Ubiquitylation of the intracellular domain of Drosophila Delta is necessary for Notch activation.
Abstract
DSL proteins are transmembrane ligands of the Notch receptor. They associate with a RING (really interesting new gene) family E3 ubiquitin ligase, either Neuralized (Neur) or Mindbomb 1 (Mib1), as a prerequisite to signaling. Although Neur and Mib1 stimulate internalization of DSL ligands, it is not known how ubiquitylation contributes to signaling. We present a molecular dissection of the intracellular domain (ICD) of Drosophila melanogaster Delta (Dl), a prototype DSL protein. Using a cell-based assay, we detected ubiquitylation of Dl by both Neur and Mib1. The two enzymes use distinct docking sites and displayed different acceptor lysine preferences on the Dl ICD. We generated Dl variants that selectively perturb its interactions with Neur or Mib1 and analyzed their signaling activity in two in vivo contexts. We found an excellent correlation between the ability to undergo ubiquitylation and signaling. Therefore, ubiquitylation of the DSL ICD seems to be a necessary step in the activation of Notch.
Introduction
The Notch transmembrane receptors are activated by transmembrane ligands of the DSL family, which is subdivided into the Delta (Dl) and Serrate (Ser)/Jagged subfamilies in higher metazoans (Kopan and Ilagan, 2009). Contact of Notch and DSL, however, is not sufficient for eliciting intracellular signal transduction. Signaling is productive only when Notch and DSL are engaged in trans, namely from adjacent cells, whereas cis-binding (Notch and DSL on the same cell) is usually inhibitory to signaling (de Celis and Bray, 1997; Klein et al., 1997; Micchelli et al., 1997; Miller et al., 2009; Sprinzak et al., 2010). Even when Notch and DSL are engaged in trans, signaling ensues only when DSL proteins are coexpressed with a ubiquitin (Ub) E3 ligase (Pitsouli and Delidakis, 2005; Le Borgne, 2006). Work from our laboratory and others over the past decade has characterized two families of RING (really interesting new gene) domain E3 ligases, which have the ability to activate the DSL proteins Neuralized (Neur; Deblandre et al., 2001; Lai et al., 2001; Pavlopoulos et al., 2001; Yeh et al., 2001) and Mindbomb 1 (Mib1; Itoh et al., 2003; Barsi et al., 2005; Koo et al., 2005a; Lai et al., 2005; Le Borgne et al., 2005; Pitsouli and Delidakis, 2005; Wang and Struhl, 2005). RING domains catalyze Ub transfer from an E2 intermediate (Ub-conjugating enzyme) to the substrate protein (Deshaies and Joazeiro, 2009). Coexpression of DSL proteins with a Neur or Mib1 E3 ligase stimulates DSL clearance from the cell surface and its relocalization into endosomes (Lai et al., 2001, 2005; Pavlopoulos et al., 2001; Le Borgne et al., 2005). Ubiquitylation of plasma membrane proteins is a signal for endocytosis as well as further sorting steps in intracellular trafficking (Acconcia et al., 2009; Clague and Urbé, 2010), raising the possibility that Neur and Mib1 proteins ubiquitylate DSL ligands to trigger their endocytosis. Indeed, DSL activity seems to depend on a select set of endocytic proteins, namely dynamin (Seugnet et al., 1997), epsin (Overstreet et al., 2004; Wang and Struhl, 2004), and auxilin (Eun et al., 2008; Kandachar et al., 2008; Banks et al., 2011).
The correlation between E3 ligase expression, DSL internalization, and signaling has given rise to several (nonmutually exclusive) hypotheses regarding the mechanism of DSL signal emission (Le Borgne, 2006; Weinmaster and Fischer, 2011). The mechanical force hypothesis proposes that DSL endocytosis pulls on the trans-bound Notch molecule, thus deforming its extracellular juxtamembrane domain and exposing a buried juxtamembrane metalloprotease cleavage site (Parks et al., 2000; Nichols et al., 2007; Gordon et al., 2008). This promotes Notch cleavage, which is a prerequisite for receptor activation. The recycling hypothesis proposes that endocytosis of DSL, which is synthesized as an inactive molecule, is followed by its recycling to the plasma membrane after it has been modified (in a yet uncharacterized manner) in an endosomal compartment, such that it is now competent to engage in productive signaling (Wang and Struhl, 2004; Emery et al., 2005). Recycling may mediate relocalization of DSL to a plasma membrane microdomain conducive to signaling (Heuss et al., 2008; Rajan et al., 2009; Benhra et al., 2010). All hypotheses emphasize internalization rather than ubiquitylation, assuming that the former is a direct consequence of the latter. Yet, there are still many open questions. The cargo complex, which undergoes ubiquitylation, is only rather poorly characterized. Is the DSL protein itself ubiquitylated or does the Ub tag mark another adaptor protein, perhaps even the E3 ligase itself? The little data on DSL ubiquitylation by Neur and Mib1 are based mostly on in vitro reconstitutions (Deblandre et al., 2001; Koutelou et al., 2008). Ubiquitylation using cell-based assays has also been reported (Itoh et al., 2003; Chen and Casey Corliss, 2004; Koo et al., 2005b; Song et al., 2006; Skwarek et al., 2007). However, these assays used native immunoprecipitation conditions, leaving open the possibility that additional proteins, besides Dl itself, may have been detected bearing the Ub modification, whereas the molecular masses (MMs) detected are consistent with proteins in a size range similar to Dl.
DSL intracellular domains (ICDs) should play a central role in assembling the cargo recognition complexes in the process of DSL trafficking. Consistent with a trafficking–signaling connection, removal of the ICD has been shown to disable DSL proteins, even to convert them to signaling antagonists (Chitnis et al., 1995; Hukriede et al., 1997; Sun and Artavanis-Tsakonas, 1997). Replacement of the Dl ICD with a heterologous non-Ub–mediated endocytic motif from the low density lipoprotein (LDL) receptor (Wang and Struhl, 2004) was able to restore signaling, albeit only partially; this rescue required the integrity of the LDL receptor endocytic motif, suggesting a causal role of endocytosis in signal emission rather than the converse (endocytosis being a consequence of productive signaling). Dissection of the Drosophila melanogaster Dl and Ser ICDs has identified three endocytic motifs, an Asn-based peptide on each protein, and a dileucine motif on Ser (Glittenberg et al., 2006; Fontana and Posakony, 2009). The Ser Asn motif mediates interaction with both Neur and Mib1, whereas the similar Dl motif was shown to be necessary for Neur binding (Mib1 was not tested). Furthermore, the Ser Asn motif was shown to be absolutely necessary for signaling activity, whereas the LL motif was dispensable. It is likely that additional endocytic motifs are found on DSL proteins. Vertebrate DSL proteins, whose ICDs have diverged significantly from insect ones, do not contain the aforementioned Asn or LL motifs. Two mouse Dl paralogues, Dll1 and Dll3, display distinct modes of endocytosis: Dll1 requires ubiquitylation, whereas Dll3, which is not ubiquitylated, as it contains no lysines in its ICD, can internalize and recycle just as efficiently (Heuss et al., 2008). However, only Dll1 can signal efficiently, suggesting that internalization alone is not sufficient for signaling. In Drosophila too, the correlation between DSL internalization and signaling is not perfect. In liquid facets mutant tissue (lacking the endocytic adaptor epsin), bulk Dl internalization occurs normally, but signaling is abolished (Wang and Struhl, 2004). Conversely, the SerLL variant, which lacks the dileucine internalization motif, displays defective internalization but signals efficiently (Glittenberg et al., 2006).
In summary, DSL proteins have been variously shown to interact with E3 ligases and to be actively endocytosed. However, the mechanistic relation between these events and DSL signaling is still largely unknown, owing to the complexity of transmembrane protein trafficking and the inability to distinguish the signaling pool of DSL proteins from the bulk. Here, we have molecularly dissected the Drosophila Dl ICD and have tested five parameters: (1) interaction with E3 ligases Neur and Mib1, (2) ubiquitylation by these enzymes, (3) ability to signal in two different contexts, (4) subcellular distribution, and (5) efficiency of endocytosis. We identify distinct motifs of the Dl ICD that are required for physical interaction with Neur and Mib1. We find that Dl is ubiquitylated and that physical interactions between Dl and the E3 ligases via the Dl ICD motifs are a prerequisite for its ubiquitylation. Therefore, we propose that both Mib1 and Neur can directly ubiquitylate Dl and that this enhances Dl endocytosis. More importantly, we find an excellent correlation between the ability of Dl to undergo ubiquitylation and its ability to signal. Activity also correlates quite well with endocytic efficiency but not with subcellular localization.
Results
Dl ICD conserved motifs are interaction domains for Neur and Mib1
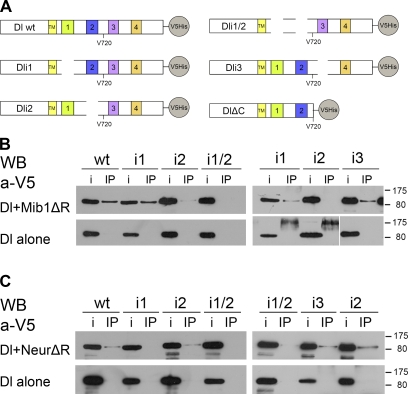
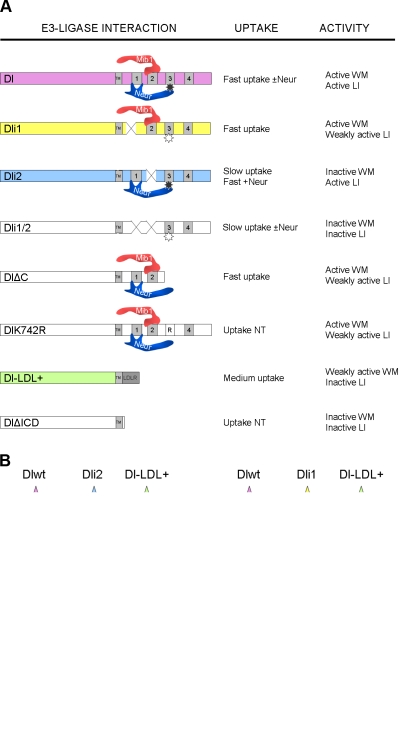
To identify important functional elements in the Dl ICD, we searched for conserved motifs among distantly related insect species. We retrieved Dl orthologue sequences from a basal dipteran (Aedes aegypti) as well as from representatives of six different insect families (Bombyx mori, Tribolium castaneum, Apis mellifera, Periplaneta americana, Acyrthosiphon pisum, and Pediculus humanus corporis). When we aligned Dl ICDs (Fig. S1), four short conserved motifs were identified; a fifth motif was noticed by visual inspection of the alignment, although it had not been deemed significant enough by the ClustalW2 algorithm. The first motif is the stop transfer sequence at the very N terminus of the ICD. We named the remaining four Dls ICD1, ICD2, ICD3, and ICD4. To investigate the functional importance of these motifs, we generated mutated forms of Dl by deleting each of the three motifs ICD1–3: Dli1 lacks ICD1, Dli2 lacks ICD2, and Dli3 lacks ICD3 (Fig. 1 A). We further constructed Dli1/2, which lacks both ICD1 and ICD2, and we obtained the C-terminal truncation DlΔC, which inserts a stop codon after amino acids 720 and removes ICD3, ICD4, and beyond (Rand, M.D., personal communication). All mutants and the wild-type (wt) control were C-terminally tagged with a 6×His-V5 epitope to facilitate biochemical detection (see Materials and methods and Fig. 1 A).
