table 1) are highlighted (bold).
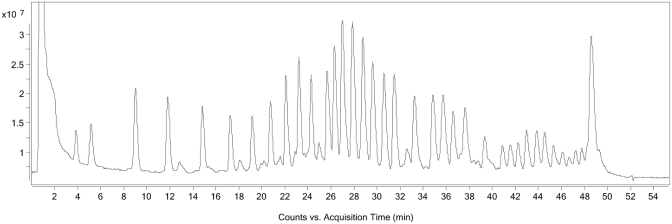
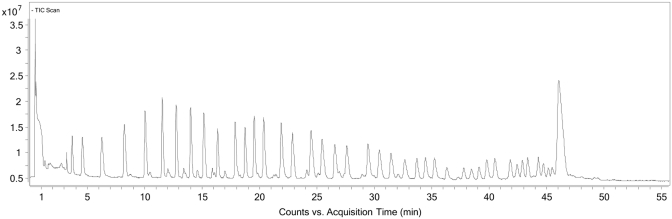
The method was applied to another l-RNA oligonucleotide, Spiegelmer NOX-A12 Int (for sequence, see Table 1). Here, the protocol needed to be altered slightly to accommodate the new sequence. Firstly, it was found that NOX-A12 required a longer fragmentation time (20 min. versus 12.5 min. for NOX-E36). Secondly, the bead-washing protocol after immobilization of the biotin-labelled fragments also needed to be altered as it was found that washing the beads with water and subsequent fragment release did not remove all unlabelled fragments. This phenomenon was probably caused by self-aggregation of the molecule whereby non-labelled fragments aggregated with labelled immobilized fragments and were therefore not removed by the washing step (Figure 5). By using a chaotrope, in this case 8 M urea aqueous solution, to disrupt this hydrogen bonding, the non-labelled fragments could be successfully removed (Figure 6). A sequencing ladder and subsequent elucidation were readily obtained (see Supplementary Table 1).
Figure 5.
TIC of NOX-A12 sequencing using NOX-E36 washing protocol reveals non-labelled fragments have been co-immobilized with the labelled fragments.
Figure 6.
TIC of NOX-A12 sequencing using improved chaotropic washing protocol with 8 M urea. Chromatogram reveals non-labelled fragments that in previous protocol were bound to the immobilized 5′–fragments and therefore co-isolated have been successfully washed away.
It occurred to us that the presumption that physical isolation of a set of fragments (e.g. via immobilization) is a critical element for a successful sequencing process could be challenged. The origins of this presumption lies in the knowledge that all fragments, be they 5′-, 3′- or internal fragments, provide mass data, which would confound the generation of a sequencing ladder. However, by attaching a label to the NOX-E36 Int. such as a fluorescein dye label, 5′-fragments could be resolved by using an alternative detection wavelength. At this wavelength, a ladder depicting all possible 5′-fragments can be selectively observed and mass data at the corresponding retention times in the (TIC) can be generated.
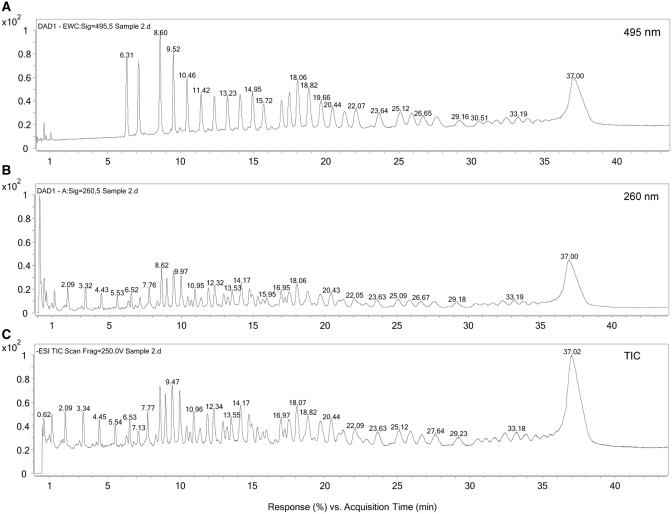
To demonstrate the feasibility of this method, a fluorescein-5-isothiocyanate (FITC Isomer I) label was attached to NOX-E36 Int. The FITC-labelled NOX-E36 Int. was then subjected to controlled fragmentation and the sample analysed by LC-MS. The label has a selective wavelength absorbance at 495 nm. Therefore, at 495 nm, only nucleic acid molecules containing an intact 5′-end (5′-fragments of the nucleic acid molecule and the intact-labelled NOX-E36 Int.) will be observed. As can be seen from Figure 7A, the chromatogram looks very similar to that observed from the biotin-immobilization method (Figures 2, 4 and 6). However, comparing this UV chromatogram extracted at 495 nm with the corresponding UV chromatogram extracted at 260 nm (Figure 7B) it can be clearly seen that there are many other fragments present. Moreover, a comparison of the UV chromatogram extracted at 260 nm with the TIC (Figure 7C) shows that the two chromatograms are very similar, thus all fragments either with or without label generate mass data.
