Abstract
Host cell death is an intrinsic immune defense mechanism in response to microbial infection. However, bacterial pathogens use many strategies to manipulate the host cell death and survival pathways to enhance their replication and survival. This manipulation is quite intricate, with pathogens often suppressing cell death to allow replication and then promoting it for dissemination. Frequently, these effects are exerted through modulation of the mitochondrial pro-death, NF-κB–dependent pro-survival, and inflammasome-dependent host cell death pathways during infection. Understanding the molecular details by which bacterial pathogens manipulate cell death pathways will provide insight into new therapeutic approaches to control infection.
Introduction
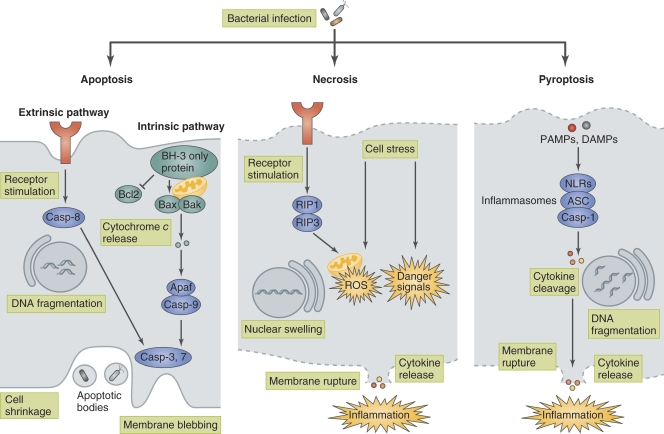
One of the most intrinsic immune defense mechanisms of multicellular organisms is to sacrifice infected cells for the benefit of the remaining cells. The major cell death modalities, including apoptotic cell death, necrotic cell death, and pyroptotic cell death, are critical defense mechanisms against microbial infection (Fig. 1; Bergsbaken et al., 2009; Lamkanfi and Dixit, 2010; Zitvogel et al., 2010). For instance, in response to bacterial infection, programmed cell death, such as apoptosis, is induced as a host innate immune response. This cell death plays two defensive roles in infection. One is to eliminate pathogens at the early stage of infection without emitting alarm signals. The other role is to induce dendritic cells (DCs) to engulf apoptotic bodies containing infected microbes, which allows extracellular antigens to access MHC I molecules and subsequently induce a protective immune response (Elliott and Ravichandran, 2010). Cell death can also benefit pathogens: one prominent strategy of many bacterial pathogens is to induce the demise of infected host cells, which allows the bacteria to efficiently exit the host cell, spread to neighboring cells, evade immune cells, and/or gain nutrients. Meanwhile, many bacterial pathogens, especially those capable of invading and multiplying within host cells, use multiple mechanisms to manipulate host cell death and survival pathways in order to maintain their replicative compartment (Behar et al., 2010; Kim et al., 2010; Lamkanfi and Dixit, 2010).
Figure 1.
Bacteria-induced host cell death. Bacteria induce host cell death through several distinct modalities, including apoptosis, necrosis, and pyroptosis. Apoptosis is a type of noninflammatory programmed cell death that is triggered by two different pathways, the intrinsic (mitochondria-mediated) pathway and extrinsic (receptor-mediated) pathway. Apoptosis is morphologically characterized by membrane blebbing, cell shrinkage, DNA fragmentation, mitochondrial permeability, and caspase (except for caspase-1) activation. In apoptosis, bacteria are retained within apoptotic bodies and engulfed by phagocytic cells. Necrosis is characterized by membrane rupture, nuclear swelling, and the release of cellular contents and is accompanied by caspase-independent inflammation. Necrosis is triggered by ROS production or danger signals, such as lysosomal destabilization, calpain release, and depletion of ATP, that are induced upon bacterial infection or physical damage. Pyroptosis is a type of programmed cell death that is coordinated by inflammasome-mediated caspase-1 activation and accompanied by membrane rupture, DNA fragmentation, and the release of pro-inflammatory cytokines, including IL-1β and IL-18. PAMPs and DAMPs are recognized by NLR proteins, which assemble the inflammasome to activate caspase-1and trigger pyroptosis.
Professional phagocytes play pivotal roles in sensing bacteria through pathogen-associated molecular patterns (PAMPs) or danger-associated molecular patterns (DAMPs) by various pathogen recognition receptors (PRRs), including Toll-like receptors (TLRs), NOD-like receptors (NLRs), RIG-I–like receptors, C-type lectin receptors, and absence in melanoma 2 (AIM2)–like receptors (Chen and Nuñez, 2010; Davis et al., 2011). These sensors trigger innate immune responses and enhance antimicrobial defense responses. Phagocytes activated in response to infection efficiently phagocytose target microbes and transport them to lysosomes, where they are ultimately degraded. The phagocytes also emit various alarms to further amplify innate immune responses (Mosser and Edwards, 2008). In addition to acting as front-line defense executioners, macrophages provide a safe niche for some bacterial pathogens, such as Legionella pneumophila, Mycobacterium tuberculosis, Coxiella burnetii, and Brucella spp. These bacteria have evolved mechanisms to manipulate host membrane trafficking, remodel bacteria-containing vacuoles, modulate cell death signaling, and prolong the longevity of the replicative compartment so they can survive and multiply therein (Kumar and Valdivia, 2009; Behar et al., 2010). Non-professional phagocytic cells such as epithelial cells also act as front-line defense executioners against microbial intrusion by using various PRRs to recognize PAMPs and DAMPs. Epithelial cells that sense microbial infection trigger oxidative stress, ER stress, mitochondrial stress, DNA stress, inflammatory responses, and autophagy, subsequently activate various antimicrobial defense systems including cell death and cell exfoliation (Lamkanfi and Dixit, 2010; Zitvogel et al., 2010). In addition, many epithelial cells, such as gut epithelial cells, are exploited as an infectious foothold by various gastrointestinal pathogens such as enteropathogenic E. coli (EPEC), enterohemorrhagic E. coli (EHEC), Salmonella Typhimurium, Yersinia, Shigella flexneri, and Helicobacter pylori. These pathogenic bacteria have highly evolved mechanisms to manipulate the host cell membrane architecture, cell–cell junctions, cytoskeletal dynamics, membrane trafficking, cell cycle progression, and cell death signaling to promote bacterial colonization of the gut epithelium (Ray et al., 2009; Kim et al., 2010).
Although the mechanisms whereby bacterial pathogens manipulate the host cell death pathway vary among different pathogens and host cell types, the following three pathways are widely used as targets for bacterial pathogens to enhance pathogenesis: the mitochondria-dependent pro-death pathway, NF-κB–dependent pro-survival pathway, and inflammasome-dependent cell death pathway (Bergsbaken et al., 2009; Lamkanfi and Dixit, 2010; Rudel et al., 2010). In this review, we highlight some selected examples of the strategies that are used by human bacterial pathogens to manipulate these host cell pathways during infection (Table I).
Table I.
