The yeast Kes1p/Osh4p protein functions as a sterol/PI(4)P exchanger between lipid membranes, which suggests the possibility of creating a sterol gradient via phosphoinositide metabolism.
Abstract
Osh/Orp proteins transport sterols between organelles and are involved in phosphoinositide metabolism. The link between these two aspects remains elusive. Using novel assays, we address the influence of membrane composition on the ability of Osh4p/Kes1p to extract, deliver, or transport dehydroergosterol (DHE). Surprisingly, phosphatidylinositol 4-phosphate (PI(4)P) specifically inhibited DHE extraction because PI(4)P was itself efficiently extracted by Osh4p. We solve the structure of the Osh4p–PI(4)P complex and reveal how Osh4p selectively substitutes PI(4)P for sterol. Last, we show that Osh4p quickly exchanges DHE for PI(4)P and, thereby, can transport these two lipids between membranes along opposite routes. These results suggest a model in which Osh4p transports sterol from the ER to late compartments pinpointed by PI(4)P and, in turn, transports PI(4)P backward. Coupled to PI(4)P metabolism, this transport cycle would create sterol gradients. Because the residues that recognize PI(4)P are conserved in Osh4p homologues, other Osh/Orp are potential sterol/phosphoinositol phosphate exchangers.
Introduction
In eukaryotic cells, sterols reside in all compartments but are unevenly distributed. Cholesterol (ergosterol in yeast) is synthesized in the ER, but is rare in this compartment and more concentrated at the trans Golgi, endosomes, and the plasma membrane (PM). Sterols are transferred between organelles either by transport vesicles or by soluble carriers (Maxfield and Tabas, 2005; Prinz, 2007; Mesmin and Maxfield, 2009). In yeast, ergosterol moves from the ER to the PM mostly via protein-mediated nonvesicular routes (Baumann et al., 2005). Little is known about the identity of the carriers, but oxysterol-binding homology protein (Osh) are plausible candidates (Prinz, 2007; Fairn and McMaster, 2008; Lev, 2010). Suppression of the entire Osh family is lethal, depriving the PM in ergosterol and stopping endocytosis. Conversely, expressing any of the seven Osh restores cell viability (Beh and Rine, 2004).
Osh4p, also known as Kes1p, is the simplest and best characterized Osh. Its structure consists of a β-barrel forming a sterol-binding pocket (Im et al., 2005). When Osh4p is empty, the N-terminal segment (29 amino-acids) is unfolded and leaves the pocket open. Upon sterol binding, this segment forms a lid that blocks the sterol molecule in the pocket. Thus, Osh4p seems adapted to transport sterols (Levine, 2005) and, in accord with this, was found to move sterol between artificial membranes (Raychaudhuri et al., 2006). Because all Osh proteins and the human oxysterol-binding protein-related protein (Orp; Ridgway, 2010) contain a sterol-binding domain akin to Osh4p, better understanding the sterol transport activity of Osh4p should have general implications.
To properly allocate sterol in the cell, Osh/Orp must ensure a one-way transport of sterol between defined compartments. This seems the simplest way to enrich one membrane with sterol at the expense of another. Such transport must involve transient and sequential interactions between the protein, the sterol molecule, and two lipid bilayers, one acting as a donor membrane and one as an acceptor (Lev, 2010). Osh4p adopts two different structures depending on its state (empty or sterol-bound), and this is likely critical for proper protein targeting to donor and acceptor membranes during a cycle of transport. In line with this hypothesis, Osh4p has been proposed to transport sterol from the PM to the ER (Raychaudhuri et al., 2006). A key argument is that sterol uptake in vitro is facilitated by phosphatidylinositol (PI) 4,5 bisphosphate (PI(4,5)P2), a PM lipid.
However, control of Osh4p by PI(4,5)P2 is hard to reconcile with other observations. First, the level of ergosterol at the PM increases upon Osh4p expression (Beh et al., 2001). Second, genetic and cellular studies suggest that Osh4p has a specific function at the Golgi, and that this function depends on another phosphoinositide, PI 4-phosphate (PI(4)P), present at the trans side. Osh4p is the only Osh whose expression is lethal in Sec14p-deficient yeast (Fang et al., 1996; Li et al., 2002). Sec14p is a key protein, which maintains a lipid composition permissive for vesicular transport (Bankaitis et al., 2005). In the absence of Sec14p, the amount of PI(4)P is limiting. Osh4p, whose endogenous expression is high, monopolizes all remaining PI(4)P molecules at the expense of essential proteins of vesicular transport (Fairn et al., 2007; LeBlanc and McMaster, 2010). How Osh4p binds PI(4)P is not well understood. Osh4p does not have any known phosphoinositol phosphate–binding domain, and in vitro binding assays failed to prove that Osh4p could distinguish a PI(4)P- from a PI(4,5)P2-containing membrane (Li et al., 2002; Fairn and McMaster, 2005; Schulz et al., 2009). Moreover, no link between PI(4)P and sterol transport by Osh4p has been found. Here, using a combination of assays for lipid exchange and x-ray crystallography, we solve this issue by showing that Osh4p acts as a sterol/PI(4)P exchanger.
Results
Osh4p quickly extracts and solubilizes the fluorescent ergosterol dehydroergosterol (DHE) from neutral liposomes
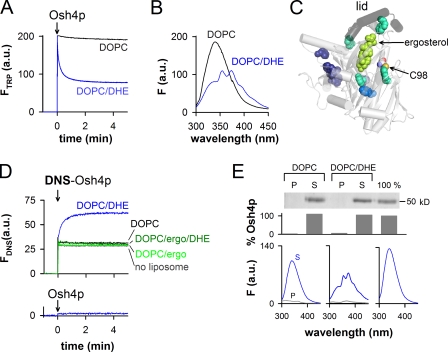
So far, the ability of Osh4p to extract, deliver, or exchange sterols has been mostly tested using radiolabeled sterols (Raychaudhuri et al., 2006; Schulz et al., 2009). However, the separation procedures required for determining the sterol distribution between the donor liposomes, the acceptor liposomes, and the protein do not permit precise kinetics measurements. To circumvent these limitations, we took advantage of the fluorescence properties of DHE, a natural ergosterol analogue. We observed that the intrinsic fluorescence of Osh4p quickly diminished once added to large liposomes made of dioleoyl phosphatidylcholine (DOPC) doped with 0.5% DHE (Fig. 1 A). Spectral analysis revealed that tryptophan quenching at 340 nm was concomitant with the apparition of three emission peaks at 354, 373, and 393 nm (Fig. 1 B). These peaks are characteristic of DHE, which suggests fluorescence resonance energy transfer (FRET) from Osh4p to DHE (Schroeder et al., 1990; Holt et al., 2008; Liu et al., 2009). Osh4p contains several tryptophans close to the ergosterol-binding pocket, which should be responsible for the observed FRET signal (Fig. 1 C).
Figure 1.
Real-time measurement of DHE loading and extraction by Osh4p from large DOPC liposomes. (A) Intrinsic fluorescence at 340 nm of Osh4p (0.5 µM) upon addition to DOPC/DHE (99.5:0.5 mol/mol) liposomes (blue trace) or to DOPC liposomes (black trace). The final lipid concentration is 0.5 mM. (B) Fluorescence emission spectra (λex = 285 nm) of the samples shown in A at the end of the kinetics. (C) Close-up view of the Osh4p structure (PDB accession no. 1ZHZ) showing the lid (segment 1–29, black) covering ergosterol (bright green). Tryptophans located at ≤15 Å, between 15 and 20 Å, or at ≥20 Å from ergosterol are colored in cyan, blue, and purple, respectively. Cystein C98 is indicated. (D) Fluorescence at 510 nm (λex = 310 nm) of 0.5 µM DNS-Osh4p added to buffer alone (gray trace) or to DOPC/DHE (99.5:0.5 mol/mol, blue trace), DOPC (black trace), DOPC/ergosterol (95:5, light green trace), or DOPC/ergosterol/DHE (94.5:5:0.5, dark green trace) liposomes. Bottom trace, unlabeled Osh4p added to DOPC/DHE liposomes. (E) 0.5 µM Osh4p was incubated with DOPC or DOPC/DHE (99.5:0.5) liposomes (0.5 mM lipids) filled with sucrose. A reference experiment (100%) was done without liposomes. After centrifugation, the percentage of Osh4p in the pellet (P) and supernatant (S) was determined by SDS-PAGE. The fluorescence spectrum of each sample was recorded (λex = 285 nm). Control spectra corresponding to buffer or liposome alone (0.5 mM lipids) were subtracted from the spectra of the supernatant and pellet, respectively. Data in E are from a single representative experiment out of two independent experiments.
Next, we created a more defined FRET pair by labeling an endogenous cysteine (C98) with a dansyl (DNS) probe. C98 is exposed on the external side of Osh4p, at 12.8 Å from the center of the sterol-binding pocket. Once DNS-Osh4p was added to DHE-containing liposomes, the fluorescence of DNS at 510 nm upon DHE excitation increased with kinetics similar to tryptophan quenching by DHE (1/t1/2 of 0.13 and 0.18 s−1, respectively; Fig. 1, A and D). No FRET was observed when Osh4p or DNS-Osh4p was mixed with liposomes lacking DHE or combining DHE and an excess of nonfluorescent ergosterol (Fig. 1, A and D). Therefore, the two FRET signals presented in Fig. 1 monitor the loading of DHE by Osh4p.
The experiments shown in Fig. 1 (A and D) did not allow us to determine whether Osh4p remained bound to the liposomes or returned in solution after DHE extraction. To distinguish between these two possibilities, we incubated Osh4p with sucrose-loaded DOPC liposomes containing DHE. After 15 min, the liposomes were collected by centrifugation. Gel and spectral analysis indicated that the protein was mostly recovered in the supernatant and quenched by DHE (Fig. 1 E). No spectral change was observed with DHE-free liposomes (Fig. 1 E) or when a “lidless” form of Osh4p lacking the amphipathic lipid packing sensor (ALPS)/lid motif and deficient in sterol binding was used (Fig. S1; Im et al., 2005; Raychaudhuri et al., 2006). We concluded that Osh4p interacts transiently with neutral liposomes, traps sterol, and goes back in solution in a sterol-loaded form stabilized by the lid. “Extraction” will be used to indicate DHE loading regardless of the retention of Osh4p on membranes, whereas “solubilization” will be used to indicate that Osh4p not only extracts DHE but also dissociates from membranes.