Figure 7.
LC-MS analysis of FITC-labelled NOX-E36 Int. after controlled fragmentation. (A) UV chromatogram extracted at 495 nm revealing just 5′–fragments; (B) UV chromatogram extracted at 260 nm revealing all nucleic acid fragments; (C) TIC showing all ions present.
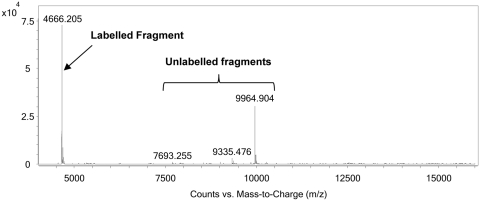
As determined through the chromatogram extracted at 495 nm, the first 5′-fragment [representing in this example FITC-NH-(CH2)6-OP(O)(OH)-Gcp where cp is 2′,3′-cyclic phosphate] can be readily identified to be that eluting at 6.31 min (Figure 7A and enlargement Figure 8). It can also be clearly seen by comparing the TIC and the extracted 495 nm chromatogram that there are many non-labelled fragments that elute earlier than this peak; however, these represent fragments of between 8 and 14 nt in length as estimated according to their observed masses. Therefore, due to the lipophilicity afforded to the labelled fragments, any non-labelled fragment that co-elutes with the labelled fragments can be disregarded due to the significant difference in mass to that expected for a particular fragment size (>2 kDa). This is clearly illustrated in Figure 9, where the extracted wavelength of 495 nm represents the FITC-labelled 5′-fragments; the extracted wavelength of 260 nm represents all nucleic acid fragments, as does the TIC. By obtaining a mass spectrum for the ions within the range marked by the two dotted lines in Figure 9 that represent a typical ion window for obtaining the mass spectrum of the labelled fragment (Figure 10), more than one peak is obtained upon deconvolution. As the peaks vary greatly in mass value, only one is feasible as coming from the labelled fragment, in this case the average molecular mass deconvoluted value of 4666 Da. This ability to discount spurious masses enables the generation of the sequencing ladder required for Spiegelmer identification, and can therefore be also used for de novo sequencing (see Supplementary Tables 2 and 3).
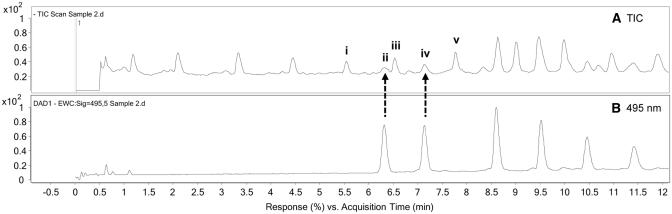
Figure 8.
Zoomed view of Figure 7 showing the location of the first two labelled fragments in the TIC (A), as revealed in the extracted wavelength chromatogram at 495 nm (B), as highlighted with the dotted-line arrows. Fragments i, iii, and v with deconvoluted isotopic masses of 4106.55, 4451.60 and 4780.64 Da, respectively, have molecular masses far greater than those expected for the first two labelled fragments with similar retention time ii and iv, which have the expected values of 913.15 and 1218.19 Da, respectively.
Figure 9.
