A novel membrane structure called CUPS is assembled during the secretion of unconventional cargo such as Acb1.
Abstract
The endoplasmic reticulum (ER)–Golgi-independent, unconventional secretion of Acb1 requires many different proteins. They include proteins necessary for the formation of autophagosomes, proteins necessary for the fusion of membranes with the endosomes, proteins of the multivesicular body pathway, and the cell surface target membrane SNARE Sso1, thereby raising the question of what achieves the connection between these diverse proteins and Acb1 secretion. In the present study, we now report that, upon starvation in Saccharomyces cerevisiae, Grh1 is collected into unique membrane structures near Sec13-containing ER exit sites. Phosphatidylinositol 3 phosphate, the ESCRT (endosomal sorting complex required for transport) protein Vps23, and the autophagy-related proteins Atg8 and Atg9 are recruited to these Grh1-containing membranes, which lack components of the Golgi apparatus and the endosomes, and which we call a novel compartment for unconventional protein secretion (CUPS). We describe the cellular proteins required for the biogenesis of CUPS, which we believe is the sorting station for Acb1’s release from the cells.
Introduction
Proteins that contain a signal sequence are targeted to the ER. In the ER, the signal sequence is cleaved and, when permitted, the secretory proteins are exported to the Golgi apparatus. Within the Golgi, the cargo is sorted and then transported to various cellular destinations, including the extracellular space. Several key features of this conventional secretory pathway are well understood (Pfeffer, 2007). However, eukaryotic cells use another, unconventional, mode of protein secretion. Proteins following this route lack a classical signal sequence for entering the ER, and their secretion is independent of traffic through the Golgi membranes (Nickel and Rabouille, 2009). The best example of this mode of secretion is the release of a-factor in yeast, which is mediated by the cell surface transporter Ste6 (Kuchler et al., 1989; McGrath and Varshavsky, 1989). Unlike this nonvesicular transport of a-factor, the ER–Golgi-independent secretion of IL-1β (interleukin 1β) involves a membrane compartment (Rubartelli et al., 1990). Secretion of FGF-2, for example, is considered to occur directly from the cytoplasm across the plasma membrane in a phosphatidylinositol 4,5-bisphosphate–dependent manner (Schäfer et al., 2004; Seelenmeyer et al., 2008; Temmerman et al., 2008). Although the repertoire of unconventionally secreted proteins has increased considerably over the years (Grundmann et al., 1988; Lutomski et al., 1997; Joliot et al., 1998; Menon and Hughes, 1999; Flieger et al., 2003; Loomis et al., 2010), the mechanism of their release into the extracellular space remains poorly understood (Nickel and Seedorf, 2008; Nickel and Rabouille, 2009; Nickel, 2010). However, the starvation-induced secretion of the Acyl-CoA binding protein Acb1 in yeast and AcbA in Dictyostelium discoideum, like IL-1β in mammalian cells, occurs via a membrane-bound compartment (Cabral et al., 2010; Duran et al., 2010). The orthologue of the Golgi-associated proteins GRASP65 and GRASP55 of mammalian cells (Grh1 in yeast/GrpA in D. discoideum) is essential for Acb1 as well as AcbA secretion but dispensable for the secretion of a-factor (Kinseth et al., 2007; Duran et al., 2010; Manjithaya et al., 2010). Knockdown of the single GRASP protein in Drosophila melanogaster tissue-culture cells by double-stranded RNA also inhibited the Golgi-independent trafficking of α-PS1 integrin from the ER to the cell surface (Schotman et al., 2008). More recent studies have revealed that secretion of Acb1 requires proteins needed for the formation of autophagosomes, trafficking to the endosomes, and for vesicle fusion processes at the cell surface (Duran et al., 2010; Manjithaya et al., 2010). These findings raise several important questions: (a) why do signal sequence–lacking proteins follow different routes for their export; (b) how are these cargoes recognized for secretion; (c) what is the source of the membranes for the generation of the transport carrier; and (d) where do the components for autophagosome formation fit into this scheme?
In this present study, we now provide evidence that Grh1 assembles into a novel compartment in the vicinity of the ER exit site in yeast upon starvation. This novel compartment called compartment for unconventional protein secretion (CUPS) contains many of the proteins compulsory for Acb1 secretion. We also discuss the requirements for the biogenesis of CUPS.
Results
Relocalization of Grh1 during starvation
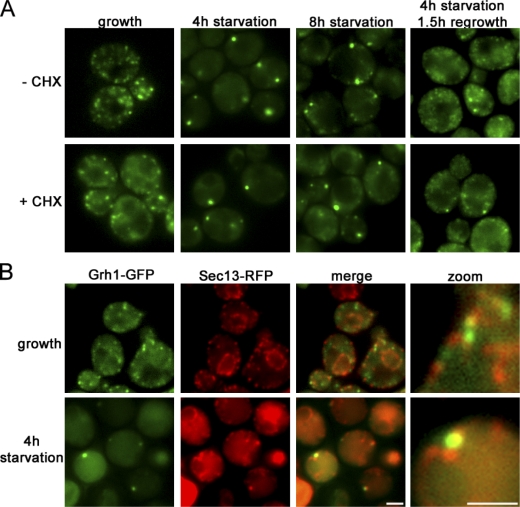
Grh1 is essential for Acb1 secretion during starvation but dispensable for general autophagy (Duran et al., 2010). To monitor its localization, we tagged endogenous Grh1 with GFP. In yeast cells grown in normal medium, Grh1-GFP was localized to several small punctate elements. Surprisingly, upon nutrient (glucose and nitrogen) starvation, Grh1 was found in one to three larger structures. The Grh1-containing compartment was clearly visible after 2 h of culturing in starvation medium and stable for ≤8 h. When starved yeast cells (4 h) were collected and further cultured in normal growth medium, Grh1-GFP was found to redistribute into numerous small elements. These events were independent of new protein synthesis, as cycloheximide treatment had no effect on the starvation-induced relocalization of Grh1 and its recovery upon culturing in growth medium (Fig. 1 A).
Figure 1.
Relocalization of Grh1-GFP upon starvation. (A) Yeast expressing Grh1-GFP grown in normal growth medium were washed and cultured in starvation medium with or without cycloheximide (CHX) for 4 or 8 h. The cells cultured in starvation medium for 4 h were cultured in growth medium for 1.5 h. The cells were visualized by fluorescence microscopy to monitor the localization of Grh1-GFP. (B) Yeast coexpressing Sec13-RFP and Grh1-GFP cultured in growth conditions or starved for 4 h were visualized by fluorescence microscopy. (right) Image at higher magnification. Bars, 2 µm.