Effect of anionic lipids on DHE solubilization by Osh4p
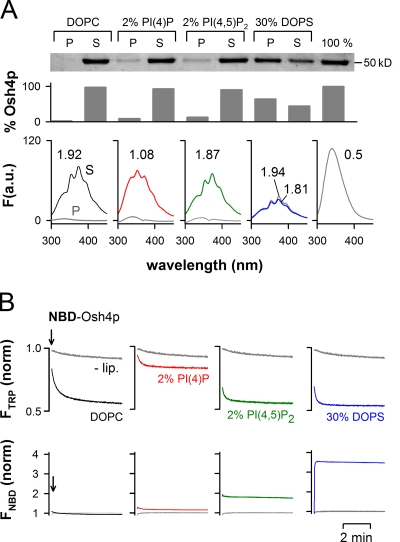
It has been reported that PI(4,5)P2 specifically stimulates sterol extraction by Osh4p (Raychaudhuri et al., 2006). We used our assay combining sedimentation and fluorescence analysis to examine the effect of various anionic lipids on the ability of Osh4p to extract and solubilize DHE (Fig. 2 A). To have an index of DHE binding by Osh4p, we calculated for both the liposome pellet and the supernatant an intensity ratio I372/I340, which reflects the extent of FRET between Osh4p and DHE in these fractions (from 0.5 for DHE-free Osh4p to 1.9 for Osh4p saturated with DHE).
Figure 2.
Influence of membrane composition on the solubilization of DHE by Osh4p. (A) Solubilization of DHE by Osh4p from DOPC liposomes doped with the indicated anionic lipid using the same protocol as in Fig. 1 E. When the fluorescence intensity was high enough, the I372/I340 ratio, which reflects the amount of DHE in complex with Osh4p in the supernatant (S) or in the pellet (P), was determined. (B) 0.5 µM NBD-Osh4p was added to buffer (−lip) or to liposomes (0.5 mM lipids) of various compositions. Trp fluorescence was monitored at 340 nm (λex = 285 nm; top), whereas NBD fluorescence was followed at 525 nm (λex = 495 nm; bottom). The contribution of the liposomes to the fluorescence signal was subtracted for each curve, and the fluorescence in buffer at t = 0 was used as a reference for normalization. Data shown in A are from a single representative experiment out of two independent experiments.
Anionic lipids had contrasting effects on the extraction and solubilization of DHE by Osh4p. First, dioleoyl phosphatidylserine (DOPS), which was used at 30 mol%, caused strong retention of Osh4p to the liposomes but did not modify the extent of DHE extraction. Both the soluble and the membrane-bound forms of Osh4p were loaded with DHE (I372/I340 = 1.81 and 1.94). PI(4)P or PI(4,5)P2, which were both used at 2 mol%, caused much less retention of Osh4p on the liposomes (∼8–10%). Surprisingly, the fluorescence spectra of the supernatants revealed a major difference: with PI(4)P, Osh4p failed to fully extract DHE (I372/I340 = 1.08). In contrast, DHE loading in the presence of PI(4,5)P2 was complete and similar to that observed on pure DOPC liposomes. This suggested that PI(4)P specifically inhibits DHE extraction.
The fact that PI(4)P had a specific inhibitory effect on Osh4p was striking in real-time measurements. As mentioned previously (Fig. 1 A), we recorded the rate of DHE extraction by Osh4p by measuring the quenching of tryptophan at 340 nm. In addition, we measured Osh4p binding to the liposomes through a second and independent fluorescence signal at 525 nm. To this end, we labeled the ALPS/lid motif with 7-nitrobenz-2-oxa-1,3-diazol (NBD), a probe whose fluorescence increases upon insertion between lipids. Control experiments indicated that this probe was well positioned to measure liposome binding of Osh4p (Fig. S2). As shown in Fig. 2 B, PI(4)P at 2 mol% almost completely blocked DHE extraction by NBD-Osh4p (red trace). In contrast, DOPS and PI(4,5)P2 had no effect on the extent of DHE loading and, instead, accelerated the rate of sterol extraction. Intriguingly, when the binding of Osh4p to the liposomes was measured by NBD fluorescence, PI(4)P did not contrast with other anionic lipids. In agreement with the sedimentation assay (Fig. 2 A), the NBD intensity was minimal with neutral liposomes, slightly increased with 2 mol% PI(4)P or 2 mol% PI(4,5)P2, and became maximal with liposomes containing a large amount of DOPS (30 mol%).
In conclusion, anionic lipids affect DHE extraction/solubilization by Osh4p in very different ways: by stimulating the rate of sterol extraction (2% PI(4,5)P2; 30% DOPS), by retaining the protein on the liposome surface (30% DOPS), or by inhibiting DHE extraction (2% PI(4)P). The most surprising observation was the inhibitory effect of PI(4)P.
Osh4p specifically extracts PI(4)P from liposomes
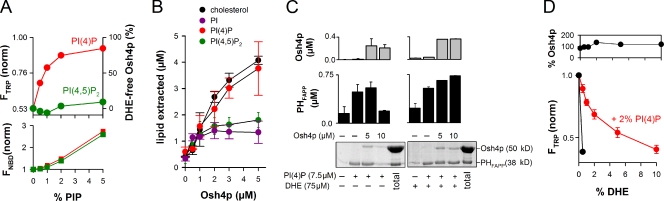
To further study the inhibitory effect of PI(4)P on DHE extraction by Osh4p, we performed experiments at increasing surface density of PI(4)P. The amount of DHE was kept at 0.5 mol%. As shown in Fig. 3 A, PI(4)P inhibited DHE extraction by NBD-Osh4p with a half-maximal effect of 0.5–1 mol%. No inhibition was seen with PI(4,5)P2, even at 5 mol%. Interestingly, NBD fluorescence indicated negligible protein retention on the liposomes when the density of PI(4)P did not exceed 1 mol%. At higher density (1–5 mol%), PI(4)P recruited Osh4p on membranes with similar efficiency as PI(4,5)P2, which suggests nonspecific electrostatic interactions. The lack of correlation between the concentration range at which PI(4)P inhibited DHE loading (0–2 mol%) and the concentration range at which PI(4)P caused Osh4p liposome binding (1–5 mol%) indicated that PI(4)P blocked DHE uptake by a mechanism different from membrane retention.
Figure 3.
DHE and PI(4)P compete for extraction by Osh4p. (A) 0.5 µM NBD-Osh4p was added to buffer alone or to DOPC liposome (0.5 mM lipids) containing 0.5 mol% DHE and increasing amounts of PI(4)P or PI(4,5)P2. The changes in Trp and NBD fluorescence, which reflect DHE extraction and liposome binding, respectively, were monitored as in Fig. 2 B. The plots show the fluorescence levels 9 min after the addition of Osh4p. The amount of DHE-free Osh4p reported on the right axis was determined from the Trp intensity. (B) Lipid extraction assay. Sucrose-loaded DOPC liposomes (0.5 mM lipids) containing 2% PI(4)P (red curve), 2% PI(4,5)P2 (green curve), 2% PI (purple curve), or 2% cholesterol (black curve) and doped, respectively, with [32P]PI(4)P, [3H]PI(4,5)P2, [3H]PI, or [3H]cholesterol were incubated with Osh4p (0–5 µM) for 20 min (room temperature). After centrifugation, radioactivity in the supernatant and in the pellet was counted to calculate the fraction of extracted lipid. Data are represented as mean ± SEM (error bars; n = 2–3). (C) Two-stage flotation assay. DOPC liposomes containing the indicated amounts of PI(4)P and DHE were mixed with 0, 5, or 10 µM Osh4p, collected by flotation, and then incubated with 0.75 µM PHFAPP (GST-tagged). The final amount of liposome-bound Osh4p and PHFAPP after the second stage was determined by SDS-PAGE. (D) Solubilization of DHE by Osh4p from DOPC liposomes containing 2% PI(4)P and increasing amounts of DHE (0–10%). After centrifugation, the amount of protein in the supernatant (top) was assessed by SDS-PAGE. The bottom shows Osh4p fluorescence at 340 nm (λex = 285 nm). Data are represented as mean ± SEM (error bars; n = 2). For comparison, an experiment was performed with liposomes containing 0.5% DHE but no PI(4)P (black curve).
Consequently, we hypothesized that PI(4)P could be itself extracted by Osh4p. As such, PI(4)P would act as a competing inhibitor of DHE. This mechanism would explain why Osh4p was recovered in the supernatant in the presence of PI(4)P, but did not show strong DHE loading (Fig. 2 A). To test this hypothesis, we incubated sucrose-loaded DOPC liposomes containing radiolabeled PI(4)P with an increasing amount of Osh4p. At 5 µM, Osh4p extracted ∼40%, i.e., 4 µM of PI(4)P (Fig. 3 B). Considering the amount of lipids (10 µM PI(4)P) and the fact that only half was accessible (in the liposome outer leaflet), this suggested that Osh4p extracted a stoichiometric amount of PI(4)P. In line with this, the dose response curves of cholesterol and PI(4)P extraction were very similar. In contrast, less PI or PI(4,5)P2 was extracted (Fig. 3 B).
In yeast, Osh4p inhibits the recruitment at the Golgi of a reporter protein for PI(4)P under conditions where this lipid is limiting (Fairn et al., 2007). Extraction of PI(4)P by Osh4p could explain this observation. We tested this model using a two-stage assay. In the first stage, liposomes containing 2 mol% PI(4)P were incubated with Osh4p and recovered at the top of a sucrose gradient by centrifugation. In the second stage, the liposomes were incubated with a GST-tagged Pleckstrin homology domain that targets PI(4)P (PHFAPP, 0.75 µM). In the absence of Osh4p during the first stage, almost all PHFAPP was recovered on the liposomes. When Osh4p was included at a concentration (10 µM) exceeding the amount of accessible PI(4)P (7.5 µM), binding of PHFAPP was low and undistinguishable from that observed on pure DOPC liposomes (Fig. 3 C). When the concentration of Osh4p (5 µM) was below the concentration of accessible PI(4)P, PHFAPP bound efficiently to the liposomes (Fig. 3 C). Importantly, under all conditions, the final amount of liposome-bound Osh4p was low and nearly identical. These observations suggested that Osh4p can extract a stoichiometric amount of PI(4)P, thereby preventing the subsequent binding of PHFAPP to the liposomes. Last, when 10% mol DHE was included in the liposome formulation, binding of PHFAPP to the liposomes was no longer affected by preincubation with Osh4p. By competing with PI(4)P, DHE in excess likely prevented Osh4p from extracting PI(4)P.