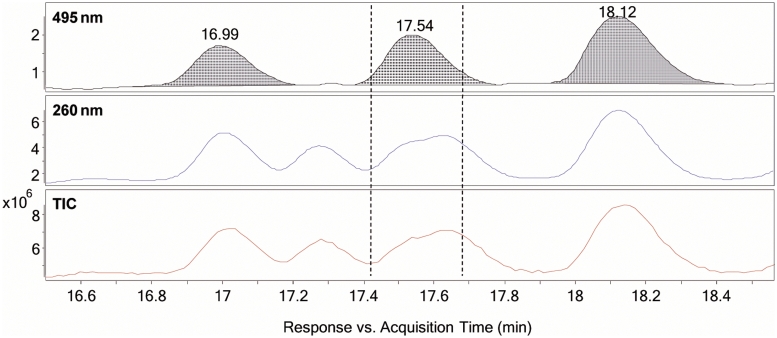
Zoomed view of Figure 7 (16.6–18.5 min) illustrating FITC-labelled and non-FITC-labelled fragments co-eluting. The extracted wavelength chromatogram at 495 nm shows FITC-labelled nucleic acid fragments, the extracted wavelength chromatogram at 260 nm indicates all nucleic acid fragments (labelled and non-labelled) and the TIC displays all ions in the sample material. It is clear to see that in the marked area between the dotted lines there are other species present that produce ions alongside the labelled fragment with the retention time of 17.54 min.
Figure 10.
Deconvoluted mass spectrum of the area in the TIC in Figure 9 between the dotted lines. Out of the masses observed, only one peak has a mass value that would be plausible for the following fragment in the sequencing ladder (Δ range: 305–345 Da). Nonsensical masses can therefore be identified as non-labelled fragments.
CONCLUSION
In conclusion, we have developed effective and robust methods for the elucidation of mirror-image RNA-oligonucleotide (Spiegelmer) primary structures. The methods employed take advantage of the amino-handle pre-installed on the Spiegelmer intermediate by attaching either an affinity tag or a dye label to it. This mild procedure can be done on purified and also crude material if required, and can be used for sequencing after a rudimentary desalting procedure. Chemical fragmentation of the labelled molecule under mild basic conditions serves to reduce the chance of artefactual fragments being produced, and either isolating or resolving the labelled fragments creates the sequencing ladder needed for structure identification. In the case of immobilization, it was found that washing immobilized fragments with a chaotropic solution served to remove co-isolated unlabelled fragments, thus suppressing contamination arising from this phenomenon, the extent of which was sequence dependent.
The methods described herein were proven to meet selectivity and accuracy requirements as shown by successfully testing the method on a mutant (isomeric) NOX-E36 Int. derivative with two CU switches compared to the parent compound. Consequently, the method can be readily applied to the de novo sequencing of mirror-image RNA-oligonucleotides. It should also be noted that the ability to generate all desired fragments in one step and for those fragments to be resolved in one LC-MS measurement serves to enhance the robustness and convenience of the method, as does the avoidance of MS/MS experiments and their subsequent interpretation. Furthermore, the method is also applicable to other RNA molecules with the ability to install reactive handles and linkers onto unmodified nucleic acids (23), making the technique more widely applicable.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: NOXXON Pharma AG. Max-Dohrn-Str. 8–10, D-10589 Berlin, Germany.
Conflict of interest statement. All authors are employed by NOXXON Pharma AG.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Christian Mihm for the preparation of some figures.
REFERENCES
- 1.Eckstein F, Krieg AM, Stein CA, Agrawal S, Beaucage S, Cook PD, Crooke S, Gait MJ, Gewirtz A, Helene C, et al. On the quality control of antisense oligonucleotides [editorial] Antisense Nucleic Acid Drug Dev. 1996;6:149. doi: 10.1089/oli.1.1996.6.149. [DOI] [PubMed] [Google Scholar]
- 2.Srivasta GS. In: Handbook of Analysis of Oligonucleotides and Related Products. Bonilla JV, Srivasta GS, editors. Boca Raton, FL: CRC Press; 2011. pp. 465–486. [Google Scholar]
- 3.Timar Z. In: Handbook of Analysis of Oligonucleotides and Related Products. Bonilla JV, Srivasta GS, editors. Boca Raton, FL: CRC Press; 2011. pp. 167–218. [Google Scholar]
- 4.Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, Hartsough K, Machemer L, Radka S, Jadhav V, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 5.Heidenreich O, Pieken W, Eckstein F. Chemically modified RNA: approaches and applications. FASEB J. 1993;7:90–96. doi: 10.1096/fasebj.7.1.7678566. [DOI] [PubMed] [Google Scholar]
