Abstract
Fungal pathogens of plants and insects infect their hosts by direct penetration of the cuticle. Plant and insect cuticles are covered by a hydrocarbon-rich waxy outer layer that represents the first barrier against infection. However, the fungal genes that underlie insect waxy layer degradation have received little attention. Here we characterize the single cytochrome P450 monoxygenase family 52 (MrCYP52) gene of the insect pathogen Metarhizium robertsii, and demonstrate that it encodes an enzyme required for efficient utilization of host hydrocarbons. Expressing a green florescent protein gene under control of the MrCYP52 promoter confirmed that MrCYP52 is up regulated on insect cuticle as well as by artificial media containing decane (C10), extracted cuticle hydrocarbons, and to a lesser extent long chain alkanes. Disrupting MrCYP52 resulted in reduced growth on epicuticular hydrocarbons and delayed developmental processes on insect cuticle, including germination and production of appressoria (infection structures). Extraction of alkanes from cuticle prevented induction of MrCYP52 and reduced growth. Insect bioassays against caterpillars (Galleria mellonella) confirmed that disruption of MrCYP52 significantly reduces virulence. However, MrCYP52 was dispensable for normal germination and appressorial formation in vitro when the fungus was supplied with nitrogenous nutrients. We conclude therefore that MrCYP52 mediates degradation of epicuticular hydrocarbons and these are an important nutrient source, but not a source of chemical signals that trigger infection processes.
Introduction
Although hydrocarbon degrading enzymes are generally associated with oil-polluted environments, alkanes are persistent molecules and this makes them useful components for natural barriers. Thus, both plant and insect cuticles are covered by a waxy epicuticular layer comprising a mixture of long chain alkanes and related chemicals that represent the first barrier to environmental threats [1]. Most plant and insect diseases are caused by fungi that infect their hosts by direct penetration of the cuticle [2], but the fungal genes responsible for waxy layer degradation remain almost completely unexplored. In the rice blast fungus Magnaporthe oryzae, a putative alkane degrading cytochrome P450 is up-regulated upon the first stages of infection [3], which implies that alkane degradation is required for breaching host defenses and/or to access nutrients.
The entomopathogens Metarhizium robertsii and Beauveria bassiana infect a broad array of insects and hence can be exploited as biological control agents of pests. They are able to grow on straight chain and branched hydrocarbons as sole carbon and energy source [4]–[6], and the specific locust pathogen Metarhizium acridum extensively hydrolyzes surface lipids during germination and pre-penetration growth on locust cuticles [7]. Pedrini et al [6] postulated that the cytochrome P450 monoxygenases (CYP) are involved in alkane and insect epicuticle degradation, and they showed that Beauveria bassiana differentially expressed CYP genes when grown on different hydrocarbons. The versatile CYP52 family (membrane bound class II) contains several enzymes with demonstrated activity towards alkanes and/or fatty acids [1]. Although the genome of M. robertsii encodes 123 highly divergent CYP genes [8], M. robertsii has only a single CYP52 (MAA_06634) compared to four in M. acridum [8].
This study focuses on M. robertsii strain ARSEF 2575 (Mr2575). Mr2575 is the type strain of M. robertsii, and is frequently employed as a model for studies on host pathogen interactions and genetic engineering. Infection by Mr2575 proceeds via spores that adhere to the host surface (epicuticle) and germinate to form germ tubes that continue undifferentiated hyphal growth if nutrient quality and quantity is not conducive to differentiation. On a host, however, apical elongation terminates and germ tubes produce infection structures, called appressoria, which promote the localized production of cuticle degrading enzymes [9]–[11]. Lipids are the main nutrient reserve in fungal spores [12]. Lipid bodies are transported to the developing appressoria and degraded to release glycerol which increases hydrostatic pressure and provides a driving force for mechanical penetration [11], [13]. Despite their intracellular lipid reserves, Mr2575 spores are nutrient deficient, as they also need extracellular nutrients in order to germinate [9].
In this study we show for the first time that a specific hydrocarbon utilizing enzyme is a virulence factor in an entomopathogenic fungus. The M. robertsii CYP52 (MrCYP52) increases differentiation of appressoria on grasshopper cuticle, and is required for full pathogenicity, but is expendable for appressorial formation in vitro when the fungus is supplied with nitrogenous nutrients. The implication is that epicuticular hydrocarbons are an important nutrient source, but not a source of chemical signals that trigger infection processes.
Materials and Methods
Fungal and bacterial strains
M. robertsii wild-type strain ARSEF2575 was obtained from the USDA/ARS Collection of Entomopathogenic Fungal Cultures, Ithaca, NY. Escherichia coli DH5α was employed for DNA manipulation. Agrobacterium tumefaciens AGL-1 was used for M. robertsii transformation.