Selected examples of bacterial pathogens and host cell death
| Bacteria | Factor | Function | Mechanism | Host target | Reference |
| Chlamydia trachomatis | CPAF | Inhibition of apoptosis | Degradation of proapoptotic proteins | BH3-only proteins | Pirbhai et al., 2006 |
| Unknown | Inhibition of apoptosis | Leads to mislocalizaton and prevents proapoptotic activity | Bad | Verbeke et al., 2006 | |
| EPEC | NleH | Inhibition of apoptosis | Inhibits caspase-3 activation | BI-1 | Hemrajani et al., 2010 |
| NleD | Inhibition of apoptosis | Cleaves JNK via metalloprotease activity and inhibits JNK signaling | JNK | Baruch et al., 2011 | |
| Helicobacter pylori | CagA | Inhibition of apoptosis | Up-regulation of anti-proapoptotic protein, Mcl-1 | MCL-1 | Mimuro et al., 2007 |
| Unknown | Inhibition of apoptosis | Activates EGFR–Akt–Bcl2 antiapoptotic pathway | EGFR | Yan et al., 2009 | |
| Legionella pneumophila | SdhA | Inhibition of cell death | Inhibits mitochondrial disruption and caspase activation | Unknown | Laguna et al., 2006 |
| SidF | Inhibition of apoptosis | Inhibits apoptosis signaling | BNIP3, Bcl-rambo | Banga et al., 2007 | |
| LegK1 | Inhibition of cell death? | Promotes host pro-survival signal via NF-κB activation | IκBα, p100 | Ge et al., 2009 | |
| LnaB | Inhibition of cell death? | Promotes host pro-survival signal via NF-κB activation | Unknown | Losick et al., 2010 | |
| Flagellin | Induction of pyroptosis | Induces the NAIP5–NLRC4–inflammasome–caspase-1 activation | NLRC4, NAIP5 | Amer et al., 2006; Ren et al., 2006; Kofoed and Vance, 2011; Zhao et al., 2011 | |
| Mycobacterium tuberculosis | Unknown | Inhibition of apoptosis | Up-regulation of anti-proapoptotic proteins, Mcl-1 and A1 | MCL-1, A1 | Sly et al., 2003; Loeuillet et al., 2006; Dhiman et al., 2008 |
| Zmp1 | Inhibition of inflammasome activation | Inhibits caspase-1 activation and IL-1β secretion | Unknown | Master et al., 2008 | |
| Salmonella Typhimurium | Flagellin, PrgJ | Induction of pyroptosis | Induces the NAIP–NLRC4–inflammasome–caspase-1 activation | NLRC4, NAIP2, NAIP5 | Franchi et al., 2006; Miao et al., 2006, 2010a; Kofoed and Vance, 2011; Zhao et al., 2011 |
| AvrA | Inhibition of apoptosis | Modifies acetyltransferase activity toward MAPKK and inhibits JNK activation | MAPKKs | Jones et al., 2008 | |
| SopB | Inhibition of apoptosis | Induction of host pro-survival activity | PI3K/Akt | Knodler et al., 2005; Kum et al., 2011 | |
| Shigella flexneri | Unknown | Induction of necrotic cell death | Induces mitochondrial dysfunction | BNIP3, CypD | Carneiro et al., 2009 |
| Unknown | Inhibit mitochondrial dysfunction | Activates Nod1–RIP2–NF-κB pro-survival signaling | Bcl2 | Carneiro et al., 2009 | |
| MxiI | Induction of pyroptosis | Induces the NLRC4–inflammasome–caspase-1 activation | NLRC4 | Suzuki et al., 2007; Miao et al., 2010a | |
| Yersinia | YopJ | Induction of apoptosis | Inhibition of NF-κB and MAPK signaling via acetyltransferase activity results in induction of apoptosis | MAPKKs, IKKβ | Monack et al., 1997, Orth et al., 1999; Mukherjee et al., 2006 |
| YopK | Inhibition of pyroptosis | Prevents inflammasome recognition by interacting with T3SS translocon | T3SS translocon (bacterial target) | Brodsky et al., 2010; Dewoody et al., 2011 |
The mitochondria-dependent cell death pathway
Apoptosis is a major cellular defense response during pathogen infection, and the mitochondria play a central role in modulating the host cell death and survival pathways (Rudel et al., 2010). Apoptosis is triggered by two different pathways: the extrinsic pathway and the intrinsic pathway. The extrinsic pathway is activated upon stimulation of the transmembrane death receptors, including FasL, TNF-R1, and Apo2/Apo3. These receptors transmit external apoptotic signaling to the death machinery, which results in the activation of the executioners caspase-3 and -7. The intrinsic pathway is initiated through the release of signaling factors from the mitochondria. Various intrinsic stimuli activate Bcl-2 homology 3 (BH3)–only proteins, which overcome the inhibitory effects of anti-apoptotic Bcl-2 proteins. The activated BH3-only proteins promote the oligomerization of pro-apoptotic proteins, such as Bax and Bak, in the mitochondrial outer membrane, which results in the release of cytochrome c into the cytoplasm. Cytochrome c stimulates formation of the apoptosome, a multimeric protein complex that serves as a scaffold for the caspase activation, which proteolytically activates procaspase-9. Caspase-9 then cleaves and activates other caspases that cleave numerous substrates to ultimately induce apoptosis (Fig. 1; Arnoult et al., 2009). Blocking or delaying cell death is a prominent strategy used by pathogens to promote intracellular survival and replication (Arnoult et al., 2009; Rudel et al., 2010).
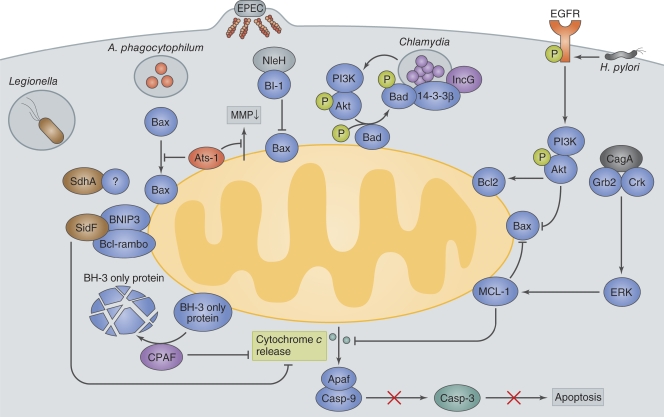
Manipulation of the mitochondrial cell death pathway by intracellular pathogens.
L. pneumophila replicates within alveolar macrophages, during which the pathogen postpones host cell death by delivering a subset of effectors, including SdhA and SidF, via a type IV secretion system (T4SS; Fig. 2). The mechanism by which SdhA inhibits macrophage cell death remains unclear (Laguna et al., 2006). SidF targets two pro-apoptotic factors, BNIP3 and Bcl-rambo, thereby inhibiting mitochondria-mediated apoptotic signaling (Banga et al., 2007). Indeed, macrophages infected with a L. pneumophila sdhA or sidF mutant undergo more apoptosis and have reduced bacterial replication than macrophages infected with wild-type Legionella, suggesting that blocking the macrophage apoptotic response is important for Legionella pathogenesis.
Figure 2.
Bacterial manipulation of mitochondrial cell death pathways. Legionella deliver SidF and SdhA. SidF interacts with two pro-apoptotic proteins, BNIP3 and Bcl-rambo, and inhibits apoptotic signaling. SdhA inhibits host cell death and enhances bacterial survival and replication by an as yet unknown mechanism. A. phagocytophilum delivers Ats-1 and prevents the loss of mitochondrial membrane potential (MMP) and Bax translocation to the mitochondria to inhibit apoptosis. EPEC delivers NleH, which interacts with Bax inhibitor 1 (BI-1) and blocks epithelial apoptosis. Chlamydia secrete CPAF, which degrades the pro-apoptotic BH3-only proteins, thereby blocking the release of cytochrome c from the mitochondria. Chlamydia infection also inhibits the function of the pro-apoptotic protein Bad by causing its phosphorylation by Akt and mislocalization to Chlamydia inclusion vacuoles via its interaction with 14-3-3β and IncG. H. pylori delivers the T4SS effector CagA to attenuate epithelial cell apoptosis. The CagA and Grb2 or Crk interaction stimulates the ERK1/2 pathway and up-regulates the production of the anti-apoptotic factor MCL-1. H. pylori activates EGFR, thereby up-regulating anti-apoptotic signaling involving Akt and Bcl2.
Chlamydia spp. use multiple mechanisms to prevent premature host cell death that involve bacterial factors that likely interact with mitochondrial components involved in apoptotic cell death (Fig. 2; Fan et al., 1998; Fischer et al., 2004; Dong et al., 2005; Ying et al., 2005). Chlamydia trachomatis inhibits apoptosis by degrading pro-apoptotic BH3-only proteins, including Bim, Puma, and Bad (Fischer et al., 2004; Dong et al., 2005; Ying et al., 2005), and this degradation is partially mediated by Chlamydia-secreted chlamydial protease-like activity factor (CPAF; Pirbhai et al., 2006). Furthermore, Chlamydia infection activates phosphoinositide-3 kinase (PI3K), which leads to Akt activation and subsequent phosphorylation of the pro-apoptotic protein, Bad (Verbeke et al., 2006). Phosphorylated Bad can interact and colocalize with 14-3-3β on the Chlamydia inclusion vacuole through an interaction between IncG, a bacterial-encoded inclusion protein, and 14-3-3β (Scidmore and Hackstadt, 2001). These interactions sequester Bad from the mitochondria, thus allowing the pathogen to prevent host cell apoptosis (Fig. 2; Verbeke et al., 2006).
Anaplasma phagocytophilum, the causative agent of human granulocytic anaplasmosis, infects neutrophils and prevents their apoptosis by inhibiting the decrease in the anti-apoptotic protein Bfl1 and the loss of mitochondrial membrane potential (Ge et al., 2005; Rikihisa, 2010). A recent study determined that the Anaplasma T4SS effector Ats-1, which contains an N-terminal mitochondria-targeting sequence, can target the mitochondria (Niu et al., 2010). Niu et al. (2010) showed that Ats-1 can prevent Bax from relocalizing from the cytoplasm into the mitochondria and inhibit cytochrome c release in neutrophils, suggesting that Anaplasma has evolved strategies to survive within neutrophils (Fig. 2).
These studies suggest that the demise of infected cells is an important host innate defense mechanism, and that bacteria counteract these host cell death responses during the early stage of infection to facilitate their intracellular replication.
Modulation of the mitochondrial cell death pathway by enteric pathogens.