Molecular composition of a starvation-induced Grh1-GFP–containing compartment
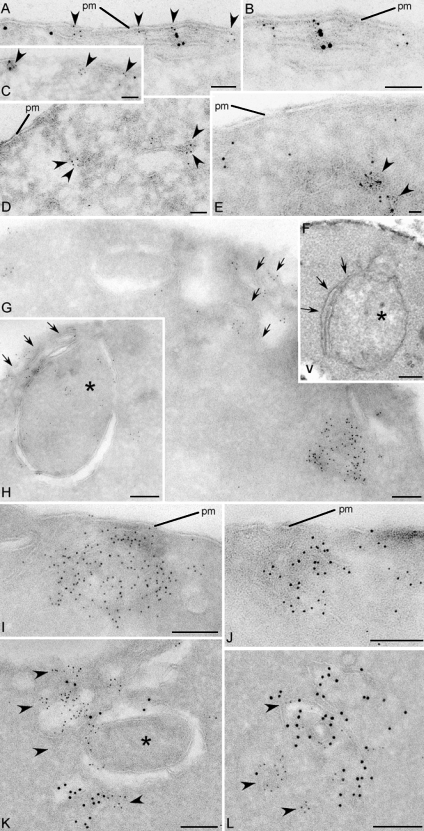
Yeast cells were grown in normal medium or starved for 4 h and visualized by fluorescence microscopy with several exocytic and endocytic compartment-specific proteins. Yeast grown in normal medium revealed Grh1-GFP to be proximal and, in some cases, colocalized with a subset of the ER exit site–specific Sec13-RFP. In cells cultured in starvation medium, Grh1-GFP was found in the vicinity of a subset of Sec13-RFP (Fig. 1 B and Fig. S1 A). Grh1 as well as its mammalian orthologues GRASP55 and GRASP65 are Golgi-associated proteins (Barr et al., 1997; Shorter et al., 1999; Behnia et al., 2007). However, Levi et al. (2010) have recently reported its localization to the ER exit site based on fluorescence microscopy. Furthermore, the Drosophila orthologue dGRASP also localizes to the Golgi and the ER exit sites under normal growth conditions (Kondylis et al., 2005). To ascertain the localization of Grh1 with respect to the ER exit site–specific protein Sec13, we visualized these proteins by immunoelectron microscopy of ultrathin cryosections (Fig. 2). In yeast cells grown in normal medium, Grh1 was found to be in close proximity but clearly segregated from the Sec13-RFP–containing ER exit sites (Fig. 2, A–C). Upon nutrient starvation, Grh1-GFP was detected closely apposed to and in rare instances colocalized with Sec13-RFP (Fig. 2, K and L). To quantitate the proximity of Sec13-RFP to Grh1-GFP, we visualized the localization of gold-conjugated antibodies directed against the tagged Sec13 (6 nm) and Grh1 (12 nm), respectively, over a distance of <10, 50, and 100 and >100 nm. 20 random images were scored and found to contain a total of 745 particles (Grh1, 12 nm) and 2,144 particles (Sec13, 6 nm). The relative distribution of these two proteins was as follows: 18 particles each of Grh1 and Sec13 were found at <10 nm from each other, 87 Grh1 and 129 Sec13 gold particles were found at <50 nm, 52 Grh1 and 89 Sec13 particles were found at <100 nm, and 588 Grh1 and 1,908 Sec13 particles were found at >100 nm. In sum, ∼80% of Grh1 particles were >100 nm removed from Sec13 particles (Fig. S1 A). This suggests that Grh1 and Sec13 are in close proximity but do not localize to the same structure during starvation. Upon starvation, the Grh1-containing membranes are fewer in number but larger in size and appear to be similar to the classical preautophagosomal structures (PASs)/forming autophagosomes (Fig. 2, F, H, and K; Baba et al., 1994; Kirisako et al., 1999).
Figure 2.
Grh1 localizes near ER exit sites and relocalizes to cup-shaped structures upon starvation. (A–L) Immunoelectron microscopy of ultrathin cryosections of yeast cells. Growth conditions (A–E), 4-h starvation conditions (G–L), and conventional morphology at 4-h starvation (F) are shown. (A–C) Double immunogold labeling of Grh1-GFP (12 nm) and Sec13-RFP (6 nm). (D) Single labeling of Grh1-GFP. (E) Single labeling of Sec13-RFP. (F) Conventional morphology. (G–J) Labeling of Grh1-GFP. (K and L) Double labeling of Grh1-GFP (12 nm) and Sec13-RFP (6 nm). Arrowheads indicate 6-nm gold particles. Arrows indicate membrane lamellae. Asterisks indicate preautophagosomal structure. pm, plasma membrane; v, vacuole. Bars: (A–D, G–I, K, and L) 200 nm; (F) 500 nm; (E and J) 100 nm.
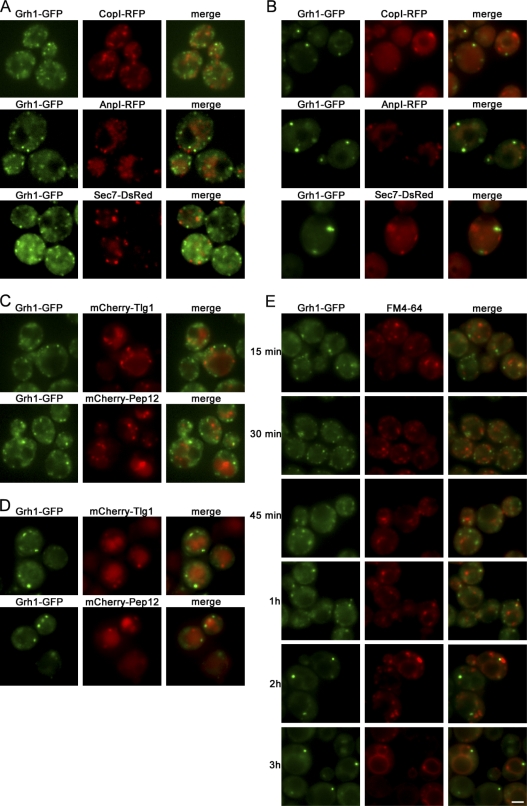
The Grh1-GFP–containing compartment did not colocalize with the early (CopI-RFP and Anp1-RFP) or the late (Sec7-DsRed) Golgi apparatus–specific marker under growth or starvation conditions (Fig. 3, A and B). As the Golgi membranes in Saccharomyces cerevisiae are neither stacked nor connected (Papanikou and Glick, 2009), the lack of colocalization with these marker proteins does not completely exclude the possible localization of Grh1 to early Golgi membranes, but given its consistent non-Golgi immunoelectron microscopy localization, it is most likely not a Golgi-associated protein.
Figure 3.
Grh1-GFP does not colocalize with marker proteins of the Golgi apparatus and endosomes under normal growth conditions or upon starvation. (A and B) Grh1-GFP was coexpressed with the early Golgi marker proteins CopI-RFP and AnpI-RFP or the late Golgi marker protein Sec7-DsRed. Cells were cultured in growth conditions (A) or nutrient starved for 4 h (B) and visualized by fluorescence microscopy. (C and D) Grh1-GFP was coexpressed with mCherry-Tlg1 or mCherry-Pep12 in growth conditions (C) or nutrient starved (D) and visualized by fluorescence microscopy. (E) Grh1-GFP–expressing cells were labeled with FM 4-64 and transferred to starvation medium for up to 3 h to visualize the localization of the internalized FM 4-64 with reference to Grh1-GFP. Bar, 2 µm.
We then tested whether endosomal membrane proteins, which are involved in the unconventional secretion of Acb1, are present in the Grh1-GFP–containing compartment upon nutrient starvation. Grh1-GFP did not colocalize with the mCherry-tagged early and late endosomal t-SNARE proteins, Tlg1 and Pep12, respectively, under normal growth conditions or upon starvation (Fig. 3, C and D). Furthermore, the Grh1-GFP–containing structure did not incorporate the endocytosed FM 4-64 dye in yeast cells that had been pulsed 15 min, subjected to starvation, and visualized by fluorescence microscopy at different time points. This further indicates a lack of mixing with the endosomal membranes under starvation conditions (Fig. 3 E).
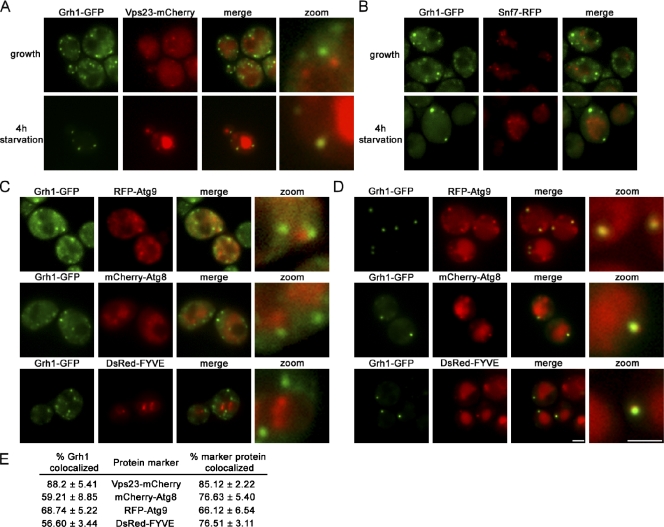
We next looked at Vps23, part of the ESCRT-I (endosomal sorting complex required for transport I) machinery, which is required for the sorting of cargo to the multivesicular body (MVB) as well as for the secretion of Acb1 in yeast under starvation conditions (Katzmann et al., 2001; Duran et al., 2010) and found that Vps23 did not colocalize with Grh1-GFP in yeast grown in normal medium. Surprisingly, upon starvation, 88% of Grh1 punctate structures were positive for Vps23-mCherry, and 87% of Vps23 punctae were positive for Grh1 (Fig. 4 A). This prompted us to test Snf7 of the ESCRT-III complex (Teis et al., 2008) for its presence in this starvation-specific ER exit site–associated compartment. Surprisingly, Snf7-RFP did not colocalize with Grh1-GFP in yeast cells grown in normal or starvation medium (Fig. 4 B).