As previously mentioned, PI(4)P inhibited the loading of 0.5 mol% DHE by Osh4p with a half-maximal effect of 0.5–1 mol% (Fig. 3 A). Conversely, when the surface density of PI(4)P was kept at 2 mol%, the dose response curve of DHE loading was shifted to higher density values (half-maximal effect = 2 mol%; Fig. 3 D). Therefore, PI(4)P and DHE act not only as competing ligands for extraction by Osh4p, but they also display similar affinities for the protein.
Structural basis of PI(4)P-specific recognition by Osh4p
Because PI(4)P competes with DHE, the PI(4)P-binding site in Osh4p should overlap with the sterol-binding pocket. Using in silico approaches, we noticed that the sterol-binding site may also house a PI(4)P molecule. This prompted us to perform structural studies.
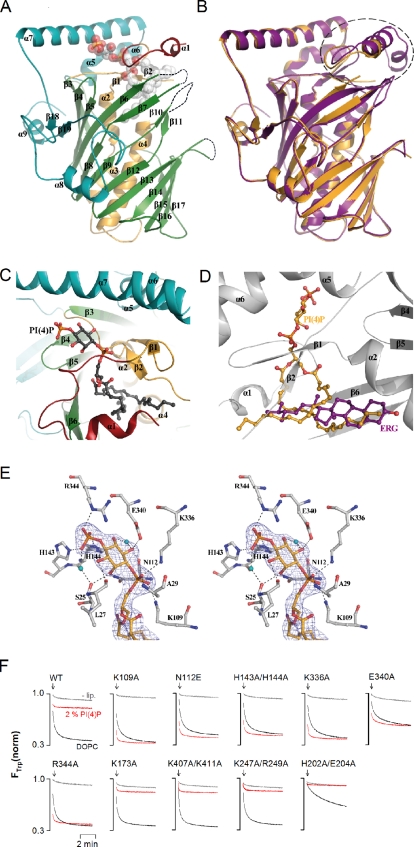
We crystallized Osh4p with PI(4)P and solved the structure of the complex by molecular replacement at 2.6-Å resolution (Table S1). The overall structure of Osh4p (Fig. 4 A) displays a central near-complete β-barrel (residues 117–307) forming a mainly hydrophobic tunnel flanked by an N-terminal domain (residues 30–116), which consists of a two-stranded β-sheet and three α-helices that close the incomplete β-barrel, and a large C-terminal region (residues 308–434). With a root mean square deviation (rmsd) of 1.14 Å for 382 residues, the structure of the complex with PI(4)P is highly similar to that obtained in the presence of ergosterol (PDB accession no. 1ZHZ; Fig. 4 B; Im et al., 2005). However, the lid (residues 1–29) adopts another conformation to accommodate the PI(4)P molecule (Fig. 4 B). In addition, several regions of the protein display poor electron density and could not be modeled reliably. These unresolved segments in the PI(4)P-bound conformation correspond to loops 99–105, 172–175, 204–212, and 237–242, and to the N-terminal part of the lid (1–12; Fig. 4, A and B).
Figure 4.
Crystal structure of Osh4p in complex with PI(4)P. (A) Overall structure. The lid region is shown in red (residues 13–29), the N-terminal domain in orange (30–116), the β-barrel in green (117–307), and the C-terminal domain in cyan (308–434). PI(4)P is shown as spheres with carbon atoms colored in white, oxygen in red, and phosphorus in orange. The missing loops are represented by dashed lines. (B) Structure superposition of Osh4p in complex with PI(4)P (in orange) or ergosterol (in purple; PDB accession no. 1ZHZ). The black circle indicates the region that differs significantly in the two structures. (C) Close-up view of the PI(4)P binding site. PI(4)P is shown in black with oxygen in red and phosphorus in orange. (D) Superposition of PI(4)P (colored in orange) and ergosterol (purple) molecules in Osh4p. The backbone of Osh4p is shown in light gray. (E) Stereo view of the interaction network of the PI(4)P polar head. The electron density colored in blue represents the Fo-Fc–simulated annealing omit map contoured at 2.5 σ. Nitrogen, oxygen, and phosphorus atoms are shown in blue, red, and orange, respectively. Key water molecules contacting PI(4)P and the protein are displayed as cyan spheres. (F) Effect of mutations on the ability of Osh4p to extract DHE from DOPC liposomes doped or not doped with 2% PI(4)P. The experimental conditions were the same as in Fig. 2 B.
PI(4)P binds to Osh4p by inserting its two acyl chains into the central tunnel of the β-barrel while the phosphorylated inositol ring lies near the protein surface in a shallow pocket formed by residues from the C-terminal loop of the lid, strand β2, the tip of the turn β4-β5, and helix α7 (Fig. 4, A and C). The acyl chains of PI(4)P interact loosely with the sterol-binding site in a rather nonspecific manner, as suggested by the faint electron density observed for this part of the ligand (Fig. 4 D and Fig. S3 A). At the opposite, the polar head group is involved in many direct and water-mediated contacts with the protein, accounting for the specific recognition of PI(4)P by Osh4p and the well-defined electron density of this part of the ligand (Fig. 4 E). The 4-phosphate group makes direct hydrogen-bonds with the side-chains of H143, H144 (β4-β5), and R344 (α7), and a water-mediated interaction with the main chain oxygen atom of S25 from the lid. The 1-phosphate group bridging the inositol ring and the glycerol moiety is hydrogen bonded to K109 (β2), K336 (α7), and the backbone amide of A29 in the lid (Fig. 4 E). Finally, the hydroxyl groups of the inositol ring are engaged in direct or water-mediated hydrogen bonds with the main chain oxygen atoms of S25, L27 (lid), and N112 (β2-β3 loop), and with the side chain of E340 (α7). Molecular modeling suggested that Osh4p could not bind PI(4,5)P2 as the 5-phosphate group should sterically clash with the side-chain of H143 (Fig. S3 B). A phosphate at position 3 seems also unfavorable.
We noted that several residues making contact with the PI(4)P headgroup are strictly conserved in Osh/Orp proteins. We tested their functional role by examining the inhibitory effect of PI(4)P on DHE loading by Osh4p mutants (Fig. 4 F). The mutants K109A, N112E, H143A/H144A, E340A, R344A, and K336A loaded DHE from liposomes as efficiently as the wild-type form but were not inhibited by PI(4)P. As a control, mutating basic residues (K174A, K407A/K411A, and K247A/R249A) on the other side of the sterol pocket preserved both the ability of Osh4p to extract DHE and its inhibition by PI(4)P. Finally, we noted that loop 204–212, which is defined in the sterol-bound form of Osh4p but not in the Osh4p–PI(4)P complex, is likely key for sterol binding but not for PI(4)P: the H202A/E204A mutant loaded DHE very slowly but remained sensitive to PI(4)P.
Osh4p delivers DHE to PI(4)P-containing acceptor liposomes
The crystal structures of Osh4p demonstrate that sterols and PI(4)P are mutually exclusive ligands, which explains why they compete for Osh4p extraction when present in the same membrane. However, this observation does not necessarily imply that sterol and PI(4)P act in an antagonist manner. In the context of lipid transport, Osh4p might exchange sterol for PI(4)P such that forward transport of sterol between two membranes would be coupled to backward transport of PI(4)P.
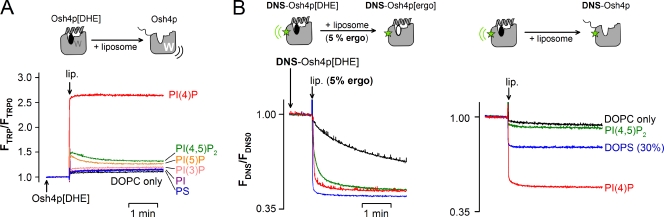
As a first step toward testing this hypothesis, we performed DHE delivery assays. We prepared Osh4p loaded with DHE (Osh4p[DHE]), then followed the kinetics of DHE release upon the addition of acceptor liposomes by measuring the increase in tryptophan fluorescence of Osh4p that resulted from the loss of FRET with DHE. With liposomes containing 2% PI(4)P, a jump in the fluorescence of Osh4p[DHE] was observed. The signal increase (2.6×) mirrored the signal decrease observed when Osh4p extracted DHE (1/2.5×; Fig. 1 A), which suggests the complete release of DHE. The effect of PI(4)P was very specific. Substituting other anionic lipids for PI(4)P such as DOPS, PI, PI(3)P, PI(5)P, or PI(4,5)P2 resulted in minor DHE release (Fig. 5 A).
Figure 5.
Osh4p delivers DHE to PI(4)P-containing liposomes. (A) Osh4p loaded with DHE (∼0.5 µM Osh4p[DHE]) was incubated at 30°C in buffer. At the indicated time, 30 µl of a stock suspension of liposomes (5 mM lipids) was injected (final concentration = 240 µM). The release of DHE was followed by measuring the increase in Osh4p fluorescence at 340 nm (λex = 285 nm). The liposomes contained DOPC and, when indicated, 2 mol% of DOPS, PI, PI(3)P, PI(4)P, PI(5)P, or PI(4,5)P2. (B) DNS-Osh4p loaded with DHE (∼0.5 µM) was mixed with 240 µM liposomes with (left) or without (right) 5 mol% ergosterol at 30°C. The release of DHE was followed by measuring the diminution in FRET between DHE and DNS (λex = 310 nm; λem = 510 nm). In all assays, the recordings were corrected for the light-scattering signal and the dilution effect was due to liposome addition.