- 6.Kawasaki AM, Casper MD, Freier SM, Lesnik EA, Zounes MC, Cummins LL, Gonzalez C, Cook PD. Uniformly modified 2′-deoxy-2′-fluoro phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high affinity and specificity for RNA targets. J. Med. Chem. 1993;36:831–841. doi: 10.1021/jm00059a007. [DOI] [PubMed] [Google Scholar]
- 7.Shaw JP, Kent K, Bird J, Fishback J, Froehler B. Modified deoxyoligonucleotides stable to exonuclease degradation in serum. Nucleic Acids Res. 1991;19:747–750. doi: 10.1093/nar/19.4.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank R, Koster H. DNA chain length markers and the influence of base composition on electrophoretic mobility of oligodeoxyribonucleotides in polyacrylamide-gels. Nucleic Acids Res. 1979;6:2069–2087. doi: 10.1093/nar/6.6.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirpekar F, Nordhoff E, Larsen LK, Kristiansen K, Roepstorff P, Hillenkamp F. DNA sequence analysis by MALDI mass spectrometry. Nucleic Acids Res. 1998;26:2554–2559. doi: 10.1093/nar/26.11.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kramer FR, Mills DR. RNA sequencing with radioactive chain-terminating ribonucleotides. Proc. Natl Acad. Sci. USA. 1978;75:5334–5338. doi: 10.1073/pnas.75.11.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li S, Haces A, Stupar L, Gebeyehu G, Pless RC. Elimination of band compression in sequencing gels by the use of N4-methyl-2′-deoxycytidine 5′-triphosphate. Nucleic Acids Res. 1993;21:2709–2714. doi: 10.1093/nar/21.11.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taucher M, Rieder U, Breuker K. Minimizing base loss and internal fragmentation in collisionally activated dissociation of multiply deprotonated RNA. J. Am. Soc. Mass Spectrom. 2010;21:278–285. doi: 10.1016/j.jasms.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 13.Eulberg D, Jarosch F, Vonhoff S, Klussmann S. In: The Aptamer Handbook, Functional Oligonucleotides and Their Applications. Klussmann S, editor. Weinheim: Wiley-VCH; 2006. pp. 417–439. [Google Scholar]
- 14.Eulberg D, Klussmann S. Spiegelmers: biostable aptamers. Chembiochem. 2003;4:979–983. doi: 10.1002/cbic.200300663. [DOI] [PubMed] [Google Scholar]
- 15.Klussmann S, Nolte A, Bald R, Erdmann VA, Furste JP. Mirror-image RNA that binds D-adenosine. Nat. Biotechnol. 1996;14:1112–1115. doi: 10.1038/nbt0996-1112. [DOI] [PubMed] [Google Scholar]
- 16.Damha MJ, Ogilvie KK. Oligoribonucleotide synthesis. The silyl-phosphoramidite method. Methods Mol. Biol. 1993;20:81–114. doi: 10.1385/0-89603-281-7:81. [DOI] [PubMed] [Google Scholar]
- 17.Wincott F, DiRenzo A, Shaffer C, Grimm S, Tracz D, Workman C, Sweedler D, Gonzalez C, Scaringe S, Usman N. Synthesis, deprotection, analysis and purification of RNA and ribozymes. Nucleic Acids Res. 1995;23:2677–2684. doi: 10.1093/nar/23.14.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moyroud E, Biala E, Strazewski P. Synthesis and Enzymatic Digestion of an RNA nonamer in both enantiomeric forms. Tetrahedron. 2000;56:1475–1484. [Google Scholar]
- 19.Tolson DA, Nicholson NH. Sequencing RNA by a combination of exonuclease digestion and uridine specific chemical cleavage using MALDI-TOF. Nucleic Acids Res. 1998;26:446–451. doi: 10.1093/nar/26.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peattie DA. Direct chemical method for sequencing RNA. Proc. Natl Acad. Sci. USA. 1979;76:1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farand J, Beverly M. Sequence confirmation of modified oligonucleotides using chemical degradation, electrospray ionization, time-of-flight, and tandem mass spectrometry. Anal. Chem. 2008;80:7414–7421. doi: 10.1021/ac8011158. [DOI] [PubMed] [Google Scholar]
- 22.Farand J, Gosselin F. De novo sequence determination of modified oligonucleotides. Anal. Chem. 2009;81:3723–3730. doi: 10.1021/ac802452p. [DOI] [PubMed] [Google Scholar]
- 23.Proudnikov D, Mirzabekov A. Chemical methods of DNA and RNA fluorescent labeling. Nucleic Acids Res. 1996;24:4535–4542. doi: 10.1093/nar/24.22.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.