Figure 4.
Grh1 colocalizes with Vps23 (ESCRT-I), PI3P, and components of autophagy machinery upon starvation. (A and B) Grh1-GFP was coexpressed with Vps23-mCherry (A) or with Snf7-RFP (B), cultured in growth conditions or nutrient starved for 4 h, and visualized by fluorescence microscopy. The zoom images of A represent high magnification images. (C and D) Grh1-GFP was coexpressed with RFP-Atg9, mCherry-Atg8, and a DsRed-tagged FYVE domain and cultured in growth conditions (C) or nutrient starved (D) and visualized by fluorescence microscopy. The right images represent one of the CUPS at high magnification. (E) The percentages of colocalization were quantified with respect to Grh1 or the indicated marker. At least 60 cells per marker were assessed, and errors are represented as SEM. Bars, 2 µm.
Because the deletion of various autophagy genes impaired the starvation-specific secretion of Acb1, we asked whether two marker proteins of the PAS, Atg8 and Atg9, are present in the Grh1-GFP–containing compartment. In yeast grown in normal medium, Grh1-GFP did not colocalize with Atg8 and Atg9 (Fig. 4 C). However, a starvation-specific colocalization of Grh1-GFP was readily evident for both marker proteins, as 59% (Atg8) and 68% (Atg9) of Grh1-GFP punctae showed colocalization (Fig. 4 D).
It has recently been shown that phosphatidylinositol 3 phosphate (PI3P) accumulates in large elements in close proximity to the ER during amino acid starvation of mammalian cells (Axe et al., 2008; Matsunaga et al., 2010). We therefore tested whether the Grh1-GFP compartments were enriched in PI3P by coexpressing a DsRed-tagged FYVE domain that binds PI3P (Stenmark et al., 2002). Under normal growth conditions, Grh1-GFP–containing structures were devoid of PI3P (Fig. 4 C); however, upon starvation, colocalization of Grh1 with DsRed-FYVE was observed for 56% of Grh1-GFP and 76% of DsRed-FYVE punctae, indicating the presence of PI3P (Fig. 4 D).
Collectively, these findings support the conclusion that Grh1-GFP localizes to a compartment close to the Sec13-containing ER exit sites in yeast grown in normal medium, whereas, upon starvation, Grh1-GFP concentrates in one to three larger punctae that are enriched in PI3P and contain Vps23 of the ESCRT-I complex as well as the autophagy gene products Atg8 and Atg9. Interestingly, this starvation-specific compartment does not contain membranes or proteins of the endosomes or the Golgi. Based on these findings, we believe that we have identified a new compartment, which we have named CUPS. This Grh1-GFP–containing compartment is generated upon starvation, has a characteristic cup shape, and is distinguished from its morphologically similar Berkeley body by the absence of Golgi/endosomal membranes (Novick et al., 1981).
CUPS is separated from the ER and the Golgi membranes
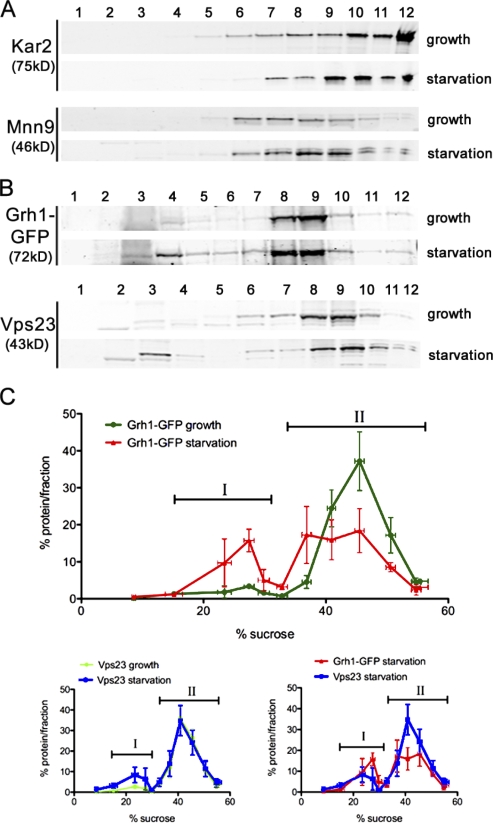
Yeast cells grown in normal medium or starved for 3 h were homogenized, and the membranes were fractionated by sucrose density gradients. The fractions were probed with several antibodies to secretory and endocytic membrane-specific proteins. Western blotting with antisera against the ER protein Kar2 and the Golgi protein Mnn9 revealed a similar distribution pattern when grown in normal or starvation medium (Fig. 5 A). Grh1-GFP was contained in membranes that fractionated with Kar2 and Mnn9 under normal growth conditions (Fig. 5, B and C, II). However, upon starvation, a pool of Grh1-GFP was found in lighter fractions that were well separated from the ER and the Golgi membranes (Fig. 5, B and C, I). We presume that these Grh1-containing fractions are enriched in CUPS. To further ascertain the identity of these fractions, we tested whether they contained Vps23. Importantly, fractionation of membranes from starved yeast cells revealed the presence of Vps23 in membrane fractions enriched in Grh1 (Fig. 5, B and C, I). Thus, these results support the microscopy-based localization of Grh1 and Vps23 to CUPS in yeast cultured in starvation medium.
Figure 5.
CUPS is separated from the ER and Golgi membranes. (A and B) Yeast cells cultured in growth conditions or starved for 3 h were fractionated on a continuous 15–60% sucrose gradient for 18 h. The gradient fractions were Western blotted to detect the ER protein Kar2 and the Golgi protein Mnn9 (A) or Vps23 and GFP to monitor Grh1-GFP (B). The numbers indicate the fractions. (C) The percentage of protein contained in each fraction was plotted against the sucrose concentration, and the error bars represent the results and SEM from three independent experiments. The fractions marked ‘I’ represent the starvation-specific pool of Grh1 and Vps23, and the denser fractions ‘II’ represent the ER–Golgi pool of Grh1-GFP and the endosomal pool of Vps23.
Molecular requirements for the biogenesis of CUPS
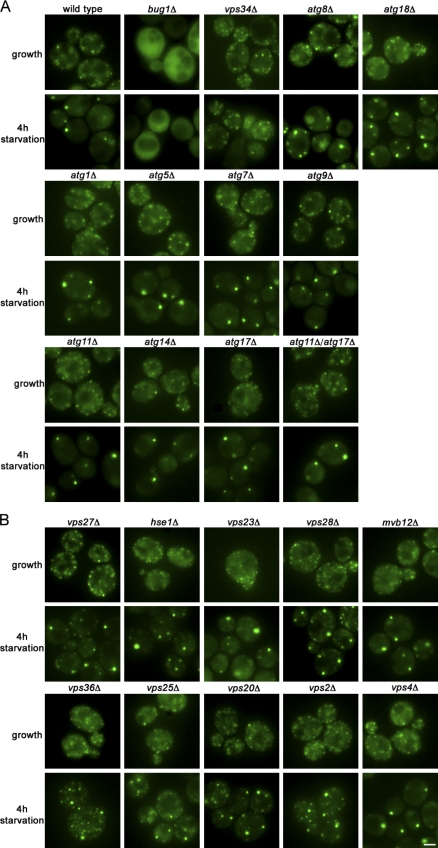
The association of Grh1 to membranes depends on an N-terminal acetylation and the interaction with a protein called Bug1, which is also essential for Acb1 secretion (Behnia et al., 2007; Duran et al., 2010). We therefore tested whether Bug1 was required for the recruitment of Grh1-GFP to CUPS. Deletion of BUG1 abolished the localization of Grh1-GFP to any membranous structures under normal growth and to CUPS upon starvation (Fig. 6 A). As CUPS is enriched in PI3P, we asked whether its biogenesis requires the sole PI3P kinase Vps34. A VPS34 deletion strain was unaffected with respect to the localization of Grh1-GFP when grown in normal medium. However, deletion of VPS34 inhibited the starvation-induced relocalization of Grh1-GFP to CUPS (Fig. 6 A).
Figure 6.