DHE delivery probably involves sequential steps: binding of Osh4p[DHE] to the liposome surface, opening of the lid, and the exit of DHE. To better define this sequence of events and the role of PI(4)P, we compared experiments in which DNS-Osh4p[DHE] was mixed with acceptor liposomes with or without an excess of ergosterol (Fig. 5 B). With DOPC liposomes, the decay in FRET between DHE and DNS-Osh4p indicated a slow (t1/2 = 188 s) replacement of DHE by ergosterol. In contrast, Osh4p rapidly exchanged DHE for ergosterol (t1/2 < 5 s) on liposomes containing 2% PI(4)P, PI(4,5)P2, or 30% DOPS. This implied that Osh4p was able to quickly release DHE on liposomes containing anionic lipids. However, when we repeated these experiments with liposomes devoid of ergosterol, DHE appeared poorly delivered, except when PI(4)P was present. These results indicate that DHE release on anionic liposomes can be followed by recapture of another sterol molecule, leading to futile exchange. What makes PI(4)P unique compared with other anionic lipids is its ability to block sterol re-extraction by being loaded by Osh4p. As such, PI(4)P ensures net delivery of sterol.
Fast DHE transport from neutral membranes to PI(4)P-containing membranes
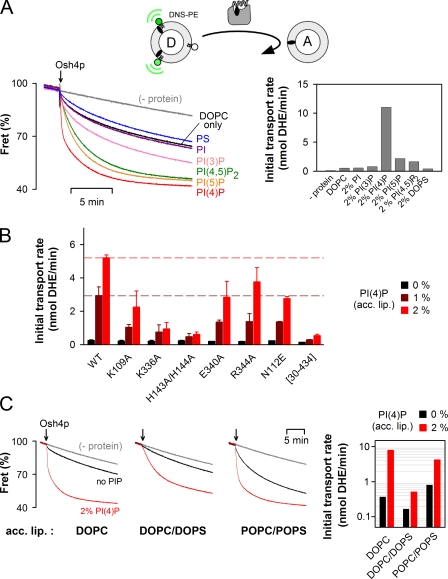
Osh4p optimally solubilized DHE from neutral membranes and fully delivered DHE to PI(4)P-containing membranes. These results prompted us to recapitulate sterol transport by Osh4p from the first to the second type of liposome. For this, donor liposomes made of DOPC and containing 10% DHE and 2.5% DNS-phosphatidylethanolamine (DNS-PE) were incubated with a ninefold excess of acceptor liposomes and with Osh4p. Although the FRET decrease between DHE and DNS-PE monitored DHE extraction from the donor liposomes (John et al., 2002), a decrease of ∼60% as seen in Fig. 6 A necessarily implies DHE delivery to the acceptor liposomes and Osh4p recycling through multiple cycles of transport. Indeed, Osh4p was used in a substoichiometric amount compared with DHE (ratio 1:20). Without acceptor liposomes, only minor FRET decrease was observed (see Materials and methods).
Figure 6.
Osh4p optimally transports DHE from neutral to PI(4)P-containing membranes. (A) Effect of anionic lipids in the acceptor liposomes. DOPC/DNS-PE/DHE liposomes (87.5:2.5:10 mol/mol, 100 µM lipids) were mixed with DOPC liposomes (900 µM lipids) containing 2 mol% of the indicated lipid. After 3 min, 0.5 µM Osh4p was added. FRET between DHE and DNS-PE in the donor liposomes diminishes as DHE is transported to the acceptor liposomes. The initial fluorescence signal was set at 100%. The slow decay observed without Osh4p is caused by spontaneous DHE transfer between the liposomes. (A, right) Initial transport rates. (B) Osh4p WT or mutants (0.5 µM) were added to DOPC/DNS-PE/DHE liposomes (87.5:2.5:10 mol/mol, 100 µM lipids) mixed with DOPC liposomes (900 µM lipids) containing 0, 1, or 2% PI(4)P. The initial transport rates are indicated on a linear scale for each mutant (black bars, 0% PI(4)P; dark red bars, 1% PI(4)P; red bars, 2% PI(4)P). The broken lines correspond to the value of the initial transport rate measured for the wild-type Osh4p with acceptor liposomes containing 1% or 2% PI(4)P. Data are represented as mean ± SEM (error bars; n = 2). (C) Transport was measured by mixing 0.5 µM Osh4p with DOPC/DNS-PE/DHE liposomes (87.5:2.5:10 mol/mol, 100 µM lipids) and acceptor liposomes (900 µM lipids) of different compositions (DOPC, DOPC/DOPS or POPC/POPS 70:30 mol/mol) with 2% PI(4)P (red traces) or without PI(4)P (black traces). (C, right) Initial transport rate. Black bars, without PI(4)P; red bars, with 2% PI(4)P. Experiments in A and C were completed once but include common conditions.
Fig. 6 A shows DHE transport experiments where we varied the composition of the acceptor liposomes. When these liposomes contained only DOPC or were doped with 2 mol% phosphatidylserine, PI, or PI(3)P, DHE transport was very slow (<1 nmol/min). In the presence of 2 mol% PI(4,5)P2 or PI(5)P, the rate increased approximately fourfold. Finally, with acceptor liposomes containing 2 mol% PI(4)P, we observed the fastest initial rate of transport (11 nmol /min; Fig. 6 A). Thus, there was a good correlation between the transport rate under multiple turnover conditions and the ability of Osh4p to deliver DHE to acceptor liposomes (Fig. 5 A).
Next we tested the DHE transfer activity of several Osh4p mutants with donor liposomes made of DOPC and acceptor liposomes containing DOPC and 0, 1, or 2 mol% PI(4)P (Fig. 6 B). In the absence of PI(4)P, the transport activity of all mutants was as low as that of wild-type Osh4p. Marked differences emerged when the acceptor membranes included PI(4)P. Some mutants (K109A, E340A, R344A, and N112E) showed a modest decrease in activity (two- to threefold) compared with the wild-type form, whereas the K336A and H143A/H144A mutant showed a much stronger defect (five- and ninefold, respectively). Remarkably, these two mutants were almost as inactive as lidless Osh4p ([30–434]Osh4p) defective in sterol binding. Because K336A and H143A/H144A extracted DHE normally but did not respond to PI(4)P (Fig. 4 F), these results highlighted the importance of PI(4)P recognition in the sterol transport activity of Osh4p.
In yeast, PI(4)P is synthesized by two PI-4 kinases: Pik1p at the trans Golgi, and Stt4p at the PM (Strahl and Thorner, 2007). The bulk lipid composition of these compartments is complex and obviously not properly mimicked by acceptor liposomes containing only DOPC and PI(4)P. Compared with the ER, the PM is highly enriched in phosphatidylserine (Yeung et al., 2008) and contains more saturated lipids (Zinser et al., 1991; Schneiter et al., 1999; Pichler et al., 2001; Klemm et al., 2009). The trans Golgi probably displays intermediate features between the ER and the PM. To address the influence of lipid packing and membrane electrostatics, we compared three acceptor liposome compositions: DOPC (low packing), DOPC/DOPS (low packing and high charge density), and 1-palmitoyl-2-oleoyl-phosphatidylcholine (POPC)/1-palmitoyl-2-oleoyl-phosphatidylserine (POPS; medium packing and high charge density). In all cases, we observed a strong stimulatory effect of 2% PI(4)P on DHE transport, which suggests that PI(4)P retains a specific effect on Osh4p regardless of the bulk properties of the acceptor membrane (Fig. 6 C). However, the rate of DHE transport decreased by one order of magnitude when the acceptor liposomes combined low lipid packing and high charge density. Because the PI(4)P-dependant sterol delivery step was marginally affected by these changes (Fig. S4), the following step of the cycle, recycling of Osh4p from DOPC/DOPS liposomes, should be rate limiting. This effect is explained by the fact that Osh4p is strongly retained on these liposomes, as previously observed (Figs. 2 and S2). Finally, we noticed that transport occurred in all conditions without liposome aggregation (Fig. S4, B and C). Thus, Osh4p efficiently transfers sterol from neutral membranes to membranes containing PI(4)P and a high density of other anionic lipids without membrane bridging.
Counter exchange of PI(4)P and DHE by Osh4p
PI(4)P could be very efficiently extracted by Osh4p (Fig. 3), had a stimulatory effect on DHE delivery (Fig. 5), and strongly accelerated the rate of DHE transport under multiple turnover conditions (Fig. 6). Together, these observations suggested that DHE transport from donor to acceptor liposomes may be coupled to transport of PI(4)P in the opposite direction. Interestingly, this prediction could explain the atypical shape of DHE transport kinetics: in the presence of PI(4)P in the acceptor liposomes, the time courses slowed down abruptly once half of DHE had been transferred (Fig. 6, A and C). PI(4)P, which promotes DHE delivery on the acceptor liposomes, should, if transported back to the donor liposomes, become inhibitory by competing with DHE for Osh4p extraction.
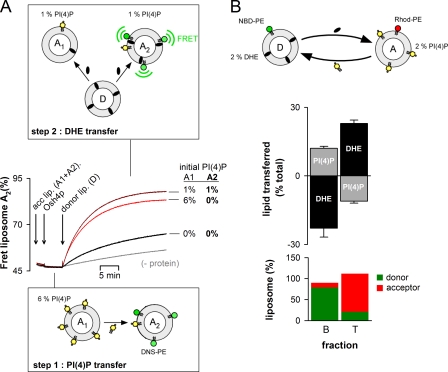
We set up an indirect assay for PI(4)P transport. In a first step, we mixed Osh4p with two liposome populations: “A1” containing 6% PI(4)P, and “A2” without PI(4)P. Given the ratio between these liposomes (1:5), transport and equilibration of PI(4)P between A1 and A2 by Osh4p should lead to a final density of 1 mol% accessible PI(4)P. Thereafter, donor liposomes (D) containing DHE were added. Because only A2 contained the fluorescent lipid DNS-PE, the FRET signal monitored DHE transport from D to A2. As shown in Fig. 7 A, the kinetic of DHE transport was almost as fast as in a control reaction performed with 1 mol% PI(4)P directly included in liposomes A2. In contrast, when A1 liposomes were devoid of PI(4)P, the delivery of DHE to A2 was very slow. This assay suggested that Osh4p had efficiently transported PI(4)P from A1 to A2 during the first step.
Figure 7.