Gene cloning and disruption
We modified the master Ti vector pFBARGFP used for gene disruption in M. robertsii [13], by removing the EcoR I site. The new bar cassette without the lox-NotI-lox site was obtained by PCR using the primers Bar5 and Bar3 (Table S1). The primers were designed to include unique Xba I/EcoR I and Spe I/EcoR V sites at the 5′-end and 3′-end of the bar gene. The resultant fragment was digested with Xba I and EcoR V, and cloned into pFBARGFP to produce pPK2BarGFPD. To construct the vector for MrCYP52 disruption, the 5′ end and 3′ end of MrCYP52 were cloned by PCR and inserted into the EcoR I and Spe I sites, respectively, of pPK2BarGFPD. The disruption mutant (ΔMrCYP52) was obtained utilizing Agrobacterium tumefaciens AGL-1 as described [14]. To complement ΔMrCYP52, the genomic sequence of MrCYP52 including its promoter (2215 bp) and terminator (427 bp) was cloned, inserted into the EcoR V site of pPK2SurGFPD [15], and transformed into ΔMrCYP52. Successful disruptant and complementation of strains were confirmed by PCR and Southern blotting (Figure S1). To overexpress of MrCYP52, the ORF of the MrCYP52 gene was amplified by reverse transcription (RT)-PCR with primers MrCYP52_ORF_F and MrCYP52_ORF_R. The MrCYP52 ORF was digested with Spe I and EcoR I and ligated into pBARGPE1 downstream of the constitutive gpd promoter, following which the expression cassette was excised with Not I and Nde I, blunt-end digested with T4 DNA polymerase (New England Biolabs, Pickering, Ontario, Canada), and inserted into the EocR V site of pPK2BarGFPD. The resulting plasmid (pPK2BarGFPD- MrCYP52) was transformed into wild type M. robertsii. The primers used in this study were listed in Table S1. All PCR products in this study were cloned into the pGEM-T vector (Promega) and sequenced for confirmation.
Southern blotting
Southern blotting was performed with 20 µg of DNA for each sample. Genomic DNA was extracted as described previously [16] and digested with EcoR V. Agarose gel electrophoresis and DNA transfers were performed as described [17]. The fragment used as the probe was generated using primers MrCYP52_UR and MrCYP52_CF (Table S1). Probe preparation, membrane hybridization and visualization were according to the manufacturer's instructions (DIG High Prime DNA Labeling and Detection Starter Kit II, Roach).
Insect bioassay
M. robertsii was bioassayed using Galleria mellonella larvae from Big Apple Herpetological (Hauppauge, NY) as described [13]. Briefly, insects were inoculated by a 10 second immersion in conidial suspensions (1×107 conidial ml−1). All bioassays were repeated three times with 36 insects per replicate. Mortality was recorded every day.
Utilization of different alkanes
Germination rates of the wild type Mr2575 were compared with ΔMrCYP52 in basal medium (BS) (0.1% KH2PO4, 0.025% Na2SO4, 0.05% KCl, 0.0125% MgSO4·7H2O, 0.00625% CaCl2, and 0.3% NaNO3) supplemented with the alkanes listed in Table 1. To obtain insect-derived hydrocarbons, we extracted locust wings using hexane [12], [18]. The extract was dried and the precipitate resuspended in hexane. One hundred microliter hexane extract, the equivalent to 4 locust wings, was evaporated per slide on sterilized glass microscope cavity slides. The slides were inoculated with 40 µl drops of 1×106 spores ml−1 and incubated at 27°C. Germination rates were recorded as described above. Mycelial biomasses were determined by inoculating 1.5×107 spores into 50 ml of autoclaved BS medium supplemented with filter sterilized hydrocarbons. Cultures were shaken (200 rpm) at 27°C and harvested by vacuum filtration on pre-weighed glass-fiber filters five days after the first appearance of visible growth. The filters were washed in water and acetone, and dried to constant weight at 80°C [4].
Table 1. Germination rate of WT and ΔMrCYP52 on 1% alkanes and cuticular hydrocarbons (20 hrs).
| Wild type | ΔMrCYP52 | % reduction in alkane induced growth | |
| BS | 10.7±0.8% | 10.9±1.1% | |
| n-alkanes | |||
| Nonane (C9) | 16.7±0.6% | 15.8±2.3% | 18.3% |
| Decane (C10) | 18.9±0.8% | 12.9±0.8% | 75.6% |
| Dodecane (C12) | 33.3±1.5% | 32.9±1.8% | 2.6% |
| Tridecane (C13) | 33.0±1.3% | 32.6±1.7% | 2.7% |
| Tetradecane (C14) | 18.2±1.6% | 16.3±1.3% | 28% |
| Pentadecane (C15) | 23.3±2.9% | 19.3±0.3% | 33.3% |
| Hexadecane (C16) | 31.9±0.6% | 25.9±1.3% | 28.7% |
| Hexacosane (C26) | 39.4±1.0% | 32.3±1.4% | 25.4% |
| Octacosane (C28) | 50.8±2.1% | 29.2±2.0% | 54.4% |
| Cuticular hydrocarbons | |||
| Hexane extract | 57.7±2.1% | 35.6±1.3% | 47.4% |
| Branched alkanes | |||
| Pristane | 24.4±1.1% | 23.6±2.6% | 7.3% |
| Squalane | 24.9±1.6% | 24.2±1.8% | 6.3% |
Conidial germination and appressorial differentiation
The germination rate of conidia was measured by inoculating 20 µl of spore suspension (2×107 spores ml−1) into 3.5 cm polystyrene petri dishes containing 2 ml of 0.0125% yeast exact (YE). Three hundred spores from each of three replicates were recorded microscopically to assess germination and appressorial differentiation against the hydrophobic surface of the Petri dish. Appressoria were also induced against locust (Schistocerca gregaria) hind wings as described previously [12], [19]. Cuticles were inoculated with 3 µl of conidia suspension (5×106 ml−1) and incubated on 1.5% water agar.
