Abstract
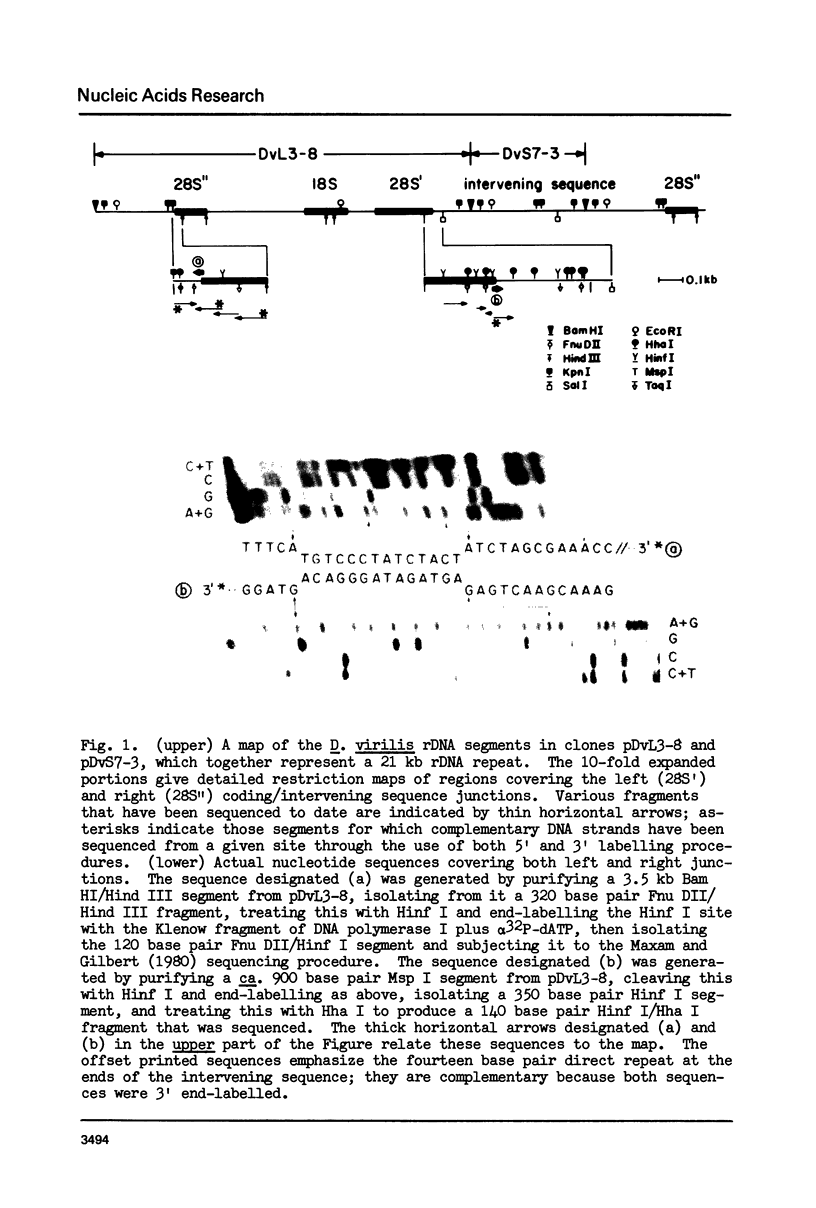
Most repeat units of rDNA in Drosophila virilis are interrupted in the 28S rRNA coding region by an intervening sequence about 10 kb in length; uninterrupted repeats have a length of about 11 kb. We have sequenced the coding/intervening sequence junctions and flanking regions in two independent clones of interrupted rDNA, and the corresponding 28S rRNA coding region in a clone of uninterrupted rDNA. The intervening sequence is terminated at both ends by a direct repeat of a fourteen nucleotide sequence that is present once in the corresponding region of an intact gene. This is a phenomenon associated with transposable elements in other eukaryotes and in prokaryotes, and the Drosophila rDNA intervening sequence is discussed in this context. We have compared more than 200 nucleotides of the D. virilis 28S rRNA gene with sequences of homologous regions of rDNA in Tetrahymena pigmentosa (Wild and Sommer, 1980) and Xenopus laevis (Gourse and Gerbi, 1980): There is 93% sequence homology among the diverse species, so that the rDNA region in question (about two-thirds of the way into the 28S rRNA coding sequence) has been very highly conserved in eukaryote evolution. The intervening sequence in T. pigmentosa is at a site 79 nucleotides upstream from the insertion site of the Drosophila intervening sequence.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barnett T., Rae P. M. A 9.6 kb intervening sequence in D. virilis rDNA, and sequence homology in rDNA interruptions of diverse species of Drosophila and other diptera. Cell. 1979 Apr;16(4):763–775. doi: 10.1016/0092-8674(79)90092-8. [DOI] [PubMed] [Google Scholar]
- Beckingham K., White R. The ribosomal DNA of Calliphora erythrocephala; and analysis of hybrid plasmids containing ribosomal DNA. J Mol Biol. 1980 Mar 15;137(4):349–373. doi: 10.1016/0022-2836(80)90162-x. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Cameron J. R., Loh E. Y., Davis R. W. Evidence for transposition of dispersed repetitive DNA families in yeast. Cell. 1979 Apr;16(4):739–751. doi: 10.1016/0092-8674(79)90090-4. [DOI] [PubMed] [Google Scholar]
- Campbell G. R., Littau V. C., Melera P. W., Allfrey V. G., Johnson E. M. Unique sequence arrangement of ribosomal genes in the palindromic rDNA molecule of Physarum polycephalum. Nucleic Acids Res. 1979 Apr;6(4):1433–1447. doi: 10.1093/nar/6.4.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chooi W. Y. The occurrence of long transcription units among the X and Y ribosomal genes of Drosophila melanogaster: transcription of insertion sequences. Chromosoma. 1979 Sep 1;74(1):57–74. doi: 10.1007/BF00344483. [DOI] [PubMed] [Google Scholar]
- Dawid I. B., Botchan P. Sequences homologous to ribosomal insertions occur in the Drosophila genome outside the nucleolus organizer. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4233–4237. doi: 10.1073/pnas.74.10.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawid I. B., Wellauer P. K., Long E. O. Ribosomal DNA in Drosophila melanogaster. I. Isolation and characterization of cloned fragments. J Mol Biol. 1978 Dec 25;126(4):749–768. doi: 10.1016/0022-2836(78)90018-9. [DOI] [PubMed] [Google Scholar]
- Din N., Engberg J. Extrachromosomal ribosomal RNA genes in Tetrahymena: structure and evolution. J Mol Biol. 1979 Nov 5;134(3):555–574. doi: 10.1016/0022-2836(79)90367-x. [DOI] [PubMed] [Google Scholar]
- Din N., Engberg J., Kaffenberger W., Eckert W. A. The intervening sequence in the 26S rRNA coding region of T. thermophila is transcribed within the largest stable precursor for rRNA. Cell. 1979 Oct;18(2):525–532. doi: 10.1016/0092-8674(79)90069-2. [DOI] [PubMed] [Google Scholar]