Osh4p transfers PI(4)P between membranes. (A) 0.5 µM Osh4p was mixed at 30°C with DOPC liposomes containing 6% PI(4)P (liposome A1, 15 µM) and DOPC liposomes doped with 2.5% DNS-PE (A2, 75 µM). In a second step, DOPC liposomes containing 18% DHE were added (donor liposome, 90 µM). Control experiments were done with A1 and A2 liposomes containing either 0 or 1% PI(4)P (black and dark red trace, respectively) or without Osh4p (gray trace). (B) Sucrose-loaded DOPC/DHE liposomes (98:2 mol/mol, 0.5 mM lipids, labeled with 0.1% mol NBD-PE) were incubated with DOPC/PI(4)P liposomes doped with [32P]PI(4)P (98:2 mol/mol, 0.5 mM lipids, labeled with 0.1 mol% Rho-PE). After centrifugation on a sucrose gradient, a bottom and top fraction were collected. The fluorescence of NBD-PE, Rho-PE, and DHE was measured and PI(4)P radioactivity was counted for each fraction. (B, bottom) The relative amount of donor and acceptor liposomes in each fraction. (B, top) The gain or loss of DHE and PI(4)P (in percentages) for each liposome population. Data are represented as mean ± SEM (n = 2).
Next, we devised a direct assay for PI(4)P transport based on liposome separation. Donor liposomes containing 2% DHE were loaded with sucrose and labeled with NBD-PE. Acceptor liposomes containing 2% radiolabeled PI(4)P were doped with rhodamine-phosphatidylethanolamine (Rhod-PE). The liposomes were mixed in a 1:1 ratio, incubated with or without Osh4p, and then separated by centrifugation. We collected a bottom and top fraction containing, respectively, mostly donor (80%) or acceptor liposomes (90%), as estimated by the relative fluorescence of NBD-PE and Rhod-PE. We determined that the donor liposomes lost 25% DHE and gained 12–13% PI(4)P when incubated with Osh4p. Conversely, the acceptor liposomes showed an increase in DHE and a decrease in PI(4)P (Fig. 7 B). If forward transport of sterol was strictly coupled with backward transport of PI(4)P, the same amount of PI(4)P and DHE should have been displaced. However, DHE also spontaneously exchanges between liposomes (see, e.g., the gray trace in Fig. 6 A), making quantitative comparisons difficult with this assay, which also lacks time resolution. Notwithstanding these limitations, these experiments indicate that Osh4p can transport DHE and PI(4)P in opposite ways.
Discussion
Identifying the mechanisms by which lipid transfer proteins selectively convey lipids between membranes is crucial for understanding how organelles maintain their composition (Lev, 2010). Osh4p has been suggested to transport sterol from the PM, marked by PI(4,5)P2, to the ER (Raychaudhuri et al., 2006). We propose a different model in which Osh4p physically exchanges sterols for PI(4)P between membranes. Based on structural and biochemical data, this model reconciles several cellular observations on Osh4p and suggests that the distribution of sterol may be controlled by phosphoinositide metabolism.
Osh4p efficiently solubilizes PI(4)P from membranes and, for this, uses roughly the same binding site as for sterol extraction. This unexpected finding is nicely explained by the structure of Osh4p bound to PI(4)P. First, the sterol-binding pocket accommodates the PI(4)P acyl chains. Second, a shallow pocket at the entrance of the tunnel, which contains critical residues such as K336 and the H143/H144 pair, selects the polar head of PI(4)P with high specificity. This interaction is probably essential for compensating loose binding of the PI(4)P acyl chains. Third, the N-terminal lid secures the bound PI(4)P molecule by wrapping its glycerol moiety. Overall, the result of these three interactions is functionally very important: Osh4p displays a similar affinity for PI(4)P and sterols.
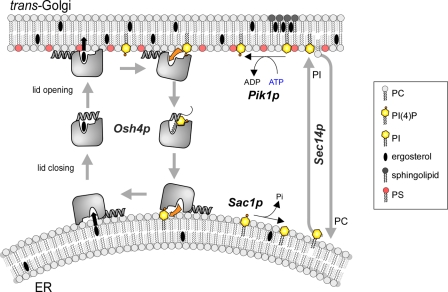
Our kinetic analysis suggests that Osh4p is designed to promote rapid exchange of sterols for PI(4)P between membranes according to the cycle shown in Fig. 8. In this scheme, sterol release in the acceptor membrane is followed by PI(4)P extraction; conversely, PI(4)P release in the donor membrane precedes sterol extraction. When integrated in a more general view including other lipid-modifying enzymes and lipid transporters at the ER–Golgi interface, the proposed cycle explains several genetics and cellular observations as detailed (Fig. 8).
Figure 8.
Model for the coupling between sterol and PI(4)P exchange by Osh4p. Osh4p solubilizes sterols efficiently from neutral and loosely packed membranes. The sterol is locked by the lid. The interaction of Osh4p with anionic membranes promotes the opening of the lid and sterol release. Only PI(4)P promotes net sterol delivery because PI(4)P is extracted by Osh4p at the expense of sterol re-extraction. Tight lipid packing in the acceptor membrane limits the retention of Osh4p through its ALPS motif. These observation suggest that Osh4p is suited to transport sterols preferentially from ER to the trans Golgi marked by PI(4)P. Conversely, Osh4p transports PI(4)P from the trans Golgi to the ER, and another round of transport can resume. Although each step of the cycle is reversible by itself, the presence of a PI-4 kinase at the TGN (Pik1p) and of a PI(4)P phosphatase at the ER (Sac1p) favors directionality of lipid transport. The sterol/PI(4)P exchange activity of Osh4p is also coupled to that of Sec14p, a PC/PI exchanger.
Osh4p rescues yeast lacking the entire Osh family by restoring the level of sterol at the PM (Beh and Rine, 2004). By exchanging sterol for PI(4)P, Osh4p might supply the trans Golgi and PM, pinpointed by this phosphoinositol phosphate, with sterol. The coupling between sterol delivery and PI(4)P extraction also explains why mutants such as K336A and H143A/H144A, although fully competent for sterol extraction, do not rescue Osh-depleted yeast (Im et al., 2005). These mutants do not promote net transfer of sterol to the PM because they are unable to capture, in turn, PI(4)P.
Osh4p overexpression reduces both the cellular level of PI(4)P and the Golgi targeting of a PI(4)P reporter (Fairn et al., 2007). These observations can be explained by retrograde transport of PI(4)P from the Golgi to the ER by Osh4p, where PI(4)P is metabolized by Sac1p, an ER-resident PI(4)P-phosphatase. We note that this mechanism offers an alternative explanation for the interplay between Osh4p and Sac1p. Osh proteins have been proposed to facilitate the hydrolysis of PI(4)P in “trans” by Sac1p by forming membrane contact sites between organelles (Stefan et al., 2011). This mechanism is appealing in the case of Osh proteins with additional domains that can effectively bridge two membranes. However, Osh4p lacks such domains and efficiently exchanges sterol for PI(4)P between liposomes without tethering them. Therefore, we hypothesize that Osh4p supplies the ER with PI(4)P, which is then hydrolyzed by Sac1p.
Finally, the sterol/PI(4)P transfer activity of Osh4p gives a straightforward explanation for the long-established genetic interactions between OSH4, SEC14, SAC1, and PIK1 genes (Fang et al., 1996; Li et al., 2002; Fairn et al., 2007). SEC14 encodes for a PI/phosphatidylcholine (PC) transfer protein and is an essential gene. However, yeast cells can survive when SEC14 is inactivated together with OSH4. As long as Osh4p was considered as a mere sterol transporter, the functional antagonism between Osh4p and Sec14p remained difficult to explain. The fact that Osh4p also acts as a PI(4)P transporter suggests that cell survival upon deletion of OSH4 and SEC14 is caused by reduced backward transport of PI(4)P from the trans Golgi to the ER. Elimination of this transport pathway probably compensates for the limiting amount of PI(4)P that can be synthesized by Pik1p when forward transport of PI by Sec14p is impaired (Fairn et al., 2007). Again, this model better explains mutagenesis data: Osh4p mutants that are unable to extract PI(4)P but behave normally for sterol extraction (K336A, H143A/H144A) are not lethal in Sec14p-deficient yeast, in contrast to the wild-type form (Im et al., 2005).
In conclusion, we propose a phosphoinositide cycle in which Sec14p and Osh4p transport PI and PI(4)P along opposite routes, PI(4)P being converted into PI at the ER by Sac1p and PI being phosphorylated into PI(4)P at the trans Golgi by Pik1p (Fig. 8). When considered from the sole “point of view” of phosphoinositides, this cycle seems futile: ATP is used to continually regenerate PI(4)P. However, from a sterol-centric point of view, the consumption of ATP by Pik1p ensures net transfer of sterol in the forward direction and PC in the backward direction. As such, Osh4p could drive sterols up a concentration gradient and promote sterol enrichment of membranes of the late secretory pathway (TGN, PM) at the expense of the ER, where sterol is synthesized.
Some aspects of the proposed Osh4p cycle warrant further examination. Notably, the mechanism by which Osh4p targets the ER remains to be addressed. Osh4p quickly (t1/2 of ∼7 s) and totally solubilizes DHE from neutral (DOPC) liposomes under conditions where DHE was present at only 0.5 mol%. Thus, Osh4p should be able to efficiently pick up sterol from the ER, wherein this lipid is rare. Interestingly, we found previously that the lid of Osh4p corresponds to an ALPS motif (Drin et al., 2007). ALPS are unstructured sequences that fold into amphipathic helices upon binding to neutral membranes whose composition and high curvature result in loose lipid packing. The ER, which is characterized by a low content of anionic lipids, a high content of unsaturated lipids, and a highly tubulated structure, seems well adapted to the transient adsorption of the Osh4p ALPS/lid region. In the future, it will be important to study in a systematic manner the influence of membrane curvature and composition on the ability of Osh4p to exchange sterol and PI(4)P between membranes. In fact, one reason for the conflicting results between this study and previous studies may be related to membrane curvature. In several in vitro assays for Osh4p transport, very small liposomes obtained by sonication and containing high amounts of anionic lipids have been used (Raychaudhuri et al., 2006; Schulz et al., 2009; Stefan et al., 2011). It is likely that the combination of extreme curvature and high electrostatic favors Osh4p retention on the liposomes, impairs lipid transport, and causes membrane aggregation through the multiple potential membrane-binding sites of Osh4p (ALPS motif, basic patches).