Some gastrointestinal bacterial pathogens use countermeasures against the rapid epithelial cell death response by interfering with the mitochondria-dependent cell death pathway (and enhancing the NF-κB pro-survival pathway). EPEC, EHEC, and Citrobacter rodentium use a type III secretion system (T3SS) to deliver a subset of effectors into the intestinal epithelium. One of these effectors, EspF, has multiple functions that manipulate a variety of host signaling pathways that are involved in cytoskeletal rearrangement, disruption of cell–cell junctions, epithelial cell death, and immune modulation (Holmes et al., 2010). EspF, which contains an N-terminal mitochondrial targeting sequence, targets the mitochondria and stimulates cell death by disrupting the mitochondrial membrane potential (Nougayrède and Donnenberg, 2004; Nagai et al., 2005). These EspF-dependent cellular responses appear to benefit pathogenesis because mice infected with a C. rodentium espF mutant have reduced lethality, intestinal hyperplasia, and bacterial loads in the intestine (Deng et al., 2004; Nagai et al., 2005). Earlier studies showed that EPEC-infected epithelial cells expressed phosphatidylserine on their surface and had DNA fragmentation, but showed no signs of cell shrinkage, membrane blebbing, or nuclear condensation, which are characteristic of late-stage apoptotic cell death (Crane et al., 1999, 2001). Thus, these findings indicate that EPEC can inhibit the host cell death response despite initially triggering it through EspF. A recent study provided insight into the mechanisms by which EPEC inhibits early apoptosis. The EPEC NleH effector targets the ER six-transmembrane protein Bax inhibitor-1 (BI-1) to prevent elevated cytoplasmic Ca2+ levels, nuclear condensation, caspase-3 activation, and membrane blebbing of infected epithelial cells (Hemrajani et al., 2010). Infection of BI-1 knockdown cells with the EPEC-related mouse pathogen, C. rodentium, resulted in the loss of NleH-dependent anti-apoptotic activity, and mice infected with a C. rodentium nleH mutant exhibited increased caspase-3 cleavage in the intestinal epithelium compared with mice infected with wild-type C. rodentium (Hemrajani et al., 2010). Thus, NleH may promote colonization by slowing down enterocyte loss from the intestine.
H. pylori can manipulate the gastric epithelial mitochondria-dependent and -independent cell death pathways to promote persistent colonization of the stomach mucosa (Fischer et al., 2009). H. pylori feeds on nutrients in the mucin layer and exudates from epithelial cells that are damaged as a result of gastric inflammation, reactive oxygen species (ROS), apoptosis, and compromised cell–cell junctions (Fischer et al., 2009). H. pylori delivers CagA, a major virulence factor, via a T4SS into the gastric epithelium. CagA has multiple activities that usurp signaling pathways involved in gastric epithelial cell proliferation, cell death, cell–cell junctions, cell scattering, inflammatory responses, and gene regulation, all of which contribute to bacterial colonization and gastritis (Fischer et al., 2009). For instance, CagA interacts with many cellular proteins that activate a variety of transcriptional factors such as NF-κB, nuclear factor of activated T cells (NFAT), serum response factor (SRF), and T cell factor/lymphoid enhancer factor (TCF/LEF; Amieva and El-Omar, 2008; Wessler and Backert, 2008). CagA is also responsible for dampening apoptosis of matured gastric epithelial cells (Mimuro et al., 2007). When etoposide, an apoptosis inducer, was orally administered to Mongolian gerbils to induce intrinsic apoptotic cell death, H. pylori infection of the stomach up-regulated the production of ERK, a pro-survival factor, and Mcl-1, an anti-apoptotic factor, in a CagA-dependent manner, which promoted H. pylori colonization of the gerbil stomach (Mimuro et al., 2007). Yan et al. (2009) infected wild-type and kinase-defective epidermal growth factor receptor (EGFR) mice with H. pylori and demonstrated that H. pylori–induced EGFR phosphorylation stimulated PI3K-dependent activation of the anti-apoptotic factor Akt, increased expression of the anti-apoptotic factor Bcl-2, and decreased expression of the pro-apoptotic factor Bax, suggesting that H. pylori uses multiple mechanisms to suppress gastric epithelial cell death and promote infection (Fig. 2).
Mitochondria-dependent cell death pathways in bacteria–host interactions have been extensively documented and are well recognized as major pathogenic strategies for many bacterial pathogens (Böhme and Rudel, 2009; Lamkanfi and Dixit, 2010). However, the underlying molecular mechanisms of cell death that result from interactions between bacterial factors and host factors as well as the biological significance of these interactions in vivo are still unknown. Further studies using animal models will be important to evaluate the impact of mitochondria-dependent and -independent cell death responses on bacterial pathogenesis and disease progression.
Manipulation of the NF-κB pro-survival pathway by bacterial pathogens
NF-κB is one of the master regulators of the innate immune response and it functions by activating the host inflammatory signaling pathway and cell survival pathway. For example, NF-κB has been shown to protect against TNF-R1–induced cell death by up-regulating NF-κB–targeted genes, such as A20, FHC, Mn-SOD, XIAP, and Gadd45b. These genes suppress signaling events downstream of TNF-R1 activation, which are ROS production, MAP kinase kinase (MAPKK) activation, and induction of the Jun–N-terminal kinase (JNK) cascade (Bubici et al., 2006). Many bacterial pathogens use a variety of mechanisms to manipulate the NF-κB–regulated survival pathway in order to modulate the host cell death response and thus promote intracellular replication and pathogenicity.
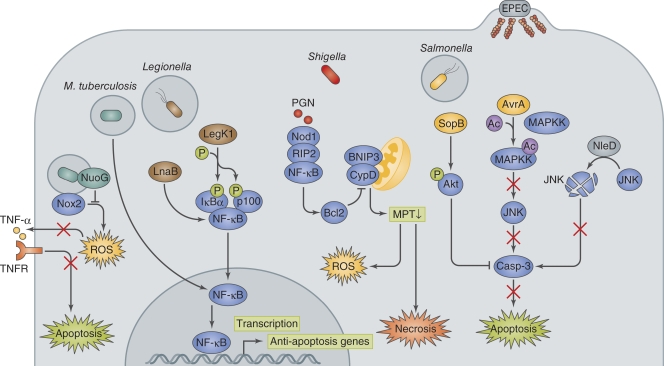
L. pneumophila delivers T4SS effectors to manipulate the NF-κB pro-survival pathway and delay macrophage cell death (Fig. 3; Losick and Isberg, 2006; Abu-Zant et al., 2007; Bartfeld et al., 2009). The L. pneumophila effector LegK1 phosphorylates and inactivates IκBα and p100, inhibitor proteins of NF-κB, thus enhancing the activation of the NF-κB pro-survival pathway (Ge et al., 2009). Legionella also deliver the LnaB effector to activate NF-κB. Although the functions of LnaB are still unclear, introduction of LnaB and an NF-κB reporter into HEK293T cells showed that LnaB enhanced NF-κB activity compared with the mock control. In addition, macrophages that are infected with a L. pneumophila lnaB mutant had reduced NF-κB activity (Losick et al., 2010). Despite this evidence showing the enhancement of the NF-κB–dependent pro-survival pathway by these effectors, additional in vivo studies are required because L. pneumophila lacking either LegK1 or LnaB did not show a significant replication defect in macrophages (Losick et al., 2010).
Figure 3.
Bacterial manipulation of host pro-survival pathway. M. tuberculosis activates host anti-apoptotic signaling by up-regulating the anti-apoptosis genes Mcl-1, bfl1, and FLIP. NuoG dampens Nox2-mediated host signaling, resulting in the inhibition of apoptosis. Legionella deliver LegK1 and LnaB, which enhance the pro-survival activity of NF-κB. Shigella invasion of the epithelium results in mitochondria permeability transition (MPT)−dependent ROS production and necrosis-like cell death, which is counterbalanced by the NOD1−RIP2−NF-κB−Bcl2 pro-survival pathway. PGN, peptidoglycan. Salmonella deliver SopB and AvrA via the T3SS, and they inhibit apoptosis by activating the PI3K–Akt pro-survival pathway via inositol phosphate activity and by preventing JNK pro-cell death signaling via acetyltransferase activity, respectively. EPEC also targets JNK signaling by delivering NleD, which uses its metalloprotease activity to cleave JNK, thereby inhibiting cell death.
M. tuberculosis uses two distinctive strategies during macrophage infection to control cell death; the pathogen postpones cell death early during infection, but induces cell death in the later stage of infection to promote bacterial egress from infected macrophages. Studies with monocyte-derived THP-1 cells showed that M. tuberculosis infection up-regulates TLR2/NF-κB signaling, which results in the up-regulation of FLIP, a powerful inhibitor of death receptor signaling (Loeuillet et al., 2006). A search for anti-apoptosis–related genes in the M. tuberculosis genome led to the identification of the nuoG gene, which encodes the NuoG subunit of the type I NADH-hydrogenase, NDH-1, and can neutralize the NOX-2–mediated increase in phagosomal ROS and TNF production and thus inhibit apoptosis (Fig. 3; Velmurugan et al., 2007; Miller et al., 2010). A M. tuberculosis nuoG mutant decreased the ability to inhibit macrophage apoptosis and significantly reduced virulence in mice (Velmurugan et al., 2007). The apoptotic phenotype of the nuoG mutant was significantly reduced in human macrophages treated with a TNF-neutralizing antibody as well as in TNF−/− murine macrophages (Miller et al., 2010). The bacterial pathogenic strategy to counteract host cell apoptosis was also verified by infecting either macrophages with reduced Mcl-1 expression (by siRNA knockdown) or bfl1 knockout mice. Both of these models exhibited decreased mycobacterial pathogenesis (Sly et al., 2003; Dhiman et al., 2008). Recent studies reported that M. tuberculosis uses a different strategy at the late stage of infection in order to augment macrophage cell death and facilitate the egress of intracellular bacteria and further infection of other cells via a yet-uncharacterized caspase-independent pathway that shares features of necrosis (Behar et al., 2010).