The role of Bug1, autophagy-related proteins, and the ESCRT machinery in CUPS formation. (A and B) Wild-type yeast, yeast strains deleted for BUG1, VPS34, ATG1, 5, 7, 8, 9, 11, 14, 17, 18, and 11/17 (A) or yeast cells deleted for components of the ESCRT machinery (VPS27, HSE1, VPS23, VPS28, MVB12, VPS36, VPS25, VPS20, VPS2, and VPS4; B) expressing Grh1-GFP were cultured in growth medium or starved for 4 h, and the formation of CUPS was visualized by fluorescence microscopy. Bar, 2 µm.
As Atg8 and Atg9 are present in CUPS and required for the secretion of Acb1, we tested whether their deletion affected the starvation-specific relocalization of Grh1-GFP. Surprisingly, deletion of various autophagy genes, which are involved in different steps of autophagy, e.g., regulation of autophagy, vesicle nucleation, expansion, and completion (atg1Δ, atg5Δ, atg7Δ, atg9Δ, atg11Δ, atg14Δ, atg17Δ, atg18Δ, and atg11Δ/atg17Δ; Yang and Klionsky, 2009), did not impair the relocalization of Grh1 to CUPS. Interestingly, in the case of an atg8Δ strain, larger punctate elements representing CUPS were formed, but several Grh1-GFP–containing smaller elements were still present. The same phenotype was observed upon deletion of ATG18, which is known to bind PI3P and to be essential for autophagosome formation (Fig. 6 A and Fig. S1 C; Dove et al., 2004).
Next, we investigated whether proteins of the ESCRT machinery are involved in the formation of CUPS as Vps23 is contained in this structure and also required for the secretion of Acb1 (Duran et al., 2010). Vps27 is a component of the ESCRT-0 complex, contains a FYVE domain for PI3P binding, and has been shown to interact with Vps23 (Katzmann et al., 2003). Deletion of VPS27 did not impair the formation of CUPS, but a large number of smaller structures of Grh1-GFP was also visible, implying that there is a kinetic delay in the formation of CUPS in vps27Δ cells. Deletion of the second member of the ESCRT-0 complex, HSE1 (Bilodeau et al., 2002), and components of the ESCRT-I complex (VPS23, MVB12, and VPS28; Katzmann et al., 2003; Curtiss et al., 2007) revealed no obvious defect in the relocalization of Grh1-GFP to CUPS. Deletion of ESCRT-II components (VPS25 and VPS36; Babst et al., 2002b) as well as ESCRT-III (VPS20 and VPS2; Babst et al., 2002a) also appeared to affect the kinetics of the localization of Grh1-GFP to CUPS similar to deletion of VPS27, as residual Grh1-GFP was present in smaller structures. Finally, deletion of VPS4 did not affect the relocalization of Grh1-GFP to CUPS (Fig. 6 B and Fig. S1 C).
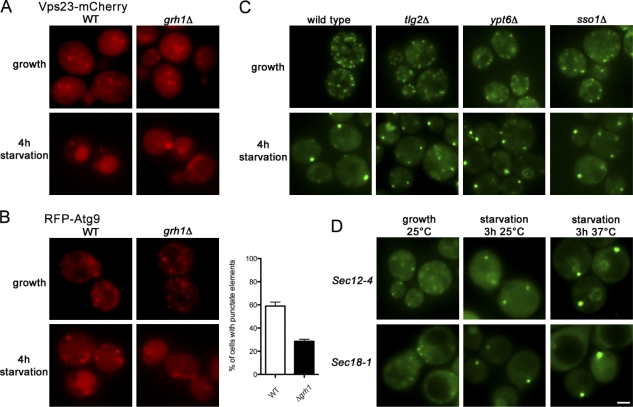
We then tested whether Grh1 has a role in the recruitment of other proteins to CUPS. Upon starvation, Vps23 localizes to CUPS and in the vacuole. However, GRH1 deletion completely abrogated the recruitment of Vps23-mCherry to CUPS and its trafficking to the vacuole, whereas deletion of VPS23 did not affect the relocalization of Grh1 (Fig. 6 B and Fig. 7 A). RFP-Atg9 under normal growth conditions is localized to punctate elements that are considered a reservoir for this protein in the vicinity of or close proximity to the mitochondria (Mari et al., 2010). Upon starvation, 59% of the wild-type cells revealed RFP-Atg9 in punctate elements that are clearly distinct from the reservoir. However, in grh1Δ yeast, only 28% of the cells revealed a punctate staining of RFP-Atg9 (Fig. 7 B). Thus, Grh1 appears to be essential for the recruitment and trafficking of Vps23 to CUPS and the vacuole and plays a significant role in the starvation-induced relocalization of Atg9.
Figure 7.
NSF is not essential for CUPS biogenesis. (A) Wild-type (WT) and grh1Δ yeast expressing Vps23-mCherry were cultured in growth conditions or starved for 4 h and analyzed by fluorescence microscopy. (B) Wild-type and grh1Δ strains expressing RFP-Atg9 were cultured in growth conditions or starved and visualized by fluorescence microscopy. Starved cells showing RFP-Atg9–specific punctate elements representing the CUPS were counted in wild-type and grh1Δ cells. 59 ± 3.3% of wild-type cells showed a localization of Atg9 to CUPS compared with only 28.5 ± 1.8% upon deletion of GRH1 (statistically significant with P < 0.0001; error bars represent SEM). (C) Grh1-GFP expressing wild-type yeast and deleted for TLG2, YPT6, and SSO1 were cultured in growth or starvation medium and visualized by fluorescence microscopy. (D) Grh1-GFP was expressed in sec12-4 and sec18-1 strains. Yeast cells were grown at permissive temperature in growth medium and starved for 3 h at either the permissive or the nonpermissive temperature and visualized by fluorescence microscopy. Bar, 2 µm.
Subsequently, we tested whether the formation of CUPS was mediated by the fusion of Grh1-GFP–containing membranes or growth of a subset of preexisting sites close to the ER. We first tested whether Sec12, a guanine nucleotide exchange factor for Sar1 and therefore necessary for the biogenesis of COPII-coated vesicles at the ER exit sites, was required for the biogenesis of CUPS (Barlowe and Schekman, 1993). The temperature-sensitive sec12-4 yeast strain expressing Grh1-GFP was grown in the normal growth medium and then shifted to the starvation medium at either the permissive (25°C) or the nonpermissive (37°C) temperature. The cells were visualized by fluorescence microscopy, and the data revealed that a temperature-sensitive mutation in SEC12 did not affect the starvation-induced relocalization of Grh1. In other words, Sec12 activity is not required for the biogenesis of CUPS (Fig. 7 D and Fig. S1 B).
The N-ethylmaleimide–sensitive protein Sec18 is required for all postmembrane fusion events, including the events leading to the secretion of Acb1 (Graham and Emr, 1991; Duran et al., 2010). A yeast strain carrying the sec18-1 temperature-sensitive allele was grown in starvation medium at the permissive (25°C) or the nonpermissive (37°C) temperature and analyzed by fluorescence microscopy for the presence of CUPS. No defect in the formation of CUPS was observed at either temperature (Fig. 7 D and Fig. S1 B). Ypt6 and Tlg2, which are required for the transport of membranes into the endosomes (by fusion; Nichols et al., 1998; Luo and Gallwitz, 2003), are essential for the unconventional secretion of Acb1 (Duran et al., 2010). Interestingly, deletion of YPT6 and TLG2 did not impair the starvation-induced relocalization of Grh1-GFP to CUPS, but they have a slight effect as shown for atg8Δ, as smaller elements were present in addition to larger punctae representing the CUPS (Fig. 7 C and Fig. S1 B). Similarly, deletion of SSO1, a plasma membrane t-SNARE that is also required for the secretion of Acb1 (Duran et al., 2010), did not affect the localization of Grh1-GFP to CUPS (Fig. 7 C and Fig. S1 B).