Most of residues that contact the PI(4)P headgroup are conserved in Osh/Orp proteins, which suggests that extraction of PI(4)P is a general hallmark of this family. Determining how this biochemical feature translates to a function will require further investigation for each Osh/Orp.
Materials and methods
Proteins
Osh4p and Osh4p[30–434] were cloned in a pGEX-4T-3 vector. Site-specific mutations were introduced using the QuikChange mutagenesis protocol and checked by DNA sequencing. Osh4p was expressed in E. coli at 30°C overnight upon induction with 1 mM IPTG (OD600 = 0.6). All purification steps were conducted in 50 mM Tris, pH 7.4, 120 mM NaCl, 1 mM MgCl2, and 1 mM DTT supplemented, during the first purification steps, with 1 mM PMSF, 1 µM bestatine, 10 µM pepstatine, 10 µM phosphoramidon, and protease inhibitor tablets (Roche). Cells were lysed with a French press and the lysate was centrifuged at 200,000 g for 1 h. The supernatant was applied to Glutathione Sepharose 4B beads. After three washing steps, the beads were incubated with thrombin to cleave the GST fusion and allow the release of Osh4p. The purity of the proteins was >95% as assessed by SDS-PAGE. The protein concentration was determined by a Bradford assay and by SDS-PAGE analysis using a BSA standard curve. All constructs contain an N-terminal GS sequence from the thrombin cleavage site.
5-((((2-iodoacetyl)amino)ethyl)amino)naphthalene-1-sulfonic acid (IAEDANS) labeling of Osh4p was performed by mixing the protein (after DTT removal by dialysis) with a 10-fold molar excess of IAEDANS (Invitrogen). After 90 min on ice, the reaction was stopped by adding a 10-fold excess of l-cysteine over the probe. The free probe was removed by gel filtration on a Sephacryl S200 HR XK16-50 column (GE Healthcare). The labeled protein was analyzed by SDS-PAGE and UV-visible spectroscopy. The gel was directly visualized in a fluorescence imaging system (U-GENIUS; Syngene) to detect DNS-labeled Osh4p excited in near-UV and then stained with Sypro Orange to determine the purity of DNS-Osh4p. The percentage of labeling was estimated from the optical density (OD) of tryptophan at 280 nm (ε = 73,900 M−1cm−1) and DNS at 340 nm (ε = 5,700 M−1cm−1). In a control experiment, we observed no labeling of the Osh4p(C98S) mutant, which indicates that CYS-98 was the sole endogenous cysteine that could be labeled by a thiol-reactive probe.
NBD labeling of Osh4p was performed on a double mutant lacking the solvent-exposed endogenous cysteine (C98S) and including a cysteine in the N-terminal lid (A5C). After DTT removal, this mutant was labeled with a 10-fold excess of N,N’-dimethyl-N-(iodoacetyl)-N’-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)ethylenediamine (IANBD-amide; Invitrogen) and purified according to the instructions used for DNS-Osh4p. The percentage of labeling was estimated from the optical density at 280 and 495 nm (ε = 25,000 M−1cm−1 for NBD according to the manufacturer). The PH domain of FAPP in fusion with GST was provided by J. Bigay (Centre National de la Recherche Scientifique, Institut de Pharmacologie Moléculaire et Cellulaire, Valbonne, France).
Lipids
DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine), POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine), DOPS (1,2-dioleoyl-sn-glycero-3-phospho-l-serine), POPS (1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine), brain PI(4)P (l-α-PI-4-phosphate), brain PI(4,5)P2 (l-α-PI-4,5-bisphosphate), liver PI (l-α-PI), DNS-PE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(5-dimethylamino-1-naphthalenesulfonyl)), NBD-PE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl)), and Rhod-PE (1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl)) were obtained from Avanti Polar Lipids, Inc. PI(3)P (1,2-dioleoyl-sn-glycero-3-inositol-3-phosphate) and PI(5)P (1,2-dioleoyl-sn-glycero-3-inositol-5-phosphate) were obtained from Echelon. Ergosterol and DHE were obtained from Sigma-Aldrich. The concentration of DHE in stock solution in methanol was carefully determined by UV spectroscopy using an extinction coefficient of 13,000 M−1cm−1.
The radioactive lipids [3H]cholesterol, [3H]PI, and [3H]PI(4,5)P2 were purchased from PerkinElmer. To prepare radioactive [32P]PI(4)P, BHK cells were labeled with 1 mCi/ml ortho[32P]phosphate during 36 h in phosphate-free DME containing 2% FCS. After one washing step in cold PBS, cells were scrapped off and recovered in 3 ml of HCl (1.6 N). A mixture of methanol and chloroform (6 ml, vol/vol) was immediately added and the sample was vortexed vigorously during 2 min and centrifuged at 300 g for 5 min. The organic phase was collected and dried under a nitrogen stream at 37°C. Lipids were resolved by thin layer chromatography (TLC silica gel 60; Merck) using CHCl3/CH3OH/NH4OH 4N (90/70/20, vol/vol) as a solvent and identified using appropriate standards. The spot of [32P]PI(4)P was scraped off and the lipid was extracted from the silica by a modified Blight and Dyer procedure (Bligh and Dyer, 1959). The purity of the PI(4)P preparation was analyzed by HPLC. In brief, an aliquot of the preparation was deacylated by adding 1 ml of methylamine reagent composed of 26.8% (vol/vol) of 40% methylamine, 45.7% (vol/vol) methanol, 11.4% (vol/vol) n-butanol, and 16% (vol/vol) H2O. After incubation at 53°C for 50 min, the methylamine reagent was completely evaporated under a nitrogen stream at 37°C. The samples were resuspended in 1.2 ml H2O and separated by HPLC using a Whatman Partisphere 5 SAX column 4.6 mm × 125 mm with guard-cartridge anion exchanger units (Ref. 4641 0005; GE Healthcare) and a gradient of 1 M (NH4)2HPO4, pH 3.8, and bi-distilled water (Payrastre, 2004). The amount of purified PI(4)P was quantified by phosphorus measurement (Fiske and Subbarow, 1925).
Liposomes
Lipids in stock solutions in chloroform were mixed at the desired molar ratio, and the solvent was removed in a rotary evaporator. For formulations containing phosphoinositides, the mixture was warmed at 30–33°C for 5 min prior to drying under vacuum. For lipid films containing radioactive lipids, the mixture was dried in a hemolysis tube under argon. The lipid film was hydrated in 50 mM Hepes, pH 7.2, and 120 mM potassium acetate (HK buffer) to give a suspension of large multilamellar liposomes (lipid concentration: 1–5 mM). The suspension was then frozen and thawed five times (using liquid nitrogen and a water bath) and extruded through polycarbonate filters of 0.2-µm pore size using a mini-extruder (Avanti Polar Lipids, Inc.). For experiments with liposomes of varying curvature, the liposomes were extruded sequentially through 0.4-, 0.2-, 0.1-, 0.05-, and 0.03-µm (pore size) polycarbonate filters. The liposome hydrodynamic radius (RH) was estimated by dynamic light scattering in a DynaPro instrument (Wyatt Technology). Liposomes were stored at 4°C and in the dark when containing light-sensitive lipids (NBD-PE, DNS-PE, DHE, or ergosterol) and used within 2 d.
DHE loading assay
For kinetics measurements, Trp fluorescence was measured at 340 nm (bandwidth 5 nm) upon excitation at 286 nm (bandwidth 1.5 nm) in a fluorometer (RF 5301-PC; Shimadzu). The sample initially contained liposomes (500 µM total lipids) in HKMD buffer (50 mM Hepes, pH 7.2, 120 mM potassium-acetate, 1 mM MgCl2, and 1 mM DTT). The liposomes used in this assay were extruded on a 0.2-µm filter. The sample (volume 600 µl) was placed in a cylindrical quartz cell, continuously stirred with a small magnetic bar, and thermostated at 30°C. At the indicated time, Osh4p or NBD-Osh4p was injected from stock solutions through a guide in the cover of the fluorometer adapted to Hamilton syringes, such as to not interrupt the fluorescence recording. Emission spectra were recorded at the end of the kinetics upon excitation at 285 nm. Loading experiments with DNS-Osh4p were performed under the same conditions except that DHE was directly excited at 310 nm (bandwidth 1.5 nm) and the fluorescence of DNS was followed at 510 nm (bandwidth 10 nm). We assessed, under conditions where Osh4p was strongly bound to the liposomes (i.e., with DOPC/DOPS liposome), that the FRET signal reflected only DHE loading by Osh4p and not the proximity between the protein and DHE embedded in the membrane. Indeed, no FRET occurred when we incubated DNS-Osh4p with DOPS/DOPS liposomes containing DHE (0.5%) and an excess of ergosterol (5%).
DHE extraction assay
0.5 µM Osh4p was mixed with 500 µM (total lipids) of large sucrose-loaded liposomes (210 mM sucrose and 50 mM Hepes, pH 7.2, extruded on a 0.2-µm filter) for 15 min at 30°C under stirring in HKMD. The sample was centrifuged at 400,000 g for 20 min at 20°C in a fixed-angle rotor (TLA 100.1; Beckman). The supernatant (600 µl) was recovered and the liposome pellet was resuspended in 600 µl HKMD. A fluorescence spectrum of each fraction was recorded between 300 and 450 nm (bandwidth 5 nm) upon excitation at 285 nm (bandwidth 1.5 nm). The background signal was determined by measuring spectra of buffer with or without liposomes (500 µM total lipids). In addition, each sample was analyzed by SDS-PAGE, using Sypro Orange staining and a fluorescence imaging system (LAS 3000; Fujifilm). Control centrifugations performed without liposomes indicated that Osh4p did not sediment.