RT-PCR analysis
To monitor the expression of MrCYP52 in different growth conditions, mycelial inoculums from 36 h Sabouraud dextrose broth (SDB) (Difco) cultures [11] were incubated (6 hours) in 50 ml of either water, Sabouraud dextrose broth (SDB), hemolymph or basal medium (BS) containing either 1% glucose, 1% decane or 1% Manduca sexta cuticle. The M. sexta cuticle and hemolymph were prepared as described previously [12]. RNA was extracted from mycelia using a QIAGEN RNase Plant Mini kit, treated with DNase I, and 1 µg was converted into single-stranded cDNA using Verso™ TR-PCR kit (Thermo Scientific).
Expression pattern of MrCYP52in M. robertsii
The expression pattern of MrCYP52 was investigated in transformants expressing GFP driven by the promoter region (2200 bp) of MrCYP52. The promoter region was cloned by PCR using primers MrCYP52_PF and MrCYP52_PR (Table S1), digested with Bgl II and EcoR I and inserted into BamH I/EcoR I sites of pBarGPE1-GFP [20]. The egfp cassette was released by Hpa I and Spe I, blunted with T4 DNA polymerase, and inserted into the EcoR V site of pPK2Bar to form pPMrCYP52:GFP. This Ti plasmid was transformed utilizing A. tumefaciens into the wild type M. robertsii ARSEF2575, producing WT-PMrCYP52: GFP. To study the time course of MrCYP52 expression, 30 µl 2×107 ml−1 spore suspensions of WT-PMrCYP52:GFP were incubated for 10 h in 0.0125% YE and then transferred to BS supplemented with different carbon source (1% liquid alkanes, 1% glucose or 1% glycerol). For solid alkanes, 40 µl 1% long chain alkane or myristic acid solution in hexane was pipette onto glass coverslips and evaporated, leaving a white greasy layer. The coverslips were then placed in polystyrene petri dishes with the alkane layer facing up, and the dishes were inoculated with 30 µl of 1×107 ml−1 spore suspensions in BS medium. Spores of WT-PMrCYP52:GFP (5×106 ml−1) were also inoculated onto locust wings and to locust wings treated with hexane to remove hydrocarbons. GFP fluorescence was followed microscopically.
Results
Protein characteristics and phylogenetic analysis
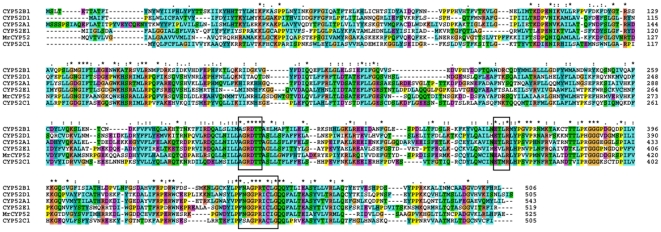
Analyses of the predicted MrCYP52 protein (EFY97851) [8] indicates that it is composed of 525 amino acid residues (59.5 kDa) with a predicted pI of 8.57. According to TMpred analysis (Figure S2), the predicted protein resembles other fungal cytochrome P450s in having a single hydrophobic transmembrane domain at the N-terminus, and it contains the signature pfam00067 Cytochrome P450 conserved domain (Figure 1). A neighbor-joining analysis of 28 fungal CYPs conducted with MEGA4 (Figure S3) showed that MrCYP52 clusters within the alkane hydroxylating CYP52 family, and is similar to CYP52 enzymes from Beauveria bassiana (ADK36660, 59% identity), Candida maltose (AAA34320, 44% identity), and Candida tropicalis (XP_002546279, 45% identity). Homologs (>40% identity) of MrCYP52 were identified in most other Ascomycete fungi, but were absent in the genomes of Basidiomycete, Chytrid and Zygomycete fungi, showing that the gene is expendable in some lifestyles.
Figure 1. Sequence alignment analysis of MrCYP52 with defined CYP52s.
CYP52A1 (AAA34354), CYP52B1 (CAA78357), CYP52C1 (CAA78358) and CYP52D1 (Q12585) from Candida tropicalis, CYP52E1 (P43083) from Candida apicola. The heme-binding regions and other conserved domains discussed in the text are marked.
Alkane utilization assay
To study the function of MrCYP52, MrCYP52 null mutants (ΔMrCYP52) were generated in Mr2575 by homologous replacement. Complementing ΔMrCYP52 with a genomic clone of MrCYP52 produced a strain indistinguishable from the wild type in all the phenotypic assays performed in this study. Consequently, the data for the complemented strains is not presented.