- Finnegan D. J., Rubin G. M., Young M. W., Hogness D. S. Repeated gene families in Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1053–1063. doi: 10.1101/sqb.1978.042.01.106. [DOI] [PubMed] [Google Scholar]
- Gubler U., Wyler T., Braun R. The gene for the 26 S rRNA in Physarum contains two insertions. FEBS Lett. 1979 Apr 15;100(2):347–350. doi: 10.1016/0014-5793(79)80366-x. [DOI] [PubMed] [Google Scholar]
- Jolly D. J., Thomas C. A., Jr Nuclear RNA transcripts from Drosophila melanogaster ribosomal RNA genes containing introns. Nucleic Acids Res. 1980 Jan 11;8(1):67–84. doi: 10.1093/nar/8.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. Translocatable elements in procaryotes. Cell. 1977 May;11(1):11–23. doi: 10.1016/0092-8674(77)90313-0. [DOI] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Expression of ribosomal DNA insertions in Drosophila melanogaster. Cell. 1979 Dec;18(4):1185–1196. doi: 10.1016/0092-8674(79)90231-9. [DOI] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Restriction analysis of spacers in ribosomal DNA of Drosophila melanogaster. Nucleic Acids Res. 1979 Sep 11;7(1):205–215. doi: 10.1093/nar/7.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Pellegrini M., Manning J., Davidson N. Sequence arrangement of the rDNA of Drosophila melanogaster. Cell. 1977 Feb;10(2):213–214. doi: 10.1016/0092-8674(77)90215-x. [DOI] [PubMed] [Google Scholar]
- Potter S. S., Brorein W. J., Jr, Dunsmuir P., Rubin G. M. Transposition of elements of the 412, copia and 297 dispersed repeated gene families in Drosophila. Cell. 1979 Jun;17(2):415–427. doi: 10.1016/0092-8674(79)90168-5. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M., Leung D. W., Gillam S., Astell C. R., Montgomery D. L., Hall B. D. Sequence of the gene for iso-1-cytochrome c in Saccharomyces cerevisiae. Cell. 1979 Apr;16(4):753–761. doi: 10.1016/0092-8674(79)90091-6. [DOI] [PubMed] [Google Scholar]
- Strobel E., Dunsmuir P., Rubin G. M. Polymorphisms in the chromosomal locations of elements of the 412, copia and 297 dispersed repeated gene families in Drosophila. Cell. 1979 Jun;17(2):429–439. doi: 10.1016/0092-8674(79)90169-7. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B. Ribosomal DNA in Drosophila melanogaster. II. Heteroduplex mapping of cloned and uncloned rDNA. J Mol Biol. 1978 Dec 25;126(4):769–782. doi: 10.1016/0022-2836(78)90019-0. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B. The structural organization of ribosomal DNA in Drosophila melanogaster. Cell. 1977 Feb;10(2):193–212. doi: 10.1016/0092-8674(77)90214-8. [DOI] [PubMed] [Google Scholar]
- White R. L., Hogness D. S. R loop mapping of the 18S and 28S sequences in the long and short repeating units of Drosophila melanogaster rDNA. Cell. 1977 Feb;10(2):177–192. doi: 10.1016/0092-8674(77)90213-6. [DOI] [PubMed] [Google Scholar]
- Wild M. A., Gall J. G. An intervening sequence in the gene coding for 25S ribosomal RNA of Tetrahymena pigmentosa. Cell. 1979 Mar;16(3):565–573. doi: 10.1016/0092-8674(79)90030-8. [DOI] [PubMed] [Google Scholar]
- Wild M. A., Sommer R. Sequence of a ribosomal RNA gene intron from Tetrahymena. Nature. 1980 Feb 14;283(5748):693–694. doi: 10.1038/283693a0. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Cech T. R. In vitro splicing of the ribosomal RNA precursor in nuclei of Tetrahymena. Cell. 1980 Feb;19(2):331–338. doi: 10.1016/0092-8674(80)90507-3. [DOI] [PubMed] [Google Scholar]