DHE delivery assay
To prepare Osh4p in complex with DHE, Osh4p was incubated in HKMD at 2 µM for 30 min at 30°C under agitation with large DOPC liposomes (200 µM total lipid) loaded with sucrose and containing 10% DHE (mol/mol lipids). The sample was centrifuged at 400,000 g for 20 min at 20°C to pellet the liposomes. The supernatant (600 µl) containing Osh4p loaded with DHE (Osh4p[DHE]) was collected. Osh4p[DHE] was diluted at 0.5 µM in HKMD in a cylindrical quartz cuvette (final volume 600 µl). Sterol delivery was monitored at 340 nm (bandwidth 10 nm) upon excitation at 285 nm (bandwidth 1.5 nm) at 30°C by adding 30 µl of large liposomes (stock solution 5 mM, final concentration ∼240 µM) to Osh4p[DHE]. The protocol used to measure DHE release from DNS-Osh4p was the same except that the signal was measured at 510 nm (bandwidth 20 nm) upon excitation at 310 nm (bandwidth 1.5 nm).
DHE transport assay
A suspension (570 µl) of acceptor liposomes (900 µM total lipids) was incubated in a cylindrical cuvette at 30°C under constant stirring. After 1 min, 30 µl donor liposomes (100 µM lipid, final concentration) containing 2.5% of DNS-PE and 10% of DHE was added. The liposomes were extruded on a 0.2-µm filter. After 3 min, Osh4p was injected at 0.5 µM. Lipid transport was followed by measuring the DNS signal at 525 nm (bandwidth 10 nm) upon DHE excitation at 310 nm (bandwidth 1.5 nm). The amplitude of the signal corresponding to maximal transport of DHE was determined by adding 1 mM of methyl-β-cyclodextrin. The linear dependency of the FRET signal on the DHE content was checked by measuring at 525 nm the fluorescence of a set of DOPC liposomes containing 2.5% of DNS-PE and 0, 2, 4, 6, 8, or 10% DHE. In the absence of acceptor liposomes, the addition of Osh4p resulted in a FRET decrease of only 5–10% as compared with −60% in the presence of acceptor liposomes, which indicates the extraction of a small amount of DHE from donor liposomes by the protein.
Lipid and cholesterol extraction assay
Sucrose-loaded liposomes (500 µM total lipids, extruded on 0.2 µm filter) containing 0.4% Rhod-PE and the indicated radiolabeled lipid were mixed with increasing amount of Osh4p (0–5 µM) for 20 min at room temperature. The sample was centrifuged at 400,000 g for 20 min at 20°C in a fixed-angle rotor (TLA 100.1). The supernatant (150 µl) was recovered and the liposome pellet, which could be easily visualized owing to the presence of Rhod-PE, was resuspended in 150 µl HKMD. Both samples were mixed with 2 ml scintillant and counted.
NBD-fluorescence lipid binding assay
0.5 µM NBD-Osh4p was incubated at 30°C for 5 min with liposomes (500 µM total lipids) in HKMD buffer. Emission fluorescence spectra were measured from 505 to 650 nm (bandwidth 5 nm) upon excitation at 495 nm (bandwidth 5 nm) in a 100-µl quartz cell. Control spectra were also recorded in the absence of protein to subtract the light-scattering signal of the liposomes from the protein spectra. To measure NBD fluorescence over time, the sample was excited at 495 nm (bandwidth 5 nm) and the emission was recorded at 525 nm (bandwidth 5 nm).
Flotation experiments
The protocol has been described in detail (Bigay and Antonny, 2005; Bigay et al., 2005). In brief, 0.75 µM of protein was incubated with NBD-PE–containing liposomes (750 µM total lipids) in 150 µl HKM buffer (HK buffer with 1 mM MgCl2) at room temperature for 5 min. The suspension was adjusted to 30% sucrose by mixing 100 µl of a 75% (wt/vol) sucrose solution in HKM buffer and overlaid with 200 µl HKM containing 25% (wt/vol) sucrose and 50 µl sucrose-free HKM. The sample was centrifuged at 240,000 g in a swing rotor (TLS 55) for 1 h. The bottom (250 µl), middle (150 µl), and top (100 µl) fractions were collected. The bottom and top fractions were analyzed by SDS-PAGE using Sypro-Orange staining and a fluorescence imaging system (LAS-3000; Fujifilm).
DHE/PI(4)P transport experiments
0.5 µM Osh4p was incubated for 30 min at 30°C under constant mixing in 125 µl HKM buffer with DOPC/DHE liposomes (98:2 mol/mol, 500 µM total lipids) doped with NBD-PE, then loaded with 50 mM Hepes, pH 7.2, 220 mM sucrose, and DOPC/PI(4)P liposomes (98:2 mol/mol, 500 µM total lipids) doped with Rhod-PE and [32P]PI(4)P. The suspension was adjusted to 7.5% sucrose by mixing 75 µl of a 20% (wt/vol) sucrose solution in HKM buffer and overlaid with three cushions of 200 µl HKM containing 5, 2.5, and 0% (wt/vol) sucrose. The sample was centrifuged at 240,000 g in a TLS 55 rotor for 1 h at 4°C. The bottom and top fractions (400 µl each) were collected. Emission fluorescence spectra of NBD-PE, Rhod-PE, and DHE were measured in a 100-µl quartz cell (DHE, from 325 to 450 nm, excitation at 310 nm; NBD-PE, from 480 to 700 nm, excitation at 470 nm; Rhod-PE, from 580 to 700 nm, excitation at 570 nm) to quantify DHE, donor liposomes, and acceptor liposomes, respectively. Samples were then mixed with 2 ml scintillant and counted to measure PI(4)P. For each fraction, we determined the percentage of transferred DHE by considering %TDHE = ΔDHE/TDHE × (FA − FD), where ΔDHE is the difference in DHE fluorescence between the samples with and without Osh4p, TDHE is the mean total DHE fluorescence measured for both experiments, and FD and FA are the relative amounts of donor and acceptor liposomes in one fraction. Because of the small contamination of one population of liposome by the other, gain of DHE in one population is compensated by loss of DHE in the second population and leads to an apparent variation in DHE that is lower than the real transfer. Similarly, the percentage of transferred PI(4)P was calculated on the basis of radioactivity considering that %TPI(4)P = ΔPI(4)P/TPI(4)P × (FD − FA).
Dynamic light scattering
Experiments were performed at 30°C in a DynaPro apparatus (Protein Solutions). The sample initially contained donor and acceptor liposomes in HKMD buffer in a small quartz cell (volume 20 µl). A first set of ∼12 autocorrelation curves was acquired to assess the size distribution of the initial liposome suspension. Then, Osh4p was added manually and mixed thoroughly, and the kinetics of aggregation was followed by acquiring one autocorrelation curve every 10 s. The data were analyzed by the Dynamics v6.1 software (Protein Solutions) by fitting the autocorrelation functions with the assumption that the size distribution is a simple Gaussian function.
Crystallization and x-ray data collection
6 mg/ml Osh4p was incubated for 2 h with large DOPC liposomes containing 2% PI(4)P and loaded with sucrose (12 mM total lipids). The sample (2 ml) was centrifuged at 400,000 g for 20 min at 4°C in a fixed-angle rotor (TLA 100.1) to separate the protein loaded with PI(4)P from liposomes. The Osh4p–PI(4)P complex recovered in the supernatant was purified by gel filtration on a Sephacryl S200 HR XK16-50 column and concentrated at 9.7 mg/ml. The Osh4p–PI(4)P complex was crystallized by the hanging drop vapor diffusion method where 1 µl of protein sample in 20 mM Tris, pH 8.0, and 100 mM NaCl was mixed to 1 µl of reservoir solution containing 100 mM MES, pH 7.0, 250 mM CaCl2, and 16% PEG at 18°C. Native diffraction data from one crystal cryoprotected with 20% glycerol were collected to 2.6-Å resolution at a wavelength of 0.9334 Å on the ID14-1 beamline at the European Synchrotron Radiation Facility. Data were processed and scaled with XDS and XSCALE (Kabsch, 2010). The PI(4)P-bound Osh4p crystallized in the space group P21 with cell parameters a = 73.46 Å, b = 54.76 Å, c = 121.86 Å, and β = 91°, with two molecules per asymmetric unit.
X-ray structure determination and refinement
The x-ray structure was determined by the molecular replacement method using PHENIX (Adams et al., 2010) and the structure of Osh4p solved in complex with 20-hydroxycholesterol (PDB accession no. 1ZHW) as the search model. The initial structure was rebuilt using COOT (Emsley and Cowtan, 2004) and refined with PHENIX and REFMAC (Murshudov et al., 1997) from the CCP4 suite (Winn et al., 2011). The R-factor of the final structure is 0.228 (Rfree = 0.269), using all data from 47.29- to 2.6-Å resolution. Data collection and refinement statistics are summarized in Table S1. The structure contains 398 residues for chain A and 390 residues for chain B, and the rmsd of the two subunits is 0.424 Å for 385 superimposed α carbons. Because both structures in the asymmetric unit are very similar and chain A is more complete, only chain A is discussed. All rmsd’s were calculated with LSQKAB (Kabsch, 1976) from the CCP4 suite, and figures were prepared with PyMOL (http://pymol.org/). Atomic coordinates and structure factors have been deposited under the PDB accession no. 3SPW.
Molecular modeling
The Osh4p/PI(4)P structure was used as the starting point for modeling studies. The 3D structure of the molecule PI(4,5)P2 has been built by using the 3D structure of PI(4)P in its crystallographic binding mode.
Online supplemental materials
Fig. S1 shows by fluorescence-based approaches that the lidless [30–434]Osh4p mutant is unable to extract DHE. Fig. S2 shows control experiments indicating that the fluorescence of NBD-Osh4p at 525 nm reflects its avidity for membranes. Fig. S3 details the interaction between PI(4)P and Osh4p, and shows that Osh4p cannot accommodate a PI(4,5)P2 headgroup. Fig. S4 shows that whatever the electrostatic and lipid-packing properties of the acceptor liposomes, Osh4p fully releases DHE into acceptor liposomes when PI(4)P is present and does not tether liposomes. Table S1 gives information on the structure determination and refinement of the Osh4p–PI(4)P complex. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201104062/DC1.
Acknowledgments
We thank N. Leroudier for DNA sequencing and the staff of European Synchrotron Radiation Facility (ID14-1 beamline, Grenoble, France).