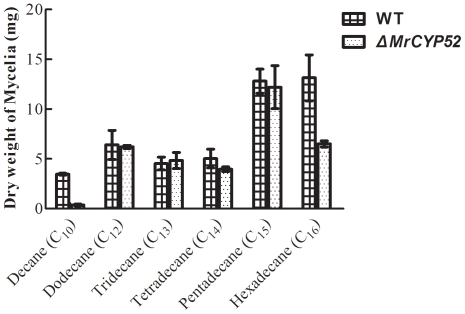
To investigate the role of MrCYP52 in utilizing alkanes, we compared the germination rate of the wild type with that of ΔMrCYP52 in BS medium (containing no carbon source) supplemented with each of the alkanes listed in Table 1. Linear alkanes, C9, C12–C15, and branched alkanes increased germination of wild type and ΔMrCYP52 spores to a similar extent. In contrast, C10 did not significantly increase germination of ΔMrCYP52 above the very low levels seen in BS medium (P>0.05). Germination rates of ΔMrCYP52 on C16, C26, C28 and locust cuticle hexane extract were significantly reduced compared with the wild type (P<0.05). ΔMrCYP52 also grew poorly in liquid cultures containing C10 and C16 (Figure 2), indicating that MrCYP52 participates in mid and long chain alkane utilization.
Figure 2. Growth of M. robertsii wild type and ΔMrCYP52 in basal salts containing 1% alkanes.
Germination behavior and appressorial formation
Pathogenicity assays involving topical application of spores (the normal route of entry) to Galleria mellonella caterpillars revealed that ΔMrCYP52 (LT50 = 8.27±0.32 d) took a significantly (P<0.05) longer time to kill than the wild type M. robertsii 2575 (LT50 = 6.85±0.37 d). Although alkane-growth adaptation of B. bassiana is correlated with increased insect host mortality [21], overproduction of the MrCYP52 gene did not increase the pathogenicity of M. robertsii (LT50 = 6.92±0.18 d). To elucidate whether MrCYP52 is needed for pre-penetration developmental processes, wild type Mr2575 and ΔMrCYP52 strains were grown in 0.0125% yeast extract (YE) which induces differentiation of appressoria by WT Mr2575 [9]. There was no significant difference in sporulation, germination, growth rates and differentiation of appressoria between M. robertsii 2575 and ΔMrCYP52, indicating that MrCYP52 does not facilitate these processes in the presence of low levels of nitrogenous nutrients (Table S2). However, the germination rate of ΔMrCYP52 on locust cuticle was significantly (P<0.001) slower than that of wild type M. robertsii (Table S2), and 6 h post inoculation only 24.6±0.7% of ΔMrCYP52 conidia had germinated as compared to 40.4±0.3% of the wild type Mr2575. Both wild type and ΔMrCYP52 had 100% germination rates 12 h post inoculation. However, 25 hours post inoculation, the wild type Mr2575 had produced ∼2.2-fold more appressoria than ΔMrCYP52. By three days post-inoculation differences were still significant, with 43.5±1.4% of ΔMrCYP52 germlings having produced appressoria as compared to 52.9±1.5% of the wild type. The germination rate of ΔMrCYP52 and the wild type were similarly low on the locust wings treated with hexane to remove cuticular hydrocarbons (Table S2). To determine if addition of exogenous nutrients overcomes the reduced ability of ΔMrCYP52 to infect locust cuticle, we inoculated cuticles with ΔMrCYP52 spores suspended in 0.0125% YE. Exogenous nutrients increased the germination rate of wild type (from 40.4±0.3 to 57.6±1.2%) and ΔMrCYP52 (from 24.6±0.7 to 54.1±2.3%). Interestingly, YE increased differentiation of infection structures by ΔMrCYP52 to the levels shown by the wild type Mr2575 in the absence of YE, but appressorial formation by the wild type was significantly (P<0.05), reduced (by 37.5±1.2%) by additional nutrition.