Figure 1.
Dl interactions with Neur and Mib1. (A) Schematic of Dl and its variants tested in this study. TM, transmembrane domain. ECDs and ICDs are not drawn to scale. For the exact extent of motifs 1–4, refer to Fig. S1. (B) C-terminally V5-tagged Dl variants, as indicated, were expressed alone (bottom) or with Myc-tagged Mib1ΔR (top). Immunoprecipitation (IP) lanes: extracts were immunoprecipitated with anti-Myc and detected with anti-V5. Input (i) lanes: 4% of the total extract from the same transfection was kept before immunoprecipitating the rest. The main band is full-length Dl (predicted MM of 90 kD). The bands visible in the bottom right near 175 kD are nonspecific cross-reacting bands. Marker sizes are shown on the right in kilodaltons. The white line indicates that intervening lanes have been spliced out. (C) The same Dl variants were expressed alone (bottom) or with NeurΔR (top). Extracts were immunoprecipitated with an anti-Neur antiserum and detected with anti-V5. Labels are the same as in B. WB, Western blot.
We coexpressed the various Dl mutants in Drosophila Schneider S2 cells with Mib1ΔR, a Mib1 variant that lacks the catalytic RING domain, to address a possible function of these motifs as Mib1 docking sites. Mib1ΔR had been shown before to physically interact with wt Dl and Ser, and deletion of the RING domain facilitated detection of these interactions, as it diminished DSL degradation (Lai et al., 2005). We observed that Dl, Dli1, and Dli3 coimmunoprecipitated with Mib1ΔR. On the contrary, coimmunoprecipitation of Dli2 and Dli1/2 was severely compromised, suggesting that motif 2 is responsible for the interaction with Mib1 (Fig. 1 B).
The same assay was used to test the ability of the mutant forms to coimmunoprecipitate with NeurΔR, a RING-less Neur variant, which was already known to interact with wt Dl and Ser (Lai et al., 2001; Pitsouli and Delidakis, 2005; Glittenberg et al., 2006). Whereas Dli2 and Dli3 were coimmunoprecipitated by an anti-Neur antibody, Dli1 and Dli1/2 coimmunoprecipitation was strongly attenuated, pointing to motif 1 as a potential interaction site for Neur (Fig. 1 C; in agreement with Fontana and Posakony, 2009). Based on these results, we propose that two of the conserved motifs in Dl ICD, ICD1 and ICD2, are docking sites for Neur and Mib1, respectively, whereas ICD3 is dispensable for the recruitment of either Ub ligase. ICD4 is also dispensable because DlΔC could be coimmunoprecipitated with either Mib1ΔR or NeurΔR (unpublished data).
Dl is ubiquitylated by Neur and Mib1
We next coexpressed the Dl variants with catalytically active (full length) E3 ligases in S2 cells to assess their ability to be ubiquitylated. Ubiquitylated species were detected after cotransfecting with an Xpress epitope–tagged version of Ub. We pulled down 6×His-tagged Dl under strong denaturing conditions to eliminate other interacting proteins and thus ensure that ubiquitylated species observed represent Dl itself. Transfected Dl displayed low background levels of ubiquitylation (anti-Xpress signal), visible only at higher exposures than those displayed in Fig. 2. For this reason, we transfected with increasing amounts of neur- or mib1-expressing plasmids and asked whether we see a corresponding increase in Dl ubiquitylated species.
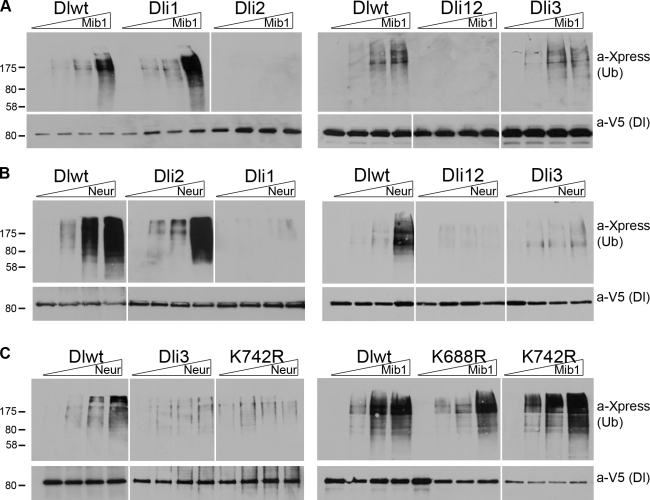
Figure 2.
Ubiquitylation of Dl variants. (A) Dl-expressing constructs were cotransfected with Xpress-Ub and increasing amounts (0, 0.5, 1, and 1.5 µg) of Mib1-expressing constructs. Dl protein was isolated by affinity purification on a Ni2+ resin under denaturing conditions. Eluates of the Ni2+ column were probed with anti-Xpress to detect ubiquitylated species (top shows 1/4 of total loaded) and anti-V5 to detect total Dl as a loading control (bottom shows 1/3 of total loaded). Note that ubiquitylated species, where present, run at much higher MMs than the expected 90 kD of unmodified Dl. (B) Dl-expressing constructs were cotransfected with Xpress-Ub and increasing amounts (0, 0.5, 1, and 1.5 µg) of Neur-expressing constructs. Note weaker levels of Ub signal in Dli3. (C) Ubiquitylation assays performed as in A and B using the Dl variants and E3 ligases shown at the top. Note that Dl-K742R is weakly ubiquitylated by Neur but resembles wt Dl in its response to Mib1. MM markers are shown in kilodaltons. White lines indicate that intervening lanes have been spliced out.
When Mib1 was expressed in increasing amounts (Fig. 2 A), we detected a concomitant increase in high MM ubiquitylated species of Dl, Dli1, or Dli3 but not of Dli2 or Dli1/2. Using Neur in the same assay, we could stimulate ubiquitylation of wt Dl as well as Dli2 but not Dli1 or Dli1/2 (Fig. 2 B). These results are in agreement with our interaction data because deletion of each docking site compromised ubiquitylation by the cognate E3 ligase; Dli1 was ubiquitylated only by Mib1, and Dli2 was ubiquitylated only by Neur. For both enzymes, ubiquitylation was dependent on the RING domain, as incubation with NeurΔR or Mib1ΔR strongly reduced the Ub signal (Fig. S2 A). In all experiments, cells were incubated with E64, a lysosomal protease inhibitor (Rock et al., 1994), before lysis. When E64 was omitted, much less ubiquitylated Dl could be detected, consistent with lysosomal clearance of Dl ubiquitylated species (Fig. S2 B). E64 did not qualitatively alter the MM pattern of Dl Ub adducts, suggesting that the modifications observed are not a secondary consequence of blocking the lysosomal pathway. Importantly, a major monoubiquitylated band at the predicted MM of 102 kD was not produced by either Mib1 or Neur. This shows that these E3 ligases catalyze preferentially the multi- or polyubiquitylation of Dl.
Dli3 gave strong ubiquitylation by Mib1 and detectable ubiquitylation by Neur, albeit at much reduced levels compared with wt (Fig. 2, A and B). This was unexpected because deletion of ICD3 had not affected recruitment of NeurΔR in our earlier coimmunoprecipitation assay (Fig. 1). We entertained the possibility that Neur docks on ICD1 but uses a lysine in ICD3 to conjugate the Ub moiety. K742, the sole lysine residue within ICD3, is in fact the most highly conserved lysine in the entire Dl ICD (Fig. S1). A mutant, Dl-K742R, converting this lysine to arginine, which cannot be ubiquitylated, behaved like Dli3, namely it was only weakly ubiquitylated by Neur, whereas it still displayed robust ubiquitylation by Mib1 (Fig. 2 C). We conclude that K742 is a preferred Ub acceptor site by Neur but not by Mib1, suggesting that the two E3 ligases produce different ubiquitylated products. Six other K → R mutations were assayed, namely K629R, K636R, K683R, K688R, K775R, as well as the double mutant K683,688R. None of these showed any defect in ubiquitylation by either E3 ligase (Fig. 2 C and not depicted). Note that K683 and K688 are the two lysines within ICD2, whereas K636 is the sole lysine within motif ICD1. None of the three is conserved across insects, although K683 and K688 show partial conservation, as do K629 and K775 (Fig. S1). Finally, we tested the C-terminal truncation DlΔC, which lacks K742 as well as K739, K762, and K775. Neur-dependent ubiquitylation was compromised, as expected from the absence of K742, but Mib1-dependent ubiquitylation was unaffected (unpublished data), eliminating the remaining C-terminal–proximal lysine residues as preferred Mib1 modification sites. It therefore appears that Neur displays strong selectivity for K742, whereas Mib1 is more promiscuous in its choice of lysine residue to be ubiquitylated.
Role of Dl ICD conserved motifs in wing dorsoventral (DV) boundary induction and sensory organ precursor (SOP) lateral inhibition
The analysis presented in the previous two sections revealed the role of ICD1 and ICD2 in binding Neur and Mib1, respectively, and that of ICD3 as a main ubiquitylation target by Neur. To address the relationship between ubiquitylation and signaling, we proceeded to ask whether these motifs play a role in Dl activity in vivo. To that end, we tested each mutant in two settings. In the first, we asked whether ectopic expression of Dl in the larval wing epithelium can induce wingless (wg), a Notch target gene normally found in the DV boundary (de Celis et al., 1996; Doherty et al., 1996; Neumann and Cohen, 1996). This event is known to be dependent on mib1 (Lai et al., 2005; Le Borgne et al., 2005; Pitsouli and Delidakis, 2005; Wang and Struhl, 2005), which is ubiquitously expressed in the wing disk. In the second setting, we asked whether transgenic Dl variants can rescue the process of SOP lateral inhibition (Bray, 1998) in a Dl Ser mutant background. Consistent with the expression of neur in proneural territories, we have previously shown that this process relies on Neur, although Mib1 also plays a minor role (Pitsouli and Delidakis, 2005).
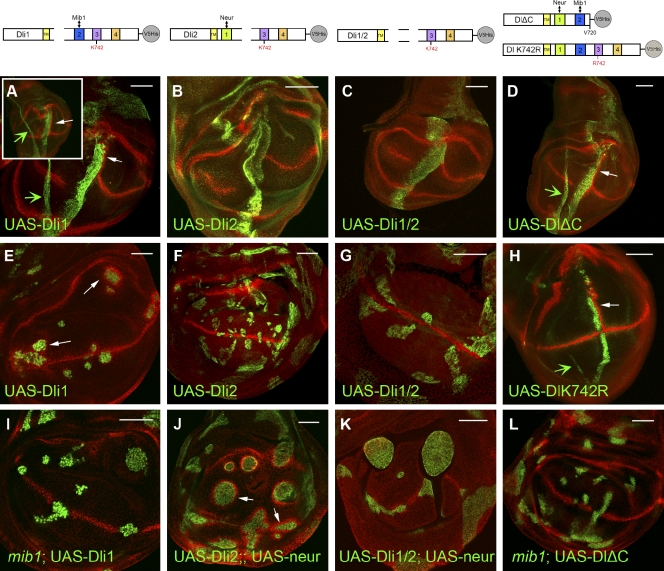
To monitor the ability of Dl mutants to signal in wing DV boundary induction, we expressed wt and mutant forms of the ligand in a stripe perpendicular to the DV boundary using ptc-Gal4. When an upstream activation sequence (UAS)-Dl wt transgene is expressed, ectopic Notch activity (reported by Wg expression) becomes apparent in cells immediately adjacent to the ptc-GAL4–expressing cells. As previously reported, Notch activation is restricted to dorsal cells, as a result of differential Notch glycosylation (Irvine and Vogt, 1997), and is excluded from high Dl-expressing cells because of the well-documented effect of Dl cis-inhibition of Notch (Doherty et al., 1996; de Celis and Bray, 1997). For this reason, Wg induction is stronger in the posterior compartment, where the ptc expression stripe ends abruptly, creating a sharp expression/nonexpression boundary, whereas it is weaker anterior to the stripe, where ectopic Dl expression levels drop more gradually (Fig. 3 A, inset). Dli1, DlΔC, and Dl-K742R were active in this assay, exhibiting robust ectopic Wg expression (Fig. 3, A, D, and H). When ICD2 was deleted (in Dli2 and Dli1/2), thus interfering with interaction with Mib1 (Fig. 1), no Wg induction was observed (Fig. 3, B and C). Thus, inability to interact with Mib1 abolishes signaling of Dl in trans to Notch but does not affect cis-inhibition in this context. Inability to interact or get ubiquitylated by Neur, on the other hand, does not abolish either trans-activation or cis-inhibition (Fig. 3, A, D, and H).