Salmonella infect the intestinal mucosa using two distinctive pathways, the Salmonella pathogenicity island 1 (SPI-1)–dependent pathway and SPI-2–dependent pathway. Salmonella infection of the gut triggers inflammation due to the delivery of effectors via the T3SS-1 and T3SS-2 encoded by SPI-1 and SPI-2, respectively (Kaiser and Hardt, 2011). In the SPI-1 pathway, Salmonella directly invades the intestinal epithelium by delivering the SopB, SopE, SopE2, and SipA effectors via the T3SS-1, in which bacterial invasion results in acute inflammatory responses. In the SPI-2 pathway, Salmonella is apprehended by intestinal DCs, which extend dendrites between epithelial cells to directly engulf luminal bacteria (Valdez et al., 2009; Kaiser and Hardt, 2011). Once Salmonella cross the epithelium, the bacteria encounter DCs, macrophages, B cells, and T cells, and some bacteria gain further access to the host circulation within CD18+ cells, which ultimately allows the bacteria to access the mesenteric lymph nodes, spleen, and liver (Valdez et al., 2009). An early report with human intestinal epithelial cells indicated that TNF and nitric oxide production, which is induced upon bacterial infection, leads to a delayed apoptosis (Kim et al., 1998). Subsequent studies indicated that infection of human intestinal epithelial cells with S. Dublin or the porcine intestine with S. Typhimurium induced caspase-3–dependent and –independent cell death, but only after prolonged exposure to the pathogens, implying that the pathogen is capable of suppressing cell death at the initial stage of infection (Paesold et al., 2002; Schauser et al., 2005). Recent studies showed that the Salmonella AvrA and SopB effectors modulate both inflammatory and cell death responses (Fig. 3). AvrA possesses acetyltransferase activity for MAPKKs and strongly inhibits the JNK signaling pathway, which helps dampen inflammatory and cell death responses (Jones et al., 2008; Du and Galán, 2009). Likewise, the EPEC NleD effector was shown to target the JNK signaling pathway to inhibit apoptosis. NleD uses its zinc metalloprotease activity to cleave JNK (and p38), and thus blocks the JNK-mediated pro-apoptotic pathway (Fig. 3; Baruch et al., 2011). SopB protects epithelial cells from apoptosis by maintaining the activation of Akt, an anti-apoptosis factor (Knodler et al., 2005). The ability of SopB to antagonize apoptosis appears to be dependent on its inositol phosphatase activity, which likely directly up-regulates Akt activity. The protective role of Akt2, an isoform of Akt, in Salmonella-induced gastroenterocolitis was recently verified using Akt2 knockout mice; Salmonella-infected Akt2−/− mice showed enhanced morbidity and mortality that was associated with increased bacterial loads in the intestine and elevated levels of proinflammatory cytokines (TNF, IFN-γ, and MCP-1), which are associated with increased apoptosis, compared with wild-type mice (Kum et al., 2011).
Shigella induce both necrosis-like and apoptosis-like cell death in a caspase-1-independent manner upon infection of epithelial cells (Clark and Maurelli, 2007; Carneiro et al., 2009; Faherty and Maurelli, 2009). However, the bacteria further manipulate the host cell survival pathway by dampening cell death signals until the late stages of infection when the bacteria have fully replicated and spread among epithelial cells. Shigella can dampen the mitochondrial dysfunction–mediated necrotic-like cell death response by activating the Nod1–RIP2–NF-κB–Bcl-2 pro-survival pathway (Fig. 3; Carneiro et al., 2009). Consistent with this model, Nod1−/− MEFs or NF-κB inhibitor–treated epithelial cells infected with Shigella have a rapid loss of mitochondria permeability transition and ROS production, leading to necrotic-like cell death (Carneiro et al., 2009). Cell death signaling in response to Shigella infection becomes predominant in the later stages of infection, and this process depends on the BH-3–only protein BNIP3 and cyclophilin D (CypD) in mitochondrial permeability and the transition to cell death. A kinetic study supports the premise that cytoprotective signaling is dominant during early bacterial replication within epithelial cells but is overwhelmed by cell death signaling at a later stage, thereby allowing the bacteria to egress and transit toward the lumen (Carneiro et al., 2009).
As discussed here, establishing a protective or replicative niche during infection is an essential pathogenic mechanism, while the demise of infected cells is an important host defense mechanism that clears the bacteria and sends alarms that activate the host immune system. In addition, the gut epithelium undergoes rapid turnover that is sustained by a balance between cell proliferation and death, and cell death plays a critical role in maintaining this tissue homeostasis. Therefore, the most simple and efficient means for bacterial pathogens to prolong cell viability, such as manipulation of NF-κB pro-survival signaling, and thus promote bacterial growth, would be a “gain in time strategy” (Kim et al., 2010).
Inflammasome activation through recognition of bacterial infection
Bacterial pathogens deliver cytotoxins and T3SS effectors (as well as T3SS components and flagellin) to the host cell membrane and cytoplasm to modify the host cell surface architecture, induce membrane damage, subvert cell signaling, and modulate cell physiology in order to promote infection. Thus, bacteria can activate the innate immune system when PAMPs and DAMPs in an atypical cellular location are recognized by cytoplasmic PRRs such as NLRs. Upon recognition of DAMPs and PAMPs, NLRs induce the assembly of the inflammasome, which is composed of NLR, ASC, and caspase-1, to drive the proximity-induced activation of caspase-1, which results in the extracellular release of IL-1β and IL-18 and induces pyroptosis. Pyroptosis is a type of inflammatory programmed cell death that is coordinated by inflammasome-mediated caspase-1 activation (Fig. 1; Bergsbaken and Cookson, 2007; Schroder and Tschopp, 2010; Davis et al., 2011). For example, NLRP3 senses damaged cell membranes, infection by Listeria monocytogenes, Shigella, S. Typhimurium, Staphylococcus aureus, Neisseria gonorrhoeae, and Chlamydia spp, as well as bacterial pore-forming toxins (Mariathasan et al., 2006; Willingham et al., 2007; Abdul-Sater et al., 2009; Duncan et al., 2009; Broz et al., 2010; Khare et al., 2010). NLRC4 detects bacterial infection with L. monocytogenes and S. Typhimurium (Mariathasan et al., 2006; Warren et al., 2008; Broz et al., 2010). In addition, NLRC4 can sense flagellin and the T3SS rod components of S. Typhimurium, L. pneumophila, Pseudomonas aeruginosa, and Shigella (Mariathasan et al., 2004; Amer et al., 2006; Franchi et al., 2006; Miao et al., 2006, 2008, 2010a; Sutterwala et al., 2007; Suzuki et al., 2007). Recent reports show that NAIP (NLR family, apoptosis inhibitory protein) family members act as pathogen sensors that determine the specificity of NLRC4 inflammasome for different bacterial ligands. In fact, NAIP5 specifically activates NLRC4 inflammasome in response to flagellin, whereas NAIP2 is required for inflammasome activation by T3SS rod components (Ren et al., 2006; Kofoed and Vance, 2011; Zhao et al., 2011). NLRP1b recognizes Bacillus anthracis lethal toxin, a major virulence factor from B. anthracis. Indeed, mutations in the Nlrp1b gene were identified as the susceptibility locus for anthrax lethal toxin-induced macrophage cell death in mice (Boyden and Dietrich, 2006). AIM2 binds to dsDNA via its C-terminal HIN200 domain, and responds to Francisella tularensis and L. monocytogenes infection in murine macrophages when these bacteria undergo spontaneous autolysis, resulting in caspase-1 activation and pyroptosis (Fernandes-Alnemri et al., 2010; Jones et al., 2010; Rathinam et al., 2010; Sauer et al., 2010; Tsuchiya et al., 2010; Warren et al., 2010). These studies indicate that NLR inflammasomes play a key role in host innate defense against a variety of microbes and in maintaining tissue homeostasis.
Modulation of inflammasome activation by bacterial pathogens.
Some bacterial pathogens use distinctive strategies to modulate inflammasome-dependent cell death, which is an important pathogenic mechanism to avoid host cell death and the release of inflammatory intracellular contents during infection (Fink and Cookson, 2007). For example, S. Typhimurium evades recognition of NLRC4 by repressing the expression of PrgJ, a T3SS-1 rod component, and flagellin after phagocytosis, whereby instead expressing T3SS-2, which is not detected by NLRC4 (Cummings et al., 2006; Miao et al., 2006, 2010a). Indeed, when macrophages are infected with a S. Typhimurium strain that constitutively expresses flagellin, the bacteria are efficiently eliminated (Miao et al., 2010b). Salmonella replicate within macrophages, and during this period the pathogen gradually switches from SPI-1 to SPI-2 expression. This change in SPI expression appears to allow the bacteria to efficiently evade NLRC4–inflammasome activation, thereby promoting bacterial replication and facilitating further infection.