Expression of MrCYP52
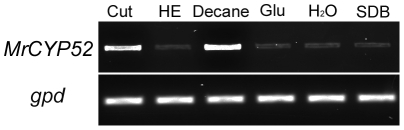
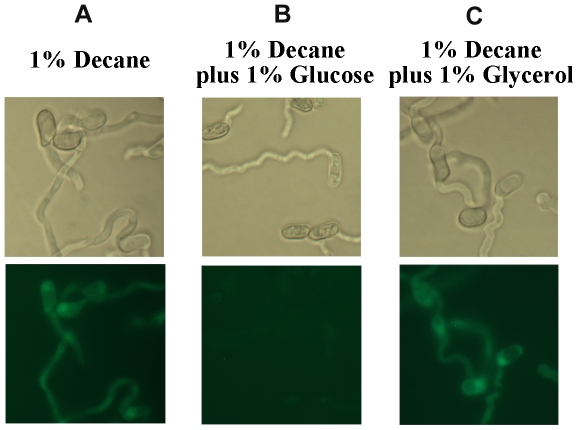
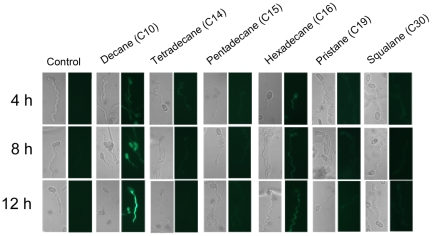
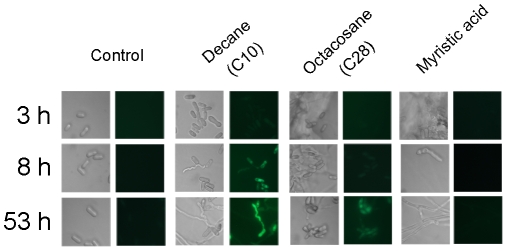
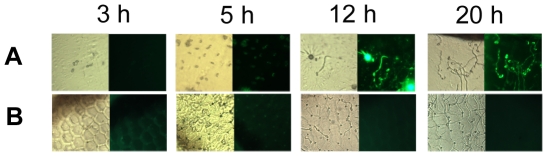
RT-PCR analyses demonstrated that MrCYP52 was expressed in diverse nutrient rich and nutrient poor media (Figure 3). The time course for MrCYP52 expression was also examined by following GFP fluorescence driven by the MrCYP52 promoter in WT-PMrCYP52:GFP. GFP fluorescence was not observed in BS medium without a carbon source. Spores of WT-PMrCYP52:GFP pre-germinated in 0.01% YE fluoresced within 1 h of transfer to BS containing 1% decane or 1% decane plus 1% glycerol. In contrast, GFP fluorescence was not observed in BS containing 1% decane plus 1% glucose (Figure 4), indicating that expression is repressed by glucose. Faint GFP fluorescence was observed within 4 hours when medium-chain alkanes (n-alkanes or branched alkanes) were used as sole carbon source (Figure 5). Spores inoculated directly into BS medium containing C28 as carbon source fluoresced faintly after 8 hours (Figure 6). However, GFP fluorescence could not be observed in BS medium with myristic acid as carbon source (Figure 6), which suggests that MrCYP52 may not have an important role in metabolizing fatty acids. WT-PMrCYP52:GFP germinated at the same rate as the wild type when inoculated onto locust cuticle. GFP fluorescence could be observed in conidia germinating 5 hours post inoculation, and the fluorescence increased in intensity as germlings grew and differentiated appressoria. In contrast, fluorescence could not be observed when WT-PMrCYP52:GFP was inoculated onto locust wings treated with hexane (Figure 7).
Figure 3. Expression of MrCYP52.
RT-PCR analysis of MrCYP52 expression by Wild type M. robertsii transferred from SDB medium to water, SDB, cell free hemolymph (HE), basal medium (BS) containing 1% Manduca larval cuticle (Cut), 1% decane or 1% glucose (MM) for 6 h.
Figure 4. Induction of WT-PMrCYP52:GFP incubated for one hour.
(A) 1% decane. (B) 1% decane supplemented with 1% glucose. (C) 1% decane supplemented 1% glycerol.
Figure 5. Time course of induction of WT-PMrCYP52:GFP in BS supplemented with different alkanes.
30 µl 2×107 ml−1 spore suspensions of WT-PMrCYP52:GFP were incubated for 10 h in 0.0125% YE and then transferred to BS supplemented with different carbon source.
Figure 6. Time course of induction of WT-PMrCYP52:GFP in BS supplemented with decane, octacosane or myristic acid.
40 µl 1% octacosane or myristic acidsolution in hexane was pipette onto glass coverslips and evaporated, leaving a white greasy layer. The coverslips were then placed in polystyrene petri dishes with the alkane layer facing up, and the dishes were inoculated with 30 µl of 1×107 ml−1 spore suspensions in BS medium.
Figure 7. Induction of WT-PMrCYP52:GFP on locust wings (A) or locust wings treated with hexane(B).