The biogenesis of CUPS is independent of rapamycin-induced autophagy
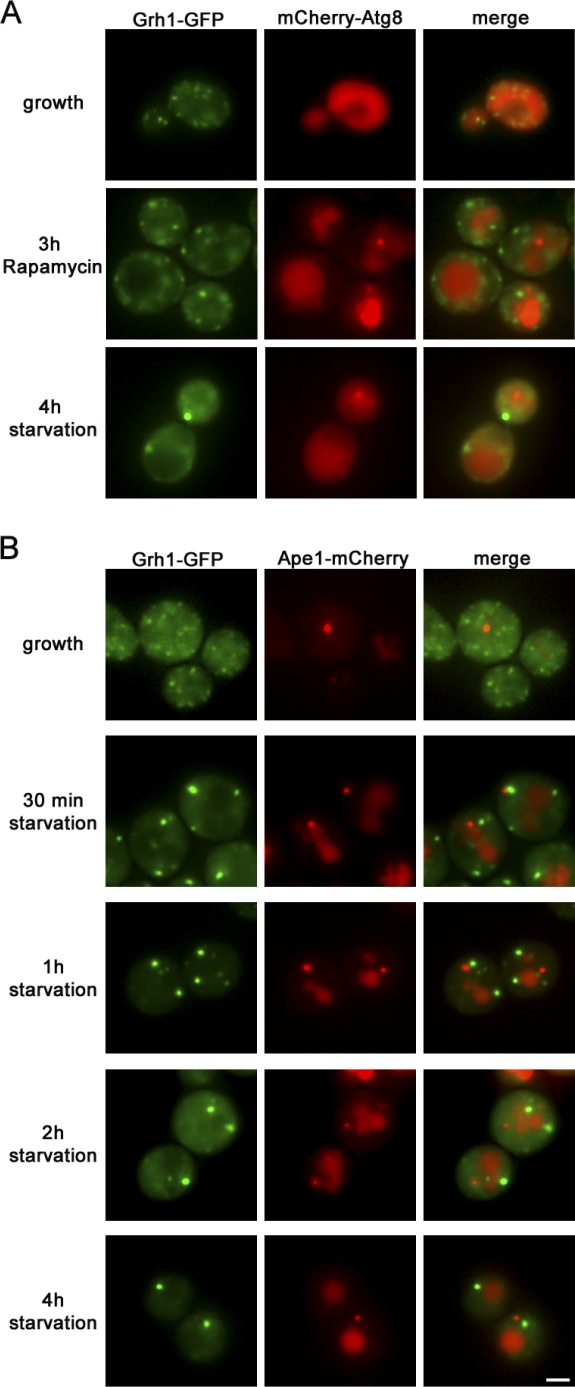
Grh1-GFP– and mCherry-Atg8–expressing yeast cells were grown in normal medium and subsequently either nutrient starved for 4 h or treated with 0.4 µg/ml rapamycin for 3 h. Rapamycin is known to form a complex with FKBP12 (Fpr1 in yeast), which inhibits TOR (target of rapamycin), thereby inducing autophagy even in nutrient-rich medium (Noda and Ohsumi, 1998). The cells were visualized by fluorescence microscopy, and the data revealed that treatment of yeast cells with rapamycin in growth medium relocalized mCherry-Atg8 from the cytosol to the newly forming autophagosome and into the vacuole. However, under these conditions, Grh1 did not relocalize to CUPS (Fig. 8 A). These data strongly suggest that the biogenesis of CUPS is independent of rapamycin-mediated autophagosome formation and its subsequent trafficking to the vacuole. In other words, CUPS and PAS differ in the requirements for their biogenesis, as treatment with rapamycin alone is not sufficient to induce CUPS formation.
Figure 8.
CUPS is distinct from PAS. (A) Yeast cells coexpressing Grh1-GFP and mCherry-Atg8 were cultured in growth medium and either nutrient starved for 4 h or treated with 0.4 µg/ml rapamycin in nutrient-rich medium for 3 h. (B) Yeast cells coexpressing Grh1-GFP and Ape1-mCherry were cultured in growth medium or starved for the indicated time points and visualized by fluorescence microscopy. Bar, 2 µm.
CUPS is devoid of the cytoplasm to vacuole (Cvt) pathway cargo Ape1
To further ascertain the relationship between CUPS and the PAS, we tested whether the vacuolar aminopeptidase Ape1, a cargo transported to the vacuole by the selective Cvt pathway, was contained in CUPS. Under nutrient-rich conditions, newly synthesized precursor of Ape1, prApe1, oligomerizes and is selectively captured into Cvt vesicles at the PAS, which subsequently fuse with the vacuole to deliver its content. Upon starvation, Ape1 is captured into autophagosomes and transported to the vacuole (Harding et al., 1995). Grh1-GFP– and Ape1-mCherry–expressing yeast cells were grown in normal and starvation medium and visualized by fluorescence microscopy (Fig. 8 B). Under growth conditions, Grh1-GFP was present in numerous punctae, whereas Ape1-mCherry was visible predominantly in a single large structure that did not colocalize with Grh1-GFP. Upon starvation, Ape1-mCherry showed an additional dispersed staining in the vacuole; however, it remained clearly visible as a single structure that still did not contain Grh1-GFP. This further indicates that CUPS is distinct from the Ape1-containing autophagosomes.
Discussion
Grh1 is essential for the secretion of the signal sequence lacking Acyl-CoA binding protein in yeast (S. cerevisiae and Pichia pastoris) and D. discoideum (Kinseth et al., 2007; Duran et al., 2010; Manjithaya et al., 2010). dGRASP is required for the Golgi-independent export of α-PS1 integrin from the ER to the cell surface in early Drosophila embryos (Schotman et al., 2008). Our new findings reveal that Grh1 relocalizes to a specific compartment near the ER exit sites under conditions that promote Acb1 secretion. Our key findings are summarized as follows:
(a) Upon culturing of yeast in starvation medium, Grh1 is concentrated in one to three large punctae that are closely apposed to Sec13 of the ER exit sites. These punctae, or CUPS, form without the involvement of new protein synthesis and the activity of Sec18. Even though transport of membranes between Golgi and the endosomes (by Ypt6 and Tlg2) and fusion with the cell surface (through Sso1) are required for the secretion of Acb1 (Duran et al., 2010), the formation of CUPS is independent of these membrane fusion events. These components of the fusion machinery therefore act after CUPS assembly for the secretion of Acb1.
(b) CUPS contain Vps23 (ESCRT-I) but are devoid of endosomal and the Golgi membrane–specific components. Because CUPS is enriched in PI3P, which is required for the relocalization of Grh1 and thus for Vps23, we suggest that Vps23 can be recruited to CUPS in addition to the endosomes through a PI3P/Grh1-dependent process. Interestingly, however, CUPS does not contain the ESCRT-III component Snf7. This suggests that not all of the machinery for the biogenesis of the MVB is recruited to CUPS during starvation.
(c) Conventional and immunoelectron microscopy has revealed that CUPS is composed of internal membranes, but their overall morphology is distinct from an MVB. CUPS are visible after 2 h and detected even after 8 h of starvation. This is consistent with the dynamics of unconventional secretion of Acb1, which was first observed after 3 h in the starvation medium and present for up to 16 h (Duran et al., 2010). Atg8 and Atg9 are contained in CUPS but not required for their formation. Based on these data, we suggest that CUPS is stable for the duration of culturing yeast in starvation medium, and Atg8 and Atg9 are presumably recruited for the biogenesis of secretory autophagosomes from CUPS.
CUPS: A novel compartment for the biogenesis of secretory autophagosomes
CUPS forms very close to the Sec13-containing ER exit sites specifically under starvation conditions. It has recently been reported that amino acid starvation of mammalian cells induces the formation of a PI3P-containing ER-associated compartments called omegasomes (Axe et al., 2008; Matsunaga et al., 2010), which is hypothesized to generate or provide membranes for the formation of autophagosomes for the purpose of autophagy. CUPS in yeast and omegasomes in mammalian cells share three common features: they are enriched in PI3P, they contain the PAS marker protein Atg8 (or the mammalian orthologue LC3), and they form close to or associated with the ER. Other than the presence of the PI3P, the molecular composition of omegasomes, the site of their assembly at the ER, and the requirements for their formation are not known. But because neither Grh1 nor Vps23 is required for degradative autophagy, it is possible that CUPS and omegasomes are separate compartments, which are specialized for the biogenesis of secretory and degradative autophagosomes, respectively.