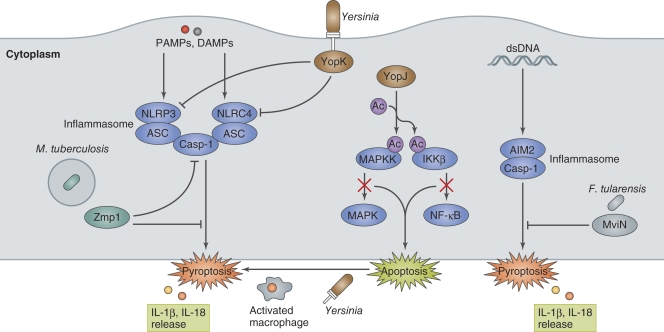
Yersinia use strategies that both inhibit and induce cell death during infection of macrophages. Some of the Yop effectors delivered via the T3SS play key roles in modulating host innate and adaptive immune responses (Dube, 2009). When Yersinia infect naive macrophages, the LPS that is released from the bacteria is recognized as a PAMP, which rapidly activates NF-κB– and MAPK-dependent pro-survival pathways. However, these signaling pathways are abrogated in the early stage of infection. One Yop effector, YopJ, interferes with these signaling pathways, allowing Yersinia to enhance apoptosis (Fig. 4; Monack et al., 1997). YopJ possesses unique acetyltransferase activity that acetylates the serine and threonine residues in host kinases, including the MAPKKs and the IκB kinase complex (IKKβ). YopJ targeting of these kinases can inhibit their activity (Orth et al., 1999; Mukherjee et al., 2006), allowing Yersinia to block pro-survival signaling in the initial stage of infection in naive macrophages, which instead leads to apoptosis. Apoptosis in the initial stage of infection by Yersinia is likely to benefit pathogens because the pathogens can kill macrophages without inflammatory responses. Nevertheless, macrophage activation followed by continuous Yersinia infection can gradually redirect the initial YopJ-dependent apoptosis to YopJ-independent, caspase-1–mediated pyroptosis (Bergsbaken and Cookson, 2007). It seems that redirection to pyroptosis in activated macrophages is the host defense strategy for bacterial clearance. To evade macrophage-directed defensive actions, Yersinia deliver the T3SS YopK effector to inhibit inflammasome activation and prevent pyroptosis (Fig. 4; Brodsky et al., 2010). Interestingly, YopK can interact with the T3SS translocon and interfere with T3SS recognition by NLRP3 and NLRC4, allowing the bacteria to evade IL-1β secretion and pyroptosis (Brodsky et al., 2010). A more recent study shows that YopK regulates effector translocation through the T3SS from inside the host cells, thereby preventing pyroptosis (Dewoody et al., 2011). When bone marrow–derived macrophages are infected with a Y. pseudotuberculosis yopK mutant, NLRP3 inflammasome activation and cell death increase and the bacterial load decreases compared with infection with wild-type Yersinia. Casp1−/− mice infected with YopK-positive or -negative mutant have similar bacterial loads (Brodsky et al., 2010), indicating that caspase-1–dependent responses against Yersinia are critical for bacterial clearance.
Figure 4.
Bacterial manipulation of inflammasome activation. PAMPs or DAMPs (such as dsDNA) generated by bacterial invasion and multiplication in macrophages trigger NLRP3-, NLRC4-, AIM2-inflammasome assembly and induce pyroptosis. Yersinia induce apoptosis by YopJ-mediated inhibition of pro-inflammatory signaling in the initial stage of infection in naive macrophages. Activated macrophages redirect YopJ-mediated apoptosis to YopJ-independent pyroptosis. However, Yersinia prevent inflammasome activation by secreting YopK, which interferes with T3SS recognition by NLRP3 and NLRC4. M. tuberculosis Zmp1, a Zn metalloprotease, and F. tularensis MviN, a putative lipid II flippase, prevent inflammasome activation and IL-1β secretion/pyroptosis by unknown mechanisms.
M. tuberculosis inhibits inflammasome activation by secreting the zinc metalloprotease Zmp1 (Fig. 4). Master et al. (2008) identified Zmp1 as a mycobacterial virulence factor using a mouse infection model as well as murine and human macrophages. The Mycobacteria zmp1 mutant failed to prevent mycobacteria-containing phagosome maturation and caspase-1 activation, which are required to clear mycobacteria from infected macrophages and from mouse lungs infected via aerosol administration. However, the zmp1 mutant was not cleared from murine macrophages when either inflammasome component expression, such as NLRC4, ASC, or caspase-1, was knocked down using siRNA (Master et al., 2008). Although it is still unclear how Zmp1 antagonizes inflammasome activation, the discovery of Zmp1 underscores the important role of inflammasome-mediated IL-1β production in mycobacterial clearance.
F. tularensis, a facultative intracellular zoonotic pathogen, is phagocytosed by macrophages, and the phagosomes temporary interact with the endocytic pathway (Clemens and Horwitz, 2007). After 1–2 h of infection, Francisella escape from the phagosome and multiply within the cytoplasm. The pathogen is recognized by AIM2 when bacterial dsDNA is released into the cytosol upon spontaneous autolysis, as bacterial DNA occasionally colocalizes with AIM2 (Fernandes-Alnemri et al., 2010; Jones et al., 2010). However, the pathogen uses the lipid/polysaccharide transporter protein, MviN, which is homologous to the E. coli putative lipid II flippase, to suppress AIM2 inflammasome activation. Compared with wild-type F. tularensis, mice infected with a F. tularensis mviN mutant have increased AIM2 inflammasome-dependent IL-1β production and pyroptosis and decreased bacterial burdens (Fig. 4; Ulland et al., 2010). Conversely, infection of Aim2−/− mice with F. tularensis (as well as L. monocytogenes and DNA viruses) results in decreased IL-1β secretion and cell death, and increased mortality and bacterial loads (Fernandes-Alnemri et al., 2010). These studies validate that the ability of Francisella to suppress AIM2 inflammasome is important in enhancing pathogenesis.
We still have a limited understanding of the ability of bacteria to modulate inflammasome activation, and the mechanisms by which bacteria manipulate the inflammasome pathway are still largely speculative (Lamkanfi and Dixit, 2010; Taxman et al., 2010). Nevertheless, because inflammasome activation as well as caspase-1 activation and subsequent IL-1β secretion and pyroptosis greatly contribute in a variety of protective host responses, we envisage that many other bacterial pathogens use yet-uncharacterized mechanisms to modulate the inflammasome for their benefit.
Conclusion
Bacterial infection elicits a diverse array of host protective and stress responses, including the cell death and proliferative responses, inflammatory response, and innate immune response. The host cell death response critically influences the fate of bacterial infection, the integrity of the host innate defense barrier, innate and acquired immunity, and disease outcome. Bacteria-triggered cell death results in various modes of cell death that greatly vary with the host cell type, stage of infection, infectious dosage, physiological condition of the cell, bacterial factors, and experimental setting. In addition, our understanding of the dynamic interplay between pathogens and host innate defense responses in vitro and in vivo are occasionally contradictory. Nevertheless, as has been described here and in other literature (Lamkanfi and Dixit, 2010; Rudel et al., 2010), future studies on the cell death response during host–pathogen interactions will provide further insight into the mechanisms by which cells undergo death as an innate defense against microbial infection. Furthermore, a better understanding of the molecular details by which bacterial pathogens manipulate cell death pathways will undoubtedly provide insight into new therapeutic approaches that can be used to control bacterial infection and inflammatory disease progression.
Acknowledgments
We thank the reviewers and members of the Sasakawa laboratory for their advice. Illustrations provided by Neil Smith, neil@neilsmithillustration.co.uk.