Figure 3.
Induction of Wg by Dl variants. Third instar wing pouches overexpressing Dl variants and stained for Wg (red) are shown. Anterior is to the left and ventral is down. (A–D and H) Dl variants as indicated are expressed with ptc-Gal4 and detected with anti-Dl. (A, inset) Disk expressing wt Dl under ptc-Gal4. Note ectopic Wg expression posterior to the Dl stripe. The narrow stripe of Dl expression sometimes seen (green arrows) comes from the overlying squamous peripodial membrane cells, which do not express wg in response to Notch. (E–G) Ectopic expression of Dl variants, as indicated, in random clones. Dli1 (E) induces Wg, whereas Dli2 (F) and Dli1/2 (G) do not. In E, the Dl transgene is driven by α-tubulin–Gal4, and the clones are visualized by coexpressed UAS–nuclear GFP (green). In F and G, the Dl transgenes are expressed by act5C-Gal4, and the clones are visualized by anti-Dl (green). (I and L) The indicated Dl variants are expressed in clones mutant for mib1. Clones are marked as in E. Compared with D and E, no ectopic Wg is produced, confirming that signaling by these variants depends on Mib1. (J and K) The indicated Dl variants are coexpressed with EGFP-Neur under act5C-Gal4 control. Clones are visualized by the presence of EGFP-Neur. Comparing F with J, we conclude that Neur, when ectopically provided, can activate Dli2, whereas it cannot activate Dli1/2 (G vs. K). The slight Wg expression close to the DV boundary in K is occasionally seen also in Dli1/2-expressing clones (without UAS-Neur), hinting at a possible residual activity of the protein. Open arrows show Wg induction. TM, transmembrane domain. Bars, 50 µm.
The activity displayed in this assay by Dli1 and DlΔC is Mib1 dependent, as it was abolished in clones expressing these proteins and simultaneously mutated for mib1 (Fig. 3, E, I, and L). The same had been shown earlier for wt Dl. Conversely, it was shown before that neur, if ectopically expressed, can compensate for loss of mib1 in this context (Pitsouli and Delidakis, 2005; Wang and Struhl, 2005). When we coexpressed Dli2 with Neur (here, we did not use a mib1− background because Dli2 is inactive in a wt background anyway), we got a strong induction of Wg (Fig. 3, F and J), suggesting that this mutant regained its ability to signal. We were not able to detect any Wg induction when we expressed Dli1/2 with or without Neur (Fig. 3, G and K), confirming that this variant, which is unable to interact with either Neur or Mib1, is inactive, other than being able to cis-inhibit Notch. This is in agreement with previous data showing that Ub ligase activity is not needed for cis-inhibition (Glittenberg et al., 2006; Miller et al., 2009).
The Dl variants behaved differently when assayed in the context of lateral inhibition. Notch signaling normally restricts the generation of SOPs from a field of tens of cells to only one or two. Clones mutant for Dl Ser give rise to clustered supernumerary SOPs, as all cells within the proneural field adopt the SOP fate. In a Dl Ser background, expression of UAS-Dl by the ubiquitous driver α-tubulin–Gal4 restored Notch signaling, and individualized SOPs were born (Pitsouli and Delidakis, 2005). UAS-Dli2 reproduced this effect (Fig. 4 C), but neither Dli1 nor Dli1/2 was able to do so (Fig. 4, B and D), pointing to the ICD1–Neur interaction as playing a major role in this context. Interestingly, DlΔC and Dl-K742R, which can interact with both Mib1 and Neur, but cannot be properly ubiquitylated by Neur, could not rescue lateral inhibition (Fig. 4, E and F). These data suggest that not only interaction (affected by Dli1 and Dli1/2) but also strong ubiquitylation by Neur (affected by DlΔC and Dl-K742R) is needed in this process. Upon close observation of clone phenotypes, we noticed that Dli1/2 and DlΔICD, a mutant in which the entire Dl ICD has been deleted (Wang and Struhl, 2004), produced clusters of adjacent SOPs (Fig. 4, A and D), the same phenotype that loss of Dl Ser would have in the absence of any transgene—this is in agreement with the complete inactivity of these two variants. In the case of Dli1, DlΔC, and Dl-K742R, on the other hand (Fig. 4, B, E, and F), although supernumerary SOPs were still produced, these were often spaced apart from each other, suggesting that a low level of lateral inhibition was taking place. As Dli1, DlΔC, and Dl-K742R are good substrates for Mib1, it is likely that endogenous Mib1 can provide partial activity in this context, which agrees with a previously noted minor role of Mib1 in lateral inhibition (Pitsouli and Delidakis, 2005).
Figure 4.
Rescue of lateral inhibition by Dl variants. (A–G′′) In all panels, notum regions of third instar wing disks are shown stained for Sens (red; shown separately in A′–G′), a marker for SOPs, which normally arise as single cells at defined positions. Dl Ser mutant clones are marked by the expression of nuclear GFP (green); clones coexpress the indicated Dl variant. Note that a singularized SOP is present only in C, whereas all other variants cannot abolish the birth of clustered supernumerary SOPs. An unrescued Dl Ser mutant would look like the clones in A or D (e.g., Pitsouli and Delidakis, 2005). (G–G′′) In G, we have costained for Wg (blue; shown separately in G′′). The edge of the wing pouch is visible at the bottom, where Dl-LDL+ is capable of inducing Wg (arrows), confirming its activity in a different context. Anterior is to the left and dorsal is up. LDLR, LDL receptor; TM, transmembrane domain. Bars, 50 µm.
In summary, of the three Dl ICD motifs, Neur-associated motifs ICD1 and 3 seem to play a major role in lateral inhibition, whereas the Mib1-associated motif ICD2 is predominant in DV boundary induction. We wondered whether lateral inhibition might be supported by a previously reported active Dl variant that cannot be ubiquitylated at all because of the lack of lysine residues in its ICD. This artificial variant has been made by fusing an NPxY-dependent endocytosis signal from a mammalian LDL receptor (Chen et al., 1990) to the Dl extracellular domain (ECD)+ transmembrane domain (Wang and Struhl, 2004). This Dl-LDL+ fusion had been shown to be active in wg induction but had not been tested in the context of lateral inhibition. In Dl Ser clones expressing UAS–Dl-LDL+, we detected no rescue of lateral inhibition, as a large number of adjacent SOPs were reproducibly detected (Fig. 4 G). Therefore, whereas a heterologous endocytosis signal can restore Dl activity in one context (DV boundary induction), it fails to do so in another (lateral inhibition).
Subcellular localization of Dl variants
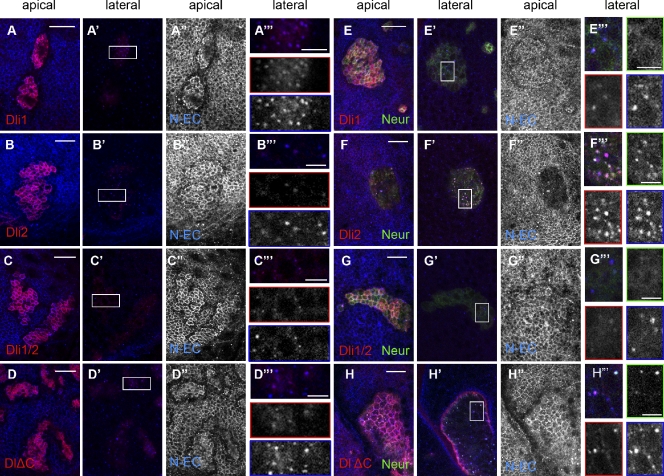
As ubiquitylation is a signal for membrane cargo trafficking, we wondered whether the inability of some Dl mutants to accept Ub moieties would alter their patterns of internalization. Most importantly, we wanted to confirm that the intracellular deletions had not adversely affected other aspects of the protein’s biogenesis, such as targeting to the cell surface. To confirm cell surface exposure for all our Dl variants, we visualized them by immunofluorescence in the absence of detergent. In all cases, strong signal was obtained on the apical plasma membrane with much weaker signal basally, suggesting that polarized exocytosis of Dl was not affected by these deletions (Fig. S3). This was even true for the inactive Dli1/2, eliminating the trivial possibility that its inactivity results from a defect in its plasma membrane localization.
When assayed in permeabilized tissue, wt Dl (either endogenous or overexpressed) is highly enriched on the apical cell surface with additional intracellular puncta, which are in large part endosomes (Pavlopoulos et al., 2001; Le Borgne and Schweisguth, 2003; Wang and Struhl, 2004). We used three representative markers of different endosomal compartments to ask whether they colocalize with intracellular Dl in an attempt to determine whether our Dl ICD mutants may affect the protein’s trafficking route. We chose Sara as an early endosome marker because of its recently reported connection with Notch–Dl signaling (Bökel et al., 2006; Coumailleau et al., 2009). Rab11, which has also been implicated in Dl function (Emery et al., 2005; Jafar-Nejad et al., 2005; Banks et al., 2011), was used as a recycling endosome marker. Hepatocyte growth factor–regulated tyrosine kinase substrate (Hrs) was used as a sorting endosome marker. It is known to bind Ub-tagged cargoes and stimulate their import into intraluminal vesicles of the multivesicular body (Lloyd et al., 2002; Jékely and Rørth, 2003). Dl wt intracellular puncta showed the best colocalization with Sara (46%). Less colocalization was evident with Hrs (37%), and even less was evident with Rab11 (15%). Dl variants showed subapical/laterobasal puncta with a similar distribution to that of the wt among the three markers used (Table 1 and Fig. S4). One exception was Dl-LDL+, which colocalized less efficiently with Sara (22%). Also, Dli1/2 and DlΔC showed decreased colocalization with Hrs, which we do not presently understand, as they have essentially complementary deletions in the Dl ICD. We conclude that the Dl ICD confers a preference to accumulate into Sara endosomes, but none of our ICD deletions seems to abolish this affinity or greatly alter Dl distribution among the endosomal compartments tested.
Table 1.