In summary, upon nutrient starvation, Grh1 concentrates to a membrane, which is in close proximity to the ER exit site. This compartment is enriched in PI3P, which is usually found in endosomal compartments and the autophagosomes. Because CUPS is devoid of the endosomal markers Tlg1 and Pep12 and is not labeled with FM 4-64, they are unlikely to be the endosomes per se. CUPS contains the autophagosome markers Atg8 and Atg9. It could, therefore, be a scaffold for the biogenesis of autophagosomes. However, the fact that CUPS contains Grh1 and Vps23, its formation is independent of rapamycin-induced autophagy, and it lacks the Cvt pathway cargo Ape1 suggests that CUPS might be involved in the formation of secretory autophagosomes exclusively.
Materials and methods
Strains
Yeast strains used in this study are summarized in Table S1. To obtain the GFP-tagged Grh1 in various deletions strains, the tagging was either induced by homologous recombination (Janke et al., 2004) or the deletion strains (BY4741 background) were mated with the Grh1-GFP strain (Matα) and sporulated in 2% potassium acetate (supplemented with amino acids as needed) at 30°C. Spore preparation was performed as described previously (Herman and Rine, 1997). Haploidity was confirmed using the halo mating type assay (Sprague, 1991).
Plasmids
To generate the Grh1-GFP fusion protein under the control of its endogenous promoter, the coding sequence of Grh1, including 1 kb upstream of the start codon without the stop codon, was amplified by PCR and cloned as a ClaI–BamHI fragment into the pRS416 plasmid. Yeast EGFP was amplified by PCR from pyM26 and cloned as a BamHI–SacI fragment into the Grh1 containing pRS416 to generate the pRS416 Grh1-GFP plasmid. The coding sequence of Ape1 including its own promoter was amplified by PCR and cloned as a Sac1–Spe1 fragment into the pRS416 plasmid containing the mCherry coding sequence to generate the pRS416 Ape1-mCherry plasmid.
mCherry-Atg8 was cloned into pRS316 plasmid under the control of its endogenous promoter and was provided by Y. Ohsumi (National Institute for Basic Biology, Okazaki, Japan). RFP-Atg9 was cloned into the pRS416 plasmid under the control of the CupI promoter (Chang and Huang, 2007) and provided by W.P. Huang (National Taiwan University, Taipei, Taiwan). Vps23-mCherry was cloned into pRS416 under the control of its endogenous promoter (Curtiss et al., 2007) and provided by M. Babst (University of Utah, Salt Lake City, UT). mCherry-Tlg1 and mCherry-Pep12 were cloned into pRS416 under the control of the Vps21 promoter and provided by D. Katzmann (Mayo Clinic, Rochester, NY). Plasmids for the expression of Sec7-DsRed (pRS316 Sec7-DsRed; Calero et al., 2003) and DsRed-FYVE (pRS425MET3 DsRed-FYVE; Katzmann et al., 2003) were provided by S. Emr (Weill Institute for Cell and Molecular Biology, Cornell University, Ithaca, NY).
Media
Yeast cells were grown in rich YPD (1% yeast extract, 2% peptone, and 2% glucose) or synthetic minimal media (synthetic complete glucose; 0.67% yeast nitrogen base, 2% glucose, amino acids, and vitamins as needed). Starvation conditions were induced by culturing yeast cells in 2% potassium acetate at 1 OD600nm. Cycloheximide treatment was performed by adding 250 µg/ml cycloheximide to growth or starvation medium, and rapamycin treatment was performed by adding 0.4 µg/ml to the nutrient-rich medium for 3 h (Suzuki et al., 2002). Unless otherwise indicated, cells were grown at 25°C.
Fluorescence microscopy
Yeast cells were grown in the appropriate medium before imaging. Cells were harvested at 500 g for 3 min in a table top centrifuge (Multifuge 3 L-R; Heraeus), spotted on a microscopy slide, and immediately live imaged with a microscope (DMI6000 B; Leica) equipped with a camera (DFC 360FX; Leica) using an HCX Plan Apochromat 100× 1.4 NA objective. Images were taken using LAS AF software (Leica) with the same exposure times for Grh1-GFP; colocalization analysis with different marker proteins was performed with lower exposure times. Figures were assembled in Photoshop (Adobe) with only linear adjustments. Statistical analysis of the localization was performed by counting ≥60 cells from three independent experiments, and the statistical significance was tested in an unpaired Student’s t test using Prism (GraphPad Software). Compared data sets were termed as statistically significant when P < 0.05.
Staining of endocytic intermediates from endosomal to vacuolar membranes with the lipophilic dye FM 4-64 was performed as described previously (Vida and Emr, 1995) with slight modifications. Cells were grown at 25°C to an OD600nm of 0.8. A 10-ml culture was centrifuged and resuspended at 30 OD600nm/ml. FM 4-64 (Invitrogen) was added at a final concentration of 30 µM, and cells were incubated for 15 min at 25°C. Cells were harvested by centrifugation, resuspended at 1 OD600nm in starvation medium, and cultured at 25°C. At different time points, cells were collected by centrifugation, spotted on a slide, and live imaged.
Conventional and immunoelectron microscopy
Yeast cells expressing Grh1-GFP or Grh1-GFP and Sec13-RFP were grown in normal growth medium or starved for 4 h at 30°C. Preparation of samples for morphology and ultrathin cryosections as well as the immunoelectron microscopy was performed as described previously (Rieder et al., 1996). In brief, for conventional electron microscopy, cells were pelleted and fixed in 3% glutaraldehyde contained in 0.1 M sodium cacodylate, pH 7.4, 5 mM CaCl2, 5 mM MgCl2, and 2.5% sucrose for 1 h at 25°C with gentle agitation, spheroplasted, embedded in 2% ultra low temperature agarose (prepared in water), cooled, and subsequently cut into small pieces (∼1 mm3). The cells were then postfixed in 1% OsO4/1% potassium ferrocyanide contained in 0.1 M cacodylate/5 mM CaCl2, pH 7.4, for 30 min at 25°C. The blocks were washed thoroughly four times with double-distilled H2O (ddH2O; 10 min in total), transferred to 1% thiocarbohydrazide at 25°C for 3 min, washed in ddH2O (four times for 1 min each), and transferred to 1% OsO4/1% potassium ferrocyanide in cacodylate buffer, pH 7.4, for an additional 3 min at 25°C. The cells were washed four times with ddH2O (15 min in total), en bloc stained in Kellenberger’s uranyl acetate for 2 h to overnight, dehydrated through a graded series of ethanol, and subsequently embedded in Spurr resin. Sections were cut on an ultramicrotome (Ultracut T; Reichert), poststained with uranyl acetate and lead citrate, and observed on a transmission electron microscope (Tecnai 12; FEI) at 100 kV. Images were recorded with a digital camera (Soft Imaging System MegaView III; Olympus), and figures were assembled in Photoshop with only linear adjustments in contrast and brightness.
For immunoelectron microscopy, cells were fixed in suspension for 15 min by adding an equal volume of freshly prepared 8% formaldehyde contained in 100 mM potassium phosphate buffer, pH 7.4. The cells were pelleted, resuspended in fresh fixative (8% formaldehyde and 100 mM PO4, pH 7.4), and incubated for an additional 18–24 h at 4°C. The cells were washed briefly in PBS and resuspended in 1% low gelling temperature agarose. The agarose blocks were trimmed into pieces of 1 mm3, cryoprotected by infiltration with 2.3 M sucrose/30% polyvinyl pyrrolidone (10,000 molecular weight)/PBS, pH 7.4, for 2 h, mounted on cryopins, and rapidly frozen in liquid nitrogen. Ultrathin cryosections were cut on an ultramicrotome (UCT; Leica) equipped with an FC-S cryoattachment and collected onto Formvar/carbon-coated nickel grids. The grids were washed through several drops of PBS containing 2.5% FCS and 10 mM glycine, pH 7.4, and then blocked in 10% FCS for 30 min and incubated overnight in chicken anti-GFP antibody (Abcam) or chicken anti-GFP plus rabbit anti-RFP (Abcam). After washing, the grids were incubated for 2 h in 12-nm gold donkey anti–chicken conjugate or a mixture of 12-nm donkey anti–chicken plus 6-nm donkey anti–rabbit (Jackson ImmunoResearch Laboratories, Inc.). The grids were washed through several drops of PBS followed by several drops of ddH2O. Grids were then embedded in an aqueous solution containing 3.2% polyvinyl alcohol (10,000 molecular weight)/0.2% methyl cellulose (400 centipoises)/0.1% uranyl acetate. The sections were examined and photographed on a transmission electron microscope (Tecnai 12) at 100 kV, and images were collected with a digital camera (Soft Imaging System MegaView III). Figures were assembled in Photoshop with only linear adjustment of contrast and brightness.