Discussion
The epicuticle (outer insect waxy layer) is the interface between the insect and its environment, and the first barrier an entomopathogenic fungi needs to cross in order to infect an insect. Hydrocarbons, mostly alkanes in the chain length range C21–C35, comprise over 90% of the epicuticular layer on the surface of grasshoppers, with the balance being composed of wax esters, free fatty acids and triacylglycerides [22]–[23]. Although several aspects of the interactions between entomopathogenic fungi and insect host epicuticular hydrocarbons have been examined, the fungal genes that underlie insect waxy layer degradation remain almost completely unexplored [6]. Cytochrome P450s constitute a superfamily of monoxygenases involved in the hydroxylation of a wide range of endogenous and xenobiotic compounds, including fatty acids and alkanes (metabolism of alkanes is coupled to fatty acid degradation as conversion of alkanes to fatty acids is an essential step in the alkane assimilation process). Besides their involvement in different physiological processes, P450s also differ in the position of the hydroxylation; this can happen close to the carboxyl group (mediated by CYP152), can occur in-chain (e.g., CYP1006) or at the terminal or subterminal end (e.g., CYP52). Blast searches of M. robertsii genome revealed that among its 123 CYP enzymes [8] there was a single CYP1006 with similarity (46% identity) to a diol synthase (CYP1006C1) from Aspergillus nidulans, and one hydroxylase belonging to the CYP52 family (membrane bound class II). There were no CYP152s. Several CYP52 enzymes have activity towards alkanes and/or fatty acids [1]. However, no induction of MrCYP52 was observed for mytistic acid, suggesting that its main function is not omega-hydroxylation of fatty acids. Alkanes are predominantly oxidized by microbes at a terminal methyl group, which represents the first and rate-limiting step in the alkane degradation pathway [1]. The importance of monoterminal oxidation to M. robertsii was shown by its ability to utilize 2-methyl but not 3-methylnonane, due to the alkyl branch at the β position causing steric inhibition of terminal oxidizing enzymes [4]. The involvement of CYP52s in terminal hydroxylation of n-alkanes implicated MrCYP52 as a potential contributor to the degradation of epicuticular lipids. Compared to ΔMrCYP52, wild type Mr2575 germinated faster with a cuticle-derived hydrocarbon extract or long chain alkanes, while expression of MrCYP52 in the wild type also boosted germination as well as appressorial differentiation on locust cuticle. These findings indicate that MrCYP52 is required by M. robertsii for rapid hydrolysis of alkanes and that this provides nutrition for germination and formation of infection structures. The expression of MrCYP52 shares common features with alkane-degrading P450 genes of Candida maltose, such as induction by alkanes and repression by glucose [24], and is consistent with alkanes playing an important role in nutrition in the absence of more readily utilized nutrients. The nutrient poor conditions that allow expression of MrCYP52 are a necessary trigger for virulence as M. robertsii strain Mr2575 only produces infection structures on cuticle surfaces with low levels of nutrients [9]. We identified three putative CREA-binding sites upstream of the MrCYP52 translation start codon (Figure S4). A trans-acting DNA-binding protein CRR1 with significant sequence similarity of A. nidulans CREA has been demonstrated to effect carbon metabolite repression in M. robertsii [25].
Utilizing an MrCYP52 promoter-reporter construct to precisely reveal the spatial and temporal pattern of MrCYP52 activity enabled us to confirm that MrCYP52 is highly expressed throughout the formation of infection structures, suggesting constant induction. Nevertheless, in vitro, long chain alkanes were comparatively poor inducers of MrCYP52, as compared to decane which is not reported to be a component of the epicuticle. ΔMrCYP52 did not grow on decane suggesting that MrCYP52 is required for its assimilation, whereas growth of ΔMrCYP52 on longer chain alkanes was reduced but not eliminated suggesting that multiple enzymes are involved in the assimilation of molecules longer than decane. CYP52 enzymes are often length specific [26], so it is possible that fungi have evolved a multitude of currently unknown enzymes that catalyze oxygenation of long chain alkanes to produce decane, and the induced MrCYP52 has a principal role in providing nutrition by oxygenation of short-chain breakdown products. Consistent with this interpretation, B. bassiana produces large amount of decane when grown on the insect-derived hydrocarbon n-octacosane [27]. MrCYP52 is able to catalyze the degradation of sufficient cuticular hydrocarbons to enhance growth, as shown by the comparatively poor performance of ΔMrCYP52. Cuticle treated with hexane to remove hydrocarbons did not induce MrCYP52 and reduced growth of the wild type fungus to the same level as ΔMrCYP52, while ΔMrCYP52 grew poorly on the extracted hydrocarbons as compared to the wild type, confirming the link between cuticular hydrocarbons, induction of MrCYP52 and growth. MrCYP52 homologs were found in many Ascomycete fungi, including species of Beauveria, Magnaporthe, Fusarium, Aspergillus, Glomerella (Colletotrichum) and Penicillium that are insect and plant pathogens or soil-dwelling saprophytes, suggesting that MrCYP52 homologs may be generally important for utilizing alkane components of the epicuticular barriers surrounding plants and insects. Homologs were absent in the plant symbionts Epichloe and Trichoderma, as well as Neurospora, which preferentially colonizes scorched dead plant material [28], showing that the gene is expendable for some Ascomycete life-styles.
Supporting Information
Disruption of MrCYP52 in M. robertsii using the Bar gene. (A) Strategy for targeted disruption of MrCYP52. Arrows indicate location of PCR primers. (B) Confirmation of the disruption of MrCYP52 by Southern blot analysis. 1: the wild type strain; 2: the MrCYP52 disruptant strain; 3: ΔMrCYP52 was complemented with the wild type MrCYP52; (C) Confirmation of the disruption of MrCYP52 by PCR analysis. M: the MrCYP52 disruptant strain; W: the wild type strain. MrCYP52_CUP+BarDCUP and MrCYP52_CUP+MrCYP52_CR indicate primer pairs used for PCR amplification.