Dl colocalization with Sara, Hrs, and Rab11
| Dl variant | Sara | Hrs | Rab11 | ||||||
| Colocalization | n | P-value | Colocalization | n | P-value | Colocalization | n | P-value | |
| % | % | % | |||||||
| Alone | |||||||||
| wt | 46 | 166 | − | 37 | 109 | − | 15 | 141 | − |
| Dli1 | 60 | 106 | 0.02 | 37 | 73 | 1.00 | 14 | 200 | 0.75 |
| Dli2 | 54 | 112 | 0.22 | 33 | 122 | 0.58 | 13 | 238 | 0.54 |
| Dli1/2 | 47 | 100 | 0.90 | 18a | 201 | <0.01a | 14 | 125 | 1.00 |
| DlΔC | 42 | 114 | 0.62 | 18a | 104 | <0.01a | 16 | 129 | 1.00 |
| Dl-LDL | 19a | 503 | <0.01a | 34 | 131 | 0.68 | 14 | 151 | 0.87 |
| +Neur | |||||||||
| wt | 45 | 317 | 0.92b | 5a | 222 | <0.01ab | 10 | 192 | 0.24b |
| Dli1 | 49 | 214 | 0.48 | 6 | 143 | 0.63 | 11 | 150 | 1.00 |
| Dli2 | 46 | 226 | 0.86 | 6 | 343 | 0.57 | 11 | 240 | 1.00 |
| Dli1/2 | 34 | 170 | 0.02 | 4 | 93 | 1.00 | 11 | 124 | 1.00 |
| DlΔC | 46 | 156 | 0.85 | 5 | 134 | 0.80 | 10 | 129 | 1.00 |
| +Mib1 | |||||||||
| wt | 50 | 92 | 0.52b | NT | − | − | 13 | 286 | 0.66b |
The percentage of colocalization of each Dl variant with Sara, Hrs, and Rab11 is shown. The numbers refer to the percentage of Dl puncta that are also positive for each endocytic marker. n = total number of Dl puncta scored. P-values were calculated using the Fisher exact test against the Dl wt control. In the +Neur and +Mib1 sections, Dl variants were coexpressed with UAS-Neur or UAS-YFP-Mib1, respectively. The variant +Neur p-values are computed against Dl wt +Neur. Minus signs indicate not applicable data. NT, not tested.
Values below 0.01 are considered significantly different than wt.
The wt +Neur/Mib1 colocalization p-values are computed against Dl wt alone.
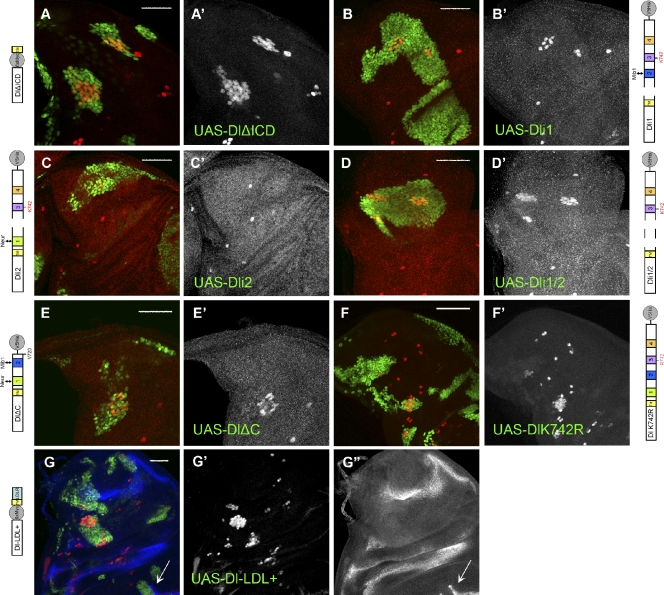
All of the aforementioned localization tests were performed in the larval wing epithelium, where mib1 is present, but neur is hardly expressed at all. We therefore repeated the assays with neur coexpression, whereupon Dl wt relocalizes dramatically as a result of stimulated endocytosis. It pulls away from the apical surface and moves into large intracellular puncta, where it often colocalizes with Neur. This apical Dl clearance is accompanied by clearance of Notch, which also accumulates on the Dl–Neur-positive intracellular puncta (Lai et al., 2001; Pavlopoulos et al., 2001). When coexpressed with Neur, dramatic differences were observed among the Dl variants (Fig. 5). Dli2 was cleared from the apical surface and massively relocalized into intracellular puncta together with Notch and Neur (Fig. 5 F), like wt Dl. Dli1, Dli1/2 (which cannot interact with Neur), and DlΔC (which interacts with Neur but cannot be properly ubiquitylated) retained their apical accumulation and were not enriched intracellularly (Fig. 5, E–H). This was mirrored in the effect on endogenous Notch, which was cleared from the apical surface only by the Dli2–Neur combination (Fig. 5, E′′–H′′). Therefore, apical clearance of Dl–Notch complexes by Neur can occur only when Dl can be ubiquitylated by Neur.
Figure 5.
Subcellular localization of Dl variants. Close ups of third instar wing epithelia stained for Dl ECD (red) and Notch ECD (N-EC). EGFP-Neur, whenever coexpressed, is detected in green. Overexpressed Dl variants are detected, whereas endogenous Dl is undetectable at the illumination level used. (A–D′′′) Ectopic expression of Dl variants, as indicated. A–D are apical single confocal sections, whereas A′–D′ are lateral single sections ∼3 µm below A–D. Strong accumulation of ectopic Dl apically is accompanied with strong endogenous Notch accumulation, shown separately in A′′–D′′. Laterally, the Dl variants accumulate in puncta that also contain Notch (A′–D′). Boxed regions of these panels are enlarged in A′′′–D′′′, in which individual channels are also shown: Dl (red borders) and Notch (blue borders). (E–H′′′) Ectopic coexpression of Dl variants, as indicated, with EGFP-Neur. E′–H′ are the corresponding lateral sections at 3 µm below E–H. E′′–H′′ show the apical Notch staining alone. (F′′) Note that only Dli2 + Neur efficiently clears Notch away from the apical surface; (F) Dl and Neur are also cleared. E′′′–H′′′ are enlarged sections of the boxed regions in E′–H′. Individual channels for Dl, Notch, and Neur are shown with red, blue, and green borders, respectively. (F′′′) Large lateral puncta of Dl are detected in the case of Dli2 colocalizing with Notch and Neur. The fewer DlΔC puncta (H′′′) also colocalize with Neur (and Notch), whereas Neur is diffusely cortical when coexpressed with Dli1 (E′′′) or Dli1/2 (G′′′). Note that, whenever overexpressed Dl accumulates apically (all panels except F), endogenous Notch seems depleted from a row of cells around the clone. This probably results from polarization of Notch in these cells toward the highly Dl-expressing cells of the clone. Bars: (A–H) 15 µm; (A′′′–H′′′) 5 µm.
We also tested the distribution of Dl variants among the three endosomal compartments, Sara, Rab11, and Hrs, upon Neur coexpression. The relative distribution of Dl in Sara and Rab11 endosomes essentially did not change (Table 1), despite the quantitative increase in endosomal puncta for the wt and Dli2. An overall decrease in Hrs colocalization was seen, but this was not Dl variant-specific, possibly hinting at a global effect of Neur, which was not studied further. A similar study cooverexpressing Dl with Mib1 failed because of lethality at prelarval stages. The few escapers that were recovered did not show a significantly altered distribution of Dl (Table 1). We conclude that the identified ICD motifs (1, 2, and 3), despite their influence on Dl ubiquitylation, do not significantly affect its trafficking route, at least as revealed by the small number of endosomal markers tested.
Endocytosis of Dl variants
As the differences in endocytosis among our Dl variants did not appear to be qualitative, we turned to a live antibody uptake assay, first described by Le Borgne and Schweisguth (2003), to gauge the efficiency of Dl internalization upon mutating the various ICD motifs. For technical reasons, this was best performed in pupal nota at 18–22 h after puparium formation. We cultured the dissected tissue in the presence of mouse anti-Dl antibody for 15 min before fixation. Using a different (guinea pig) anti-Dl antibody to detect total Dl after fixation/permeabilization, we could determine the fraction of total (guinea pig) Dl endosomal puncta that had gotten occupied by the live (mouse) antibody during the 15-min uptake window. This fraction (percentage of Dl internalized) ranged from 72 to 88% of the total Dl puncta for the wt, Dli1, and DlΔC variants (Fig. 6); the differences among these variants were statistically insignificant. However, Dli2 and Dli1/2 displayed a much slower endocytosis occupying only 38–45% of the total Dl-positive endosomes within the uptake window. Dl-LDL+ (56%) was also significantly more slowly endocytosed than wt but faster than Dli1/2. We conclude that the Dl ICD regulates the rate of Dl internalization and that motif i2, the Mib1 interaction motif, is critical for efficient internalization.
Figure 6.
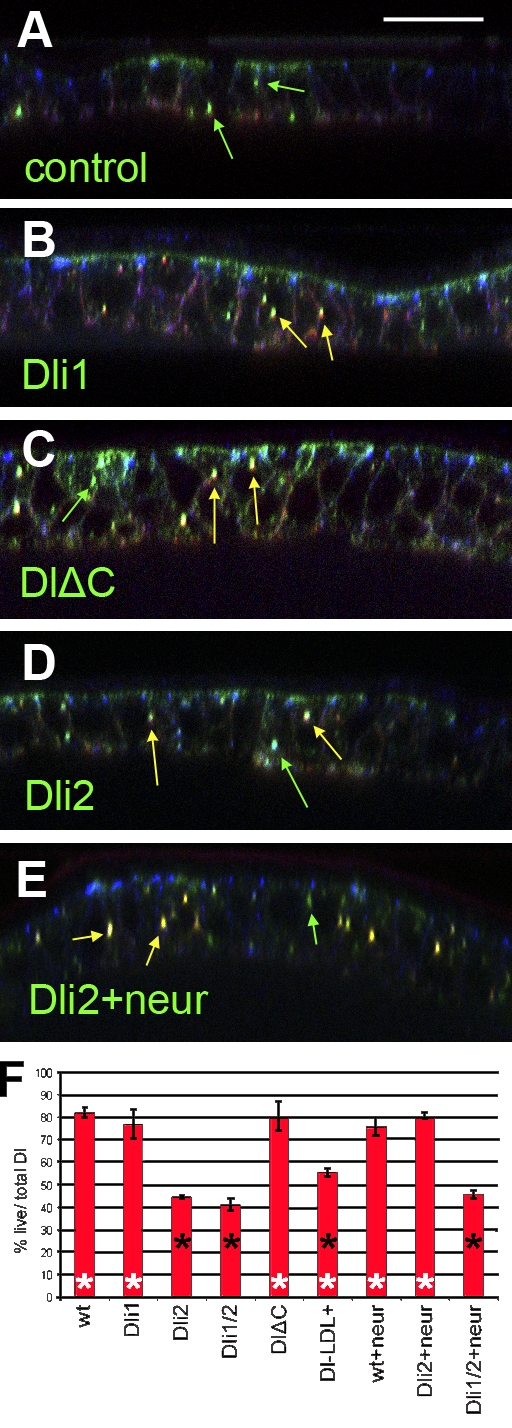
Live uptake assays for Dl variants. (A–E) Examples of pupal nota expressing the indicated Dl variant under Eq-Gal4. Optical cross sections are shown; apical is up, shown by high E-cadherin accumulation (blue). Anti-Dl taken up live for 15 min before fixation (red) and total anti-Dl (green) are shown. Yellow arrows mark Dl puncta that have been labeled by the live protocol; green arrows mark Dl puncta that did not get labeled by the live protocol. (A) The control notum expressed wt Dl but was cultured on ice to inhibit endocytosis; note the absence of yellow puncta and accumulation of the live anti-Dl (red) immunoreactivity on the basal side of the cells. In B–E, live uptake was performed at 25°C. (F) Bar graph of the percentages of total Dl puncta that were labeled by the live antibodies. The experiments were replicated two to three times, and means are shown. Error bars are standard deviations. Genotypes with significant differences from the wt (P < 0.01, Student’s t test) are indicated by filled asterisks. Genotypes with significant differences from Dli1/2 are indicated by open asterisks. Bar, 16 µm.