Subcellular fractionation
250 OD600nm of yeast expressing Grh1-GFP were grown either in normal growth medium or starved for 3 h. Cells were collected by a 5-min spin at 3,000 g and washed once with ice-cold 10 mM NaN3/NaF. Cells were resuspended at 20 OD/ml in prespheroplasting buffer (10 mM NaN3, 10 mM NaF, 100 mM Tris-H2SO4, pH 9.4, and 0.36 µl/ml β-mercaptoethanol) and incubated for 20 min at 25°C. Cells were collected and spheroplasted at 50 OD600nm/ml in spheroplasting buffer (40 mM Hepes-NaOH, pH 7.5, 1.4 M sorbitol, and 1 µl/ml β-mercaptoethanol) by treatment with 50 U/OD600nm Zymolyase for 45 min at 35°C. Spheroplasts were harvested, resuspended in 1.5 ml lysis buffer (10 mM Hepes-NaOH, 1 mM MgCl2, 0.3 M sorbitol, and protease inhibitors), and lysed by using a Dounce homogenizer (Afora). Lysates were cleared twice by centrifugation (600 g for 3 min). Equal protein concentrations were loaded on top of a continuous sucrose gradient (10 ml of 15–60% sucrose in 10 mM Hepes-NaOH and 1 mM MgCl2) and centrifuged for 18 h at 100,000 g. 1-ml fractions were taken from the top of the gradient and analyzed either directly or after TCA precipitation by Western blotting using antibodies against GFP (Santa Cruz Biotechnology, Inc.), Vps23 (a gift from S. Emr), Mnn9 (a gift from Y. Noda, University of Tokyo, Tokyo, Japan), and Kar2 (a gift from M. Rose, Princeton University, Princeton, NJ). Statistical analysis of the signal in each fraction was performed by quantifying the intensity of three independent experiments with the Odyssey 2.1 software (LI-COR Biosciences), and the percentages were plotted using Prism.
Online supplemental material
Fig. S1 shows the quantitation of the fluorescence and immunoelectron microscopy data. Table S1 indicates yeast strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201106098/DC1.
Acknowledgments
We thank all members of the Malhotra laboratory as well as Chris Stefan and Scott Emr for valuable discussions.
C. Bruns is funded by a “La Caixa” predoctoral fellowship. V. Malhotra is an Institució Catalana de Recerca i Estudis Avançats professor at the Center for Genomic Regulation, and the work in his laboratory is funded by grants from Plan Nacional (BFU2008-00414), Consolider (CSD2009-00016), Agència de Gestió d’Ajuts Universitaris i de Recerca (AGAUR) Grups de Recerca Emergents (SGR2009-1488; AGAUR-Catalan Government), and the European Research Council (268692). The project has received research funding from the European Union. This paper reflects only the author’s views. The Union is not liable for any use that may be made of the information contained therein.
Footnotes
Abbreviations used in this paper:
- CUPS
- compartment for unconventional protein secretion
- Cvt
- cytoplasm to vacuole
- ddH2O
- double-distilled H2O
- MVB
- multivesicular body
- PAS
- preautophagosomal structure
- PI3P
- phosphatidylinositol 3 phosphate
References
- Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., Ktistakis N.T. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182:685–701 10.1083/jcb.200803137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M., Takeshige K., Baba N., Ohsumi Y. 1994. Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J. Cell Biol. 124:903–913 10.1083/jcb.124.6.903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Katzmann D.J., Estepa-Sabal E.J., Meerloo T., Emr S.D. 2002a. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell. 3:271–282 10.1016/S1534-5807(02)00220-4 [DOI] [PubMed] [Google Scholar]
- Babst M., Katzmann D.J., Snyder W.B., Wendland B., Emr S.D. 2002b. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell. 3:283–289 10.1016/S1534-5807(02)00219-8 [DOI] [PubMed] [Google Scholar]
- Barlowe C., Schekman R. 1993. SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature. 365:347–349 10.1038/365347a0 [DOI] [PubMed] [Google Scholar]
- Barr F.A., Puype M., Vandekerckhove J., Warren G. 1997. GRASP65, a protein involved in the stacking of Golgi cisternae. Cell. 91:253–262 10.1016/S0092-8674(00)80407-9 [DOI] [PubMed] [Google Scholar]
- Behnia R., Barr F.A., Flanagan J.J., Barlowe C., Munro S. 2007. The yeast orthologue of GRASP65 forms a complex with a coiled-coil protein that contributes to ER to Golgi traffic. J. Cell Biol. 176:255–261 10.1083/jcb.200607151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau P.S., Urbanowski J.L., Winistorfer S.C., Piper R.C. 2002. The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat. Cell Biol. 4:534–539 [DOI] [PubMed] [Google Scholar]
- Cabral M., Anjard C., Malhotra V., Loomis W.F., Kuspa A. 2010. Unconventional secretion of AcbA in Dictyostelium discoideum through a vesicular intermediate. Eukaryot. Cell. 9:1009–1017 10.1128/EC.00337-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero M., Chen C.Z., Zhu W., Winand N., Havas K.A., Gilbert P.M., Burd C.G., Collins R.N. 2003. Dual prenylation is required for Rab protein localization and function. Mol. Biol. Cell. 14:1852–1867 10.1091/mbc.E02-11-0707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-Y., Huang W.-P. 2007. Atg19 mediates a dual interaction cargo sorting mechanism in selective autophagy. Mol. Biol. Cell. 18:919–929 10.1091/mbc.E06-08-0683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtiss M., Jones C., Babst M. 2007. Efficient cargo sorting by ESCRT-I and the subsequent release of ESCRT-I from multivesicular bodies requires the subunit Mvb12. Mol. Biol. Cell. 18:636–645 10.1091/mbc.E06-07-0588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove S.K., Piper R.C., McEwen R.K., Yu J.W., King M.C., Hughes D.C., Thuring J., Holmes A.B., Cooke F.T., Michell R.H., et al. 2004. Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. EMBO J. 23:1922–1933 10.1038/sj.emboj.7600203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran J.M., Anjard C., Stefan C., Loomis W.F., Malhotra V. 2010. Unconventional secretion of Acb1 is mediated by autophagosomes. J. Cell Biol. 188:527–536 10.1083/jcb.200911154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flieger O., Engling A., Bucala R., Lue H., Nickel W., Bernhagen J. 2003. Regulated secretion of macrophage migration inhibitory factor is mediated by a non-classical pathway involving an ABC transporter. FEBS Lett. 551:78–86 10.1016/S0014-5793(03)00900-1 [DOI] [PubMed] [Google Scholar]
- Graham T.R., Emr S.D. 1991. Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a yeast sec18 (NSF) mutant. J. Cell Biol. 114:207–218 10.1083/jcb.114.2.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundmann U., Abel K.J., Bohn H., Löbermann H., Lottspeich F., Küpper H. 1988. Characterization of cDNA encoding human placental anticoagulant protein (PP4): homology with the lipocortin family. Proc. Natl. Acad. Sci. USA. 85:3708–3712 10.1073/pnas.85.11.3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding T.M., Morano K.A., Scott S.V., Klionsky D.J. 1995. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 131:591–602 10.1083/jcb.131.3.591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman P.K., Rine J. 1997. Yeast spore germination: a requirement for Ras protein activity during re-entry into the cell cycle. EMBO J. 16:6171–6181 10.1093/emboj/16.20.6171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 21:947–962 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Joliot A., Maizel A., Rosenberg D., Trembleau A., Dupas S., Volovitch M., Prochiantz A. 1998. Identification of a signal sequence necessary for the unconventional secretion of Engrailed homeoprotein. Curr. Biol. 8:856–863 10.1016/S0960-9822(07)00346-6 [DOI] [PubMed] [Google Scholar]