This work was supported by Grant-in-Aid for Specially promoted Research (23000012); a Grant-in-Aid for Young Scientists (A) (23689027, to M. Kim); a Grant-in-Aid for Young Scientists (B) (23790471, to M. Ogawa; 23790472, to H. Ashida; and 22790403, to T. Sanada); a Grant-in-Aid for Scientific Research (B) (23390102, to H. Mimuro); a Grant-in-Aid for challenging Exploratory Research (23659220, to H. Mimuro); and Japan Initiative for Global Research Network on Infectious Diseases (to C. Sasakawa); Global COE Program “Center of Education and Research for the Advanced Genome–Based Medicine–For personalized medicine and the control of worldwide infectious diseases” (to T. Kobayashi) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT). Part of this work was supported by grants from the Takeda Science Foundation (to H. Mimuro and M. Kim) and the Naito Foundation (to H. Ashida and M. Kim).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used in this paper:
- AIM2
- absence in melanoma 2
- BH3
- Bcl-2 homology 3
- CPAF
- chlamydial protease-like activity factor
- DAMP
- danger-associated molecular pattern
- DC
- dendritic cell
- EGFR
- epidermal growth factor receptor
- EHEC
- enterohemorrhagic E. coli
- EPEC
- enteropathogenic E. coli
- JNK
- Jun N-terminal kinase
- MAPKK
- MAP kinase kinase
- NAIP
- NLR family, apoptosis inhibitory protein 5
- NLR
- NOD-like receptor
- PAMP
- pathogen-associated molecular pattern
- PI3K
- phosphoinositide-3 kinase
- PRR
- pathogen recognition receptor
- ROS
- reactive oxygen species
- SPI
- Salmonella pathogenicity island
- T3SS
- type III secretion system
- T4SS
- type IV secretion system
References
- Abdul-Sater A.A., Koo E., Häcker G., Ojcius D.M. 2009. Inflammasome-dependent caspase-1 activation in cervical epithelial cells stimulates growth of the intracellular pathogen Chlamydia trachomatis. J. Biol. Chem. 284:26789–26796 10.1074/jbc.M109.026823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Zant A., Jones S., Asare R., Suttles J., Price C., Graham J., Kwaik Y.A. 2007. Anti-apoptotic signalling by the Dot/Icm secretion system of L. pneumophila. Cell. Microbiol. 9:246–264 10.1111/j.1462-5822.2006.00785.x [DOI] [PubMed] [Google Scholar]
- Amer A., Franchi L., Kanneganti T.D., Body-Malapel M., Ozören N., Brady G., Meshinchi S., Jagirdar R., Gewirtz A., Akira S., Núñez G. 2006. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J. Biol. Chem. 281:35217–35223 10.1074/jbc.M604933200 [DOI] [PubMed] [Google Scholar]
- Amieva M.R., El-Omar E.M. 2008. Host-bacterial interactions in Helicobacter pylori infection. Gastroenterology. 134:306–323 10.1053/j.gastro.2007.11.009 [DOI] [PubMed] [Google Scholar]
- Arnoult D., Carneiro L., Tattoli I., Girardin S.E. 2009. The role of mitochondria in cellular defense against microbial infection. Semin. Immunol. 21:223–232 10.1016/j.smim.2009.05.009 [DOI] [PubMed] [Google Scholar]
- Banga S., Gao P., Shen X., Fiscus V., Zong W.X., Chen L., Luo Z.Q. 2007. Legionella pneumophila inhibits macrophage apoptosis by targeting pro-death members of the Bcl2 protein family. Proc. Natl. Acad. Sci. USA. 104:5121–5126 10.1073/pnas.0611030104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartfeld S., Engels C., Bauer B., Aurass P., Flieger A., Brüggemann H., Meyer T.F. 2009. Temporal resolution of two-tracked NF-kappaB activation by Legionella pneumophila. Cell. Microbiol. 11:1638–1651 10.1111/j.1462-5822.2009.01354.x [DOI] [PubMed] [Google Scholar]
- Baruch K., Gur-Arie L., Nadler C., Koby S., Yerushalmi G., Ben-Neriah Y., Yogev O., Shaulian E., Guttman C., Zarivach R., Rosenshine I. 2011. Metalloprotease type III effectors that specifically cleave JNK and NF-κB. EMBO J. 30:221–231 10.1038/emboj.2010.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar S.M., Divangahi M., Remold H.G. 2010. Evasion of innate immunity by Mycobacterium tuberculosis: is death an exit strategy? Nat. Rev. Microbiol. 8:668–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsbaken T., Cookson B.T. 2007. Macrophage activation redirects yersinia-infected host cell death from apoptosis to caspase-1-dependent pyroptosis. PLoS Pathog. 3:e161 10.1371/journal.ppat.0030161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsbaken T., Fink S.L., Cookson B.T. 2009. Pyroptosis: host cell death and inflammation. Nat. Rev. Microbiol. 7:99–109 10.1038/nrmicro2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhme L., Rudel T. 2009. Host cell death machinery as a target for bacterial pathogens. Microbes Infect. 11:1063–1070 10.1016/j.micinf.2009.08.014 [DOI] [PubMed] [Google Scholar]
- Boyden E.D., Dietrich W.F. 2006. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat. Genet. 38:240–244 10.1038/ng1724 [DOI] [PubMed] [Google Scholar]
- Brodsky I.E., Palm N.W., Sadanand S., Ryndak M.B., Sutterwala F.S., Flavell R.A., Bliska J.B., Medzhitov R. 2010. A Yersinia effector protein promotes virulence by preventing inflammasome recognition of the type III secretion system. Cell Host Microbe. 7:376–387 10.1016/j.chom.2010.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P., Newton K., Lamkanfi M., Mariathasan S., Dixit V.M., Monack D.M. 2010. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J. Exp. Med. 207:1745–1755 10.1084/jem.20100257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubici C., Papa S., Dean K., Franzoso G. 2006. Mutual cross-talk between reactive oxygen species and nuclear factor-kappa B: molecular basis and biological significance. Oncogene. 25:6731–6748 10.1038/sj.onc.1209936 [DOI] [PubMed] [Google Scholar]
- Carneiro L.A., Travassos L.H., Soares F., Tattoli I., Magalhaes J.G., Bozza M.T., Plotkowski M.C., Sansonetti P.J., Molkentin J.D., Philpott D.J., Girardin S.E. 2009. Shigella induces mitochondrial dysfunction and cell death in nonmyleoid cells. Cell Host Microbe. 5:123–136 10.1016/j.chom.2008.12.011 [DOI] [PubMed] [Google Scholar]
- Chen G.Y., Nuñez G. 2010. Sterile inflammation: sensing and reacting to damage. Nat. Rev. Immunol. 10:826–837 10.1038/nri2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark C.S., Maurelli A.T. 2007. Shigella flexneri inhibits staurosporine-induced apoptosis in epithelial cells. Infect. Immun. 75:2531–2539 10.1128/IAI.01866-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens D.L., Horwitz M.A. 2007. Uptake and intracellular fate of Francisella tularensis in human macrophages. Ann. N. Y. Acad. Sci. 1105:160–186 10.1196/annals.1409.001 [DOI] [PubMed] [Google Scholar]
- Crane J.K., Majumdar S., Pickhardt D.F., III 1999. Host cell death due to enteropathogenic Escherichia coli has features of apoptosis. Infect. Immun. 67:2575–2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane J.K., McNamara B.P., Donnenberg M.S. 2001. Role of EspF in host cell death induced by enteropathogenic Escherichia coli. Cell. Microbiol. 3:197–211 10.1046/j.1462-5822.2001.00103.x [DOI] [PubMed] [Google Scholar]
- Cummings L.A., Wilkerson W.D., Bergsbaken T., Cookson B.T. 2006. In vivo, fliC expression by Salmonella enterica serovar Typhimurium is heterogeneous, regulated by ClpX, and anatomically restricted. Mol. Microbiol. 61:795–809 10.1111/j.1365-2958.2006.05271.x [DOI] [PubMed] [Google Scholar]
- Davis B.K., Wen H., Ting J.P. 2011. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu. Rev. Immunol. 29:707–735 10.1146/annurev-immunol-031210-101405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Puente J.L., Gruenheid S., Li Y., Vallance B.A., Vázquez A., Barba J., Ibarra J.A., O’Donnell P., Metalnikov P., et al. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. USA. 101:3597–3602 10.1073/pnas.0400326101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewoody R., Merritt P.M., Houppert A.S., Marketon M.M. 2011. YopK regulates the Yersinia pestis type III secretion system from within host cells. Mol. Microbiol. 79:1445–1461 10.1111/j.1365-2958.2011.07534.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhiman R., Kathania M., Raje M., Majumdar S. 2008. Inhibition of bfl-1/A1 by siRNA inhibits mycobacterial growth in THP-1 cells by enhancing phagosomal acidification. Biochim. Biophys. Acta. 1780:733–742 [DOI] [PubMed] [Google Scholar]
- Dong F., Pirbhai M., Xiao Y., Zhong Y., Wu Y., Zhong G. 2005. Degradation of the proapoptotic proteins Bik, Puma, and Bim with Bcl-2 domain 3 homology in Chlamydia trachomatis-infected cells. Infect. Immun. 73:1861–1864 10.1128/IAI.73.3.1861-1864.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du F., Galán J.E. 2009. Selective inhibition of type III secretion activated signaling by the Salmonella effector AvrA. PLoS Pathog. 5:e1000595 10.1371/journal.ppat.1000595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube P. 2009. Interaction of Yersinia with the gut: mechanisms of pathogenesis and immune evasion. Curr. Top. Microbiol. Immunol. 337:61–91 10.1007/978-3-642-01846-6_3 [DOI] [PubMed] [Google Scholar]