(TIFF)
TMpred hydrophilicity analysis of MrCYP52. Shown is a plot of the MrCYP52 amino acid numbers (X coordinate) against the probability that a specific amino acid is part (positive values on the y coordinate) or not part (negative values on the y coordinate) of a transmembrane helix. Solid and dotted lines indicated the probability of a putative transmembrane helix to be oriented from the cytoplasm toward the outside (i→o) or in the opposite direction (o→i), respectively.
(TIFF)
Phylogenetic analysis of MrCYP52 along with known fungal cytochrome P450 encoding genes. MEGA4 software was used to carry out the analysis. Bootstrap values are adjacent to each internal node, representing the percentage of 1,000 bootstrap replicates.
(TIFF)
Nucleotide sequence of the 5′-upstream region of the MrCYP52 gene. The translation start point is shown in bold. The putative CREA-binding sites are underlined.
(TIFF)
Primers used in this study.
(DOCX)
Germination and appressorial formation (in brackets) against plastic or insect cuticle.
(DOCX)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by United States Department of Agriculture Cooperative State Research, Education, and Extension Service grant 2010-65106-20580. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Van Bogaert IN, Groeneboer S, Saerens K, Soetaert W. The role of cytochrome P450 monooxygenases in microbial fatty acid metabolism. FEBS J. 2011;278(2):206–221. doi: 10.1111/j.1742-4658.2010.07949.x. [DOI] [PubMed] [Google Scholar]
- 2.Roberts DW, St Leger RJ. Metarhizium spp., cosmopolitan insect-pathogenic fungi: mycological aspects. Adv Appl Microbiol. 2004;54:1–70. doi: 10.1016/S0065-2164(04)54001-7. [DOI] [PubMed] [Google Scholar]
- 3.Oh Y, Donofrio N, Pan H, Coughlan S, Brown DE, et al. Transcriptome analysis reveals new insight into appressorium formation and function in the rice blast fungus Magnaporthe oryzae. Genome Biol. 2008;9(5):R85. doi: 10.1186/gb-2008-9-5-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.St Leger RJ, Cooper RM, Charnley AK. Utilization of alkanes by entomopathogenic fungi. J Invertebr Pathol. 1988;52(2):356–359. [Google Scholar]
- 5.Crespo R, Juárez MP, Cafferata LFR. Biochemical interaction between entomopathogenous fungi and their insect-host-like hydrocarbons. Mycologia. 2000;92(3):528–536. [Google Scholar]
- 6.Pedrini N, Zhang S, Juárez MP, Keyhani NO. Molecular characterization and expression analysis of a suite of cytochrome P450 enzymes implicated in insect hydrocarbon degradation in the entomopathogenic fungus Beauveria bassiana. Microbiology. 2010;156(8):2549–2557. doi: 10.1099/mic.0.039735-0. [DOI] [PubMed] [Google Scholar]
- 7.Jarrold SL, Moore D, Potter U, Charnley AK. The contribution of surface waxes to pre-penetration growth of an entomopathogenic fungus on host cuticle. Mycol Res. 2007;111(2):240–249. doi: 10.1016/j.mycres.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 8.Gao Q, Jin K, Ying SH, Zhang Y, Xiao G, et al. Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLoS Genet. 2011;7(1):e1001264. doi: 10.1371/journal.pgen.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.St Leger RJ, Butt TM, Goettel MS, Staples RS, Roberts DW. Production in vitro of appressoria by the entomopathogenic fungus Metarhizium anisopliae. Exp Mycol. 1989;13(3):274–288. [Google Scholar]
- 10.St Leger RJ, Joshi L, Bidochka MJ, Rizzo NW, Roberts DW. Characterization and ultrastructural localization of chitinases from Metarhizium anisopliae, M. flavoviride, and Beauveria bassiana during fungal invasion of host (Manduca sexta) cuticle. Appl Environ Microbiol. 1996;62(3):907–912. doi: 10.1128/aem.62.3.907-912.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C, St Leger RJ. The Metarhizium anisopliae perilipin homolog MPL1 regulates lipid metabolism, appressorial turgor pressure, and virulence. J Biol Chem. 2007;282(29):21110–21115. doi: 10.1074/jbc.M609592200. [DOI] [PubMed] [Google Scholar]
- 12.Wang C, St Leger RJ. Developmental and transcriptional responses to host and non host cuticles by the specific locust pathogen Metarhizium anisopliae var. acridum. Eukaryot Cell. 2005;4(5):937–947. doi: 10.1128/EC.4.5.937-947.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang W, Pava-ripoll M, Wang S, St Leger RJ. Protein kinase A regulates production of virulence determinants by the entomopathogenic fungus, Metarhizium anisopliae. Fungal Genet Biol. 2009;46(3):277–285. doi: 10.1016/j.fgb.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Fang W, Pei Y, Bidochka MJ. Transformation of Metarhiziumanisopliae mediated by Agrobacterium tumefaciens. Can J Microbiol. 2006;52(7):623–626. doi: 10.1139/w06-014. [DOI] [PubMed] [Google Scholar]