The Gal4 driver used to express our Dl variants in the pupal notum expresses mostly in nonsensory epithelial cells (Fig. S5), which do not express neur, consistent with ICD2, the Mib1-interacting motif, playing an important role in the rate of internalization. We wondered what would happen if we provided Neur exogenously. When we coexpressed Dl (wt) and Neur, the percentage of Dl taken up in a 15-min window remained unchanged (Fig. 6). However, the efficiency of Dli2 endocytosis increased dramatically to wt levels. Dli1/2 uptake remained slow. Therefore, in this assay, uptake efficiency seems to be correlated with ubiquitylation, with Dli2 being endocytosed slowly in the notum epithelium but attaining a faster uptake rate when Neur is supplied.
Discussion
Mechanism of DSL ligand activation
Our dissection of the Dl ICD has revealed two discrete motifs, ICD1 and ICD2, for docking of Neur and Mib1, respectively. Moreover, we showed that Mib1 and Neur can ubiquitylate Dl in Drosophila cells. As we have used stringent denaturing conditions to isolate Dl, we are confident that Dl is the ubiquitylation substrate and have identified a major acceptor residue for Neur-mediated ubiquitylation, K742. It has been suggested in the past that Dl is usually monoubiquitylated. However, our results show that both E3 ligases produce high MM species, consistent with multi/polyubiquitylation. Whereas Neur prefers K742 as the Ub acceptor site on Dl, Mib1 does not display a lysine preference*, pointing to qualitatively different Ub modifications catalyzed by the two E3 ligases. As different Ub modifications may be recognized by different endocytic adaptors, the possibility arises that Dl modified by Mib1 versus Neur can display different trafficking behavior. From our marker colocalization and endocytic uptake assays, we could not discern any major differences among Dl ICD variants. Dl accumulated in early Sara-positive endosomes and showed lower colocalization with Hrs or Rab11. The ICD was necessary for the Sara colocalization (because Dl-LDL+ showed a significant reduction), but none of the identified ICD motifs seemed to be necessary (Table 1). We did not discern differences in uptake rate between Mib1-modified and Neur-modified Dl either. Both Ub ligases promoted a high rate of Dl uptake (∼80% of Dl-positive endosome occupancy is achieved within 15 min; Fig. 6). A slower mode of Dl uptake, independent of either Mib1 or Neur, was typified by Dli1/2 (and Dli2 in the absence of Neur) and amounted to only ∼45% endosome occupancy in the same time. We conclude that Dl contains several endocytic motifs: the conserved ICD1, 2, and 3 mediate ubiquitylation by Mib1/Neur and rapid uptake, and additional uncharacterized endocytic mechanisms must also exist to account for the slower uptake of ICD1/2. The Dl-LDL+ variant, which uses a distinct Ub-independent mechanism of endocytosis, was taken up faster than Dli1/2 but slower than wt Dl.
Can we correlate Dl signaling activity with its ubiquitylation and/or trafficking? Correlation with ubiquitylation was very good (Fig. 7 A). When ubiquitylation by Mib1 was abolished (Dli2 or Dli1/2), the ligand lost its ability to activate Notch at the wing DV boundary. Reciprocally, in cases in which we eliminated ubiquitylation by Neur (Dli1, Dli1/2, and DlΔC), the ability of the ligand to sustain lateral inhibition was compromised. As mib1 is expressed ubiquitously in the wing disk, whereas neur is limited to proneural regions, there is some residual lateral inhibition activity by Dli1 and DlΔC, variants that retain interaction with Mib1 (Fig. 7 A). The behavior of Dl-LDL+ allows us to formulate a hypothesis about the relation of endocytosis to signaling (Fig. 7 B). A high rate of internalization at the plasma membrane seems to be a prerequisite for signaling in either context tested, with two qualifications: (1) Even a moderate internalization rate (Dl-LDL+) seems sufficient to promote wg expression but not lateral inhibition, which absolutely requires high rates. (2) For lateral inhibition to be properly executed, direct interaction/ubiquitylation by Neur is required in addition to high internalization rates. This requirement was inferred from the inability of Dli1 and DlΔC to fully rescue lateral inhibition and from the fact that neur loss of function does attenuate lateral inhibition but does not completely abolish it, as in the case of the mib1 neur double mutant (Pitsouli and Delidakis, 2005). This direct requirement for Neur may be caused by some subtle modulation of Dl endocytosis or subsequent recycling, which cannot be recapitulated by Mib1. Higher resolution uptake and colocalization assays may reveal such subtle effects in the future. An additional very likely reason for the indispensability of Neur in lateral inhibition is the network of regulatory interactions in which it participates. On one hand, Notch signaling probably represses neur expression, which attains the highest levels in the Notch refractory cell, the SOP (Huang et al., 1991). On the other, Notch signaling activates transcription of the Bearded (Brd) family of genes (Castro et al., 2005), which act to inhibit Dl–Neur interaction (Bardin and Schweisguth, 2006; De Renzis et al., 2006; Fontana and Posakony, 2009) in the Notch receiving cells (non-SOPs). Neither transcription nor activity of Mib1 has in any way been shown to respond to Notch signaling, which may account for why Mib1 alone is unable to fully sustain lateral inhibition.
Figure 7.
Dl activity depends on ubiquitylation. (A) The different Dl variants used are shown interacting with Neur or Mib1, depicted as touching at their respective docking site. The star in ICD3 represents K742; filled is ubiquitylated, and open is nonubiquitylated. WM, wing margin; LI, lateral inhibition; NT, not tested; TM, transmembrane domain; LDLR, LDL receptor. (B) Behavior of indicative Dl variants in two different cell contexts. Ovals attached to the Dl ICD represent Ub moieties. In the Mib1-only cell, they are arbitrarily depicted on three positions to indicate the lack of lysine preference. See Discussion for details. +ve, positive; −ve, negative.
Divergence of intracellular motifs on DSL proteins
We have identified three conserved domains in insect Dls that seem to mediate E3 ligase recruitment and Ub ligation, explaining the necessity of the ICD for signaling. The other Notch ligand, Ser, has been previously shown to require an Asn-based motif for both Neur and Mib1 interactions. This motif is well conserved among insects (Glittenberg et al., 2006; Fontana and Posakony, 2009) and possesses features reminiscent of both Dl ICD1 and ICD2. A QNEEN stretch is similar to the QNExN stretch in Dl ICD1, and an NNL is similar to the NNI/V present in most insects in Dl ICD2. QNxxN stretches were recently shown to be direct interaction sites for Neur (Fontana and Posakony, 2009), in agreement with our data on the role of ICD1 as a Neur docking site. In fact, similar peptides are found in the Brd family of Neur inhibitors (Bardin and Schweisguth, 2006; Fontana and Posakony, 2009). This enables Brd-like proteins to outcompete DSL binding to Neur, thus decreasing signal emission. The NNL/I/V stretch of DSL proteins could be important for Mib1 docking, although this amino acid stretch is lost from the Dls of B. mori and A. pisum, which do maintain the rest of ICD2 (Fig. 1). Mutation of the Ser NNL peptide partially reduced its ability to induce wg in the wing, consistent with a role in Mib1-dependent signaling (Glittenberg et al., 2006). Although four lysine residues were found to be conserved among five insect Sers (unpublished data), none resided inside a motif reminiscent of Dl ICD3. The absence of ICD3 together with the divergence in sequence and relative arrangement of E3 ligase docking sites raise the possibility that ubiquitylation of Dls versus Sers is subject to different fine tuning.
Unlike insects, vertebrates have multiple paralogues of Dl and Ser (or Jagged). Comparing mouse (Dll1 and 4), zebrafish (DlA, DlD, and Dll4), and Xenopus laevis Dll1, we detected a conserved (L/I/V)KN(T/I)N motif, similar to the IKNTW stretch of insect Dl ICD2, as well as a nearby NNL tripeptide (unpublished data). NNL stretches were also found in Dll3 (mouse and Xenopus) and DlC (zebrafish) paralogues, which lack the (L/I/V)KN(T/I)N motif. Vertebrate Jaggeds contain a conserved NNxxxxL motif (Glittenberg et al., 2006), closely followed by an IKNxIEK motif (ICD2-like) in Jagged1 (mouse, zebrafish, and Xenopus) but not in Jagged2 (unpublished data). It is tempting to speculate that these motifs may play an important role in vertebrate DSL–Mib1 interactions, which have been documented for all aforementioned DSL paralogues (Itoh et al., 2003; Koo et al., 2005a). However, no NExN conserved stretches or other motifs similar to ICD1 (e.g., QNxxN) or ICD3 could be discerned in several vertebrate Dls and Jaggeds, whereas the looser putatively Neur-binding motif NxxN exists in some vertebrate DSLs (Fontana and Posakony, 2009). Still, mouse Dll1 was shown to respond to Neur by relocalizing from the basolateral to the apical side of polarized cultured cells via transcytosis (Benhra et al., 2010). Perhaps the molecular details of vertebrate Neur action may be different than those revealed here for Drosophila Neur.
The lack of conservation in Neur binding between insects and vertebrates is mirrored in a similar lack of conservation in function. Although vertebrate DSL proteins are putative substrates of Neur1 or 2, there is no substantiated role for either Neur paralogue in promoting Notch signaling (Song et al., 2006; Koo et al., 2007; Koutelou et al., 2008; Benhra et al., 2010). In fact, knockout of Mib1 seems to phenocopy all aspects of complete loss of Notch signaling in the mouse (Koo et al., 2007) but see also Koo et al. (2005b) and Zhang et al. (2007), suggesting that Mib1 proteins are the only E3 ligases that activate vertebrate DSLs. Neur1, on the other hand, may even act negatively on Notch signaling, as it can promote ubiquitylation-dependent degradation of Jagged1 accompanied by a decrease in signal emission (Koutelou et al., 2008). It is therefore necessary to directly test for E3 ligase–DSL interactions that may mediate ubiquitylation of vertebrate DSLs to unravel the activation mechanism of vertebrate DSL-Notch signaling.
Materials and methods
Protein sequence comparison
Dl orthologues were downloaded from the National Center for Biotechnology Information Entrez protein database. Their transmembrane domains were identified by the SMART (simple modular architecture research tool) algorithm, and their ICDs were aligned using the ClustalW2 algorithm.