- Katzmann D.J., Babst M., Emr S.D. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 106:145–155 10.1016/S0092-8674(01)00434-2 [DOI] [PubMed] [Google Scholar]
- Katzmann D.J., Stefan C.J., Babst M., Emr S.D. 2003. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J. Cell Biol. 162:413–423 10.1083/jcb.200302136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinseth M.A., Anjard C., Fuller D., Guizzunti G., Loomis W.F., Malhotra V. 2007. The Golgi-associated protein GRASP is required for unconventional protein secretion during development. Cell. 130:524–534 10.1016/j.cell.2007.06.029 [DOI] [PubMed] [Google Scholar]
- Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. 1999. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147:435–446 10.1083/jcb.147.2.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondylis V., Spoorendonk K.M., Rabouille C. 2005. dGRASP localization and function in the early exocytic pathway in Drosophila S2 cells. Mol. Biol. Cell. 16:4061–4072 10.1091/mbc.E04-10-0938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchler K., Sterne R.E., Thorner J. 1989. Saccharomyces cerevisiae STE6 gene product: a novel pathway for protein export in eukaryotic cells. EMBO J. 8:3973–3984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi S.K., Bhattacharyya D., Strack R.L., Austin J.R., II, Glick B.S. 2010. The yeast GRASP Grh1 colocalizes with COPII and is dispensable for organizing the secretory pathway. Traffic. 11:1168–1179 10.1111/j.1600-0854.2010.01089.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis W.F., Behrens M.M., Williams M.E., Anjard C. 2010. Pregnenolone sulfate and cortisol induce secretion of acyl-CoA-binding protein and its conversion into endozepines from astrocytes. J. Biol. Chem. 285:21359–21365 10.1074/jbc.M110.105858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z., Gallwitz D. 2003. Biochemical and genetic evidence for the involvement of yeast Ypt6-GTPase in protein retrieval to different Golgi compartments. J. Biol. Chem. 278:791–799 10.1074/jbc.M209120200 [DOI] [PubMed] [Google Scholar]
- Lutomski D., Fouillit M., Bourin P., Mellottée D., Denize N., Pontet M., Bladier D., Caron M., Joubert-Caron R. 1997. Externalization and binding of galectin-1 on cell surface of K562 cells upon erythroid differentiation. Glycobiology. 7:1193–1199 10.1093/glycob/7.8.1193 [DOI] [PubMed] [Google Scholar]
- Manjithaya R., Anjard C., Loomis W.F., Subramani S. 2010. Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. J. Cell Biol. 188:537–546 10.1083/jcb.200911149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari M., Griffith J., Rieter E., Krishnappa L., Klionsky D.J., Reggiori F. 2010. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J. Cell Biol. 190:1005–1022 10.1083/jcb.200912089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K., Morita E., Saitoh T., Akira S., Ktistakis N.T., Izumi T., Noda T., Yoshimori T. 2010. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J. Cell Biol. 190:511–521 10.1083/jcb.200911141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J.P., Varshavsky A. 1989. The yeast STE6 gene encodes a homologue of the mammalian multidrug resistance P-glycoprotein. Nature. 340:400–404 10.1038/340400a0 [DOI] [PubMed] [Google Scholar]
- Menon R.P., Hughes R.C. 1999. Determinants in the N-terminal domains of galectin-3 for secretion by a novel pathway circumventing the endoplasmic reticulum-Golgi complex. Eur. J. Biochem. 264:569–576 10.1046/j.1432-1327.1999.00671.x [DOI] [PubMed] [Google Scholar]
- Nichols B.J., Holthuis J.C., Pelham H.R. 1998. The Sec1p homologue Vps45p binds to the syntaxin Tlg2p. Eur. J. Cell Biol. 77:263–268 [DOI] [PubMed] [Google Scholar]
- Nickel W. 2010. Pathways of unconventional protein secretion. Curr. Opin. Biotechnol. 21:621–626 10.1016/j.copbio.2010.06.004 [DOI] [PubMed] [Google Scholar]
- Nickel W., Rabouille C. 2009. Mechanisms of regulated unconventional protein secretion. Nat. Rev. Mol. Cell Biol. 10:148–155 (published erratum appears in Nat. Rev. Mol. Cell Biol. 2009. 10:234) 10.1038/nrm2617 [DOI] [PubMed] [Google Scholar]
- Nickel W., Seedorf M. 2008. Unconventional mechanisms of protein transport to the cell surface of eukaryotic cells. Annu. Rev. Cell Dev. Biol. 24:287–308 10.1146/annurev.cellbio.24.110707.175320 [DOI] [PubMed] [Google Scholar]
- Noda T., Ohsumi Y. 1998. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273:3963–3966 10.1074/jbc.273.7.3963 [DOI] [PubMed] [Google Scholar]
- Novick P., Ferro S., Schekman R. 1981. Order of events in the yeast secretory pathway. Cell. 25:461–469 10.1016/0092-8674(81)90064-7 [DOI] [PubMed] [Google Scholar]
- Papanikou E., Glick B.S. 2009. The yeast Golgi apparatus: insights and mysteries. FEBS Lett. 583:3746–3751 10.1016/j.febslet.2009.10.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S.R. 2007. Unsolved mysteries in membrane traffic. Annu. Rev. Biochem. 76:629–645 10.1146/annurev.biochem.76.061705.130002 [DOI] [PubMed] [Google Scholar]
- Rieder S.E., Banta L.M., Köhrer K., McCaffery J.M., Emr S.D. 1996. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol. Biol. Cell. 7:985–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubartelli A., Cozzolino F., Talio M., Sitia R. 1990. A novel secretory pathway for interleukin-1 beta, a protein lacking a signal sequence. EMBO J. 9:1503–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer T., Zentgraf H., Zehe C., Brügger B., Bernhagen J., Nickel W. 2004. Unconventional secretion of fibroblast growth factor 2 is mediated by direct translocation across the plasma membrane of mammalian cells. J. Biol. Chem. 279:6244–6251 10.1074/jbc.M310500200 [DOI] [PubMed] [Google Scholar]
- Schotman H., Karhinen L., Rabouille C. 2008. dGRASP-mediated noncanonical integrin secretion is required for Drosophila epithelial remodeling. Dev. Cell. 14:171–182 10.1016/j.devcel.2007.12.006 [DOI] [PubMed] [Google Scholar]
- Seelenmeyer C., Stegmayer C., Nickel W. 2008. Unconventional secretion of fibroblast growth factor 2 and galectin-1 does not require shedding of plasma membrane-derived vesicles. FEBS Lett. 582:1362–1368 10.1016/j.febslet.2008.03.024 [DOI] [PubMed] [Google Scholar]
- Shorter J., Watson R., Giannakou M.E., Clarke M., Warren G., Barr F.A. 1999. GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. EMBO J. 18:4949–4960 10.1093/emboj/18.18.4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague G.F., Jr 1991. Assay of yeast mating reaction. Methods Enzymol. 194:77–93 10.1016/0076-6879(91)94008-Z [DOI] [PubMed] [Google Scholar]
- Stenmark H., Aasland R., Driscoll P.C. 2002. The phosphatidylinositol 3-phosphate-binding FYVE finger. FEBS Lett. 513:77–84 10.1016/S0014-5793(01)03308-7 [DOI] [PubMed] [Google Scholar]
- Suzuki K., Kamada Y., Ohsumi Y. 2002. Studies of cargo delivery to the vacuole mediated by autophagosomes in Saccharomyces cerevisiae. Dev. Cell. 3:815–824 10.1016/S1534-5807(02)00359-3 [DOI] [PubMed] [Google Scholar]
- Teis D., Saksena S., Emr S.D. 2008. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev. Cell. 15:578–589 10.1016/j.devcel.2008.08.013 [DOI] [PubMed] [Google Scholar]
- Temmerman K., Ebert A.D., Müller H.-M., Sinning I., Tews I., Nickel W. 2008. A direct role for phosphatidylinositol-4,5-bisphosphate in unconventional secretion of fibroblast growth factor 2. Traffic. 9:1204–1217 10.1111/j.1600-0854.2008.00749.x [DOI] [PubMed] [Google Scholar]
- Vida T.A., Emr S.D. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128:779–792 10.1083/jcb.128.5.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Klionsky D.J. 2009. An overview of the molecular mechanism of autophagy. Curr. Top. Microbiol. Immunol. 335:1–32 10.1007/978-3-642-00302-8_1 [DOI] [PMC free article] [PubMed] [Google Scholar]