- Duncan J.A., Gao X., Huang M.T., O’Connor B.P., Thomas C.E., Willingham S.B., Bergstralh D.T., Jarvis G.A., Sparling P.F., Ting J.P. 2009. Neisseria gonorrhoeae activates the proteinase cathepsin B to mediate the signaling activities of the NLRP3 and ASC-containing inflammasome. J. Immunol. 182:6460–6469 10.4049/jimmunol.0802696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott M.R., Ravichandran K.S. 2010. Clearance of apoptotic cells: implications in health and disease. J. Cell Biol. 189:1059–1070 10.1083/jcb.201004096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faherty C.S., Maurelli A.T. 2009. Spa15 of Shigella flexneri is secreted through the type III secretion system and prevents staurosporine-induced apoptosis. Infect. Immun. 77:5281–5290 10.1128/IAI.00800-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan T., Lu H., Hu H., Shi L., McClarty G.A., Nance D.M., Greenberg A.H., Zhong G. 1998. Inhibition of apoptosis in Chlamydia-infected cells: blockade of mitochondrial cytochrome c release and caspase activation. J. Exp. Med. 187:487–496 10.1084/jem.187.4.487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T., Yu J.W., Juliana C., Solorzano L., Kang S., Wu J., Datta P., McCormick M., Huang L., McDermott E., et al. 2010. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat. Immunol. 11:385–393 10.1038/ni.1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink S.L., Cookson B.T. 2007. Pyroptosis and host cell death responses during Salmonella infection. Cell. Microbiol. 9:2562–2570 10.1111/j.1462-5822.2007.01036.x [DOI] [PubMed] [Google Scholar]
- Fischer S.F., Vier J., Kirschnek S., Klos A., Hess S., Ying S., Häcker G. 2004. Chlamydia inhibit host cell apoptosis by degradation of proapoptotic BH3-only proteins. J. Exp. Med. 200:905–916 10.1084/jem.20040402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer W., Prassl S., Haas R. 2009. Virulence mechanisms and persistence strategies of the human gastric pathogen Helicobacter pylori. Curr. Top. Microbiol. Immunol. 337:129–171 10.1007/978-3-642-01846-6_5 [DOI] [PubMed] [Google Scholar]
- Franchi L., Amer A., Body-Malapel M., Kanneganti T.D., Ozören N., Jagirdar R., Inohara N., Vandenabeele P., Bertin J., Coyle A., et al. 2006. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1β in Salmonella-infected macrophages. Nat. Immunol. 7:576–582 10.1038/ni1346 [DOI] [PubMed] [Google Scholar]
- Ge J., Xu H., Li T., Zhou Y., Zhang Z., Li S., Liu L., Shao F. 2009. A Legionella type IV effector activates the NF-kappaB pathway by phosphorylating the IkappaB family of inhibitors. Proc. Natl. Acad. Sci. USA. 106:13725–13730 10.1073/pnas.0907200106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y., Yoshiie K., Kuribayashi F., Lin M., Rikihisa Y. 2005. Anaplasma phagocytophilum inhibits human neutrophil apoptosis via upregulation of bfl-1, maintenance of mitochondrial membrane potential and prevention of caspase 3 activation. Cell. Microbiol. 7:29–38 10.1111/j.1462-5822.2004.00427.x [DOI] [PubMed] [Google Scholar]
- Hemrajani C., Berger C.N., Robinson K.S., Marchès O., Mousnier A., Frankel G. 2010. NleH effectors interact with Bax inhibitor-1 to block apoptosis during enteropathogenic Escherichia coli infection. Proc. Natl. Acad. Sci. USA. 107:3129–3134 10.1073/pnas.0911609106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes A., Mühlen S., Roe A.J., Dean P. 2010. The EspF effector, a bacterial pathogen’s Swiss army knife. Infect. Immun. 78:4445–4453 10.1128/IAI.00635-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.W., Kayagaki N., Broz P., Henry T., Newton K., O’Rourke K., Chan S., Dong J., Qu Y., Roose-Girma M., et al. 2010. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc. Natl. Acad. Sci. USA. 107:9771–9776 10.1073/pnas.1003738107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R.M., Wu H., Wentworth C., Luo L., Collier-Hyams L., Neish A.S. 2008. Salmonella AvrA coordinates suppression of host immune and apoptotic defenses via JNK pathway blockade. Cell Host Microbe. 3:233–244 10.1016/j.chom.2008.02.016 [DOI] [PubMed] [Google Scholar]
- Kaiser P., Hardt W.D. 2011. Salmonella Typhimurium diarrhea: switching the mucosal epithelium from homeostasis to defense. Curr. Opin. Immunol. 23:456–463 10.1016/j.coi.2011.06.004 [DOI] [PubMed] [Google Scholar]
- Khare S., Luc N., Dorfleutner A., Stehlik C. 2010. Inflammasomes and their activation. Crit. Rev. Immunol. 30:463–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.M., Eckmann L., Savidge T.C., Lowe D.C., Witthöft T., Kagnoff M.F. 1998. Apoptosis of human intestinal epithelial cells after bacterial invasion. J. Clin. Invest. 102:1815–1823 10.1172/JCI2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Ashida H., Ogawa M., Yoshikawa Y., Mimuro H., Sasakawa C. 2010. Bacterial interactions with the host epithelium. Cell Host Microbe. 8:20–35 10.1016/j.chom.2010.06.006 [DOI] [PubMed] [Google Scholar]
- Knodler L.A., Finlay B.B., Steele-Mortimer O. 2005. The Salmonella effector protein SopB protects epithelial cells from apoptosis by sustained activation of Akt. J. Biol. Chem. 280:9058–9064 10.1074/jbc.M412588200 [DOI] [PubMed] [Google Scholar]
- Kofoed E.M., Vance R.E. 2011. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 477:592–595 10.1038/nature10394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kum W.W., Lo B.C., Yu H.B., Finlay B.B. 2011. Protective role of Akt2 in Salmonella enterica serovar typhimurium-induced gastroenterocolitis. Infect. Immun. 79:2554–2566 10.1128/IAI.01235-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar Y., Valdivia R.H. 2009. Leading a sheltered life: intracellular pathogens and maintenance of vacuolar compartments. Cell Host Microbe. 5:593–601 10.1016/j.chom.2009.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguna R.K., Creasey E.A., Li Z., Valtz N., Isberg R.R. 2006. A Legionella pneumophila-translocated substrate that is required for growth within macrophages and protection from host cell death. Proc. Natl. Acad. Sci. USA. 103:18745–18750 10.1073/pnas.0609012103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M., Dixit V.M. 2010. Manipulation of host cell death pathways during microbial infections. Cell Host Microbe. 8:44–54 10.1016/j.chom.2010.06.007 [DOI] [PubMed] [Google Scholar]
- Loeuillet C., Martinon F., Perez C., Munoz M., Thome M., Meylan P.R. 2006. Mycobacterium tuberculosis subverts innate immunity to evade specific effectors. J. Immunol. 177:6245–6255 [DOI] [PubMed] [Google Scholar]
- Losick V.P., Isberg R.R. 2006. NF-kappaB translocation prevents host cell death after low-dose challenge by Legionella pneumophila. J. Exp. Med. 203:2177–2189 10.1084/jem.20060766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick V.P., Haenssler E., Moy M.Y., Isberg R.R. 2010. LnaB: a Legionella pneumophila activator of NF-kappaB. Cell. Microbiol. 12:1083–1097 10.1111/j.1462-5822.2010.01452.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S., Newton K., Monack D.M., Vucic D., French D.M., Lee W.P., Roose-Girma M., Erickson S., Dixit V.M. 2004. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 430:213–218 10.1038/nature02664 [DOI] [PubMed] [Google Scholar]
- Mariathasan S., Weiss D.S., Newton K., McBride J., O’Rourke K., Roose-Girma M., Lee W.P., Weinrauch Y., Monack D.M., Dixit V.M. 2006. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 440:228–232 10.1038/nature04515 [DOI] [PubMed] [Google Scholar]
- Master S.S., Rampini S.K., Davis A.S., Keller C., Ehlers S., Springer B., Timmins G.S., Sander P., Deretic V. 2008. Mycobacterium tuberculosis prevents inflammasome activation. Cell Host Microbe. 3:224–232 10.1016/j.chom.2008.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao E.A., Alpuche-Aranda C.M., Dors M., Clark A.E., Bader M.W., Miller S.I., Aderem A. 2006. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1β via Ipaf. Nat. Immunol. 7:569–575 10.1038/ni1344 [DOI] [PubMed] [Google Scholar]
- Miao E.A., Ernst R.K., Dors M., Mao D.P., Aderem A. 2008. Pseudomonas aeruginosa activates caspase 1 through Ipaf. Proc. Natl. Acad. Sci. USA. 105:2562–2567 10.1073/pnas.0712183105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao E.A., Mao D.P., Yudkovsky N., Bonneau R., Lorang C.G., Warren S.E., Leaf I.A., Aderem A. 2010a. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl. Acad. Sci. USA. 107:3076–3080 10.1073/pnas.0913087107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao E.A., Leaf I.A., Treuting P.M., Mao D.P., Dors M., Sarkar A., Warren S.E., Wewers M.D., Aderem A. 2010b. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat. Immunol. 11:1136–1142 10.1038/ni.1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.L., Velmurugan K., Cowan M.J., Briken V. 2010. The type I NADH dehydrogenase of Mycobacterium tuberculosis counters phagosomal NOX2 activity to inhibit TNF-α-mediated host cell apoptosis. PLoS Pathog. 6:e1000864 10.1371/journal.ppat.1000864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimuro H., Suzuki T., Nagai S., Rieder G., Suzuki M., Nagai T., Fujita Y., Nagamatsu K., Ishijima N., Koyasu S., et al. 2007. Helicobacter pylori dampens gut epithelial self-renewal by inhibiting apoptosis, a bacterial strategy to enhance colonization of the stomach. Cell Host Microbe. 2:250–263 10.1016/j.chom.2007.09.005 [DOI] [PubMed] [Google Scholar]