Plasmids and transgenics
pIZ-Dl-V5-His is a plasmid expressing a C-terminally 6×His-V5 epitope–tagged wt Dl (Bland et al., 2003). It was used as a template for the generation of Dl deletion mutants i1, i2, and i3. Two PCR products were generated on either side of the motif to be deleted. Primers for the generation of pIZ-Dli1-V5-His were reaction 1, 5′-ATGAGATCTACTCCTGCGATGCC-3′ (forward) and 5′-ATGGATCCCTTTTCCTGAGCACGCTTACG-3′ (reverse), and reaction 2, 5′-ATGGATCCGCGGTGGCCACAATGC-3′ (forward) and 5′-GGTACGCGTAGAATCGAGACCGAG-3′ (reverse). Primers for the generation of pIZ-Dli2-V5-His were reaction 1, 5′-ATGAGATCTACTCCTGCGATGCC-3′ (forward) and 5′-ATGGATCCGATATTCGGGTTGCCGCC-3′ (reverse), and reaction 2, 5′-ATGGATCCTGTGCCTCAGCAGCAGC-3′ (forward) and 5′-GGTACGCGTAGAATCGAGACCGAG-3′ (reverse). Primers for the generation of pIZ-Dli3-V5-His were reaction 1, 5′-ATGAGATCTACTCCTGCGATGCC-3′ (forward) and 5′-ATGGATCCTTGCGACTTGGCTCTTTGTAG-3′ (reverse), and reaction 2, 5′-ATGGATCCCCCACGCTCATGCACCG-3′ (forward) and 5′-GGTACGCGTAGAATCGAGACCGAG-3′ (reverse). Primers for the generation of pIZ-Dli1/2-V5-His were 5′-ATAGATCTGCGGTGGCCACAATGC-3′ (forward) and 5′-GGTACGCGTAGAATCGAGACCGAG-3′ (reverse). The two fragments were joined by an artificial BamHI site and then used to replace the wt Dl coding sequence in pIZ-Dl-V5-His. In this way, each motif deleted is substituted by a Gly-Ser dipeptide. pIZ-Dli1/2-V5-His was constructed using the same strategy to delete ICD1 using pIZ-Dli2-V5-His (instead of wt Dl) as a template. pIZ-DlΔC-V5-His (Delwig, A., and M.D. Rand, personal communication) was provided by M.D. Rand (University of Vermont, Burlington, VT).
Each pUAST-Dl-V5-His deletion mutant was generated by subcloning an EcoRI–DraI fragment containing the entire V5-His–tagged Dl* coding sequence from pIZ-Dl*-V5-His into pUAST cut with EcoRI–XhoI (filled in). Transgenic flies were generated in a yw67c23 background.
To generate Dl point mutants, site-directed mutagenesis was performed using a site-directed mutagenesis kit (QuikChange; Agilent Technologies) on the P{UASDelta.Nde.Myc} vector (Parks, A.L., personal communication). An EcoRI fragment encompassing the DlNdeMyc open reading frame containing the mutation was restricted and ligated into the vector pExpUAS. A BglII–XbaI fragment from each construct (corresponding to amino acids 331–834) was then used to substitute the corresponding fragment of pIZ-Dl-V5-His. Ract-Xpress-Ub was constructed by ligating a NheI filled-in fragment from pCIneo/Ub (Koutelou, E., and J. Conaway, personal communication) into the Drosophila RactHAdh (Swevers et al., 1996) actin promoter vector cut with HincII.
Transient transfections, immunoprecipitation, and ubiquitylation assays
Transient transfections of S2 cells were performed with the calcium phosphate precipitation method. For immunoprecipitation experiments, pIZ-Dl-V5-His (Bland et al., 2003) or a deletion variant was cotransfected with pUAST-NeurΔR-GFP (Pavlopoulos et al., 2001) or UAS-HMmib1ΔR (Lai et al., 2005) and metallothionein promoter–Gal4 (inducible by 0.7 mM Cu2+). Transfected cell lysate was used for immunoprecipitation with rabbit anti-Neur antiserum or rabbit anti-Myc antibody and protein A Sepharose.
For ubiquitylation experiments, pIZ-Dl-V5-His (Bland et al., 2003) or a deletion variant was cotransfected with pUAST-EGFP-Neur (Pitsouli and Delidakis, 2005) or UAS-HMmib1 (Lai et al., 2005) and metallothionein promoter–Gal4. Ract-Xpress-Ub was included to express Xpress-tagged Ub. Transfected cells were treated with 100 µM E64 (cell-permeable lysosomal protease inhibitor; Rock et al., 1994) for 5 h before harvesting. Transfected cell lysate was used for pull-down with Ni-TED beads (Macherey-Nagel) under denaturing conditions (50 mM NaH2PO4, 300 mM NaCl, 8 M urea, and 0.2% Triton X-100, pH 8, including 10 mM N-ethylmaleimide and 1 mM PMSF). For the experiments shown in Fig. 3, an equivalent of ∼1.2 × 106 cells was loaded per lane. When 5 × 106 cells worth of extract was loaded, ubiquitylation signals became saturated, and background levels of Dl ubiquitylation became detectable, caused by endogenous S2 cell E3 ligases. HMmib1 also bears a His tag but is apparently not significantly autoubiquitylated because we get no detectable Xpress signal when we coexpress it with Dli2 or Dli1/2. Western blots were developed using HRP-coupled secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) and the SuperSignal West Pico chemiluminescent substrate obtained from Thermo Fisher Scientific.
Drosophila strains
UAS lines used in this study were UAS-Dl-V5-His, UAS-EGFP-Neur (Pitsouli and Delidakis, 2005), UAS-Dli1-V5-His, UAS-Dli2-V5-His, UAS-Dli1/2-V5-His, UAS-DlΔC-V5-His, UAS-Dl-K742R (this study), UAS-Dl-LDL+, and UAS-DlΔICD (Wang and Struhl, 2004). Driver lines used in this study were obtained as follows: Equator (Eq)-Gal4 (Pi et al., 2001) was provided by H. Bellen (Baylor College of Medicine, Houston, TX). ptc-Gal4 and hsFlp; act>CD2stop>Gal4 were obtained from the Bloomington Drosophila Stock Center. With the latter driver, larvae were incubated at 37°C for 13 min to induce clones. The MARCM (mosaic analysis with a repressible cell marker) system (Lee and Luo, 2001) was used to generate positively marked clones as follows. To express our Dl variants (Dl*) in the absence of endogenous Dl and Ser (Fig. 5), the following cross was used: y w hsFLP122 tubGal4 UAS-GFP-6xnls; FRT82B tubGal80/TM6B crossed to w; UAS-Dl*; FRT82B Dlrev10 e SerRX106/T(2;3)SM5;TM6B. To express our Dl variants (Dl*) in the absence of endogenous mib1 (Fig. 4), the following cross was used: y w hsFLP122 tubGal4 UAS-GFP-6xnls; tubG80 FRT2A/TM6B crossed to w; UAS-Dl*; mibEY9780 FRT2A/T(2;3) SM5;TM6B.
Antibodies, immunohistochemistry, and microscopy
For immunohistochemistry, dissected tissues were fixed for 20 min in 4% formaldehyde (Polysciences) in either 0.1 M Pipes, pH 6.9, 1 mM EGTA, 1 mM MgCl2, or in PBS + 1 mM CaCl2 (for the DCAD2 antibody). Antibody incubations were performed in PBS supplemented with 0.2% Triton X-100 and 0.5% BSA. Washes were performed in the same solution omitting BSA. In experiments analyzing cell surface Dl (Fig. S2), Triton X-100 was omitted from all solutions. To allow antibody access to the apical cell surface, the disk peripodial membrane was disrupted by gentle pricking with a pulled-out tungsten needle. Tissues were mounted in 80% glycerol in PBS with 0.5% N-propyl-gallate as an antibleach medium.
Antibodies used in this study were mouse anti-V5 (Invitrogen), mouse anti-Xpress (Invitrogen), mouse anti-Notch C458.2H (developed by S. Artavanis-Tsakonas [Harvard University, Boston, MA] and obtained from the Developmental Studies Hybridoma Bank), mouse anti-Wg 4D4 (obtained from the Developmental Studies Hybridoma Bank; Brook and Cohen, 1996), mouse anti-Dl (extracellular epitope) C594.9B (developed by S. Artavanis-Tsakonas and obtained from the Developmental Studies Hybridoma Bank), rat anti–E-cadherin (DCAD2; obtained from the Developmental Studies Hybridoma Bank), rabbit anti-Neur (Pitsouli and Delidakis, 2005), rabbit anti-Myc epitope (Santa Cruz Biotechnology, Inc.), rabbit anti-Sara (gift from M. González-Gaitán, University of Geneva, Geneva, Switzerland; Bökel et al., 2006), rat anti-Rab11 (gift from R. Cohen, University of Kansas, Lawrence, KS; Dollar et al., 2002), guinea pig anti-Dl 581 (extracellular epitope; Huppert et al., 1997), guinea pig anti-Senseless (Nolo et al., 2000), and guinea pig anti-Hrs full length (gift from H. Bellen; Lloyd et al., 2002). The Developmental Studies Hybridoma Bank was developed under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa Department of Biology. Fluorescent (Alexa Fluor 488, Alexa Fluor 561, and Alexa Fluor 633) secondary antibodies were obtained from Invitrogen.
Images were acquired on a confocal microscope (SP2; Leica) at the University of Crete using 20×/0.7 NA (dry), 40×/1.25 NA (oil), or 63×/1.4 NA (oil) Plan Apochromat objectives (at room temperature). They were processed with the manufacturer’s software and assembled on Photoshop (Adobe), in which some contrast adjustment was performed.
For quantifying colocalization between Dl and endosomal markers, we used the mouse anti-Dl mAb together with an antibody against endosomal markers (rat anti-Rab11, rabbit anti-Sara, or guinea pig anti-Hrs). Images were acquired at the University of Crete confocal facility (SP2) at 63× with a 3× zoom using the xz (optical cross section) mode. A computer interface developed in Matlab (MathWorks, Inc.) was used to identify and count intracellular puncta positive for each of the two markers (Dl vs. endocytic marker). The system provided an efficient methodology to facilitate image segmentation based on intensity thresholding (Tsibidis and Tavernarakis, 2007; Tsibidis et al., 2011) and offered a rapid object extraction and overview. Objects were called if they consisted of at least four contiguous square pixels (0.1 µm2) when pixel value was above a certain threshold. This depended on the quality of the staining and was (on an 8-bit scale) 38–77 for Dl, 38–51 for Rab11, 28–77 for Sara, and 38–77 for Hrs. Coalesced puncta were resolved by manual intervention. In this analysis, we excluded the extreme apical and basal domains of the wing disk epithelium, in which high levels of a contiguous Dl signal are seen. We may have therefore underestimated Dl colocalization with Sara and Rab11, as the latter also accumulated highly at the apical regions of disk cells (which were not scorable), whereas Hrs had less apical bias. The statistical significance of colocalization differences between samples was computed using Fisher’s exact test. It should be noted that the colocalizations measured by this assay reflect total Dl ECD. This will include Dl endocytosed as a full-length molecule or after ECD shedding (Delwig et al., 2006) and reuptake. Because of the inherently noisy nature of the endosomal marker antibodies, we could not consistently detect all marker-positive structures with the same confidence as we had for Dl-positive structures. We therefore present (Table 1) only the percentage of colocalization based on the total Dl-positive structures, which was more reproducible across samples. The percentage of colocalization based on the total marker-positive structures was more variable and unreliable.
Live-antibody uptake assay and image analysis
The live-antibody uptake assay was performed as previously described in Le Borgne and Schweisguth (2003). In brief, pupae 18–24 h after puparium formation were dissected in M3 tissue-culture medium supplemented with 10% fetal bovine serum and incubated in the same medium in the presence of 1:15 diluted mouse anti-Dl C594-9B supernatant, which recognizes the Dl ECD. After 15 min at room temperature, the pupal carcasses were quickly washed in M3 medium and fixed. After fixation, they were permeabilized and incubated with additional primary antibodies, namely rat anti–E-cadherin and guinea pig anti-Dl. As a control, we repeated the uptake assay with the live-tissue incubation at 4°C, in which endocytosis is blocked. Although we detected basolateral surface staining for the mouse anti-Dl, no intracellular puncta were labeled (Fig. 6 A). Also, no apical surface staining was observed, suggesting that apical access of the live antibody is blocked by the pupal cuticle; therefore, this assay measures basolateral uptake.