This work was supported by the Centre National de la Recherche Scientifique and by the Agence Nationale de la Recherche (ANR-08-BLAN-0060 and ANR-2010-1503-01). B. Antonny is supported by an advanced grant (268888) from the European Research Council program.
Footnotes
Abbreviations used in this paper:
- ALPS
- amphipathic lipid packing sensor
- DHE
- dehydroergosterol
- DNS
- dansyl
- DNS-PE
- DNS-phosphatidylethanolamine
- DOPC
- dioleoyl phosphatidylcholine
- DOPS
- dioleoyl phosphatidylserine
- FRET
- fluorescence resonance energy transfer
- NBD
- 7-nitrobenz-2-oxa-1,3-diazol
- Orp
- oxysterol-binding protein-related protein
- Osh
- oxysterol-binding homology protein
- PC
- phosphatidylcholine
- PI
- phosphatidylinositol
- PI(4)P
- PI 4-phosphate
- PI(4,5)P2
- PI 4,5-bisphosphate
- PM
- plasma membrane
- POPC
- 1-palmitoyl-2-oleoyl-PC
- POPS
- 1-palmitoyl-2-oleoyl-phosphatidylserine
- Rhod-PE
- rhodamine-phosphatidylethanolamine
- rmsd
- root mean square deviation
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W., et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis V.A., Phillips S., Yanagisawa L., Li X., Routt S., Xie Z. 2005. Phosphatidylinositol transfer protein function in the yeast Saccharomyces cerevisiae. Adv. Enzyme Regul. 45:155–170 10.1016/j.advenzreg.2005.02.014 [DOI] [PubMed] [Google Scholar]
- Baumann N.A., Sullivan D.P., Ohvo-Rekilä H., Simonot C., Pottekat A., Klaassen Z., Beh C.T., Menon A.K. 2005. Transport of newly synthesized sterol to the sterol-enriched plasma membrane occurs via nonvesicular equilibration. Biochemistry. 44:5816–5826 10.1021/bi048296z [DOI] [PubMed] [Google Scholar]
- Beh C.T., Rine J. 2004. A role for yeast oxysterol-binding protein homologs in endocytosis and in the maintenance of intracellular sterol-lipid distribution. J. Cell Sci. 117:2983–2996 10.1242/jcs.01157 [DOI] [PubMed] [Google Scholar]
- Beh C.T., Cool L., Phillips J., Rine J. 2001. Overlapping functions of the yeast oxysterol-binding protein homologues. Genetics. 157:1117–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigay J., Antonny B. 2005. Real-time assays for the assembly-disassembly cycle of COP coats on liposomes of defined size. Methods Enzymol. 404:95–107 10.1016/S0076-6879(05)04010-3 [DOI] [PubMed] [Google Scholar]
- Bigay J., Casella J.F., Drin G., Mesmin B., Antonny B. 2005. ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif. EMBO J. 24:2244–2253 10.1038/sj.emboj.7600714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh E.G., Dyer W.J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917 10.1139/o59-099 [DOI] [PubMed] [Google Scholar]
- Drin G., Casella J.F., Gautier R., Boehmer T., Schwartz T.U., Antonny B. 2007. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat. Struct. Mol. Biol. 14:138–146 10.1038/nsmb1194 [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Fairn G.D., McMaster C.R. 2005. Identification and assessment of the role of a nominal phospholipid binding region of ORP1S (oxysterol-binding-protein-related protein 1 short) in the regulation of vesicular transport. Biochem. J. 387:889–896 10.1042/BJ20041915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairn G.D., McMaster C.R. 2008. Emerging roles of the oxysterol-binding protein family in metabolism, transport, and signaling. Cell. Mol. Life Sci. 65:228–236 10.1007/s00018-007-7325-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairn G.D., Curwin A.J., Stefan C.J., McMaster C.R. 2007. The oxysterol binding protein Kes1p regulates Golgi apparatus phosphatidylinositol-4-phosphate function. Proc. Natl. Acad. Sci. USA. 104:15352–15357 10.1073/pnas.0705571104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M., Kearns B.G., Gedvilaite A., Kagiwada S., Kearns M., Fung M.K., Bankaitis V.A. 1996. Kes1p shares homology with human oxysterol binding protein and participates in a novel regulatory pathway for yeast Golgi-derived transport vesicle biogenesis. EMBO J. 15:6447–6459 [PMC free article] [PubMed] [Google Scholar]
- Fiske C.H., Subbarow Y. 1925. The colorimetric determination of phosphorus. J. Biol. Chem. 66:375–400 [Google Scholar]
- Holt A., de Almeida R.F., Nyholm T.K., Loura L.M., Daily A.E., Staffhorst R.W., Rijkers D.T., Koeppe R.E., II, Prieto M., Killian J.A. 2008. Is there a preferential interaction between cholesterol and tryptophan residues in membrane proteins? Biochemistry. 47:2638–2649 10.1021/bi702235k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im Y.J., Raychaudhuri S., Prinz W.A., Hurley J.H. 2005. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature. 437:154–158 10.1038/nature03923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John K., Kubelt J., Müller P., Wüstner D., Herrmann A. 2002. Rapid transbilayer movement of the fluorescent sterol dehydroergosterol in lipid membranes. Biophys. J. 83:1525–1534 10.1016/S0006-3495(02)73922-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. 1976. A solution for the best rotation to relate two sets of vectors. Acta Crystallogr. A. 32:922–923 10.1107/S0567739476001873 [DOI] [Google Scholar]
- Kabsch W. 2010. XDS. Acta Crystallogr. D Biol. Crystallogr. 66:125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm R.W., Ejsing C.S., Surma M.A., Kaiser H.J., Gerl M.J., Sampaio J.L., de Robillard Q., Ferguson C., Proszynski T.J., Shevchenko A., Simons K. 2009. Segregation of sphingolipids and sterols during formation of secretory vesicles at the trans-Golgi network. J. Cell Biol. 185:601–612 10.1083/jcb.200901145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc M.A., McMaster C.R. 2010. Lipid binding requirements for oxysterol-binding protein Kes1 inhibition of autophagy and endosome-trans-Golgi trafficking pathways. J. Biol. Chem. 285:33875–33884 10.1074/jbc.M110.147264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev S. 2010. Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat. Rev. Mol. Cell Biol. 11:739–750 10.1038/nrm2971 [DOI] [PubMed] [Google Scholar]
- Levine T. 2005. A new way for sterols to walk on water. Mol. Cell. 19:722–723 10.1016/j.molcel.2005.08.006 [DOI] [PubMed] [Google Scholar]
- Li X., Rivas M.P., Fang M., Marchena J., Mehrotra B., Chaudhary A., Feng L., Prestwich G.D., Bankaitis V.A. 2002. Analysis of oxysterol binding protein homologue Kes1p function in regulation of Sec14p-dependent protein transport from the yeast Golgi complex. J. Cell Biol. 157:63–77 10.1083/jcb.200201037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Lu P., Chu J.W., Sharom F.J. 2009. Characterization of fluorescent sterol binding to purified human NPC1. J. Biol. Chem. 284:1840–1852 10.1074/jbc.M803741200 [DOI] [PubMed] [Google Scholar]
- Maxfield F.R., Tabas I. 2005. Role of cholesterol and lipid organization in disease. Nature. 438:612–621 10.1038/nature04399 [DOI] [PubMed] [Google Scholar]
- Mesmin B., Maxfield F.R. 2009. Intracellular sterol dynamics. Biochim. Biophys. Acta. 1791:636–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A., Dodson E.J. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255 10.1107/S0907444996012255 [DOI] [PubMed] [Google Scholar]
- Payrastre B. 2004. Phosphoinositides: lipid kinases and phosphatases. Methods Mol. Biol. 273:201–212 [DOI] [PubMed] [Google Scholar]
- Pichler H., Gaigg B., Hrastnik C., Achleitner G., Kohlwein S.D., Zellnig G., Perktold A., Daum G. 2001. A subfraction of the yeast endoplasmic reticulum associates with the plasma membrane and has a high capacity to synthesize lipids. Eur. J. Biochem. 268:2351–2361 10.1046/j.1432-1327.2001.02116.x [DOI] [PubMed] [Google Scholar]
- Prinz W.A. 2007. Non-vesicular sterol transport in cells. Prog. Lipid Res. 46:297–314 10.1016/j.plipres.2007.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S., Im Y.J., Hurley J.H., Prinz W.A. 2006. Nonvesicular sterol movement from plasma membrane to ER requires oxysterol-binding protein-related proteins and phosphoinositides. J. Cell Biol. 173:107–119 10.1083/jcb.200510084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway N.D. 2010. Oxysterol-binding proteins. Subcell. Biochem. 51:159–182 10.1007/978-90-481-8622-8_6 [DOI] [PubMed] [Google Scholar]
- Schneiter R., Brügger B., Sandhoff R., Zellnig G., Leber A., Lampl M., Athenstaedt K., Hrastnik C., Eder S., Daum G., et al. 1999. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J. Cell Biol. 146:741–754 10.1083/jcb.146.4.741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder F., Butko P., Nemecz G., Scallen T.J. 1990. Interaction of fluorescent delta 5,7,9(11),22-ergostatetraen-3 beta-ol with sterol carrier protein-2. J. Biol. Chem. 265:151–157 [PubMed] [Google Scholar]
- Schulz T.A., Choi M.G., Raychaudhuri S., Mears J.A., Ghirlando R., Hinshaw J.E., Prinz W.A. 2009. Lipid-regulated sterol transfer between closely apposed membranes by oxysterol-binding protein homologues. J. Cell Biol. 187:889–903 10.1083/jcb.200905007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan C.J., Manford A.G., Baird D., Yamada-Hanff J., Mao Y., Emr S.D. 2011. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 144:389–401 10.1016/j.cell.2010.12.034 [DOI] [PubMed] [Google Scholar]
- Strahl T., Thorner J. 2007. Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1771:353–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G.W., McCoy A., et al. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67:235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung T., Gilbert G.E., Shi J., Silvius J., Kapus A., Grinstein S. 2008. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 319:210–213 10.1126/science.1152066 [DOI] [PubMed] [Google Scholar]
- Zinser E., Sperka-Gottlieb C.D., Fasch E.V., Kohlwein S.D., Paltauf F., Daum G. 1991. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J. Bacteriol. 173:2026–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]