- Monack D.M., Mecsas J., Ghori N., Falkow S. 1997. Yersinia signals macrophages to undergo apoptosis and YopJ is necessary for this cell death. Proc. Natl. Acad. Sci. USA. 94:10385–10390 10.1073/pnas.94.19.10385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser D.M., Edwards J.P. 2008. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 8:958–969 10.1038/nri2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Keitany G., Li Y., Wang Y., Ball H.L., Goldsmith E.J., Orth K. 2006. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science. 312:1211–1214 10.1126/science.1126867 [DOI] [PubMed] [Google Scholar]
- Nagai T., Abe A., Sasakawa C. 2005. Targeting of enteropathogenic Escherichia coli EspF to host mitochondria is essential for bacterial pathogenesis: critical role of the 16th leucine residue in EspF. J. Biol. Chem. 280:2998–3011 10.1074/jbc.M411550200 [DOI] [PubMed] [Google Scholar]
- Niu H., Kozjak-Pavlovic V., Rudel T., Rikihisa Y. 2010. Anaplasma phagocytophilum Ats-1 is imported into host cell mitochondria and interferes with apoptosis induction. PLoS Pathog. 6:e1000774 10.1371/journal.ppat.1000774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nougayrède J.P., Donnenberg M.S. 2004. Enteropathogenic Escherichia coli EspF is targeted to mitochondria and is required to initiate the mitochondrial death pathway. Cell. Microbiol. 6:1097–1111 10.1111/j.1462-5822.2004.00421.x [DOI] [PubMed] [Google Scholar]
- Orth K., Palmer L.E., Bao Z.Q., Stewart S., Rudolph A.E., Bliska J.B., Dixon J.E. 1999. Inhibition of the mitogen-activated protein kinase kinase superfamily by a Yersinia effector. Science. 285:1920–1923 10.1126/science.285.5435.1920 [DOI] [PubMed] [Google Scholar]
- Paesold G., Guiney D.G., Eckmann L., Kagnoff M.F. 2002. Genes in the Salmonella pathogenicity island 2 and the Salmonella virulence plasmid are essential for Salmonella-induced apoptosis in intestinal epithelial cells. Cell. Microbiol. 4:771–781 10.1046/j.1462-5822.2002.00233.x [DOI] [PubMed] [Google Scholar]
- Pirbhai M., Dong F., Zhong Y., Pan K.Z., Zhong G. 2006. The secreted protease factor CPAF is responsible for degrading pro-apoptotic BH3-only proteins in Chlamydia trachomatis-infected cells. J. Biol. Chem. 281:31495–31501 10.1074/jbc.M602796200 [DOI] [PubMed] [Google Scholar]
- Rathinam V.A., Jiang Z., Waggoner S.N., Sharma S., Cole L.E., Waggoner L., Vanaja S.K., Monks B.G., Ganesan S., Latz E., et al. 2010. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat. Immunol. 11:395–402 10.1038/ni.1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K., Marteyn B., Sansonetti P.J., Tang C.M. 2009. Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat. Rev. Microbiol. 7:333–340 10.1038/nrmicro2112 [DOI] [PubMed] [Google Scholar]
- Ren T., Zamboni D.S., Roy C.R., Dietrich W.F., Vance R.E. 2006. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2:e18 10.1371/journal.ppat.0020018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikihisa Y. 2010. Anaplasma phagocytophilum and Ehrlichia chaffeensis: subversive manipulators of host cells. Nat. Rev. Microbiol. 8:328–339 10.1038/nrmicro2318 [DOI] [PubMed] [Google Scholar]
- Rudel T., Kepp O., Kozjak-Pavlovic V. 2010. Interactions between bacterial pathogens and mitochondrial cell death pathways. Nat. Rev. Microbiol. 8:693–705 10.1038/nrmicro2421 [DOI] [PubMed] [Google Scholar]
- Sauer J.D., Witte C.E., Zemansky J., Hanson B., Lauer P., Portnoy D.A. 2010. Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe. 7:412–419 10.1016/j.chom.2010.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser K., Olsen J.E., Larsson L.I. 2005. Salmonella typhimurium infection in the porcine intestine: evidence for caspase-3-dependent and -independent programmed cell death. Histochem. Cell Biol. 123:43–50 10.1007/s00418-004-0731-8 [DOI] [PubMed] [Google Scholar]
- Schroder K., Tschopp J. 2010. The inflammasomes. Cell. 140:821–832 10.1016/j.cell.2010.01.040 [DOI] [PubMed] [Google Scholar]
- Scidmore M.A., Hackstadt T. 2001. Mammalian 14-3-3β associates with the Chlamydia trachomatis inclusion membrane via its interaction with IncG. Mol. Microbiol. 39:1638–1650 10.1046/j.1365-2958.2001.02355.x [DOI] [PubMed] [Google Scholar]
- Sly L.M., Hingley-Wilson S.M., Reiner N.E., McMaster W.R. 2003. Survival of Mycobacterium tuberculosis in host macrophages involves resistance to apoptosis dependent upon induction of antiapoptotic Bcl-2 family member Mcl-1. J. Immunol. 170:430–437 [DOI] [PubMed] [Google Scholar]
- Sutterwala F.S., Mijares L.A., Li L., Ogura Y., Kazmierczak B.I., Flavell R.A. 2007. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J. Exp. Med. 204:3235–3245 10.1084/jem.20071239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Franchi L., Toma C., Ashida H., Ogawa M., Yoshikawa Y., Mimuro H., Inohara N., Sasakawa C., Nuñez G. 2007. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 3:e111 10.1371/journal.ppat.0030111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxman D.J., Huang M.T., Ting J.P. 2010. Inflammasome inhibition as a pathogenic stealth mechanism. Cell Host Microbe. 8:7–11 10.1016/j.chom.2010.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya K., Hara H., Kawamura I., Nomura T., Yamamoto T., Daim S., Dewamitta S.R., Shen Y., Fang R., Mitsuyama M. 2010. Involvement of absent in melanoma 2 in inflammasome activation in macrophages infected with Listeria monocytogenes. J. Immunol. 185:1186–1195 10.4049/jimmunol.1001058 [DOI] [PubMed] [Google Scholar]
- Ulland T.K., Buchan B.W., Ketterer M.R., Fernandes-Alnemri T., Meyerholz D.K., Apicella M.A., Alnemri E.S., Jones B.D., Nauseef W.M., Sutterwala F.S. 2010. Cutting edge: mutation of Francisella tularensis mviN leads to increased macrophage absent in melanoma 2 inflammasome activation and a loss of virulence. J. Immunol. 185:2670–2674 10.4049/jimmunol.1001610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez Y., Ferreira R.B., Finlay B.B. 2009. Molecular mechanisms of Salmonella virulence and host resistance. Curr. Top. Microbiol. Immunol. 337:93–127 10.1007/978-3-642-01846-6_4 [DOI] [PubMed] [Google Scholar]
- Velmurugan K., Chen B., Miller J.L., Azogue S., Gurses S., Hsu T., Glickman M., Jacobs W.R., Jr, Porcelli S.A., Briken V. 2007. Mycobacterium tuberculosis nuoG is a virulence gene that inhibits apoptosis of infected host cells. PLoS Pathog. 3:e110 10.1371/journal.ppat.0030110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeke P., Welter-Stahl L., Ying S., Hansen J., Häcker G., Darville T., Ojcius D.M. 2006. Recruitment of BAD by the Chlamydia trachomatis vacuole correlates with host-cell survival. PLoS Pathog. 2:e45 10.1371/journal.ppat.0020045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren S.E., Mao D.P., Rodriguez A.E., Miao E.A., Aderem A. 2008. Multiple Nod-like receptors activate caspase 1 during Listeria monocytogenes infection. J. Immunol. 180:7558–7564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren S.E., Armstrong A., Hamilton M.K., Mao D.P., Leaf I.A., Miao E.A., Aderem A. 2010. Cutting edge: Cytosolic bacterial DNA activates the inflammasome via Aim2. J. Immunol. 185:818–821 10.4049/jimmunol.1000724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler S., Backert S. 2008. Molecular mechanisms of epithelial-barrier disruption by Helicobacter pylori. Trends Microbiol. 16:397–405 10.1016/j.tim.2008.05.005 [DOI] [PubMed] [Google Scholar]
- Willingham S.B., Bergstralh D.T., O’Connor W., Morrison A.C., Taxman D.J., Duncan J.A., Barnoy S., Venkatesan M.M., Flavell R.A., Deshmukh M., et al. 2007. Microbial pathogen-induced necrotic cell death mediated by the inflammasome components CIAS1/cryopyrin/NLRP3 and ASC. Cell Host Microbe. 2:147–159 10.1016/j.chom.2007.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F., Cao H., Chaturvedi R., Krishna U., Hobbs S.S., Dempsey P.J., Peek R.M., Jr, Cover T.L., Washington M.K., Wilson K.T., Polk D.B. 2009. Epidermal growth factor receptor activation protects gastric epithelial cells from Helicobacter pylori-induced apoptosis. Gastroenterology. 136:1297–1307: e1–e3 10.1053/j.gastro.2008.12.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying S., Seiffert B.M., Häcker G., Fischer S.F. 2005. Broad degradation of proapoptotic proteins with the conserved Bcl-2 homology domain 3 during infection with Chlamydia trachomatis. Infect. Immun. 73:1399–1403 10.1128/IAI.73.3.1399-1403.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Yang J., Shi J., Gong Y.N., Lu Q., Xu H., Liu L., Shao F. 2011. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 477:596–600 10.1038/nature10510 [DOI] [PubMed] [Google Scholar]
- Zitvogel L., Kepp O., Kroemer G. 2010. Decoding cell death signals in inflammation and immunity. Cell. 140:798–804 10.1016/j.cell.2010.02.015 [DOI] [PubMed] [Google Scholar]