Nota were imaged at the University of Crete confocal facility (SP2) at 63× and a 3× zoom using the xz (optical cross section) mode. This ensured that the whole height of the epithelium was imaged under identical conditions for all samples. To quantitatively estimate uptake efficiency, we calculated the percentage of total (guinea pig positive) Dl puncta that are also live uptake (mouse) positive. A total of ≥10 optical sections and ≥100 puncta were scored. Even though our optical sections were taken at 0.3-µm intervals, we scored every third image (0.9 µm apart) to avoid double scoring of large particles, which would appear in consecutive slices.
Online supplemental material
Fig. S1 presents the comparison of Dl ICDs from several insect species. Fig. S2 shows representative controls on the ubiquitylation assays of Fig. 2. Fig. S3 shows that Dl variants are exposed on the apical surface of the wing disk epithelium. Fig. S4 shows localization of Dl variants relative to Sara, Hrs, and Rab11. Fig. S5 shows the expression pattern of the Eq-Gal4 driver used for the experiments of Fig. 6. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201105166/DC1.
Acknowledgments
We are indebted to Matt Rand, Hugo Bellen, Joan Conaway, Lia Koutelou, Gary Struhl, Marcos Gonzalez-Gaitán, Bob Cohen, and Eric Lai for providing DNA constructs, fly strains, and antibodies. Special thanks go to Dan Finley and Marianthi Kiparaki for their invaluable help on establishing the ubiquitylation assay and to Nikos Giagtzoglou for his help on establishing the live uptake assay. We also thank Dina Lyka for help with statistical analysis of colocalization. Many thanks go to Savvas Christoforidis and George Diallinas for helpful discussions and also to Sarah Bray and Maura Strigini for their critical reading of the manuscript.
A. Daskalaki, K. Kux, and C. Delidakis were supported by the European Union Marie Curie Project No. 7295 and by intramural Institute of Molecular Biology and Biotechnology and University of Crete funds. Work by N.A. Shalaby and M.A.T. Muskavitch was supported by the DeLuca Professorship from Boston College.
Footnotes
We have not strictly excluded a Mib1 preference for the three lysines in the stop transfer sequence, K665 (the only other ICD lysine not mutagenized), or the single lysine residue in the V5 epitope tag. However, K665 is not conserved, and lysines 620/622/624 (stop transfer) have been tested by Wang and Struhl (2004) and shown not to be necessary for Dl activity. We therefore do not think that any of these would act as a major acceptor site for Mib1 modification.
Abbreviations used in this paper:
- Brd
- Bearded
- Dl
- Delta
- DV
- dorsoventral
- ECD
- extracellular domain
- Eq
- Equator
- Hrs
- hepatocyte growth factor–regulated tyrosine kinase substrate
- ICD
- intracellular domain
- LDL
- low density lipoprotein
- Mib1
- Mindbomb 1
- MM
- molecular mass
- Neur
- Neuralized
- Ser
- Serrate
- SOP
- sensory organ precursor
- UAS
- upstream activation sequence
- Ub
- ubiquitin
- Wg
- Wingless
- wt
- wild type
References
- Acconcia F., Sigismund S., Polo S. 2009. Ubiquitin in trafficking: the network at work. Exp. Cell Res. 315:1610–1618 10.1016/j.yexcr.2008.10.014 [DOI] [PubMed] [Google Scholar]
- Banks S.M., Cho B., Eun S.H., Lee J.H., Windler S.L., Xie X., Bilder D., Fischer J.A. 2011. The functions of auxilin and Rab11 in Drosophila suggest that the fundamental role of ligand endocytosis in notch signaling cells is not recycling. PLoS ONE. 6:e18259 10.1371/journal.pone.0018259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin A.J., Schweisguth F. 2006. Bearded family members inhibit Neuralized-mediated endocytosis and signaling activity of Delta in Drosophila. Dev. Cell. 10:245–255 10.1016/j.devcel.2005.12.017 [DOI] [PubMed] [Google Scholar]
- Barsi J.C., Rajendra R., Wu J.I., Artzt K. 2005. Mind bomb1 is a ubiquitin ligase essential for mouse embryonic development and Notch signaling. Mech. Dev. 122:1106–1117 10.1016/j.mod.2005.06.005 [DOI] [PubMed] [Google Scholar]
- Benhra N., Vignaux F., Dussert A., Schweisguth F., Le Borgne R. 2010. Neuralized promotes basal to apical transcytosis of delta in epithelial cells. Mol. Biol. Cell. 21:2078–2086 10.1091/mbc.E09-11-0926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland C.E., Kimberly P., Rand M.D. 2003. Notch-induced proteolysis and nuclear localization of the Delta ligand. J. Biol. Chem. 278:13607–13610 10.1074/jbc.C300016200 [DOI] [PubMed] [Google Scholar]
- Bökel C., Schwabedissen A., Entchev E., Renaud O., González-Gaitán M. 2006. Sara endosomes and the maintenance of Dpp signaling levels across mitosis. Science. 314:1135–1139 10.1126/science.1132524 [DOI] [PubMed] [Google Scholar]
- Bray S. 1998. Notch signalling in Drosophila: three ways to use a pathway. Semin. Cell Dev. Biol. 9:591–597 10.1006/scdb.1998.0262 [DOI] [PubMed] [Google Scholar]
- Brook W.J., Cohen S.M. 1996. Antagonistic interactions between wingless and decapentaplegic responsible for dorsal-ventral pattern in the Drosophila Leg. Science. 273:1373–1377 10.1126/science.273.5280.1373 [DOI] [PubMed] [Google Scholar]
- Castro B., Barolo S., Bailey A.M., Posakony J.W. 2005. Lateral inhibition in proneural clusters: cis-regulatory logic and default repression by Suppressor of Hairless. Development. 132:3333–3344 10.1242/dev.01920 [DOI] [PubMed] [Google Scholar]
- Chen W., Casey Corliss D. 2004. Three modules of zebrafish Mind bomb work cooperatively to promote Delta ubiquitination and endocytosis. Dev. Biol. 267:361–373 10.1016/j.ydbio.2003.11.010 [DOI] [PubMed] [Google Scholar]
- Chen W.J., Goldstein J.L., Brown M.S. 1990. NPXY, a sequence often found in cytoplasmic tails, is required for coated pit-mediated internalization of the low density lipoprotein receptor. J. Biol. Chem. 265:3116–3123 [PubMed] [Google Scholar]
- Chitnis A., Henrique D., Lewis J., Ish-Horowicz D., Kintner C. 1995. Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature. 375:761–766 10.1038/375761a0 [DOI] [PubMed] [Google Scholar]
- Clague M.J., Urbé S. 2010. Ubiquitin: same molecule, different degradation pathways. Cell. 143:682–685 10.1016/j.cell.2010.11.012 [DOI] [PubMed] [Google Scholar]
- Coumailleau F., Fürthauer M., Knoblich J.A., González-Gaitán M. 2009. Directional Delta and Notch trafficking in Sara endosomes during asymmetric cell division. Nature. 458:1051–1055 10.1038/nature07854 [DOI] [PubMed] [Google Scholar]
- Deblandre G.A., Lai E.C., Kintner C. 2001. Xenopus neuralized is a ubiquitin ligase that interacts with XDelta1 and regulates Notch signaling. Dev. Cell. 1:795–806 10.1016/S1534-5807(01)00091-0 [DOI] [PubMed] [Google Scholar]
- de Celis J.F., Bray S. 1997. Feed-back mechanisms affecting Notch activation at the dorsoventral boundary in the Drosophila wing. Development. 124:3241–3251 [DOI] [PubMed] [Google Scholar]
- de Celis J.F., Garcia-Bellido A., Bray S.J. 1996. Activation and function of Notch at the dorsal-ventral boundary of the wing imaginal disc. Development. 122:359–369 [DOI] [PubMed] [Google Scholar]
- Delwig A., Bland C., Beem-Miller M., Kimberly P., Rand M.D. 2006. Endocytosis-independent mechanisms of Delta ligand proteolysis. Exp. Cell Res. 312:1345–1360 10.1016/j.yexcr.2005.12.037 [DOI] [PubMed] [Google Scholar]
- De Renzis S., Yu J., Zinzen R., Wieschaus E. 2006. Dorsal-ventral pattern of Delta trafficking is established by a Snail-Tom-Neuralized pathway. Dev. Cell. 10:257–264 10.1016/j.devcel.2006.01.011 [DOI] [PubMed] [Google Scholar]
- Deshaies R.J., Joazeiro C.A. 2009. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78:399–434 10.1146/annurev.biochem.78.101807.093809 [DOI] [PubMed] [Google Scholar]
- Doherty D., Feger G., Younger-Shepherd S., Jan L.Y., Jan Y.N. 1996. Delta is a ventral to dorsal signal complementary to Serrate, another Notch ligand, in Drosophila wing formation. Genes Dev. 10:421–434 10.1101/gad.10.4.421 [DOI] [PubMed] [Google Scholar]
- Dollar G., Struckhoff E., Michaud J., Cohen R.S. 2002. Rab11 polarization of the Drosophila oocyte: a novel link between membrane trafficking, microtubule organization, and oskar mRNA localization and translation. Development. 129:517–526 [DOI] [PubMed] [Google Scholar]
- Emery G., Hutterer A., Berdnik D., Mayer B., Wirtz-Peitz F., Gaitan M.G., Knoblich J.A. 2005. Asymmetric Rab 11 endosomes regulate delta recycling and specify cell fate in the Drosophila nervous system. Cell. 122:763–773 10.1016/j.cell.2005.08.017 [DOI] [PubMed] [Google Scholar]
- Eun S.H., Banks S.M., Fischer J.A. 2008. Auxilin is essential for Delta signaling. Development. 135:1089–1095 10.1242/dev.009530 [DOI] [PubMed] [Google Scholar]
- Fontana J.R., Posakony J.W. 2009. Both inhibition and activation of Notch signaling rely on a conserved Neuralized-binding motif in Bearded proteins and the Notch ligand Delta. Dev. Biol. 333:373–385 10.1016/j.ydbio.2009.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glittenberg M., Pitsouli C., Garvey C., Delidakis C., Bray S. 2006. Role of conserved intracellular motifs in Serrate signalling, cis-inhibition and endocytosis. EMBO J. 25:4697–4706 10.1038/sj.emboj.7601337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon W.R., Arnett K.L., Blacklow S.C. 2008. The molecular logic of Notch signaling—a structural and biochemical perspective. J. Cell Sci. 121:3109–3119 10.1242/jcs.035683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuss S.F., Ndiaye-Lobry D., Six E.M., Israël A., Logeat F. 2008. The intracellular region of Notch ligands Dll1 and Dll3 regulates their trafficking and signaling activity. Proc. Natl. Acad. Sci. USA. 105:11212–11217 10.1073/pnas.0800695105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F., Dambly-Chaudière C., Ghysen A. 1991. The emergence of sense organs in the wing disc of Drosophila. Development. 111:1087–1095 [DOI] [PubMed] [Google Scholar]
- Hukriede N.A., Gu Y., Fleming R.J. 1997. A dominant-negative form of Serrate acts as a general antagonist of Notch activation. Development. 124:3427–3437 [DOI] [PubMed] [Google Scholar]