- 15.Lin L, Wang F, Wei D. Chlorimuron ethyl as a new selectable marker for disrupting genes in the insect-pathogenic fungus Metarhiziumrobertsii. J Microbiol Methods. 2011 doi: 10.1016/j.mimet.2011.07.018. In press. [DOI] [PubMed] [Google Scholar]
- 16.Fang W, Yang X, Zhang Y, Pei Y. Rapid extraction of DNA and RNA from fungi. Chin J Appl Environ Biol. 2002;8:305–307. [Google Scholar]
- 17.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3rd ed. Cold Spring Harbor Laboratory Press; 2001. 2344 [Google Scholar]
- 18.Carlson DA, Bernier UR, Sutton BD. Elution patterns of methyl-branched alkanes by capillary GC. J Chem Ecol. 1998;24(11):1845–1865. [Google Scholar]
- 19.St Leger RJ, Charnley AK, Cooper RM. Cuticle-degrading enzymes of entomopathogenic fungi: Synthesis in culture on cuticle. J Invertebr Pathol. 1986;48(1):85–95. [Google Scholar]
- 20.Fang W, St Leger RJ. Mrt, a gene unique to fungi, encodes an oligosaccharide transporter and facilitates rhizosphere competency in Metarhizium robertsii. Plant Physiol. 2010;154(3):1549–1557. doi: 10.1104/pp.110.163014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crespo R, Juárez MP, Dal Bello GM, Padín S, CalderónFernández G, et al. Increased mortality of Acanthoscelides obtectus by alkane-grown Beauveria bassiana. BioControl. 2002;47:685–696. [Google Scholar]
- 22.Pedrini N, Crespo R, Juárez MP. Biochemistry of insect epicuticle degradation by entomopathogenic fungi. Comp Biochem Physiol C Toxicol Pharmacol. 2007;146(1–2):124–137. doi: 10.1016/j.cbpc.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 23.Chapman RF. The insects: Structure and Function. Cambridge: Cambridge University Press; 1998. 770 [Google Scholar]
- 24.Ohkuma M, Muraoka S, Tanimoto T, Fujii M, Ohta A, et al. CYP52 (cytochrome P450alk) multigene family in Candida maltosa: identification and characterization of eight members. DNA Cell Biol. 1995;14(2):163–173. doi: 10.1089/dna.1995.14.163. [DOI] [PubMed] [Google Scholar]
- 25.Screen S, Bailey A, Charnley K, Cooper R, Clarkson J. Carbon regulation of the cuticle-degrading enzyme PR1 from Metarhizium anisopliae may involve a trans-acting DNA-binding protein CRR1, a functional equivalent of the Aspergillus nidulans CREA protein. Curr Genet. 1997;31(6):511–518. doi: 10.1007/s002940050238. [DOI] [PubMed] [Google Scholar]
- 26.Van Bogaert IN, Demey M, Develter D, Soetaert W, Vandamme EJ. Importance of the cytochrome P450 monooxygenase CYP52 family for the sophorolipid-producing yeast Candida bombicola. FEMS Yeast Res. 2009;9(1):87–94. doi: 10.1111/j.1567-1364.2008.00454.x. [DOI] [PubMed] [Google Scholar]
- 27.Crespo R, Pedrini N, Juárez MP, Dal Bello GM. Volatile organic compounds released by the entomopathogenic fungus Beauveria bassiana. Microbiol Res. 2008;163(2):148–151. doi: 10.1016/j.micres.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 28.Powell AJ, Jacobson DJ, Salter L, Natvig DO. Variation among natural isolates of Neurospora on small spatial scales. Mycologia. 2003;95(5):809–819. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Disruption of MrCYP52 in M. robertsii using the Bar gene. (A) Strategy for targeted disruption of MrCYP52. Arrows indicate location of PCR primers. (B) Confirmation of the disruption of MrCYP52 by Southern blot analysis. 1: the wild type strain; 2: the MrCYP52 disruptant strain; 3: ΔMrCYP52 was complemented with the wild type MrCYP52; (C) Confirmation of the disruption of MrCYP52 by PCR analysis. M: the MrCYP52 disruptant strain; W: the wild type strain. MrCYP52_CUP+BarDCUP and MrCYP52_CUP+MrCYP52_CR indicate primer pairs used for PCR amplification.
(TIFF)
TMpred hydrophilicity analysis of MrCYP52. Shown is a plot of the MrCYP52 amino acid numbers (X coordinate) against the probability that a specific amino acid is part (positive values on the y coordinate) or not part (negative values on the y coordinate) of a transmembrane helix. Solid and dotted lines indicated the probability of a putative transmembrane helix to be oriented from the cytoplasm toward the outside (i→o) or in the opposite direction (o→i), respectively.
(TIFF)
Phylogenetic analysis of MrCYP52 along with known fungal cytochrome P450 encoding genes. MEGA4 software was used to carry out the analysis. Bootstrap values are adjacent to each internal node, representing the percentage of 1,000 bootstrap replicates.
(TIFF)
Nucleotide sequence of the 5′-upstream region of the MrCYP52 gene. The translation start point is shown in bold. The putative CREA-binding sites are underlined.
(TIFF)
Primers used in this study.
(DOCX)
Germination and appressorial formation (in brackets) against plastic or insect cuticle.
(DOCX)