- Huppert S.S., Jacobsen T.L., Muskavitch M.A. 1997. Feedback regulation is central to Delta-Notch signalling required for Drosophila wing vein morphogenesis. Development. 124:3283–3291 [DOI] [PubMed] [Google Scholar]
- Irvine K.D., Vogt T.F. 1997. Dorsal-ventral signaling in limb development. Curr. Opin. Cell Biol. 9:867–876 10.1016/S0955-0674(97)80090-7 [DOI] [PubMed] [Google Scholar]
- Itoh M., Kim C.H., Palardy G., Oda T., Jiang Y.J., Maust D., Yeo S.Y., Lorick K., Wright G.J., Ariza-McNaughton L., et al. 2003. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev. Cell. 4:67–82 10.1016/S1534-5807(02)00409-4 [DOI] [PubMed] [Google Scholar]
- Jafar-Nejad H., Andrews H.K., Acar M., Bayat V., Wirtz-Peitz F., Mehta S.Q., Knoblich J.A., Bellen H.J. 2005. Sec15, a component of the exocyst, promotes notch signaling during the asymmetric division of Drosophila sensory organ precursors. Dev. Cell. 9:351–363 10.1016/j.devcel.2005.06.010 [DOI] [PubMed] [Google Scholar]
- Jékely G., Rørth P. 2003. Hrs mediates downregulation of multiple signalling receptors in Drosophila. EMBO Rep. 4:1163–1168 10.1038/sj.embor.7400019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandachar V., Bai T., Chang H.C. 2008. The clathrin-binding motif and the J-domain of Drosophila Auxilin are essential for facilitating Notch ligand endocytosis. BMC Dev. Biol. 8:50 10.1186/1471-213X-8-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein T., Brennan K., Arias A.M. 1997. An intrinsic dominant negative activity of serrate that is modulated during wing development in Drosophila. Dev. Biol. 189:123–134 10.1006/dbio.1997.8564 [DOI] [PubMed] [Google Scholar]
- Koo B.K., Lim H.S., Song R., Yoon M.J., Yoon K.J., Moon J.S., Kim Y.W., Kwon M.C., Yoo K.W., Kong M.P., et al. 2005a. Mind bomb 1 is essential for generating functional Notch ligands to activate Notch. Development. 132:3459–3470 10.1242/dev.01922 [DOI] [PubMed] [Google Scholar]
- Koo B.K., Yoon K.J., Yoo K.W., Lim H.S., Song R., So J.H., Kim C.H., Kong Y.Y. 2005b. Mind bomb-2 is an E3 ligase for Notch ligand. J. Biol. Chem. 280:22335–22342 10.1074/jbc.M501631200 [DOI] [PubMed] [Google Scholar]
- Koo B.K., Yoon M.J., Yoon K.J., Im S.K., Kim Y.Y., Kim C.H., Suh P.G., Jan Y.N., Kong Y.Y. 2007. An obligatory role of mind bomb-1 in notch signaling of mammalian development. PLoS ONE. 2:e1221 10.1371/journal.pone.0001221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R., Ilagan M.X. 2009. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 137:216–233 10.1016/j.cell.2009.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutelou E., Sato S., Tomomori-Sato C., Florens L., Swanson S.K., Washburn M.P., Kokkinaki M., Conaway R.C., Conaway J.W., Moschonas N.K. 2008. Neuralized-like 1 (Neurl1) targeted to the plasma membrane by N-myristoylation regulates the Notch ligand Jagged1. J. Biol. Chem. 283:3846–3853 10.1074/jbc.M706974200 [DOI] [PubMed] [Google Scholar]
- Lai E.C., Deblandre G.A., Kintner C., Rubin G.M. 2001. Drosophila neuralized is a ubiquitin ligase that promotes the internalization and degradation of delta. Dev. Cell. 1:783–794 10.1016/S1534-5807(01)00092-2 [DOI] [PubMed] [Google Scholar]
- Lai E.C., Roegiers F., Qin X., Jan Y.N., Rubin G.M. 2005. The ubiquitin ligase Drosophila Mind bomb promotes Notch signaling by regulating the localization and activity of Serrate and Delta. Development. 132:2319–2332 10.1242/dev.01825 [DOI] [PubMed] [Google Scholar]
- Le Borgne R. 2006. Regulation of Notch signalling by endocytosis and endosomal sorting. Curr. Opin. Cell Biol. 18:213–222 10.1016/j.ceb.2006.02.011 [DOI] [PubMed] [Google Scholar]
- Le Borgne R., Schweisguth F. 2003. Unequal segregation of Neuralized biases Notch activation during asymmetric cell division. Dev. Cell. 5:139–148 10.1016/S1534-5807(03)00187-4 [DOI] [PubMed] [Google Scholar]
- Le Borgne R., Remaud S., Hamel S., Schweisguth F. 2005. Two distinct E3 ubiquitin ligases have complementary functions in the regulation of delta and serrate signaling in Drosophila. PLoS Biol. 3:e96 10.1371/journal.pbio.0030096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Luo L. 2001. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 24:251–254 10.1016/S0166-2236(00)01791-4 [DOI] [PubMed] [Google Scholar]
- Lloyd T.E., Atkinson R., Wu M.N., Zhou Y., Pennetta G., Bellen H.J. 2002. Hrs regulates endosome membrane invagination and tyrosine kinase receptor signaling in Drosophila. Cell. 108:261–269 10.1016/S0092-8674(02)00611-6 [DOI] [PubMed] [Google Scholar]
- Micchelli C.A., Rulifson E.J., Blair S.S. 1997. The function and regulation of cut expression on the wing margin of Drosophila: Notch, Wingless and a dominant negative role for Delta and Serrate. Development. 124:1485–1495 [DOI] [PubMed] [Google Scholar]
- Miller A.C., Lyons E.L., Herman T.G. 2009. cis-Inhibition of Notch by endogenous Delta biases the outcome of lateral inhibition. Curr. Biol. 19:1378–1383 10.1016/j.cub.2009.06.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann C.J., Cohen S.M. 1996. A hierarchy of cross-regulation involving Notch, wingless, vestigial and cut organizes the dorsal/ventral axis of the Drosophila wing. Development. 122:3477–3485 [DOI] [PubMed] [Google Scholar]
- Nichols J.T., Miyamoto A., Olsen S.L., D’Souza B., Yao C., Weinmaster G. 2007. DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur. J. Cell Biol. 176:445–458 10.1083/jcb.200609014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolo R., Abbott L.A., Bellen H.J. 2000. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell. 102:349–362 10.1016/S0092-8674(00)00040-4 [DOI] [PubMed] [Google Scholar]
- Overstreet E., Fitch E., Fischer J.A. 2004. Fat facets and Liquid facets promote Delta endocytosis and Delta signaling in the signaling cells. Development. 131:5355–5366 10.1242/dev.01434 [DOI] [PubMed] [Google Scholar]
- Parks A.L., Klueg K.M., Stout J.R., Muskavitch M.A. 2000. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development. 127:1373–1385 [DOI] [PubMed] [Google Scholar]
- Pavlopoulos E., Pitsouli C., Klueg K.M., Muskavitch M.A., Moschonas N.K., Delidakis C. 2001. neuralized encodes a peripheral membrane protein involved in delta signaling and endocytosis. Dev. Cell. 1:807–816 10.1016/S1534-5807(01)00093-4 [DOI] [PubMed] [Google Scholar]
- Pi H., Wu H.J., Chien C.T. 2001. A dual function of phyllopod in Drosophila external sensory organ development: cell fate specification of sensory organ precursor and its progeny. Development. 128:2699–2710 [DOI] [PubMed] [Google Scholar]
- Pitsouli C., Delidakis C. 2005. The interplay between DSL proteins and ubiquitin ligases in Notch signaling. Development. 132:4041–4050 10.1242/dev.01979 [DOI] [PubMed] [Google Scholar]
- Rajan A., Tien A.C., Haueter C.M., Schulze K.L., Bellen H.J. 2009. The Arp2/3 complex and WASp are required for apical trafficking of Delta into microvilli during cell fate specification of sensory organ precursors. Nat. Cell Biol. 11:815–824 10.1038/ncb1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock K.L., Gramm C., Rothstein L., Clark K., Stein R., Dick L., Hwang D., Goldberg A.L. 1994. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 78:761–771 10.1016/S0092-8674(94)90462-6 [DOI] [PubMed] [Google Scholar]
- Seugnet L., Simpson P., Haenlin M. 1997. Requirement for dynamin during Notch signaling in Drosophila neurogenesis. Dev. Biol. 192:585–598 10.1006/dbio.1997.8723 [DOI] [PubMed] [Google Scholar]
- Skwarek L.C., Garroni M.K., Commisso C., Boulianne G.L. 2007. Neuralized contains a phosphoinositide-binding motif required downstream of ubiquitination for delta endocytosis and notch signaling. Dev. Cell. 13:783–795 10.1016/j.devcel.2007.10.020 [DOI] [PubMed] [Google Scholar]
- Song R., Koo B.K., Yoon K.J., Yoon M.J., Yoo K.W., Kim H.T., Oh H.J., Kim Y.Y., Han J.K., Kim C.H., Kong Y.Y. 2006. Neuralized-2 regulates a Notch ligand in cooperation with Mind bomb-1. J. Biol. Chem. 281:36391–36400 10.1074/jbc.M606601200 [DOI] [PubMed] [Google Scholar]
- Sprinzak D., Lakhanpal A., Lebon L., Santat L.A., Fontes M.E., Anderson G.A., Garcia-Ojalvo J., Elowitz M.B. 2010. Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature. 465:86–90 10.1038/nature08959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Artavanis-Tsakonas S. 1997. Secreted forms of DELTA and SERRATE define antagonists of Notch signaling in Drosophila. Development. 124:3439–3448 [DOI] [PubMed] [Google Scholar]
- Swevers L., Cherbas L., Cherbas P., Iatrou K. 1996. Bombyx EcR (BmEcR) and Bombyx USP (BmCF1) combine to form a functional ecdysone receptor. Insect Biochem. Mol. Biol. 26:217–221 10.1016/0965-1748(95)00097-6 [DOI] [PubMed] [Google Scholar]
- Tsibidis G.D., Tavernarakis N. 2007. Nemo: a computational tool for analyzing nematode locomotion. BMC Neurosci. 8:86 10.1186/1471-2202-8-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsibidis G.D., Burroughs N.J., Gaze W., Wellington E.M.H. 2011. Semi-automated Acanthamoeba polyphaga detection and computation of Salmonella typhimurium concentration in spatio-temporal images. Micron. 42:911–920 10.1016/j.micron.2011.06.010 [DOI] [PubMed] [Google Scholar]
- Wang W., Struhl G. 2004. Drosophila Epsin mediates a select endocytic pathway that DSL ligands must enter to activate Notch. Development. 131:5367–5380 10.1242/dev.01413 [DOI] [PubMed] [Google Scholar]
- Wang W., Struhl G. 2005. Distinct roles for Mind bomb, Neuralized and Epsin in mediating DSL endocytosis and signaling in Drosophila. Development. 132:2883–2894 10.1242/dev.01860 [DOI] [PubMed] [Google Scholar]
- Weinmaster G., Fischer J.A. 2011. Notch ligand ubiquitylation: what is it good for? Dev. Cell. 21:134–144 10.1016/j.devcel.2011.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Dermer M., Commisso C., Zhou L., McGlade C.J., Boulianne G.L. 2001. Neuralized functions as an E3 ubiquitin ligase during Drosophila development. Curr. Biol. 11:1675–1679 10.1016/S0960-9822(01)00527-9 [DOI] [PubMed] [Google Scholar]
- Zhang C., Li Q., Lim C.H., Qiu X., Jiang Y.J. 2007. The characterization of zebrafish antimorphic mib alleles reveals that Mib and Mind bomb-2 (Mib2) function redundantly. Dev. Biol. 305:14–27 10.1016/j.ydbio.2007.01.034 [DOI] [PubMed] [Google Scholar]