The ESCRT-III complex component Shrub plays a pivotal rate-limiting step in late endosomal ligand-independent Notch activation.
Abstract
The Notch signaling pathway defines a conserved mechanism that regulates cell fate decisions in metazoans. Signaling is modulated by a broad and multifaceted genetic circuitry, including members of the endocytic machinery. Several individual steps in the endocytic pathway have been linked to the positive or negative regulation of the Notch receptor. In seeking genetic elements involved in regulating the endosomal/lysosomal degradation of Notch, mediated by the molecular synergy between the ubiquitin ligase Deltex and Kurtz, the nonvisual β-arrestin in Drosophila, we identified Shrub, a core component of the ESCRT-III complex as a key modulator of this synergy. Shrub promotes the lysosomal degradation of the receptor by mediating its delivery into multivesicular bodies (MVBs). However, the interplay between Deltex, Kurtz, and Shrub can bypass this path, leading to the activation of the receptor. Our analysis shows that Shrub plays a pivotal rate-limiting step in late endosomal ligand-independent Notch activation, depending on the Deltex-dependent ubiquitinylation state of the receptor. This activation mode of the receptor emphasizes the complexity of Notch signal modulation in a cell and has significant implications for both development and disease.
Introduction
The Notch pathway is used throughout development to couple the cell fate choice of one cell to those of neighboring cells, ultimately affecting proliferation, apoptosis, and differentiation (Artavanis-Tsakonas et al., 1999; Schweisguth, 2004; Bray, 2006). Notch signaling has been associated with normal development in all organisms. In humans, alternations in Notch signaling have also been implicated in different diseases including cancer (Gridley, 2003). Notch encodes a single-pass transmembrane receptor (Wharton et al., 1985) and the classical developmental logic of the signaling pathway relies on the interaction of the receptor expressed on one cell with membrane-bound ligands expressed on the neighboring cells. The canonical signaling model has the Notch receptor being activated through a series of proteolytic events after it interacts with the ligands, Delta (Dl) or Serrate (Ser) (Bray, 2006; Kopan and Ilagan, 2009). The crucial cleavage event for signaling depends on γ-secretase and results in releasing the intracellular domain of Notch from the membrane. This allows it to translocate into the nucleus, where it directly participates in a core transcriptional complex together with DNA binding protein Suppressor of Hairless (Su(H)) and the nuclear effector Mastermind, thereby activating the transcription of target genes (Bray, 2006; Kopan and Ilagan, 2009).
Small variations of Notch signaling can profoundly affect the biology and indeed pathobiology of cells, a fact reflected by the sensitivity of development to the gene dosage of several Notch pathway components. Thus, mechanisms capable of modulating signaling are of great importance. As components of endocytic trafficking have been implicated in regulating the activity of the Notch receptor (Wilkin and Baron, 2005; Fortini, 2009; Yamamoto et al., 2010), the role and the complexity of such signal modulating mechanisms is increasingly appreciated. Several factors modulating the degradation of the Notch receptor and consequently the negative attenuation of signaling have been identified, while sorting of the receptor through the endocytic compartments has been shown to be critical for the activation of the receptor (Fortini, 2009; Yamamoto et al., 2010). Notably, such intracellular events have not only been associated with ligand-dependent (Coumailleau et al., 2009) but also with an enigmatic ligand-independent, i.e., noncanonical, activation of the receptor (Hori et al., 2004; Sakata et al., 2004; Wilkin et al., 2004, 2008; Thompson et al., 2005; Vaccari and Bilder, 2005; Childress et al., 2006; Vaccari et al., 2008, 2009). Mutations in elements of the endosomal sorting machinery were shown capable of triggering noncanonical signaling in the early endosomes (Thompson et al., 2005; Vaccari and Bilder, 2005; Vaccari et al., 2008, 2009). In addition, another distinct activation path implicates the late endosome in noncanonical activation of the receptor (Hori et al., 2004; Wilkin et al., 2008). The genetic circuitry capable of modulating such intracellular Notch signaling remains opaque, but ligand-independent activation of the receptor has been recently shown to be essential for the normal development of Drosophila blood cells (Mukherjee et al., 2011).
Here, we address these questions based on our previous study showing that Kurtz (Krz), the single nonvisual β-arrestin homologue in Drosophila together with the ubiquitin ligase Deltex (Dx), affects trafficking of the Notch receptor and regulates Notch signaling by modulating the turnover of the receptor (Mukherjee et al., 2005). To gain further insight into how Krz and Dx regulate the trafficking of the Notch receptor we performed unbiased genetic screens for modifiers of the Krz and Dx-dependent synergy, which is manifested in vivo as a typical loss of Notch function wing phenotype. We thus identified a key core component in the ESCRT (endosomal sorting complex required for transport)-III complex, Shrub, the yeast Snf7 homologue (Sweeney et al., 2006; Vaccari et al., 2008), as a modifier of Notch signaling. Our analysis gives a mechanistic insight into the role of ESCRT-III in a late endosomal ligand-independent activation of the Notch receptor. We determined that this mode of Notch regulation relies on the ubiquitinylation of the receptor, controlled by the functional association between Shrub, Dx, and Krz. The data we present emphasize both the complexity and diversity of the means used by the cell to modulate Notch signals. The Notch activation mode we uncover here has significant implications for both development and disease.
Results
shrub modulates the synergy between dx and krz
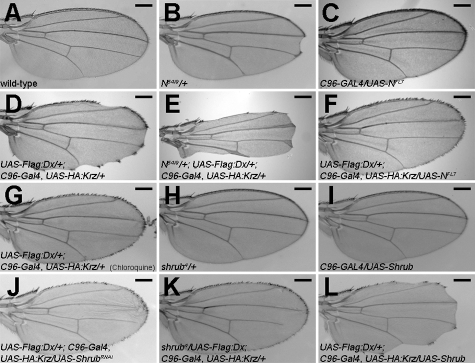
Dx, Krz, and Notch were shown to form a trimeric complex, which modulates the ubiquitinylation and trafficking of the Notch receptor, leading to its degradation (Mukherjee et al., 2005). The coexpression of Dx and Krz consequently shows a wing-nicking adult phenotype (Fig. 1 D; Mukherjee et al., 2005), a typical phenotype of Notch loss of function (Lindsley and Zimm, 1992) that is dosage sensitive as it is strongly enhanced by a heterozygous null Notch mutant, N54l9 (Fig. 1 E), and suppressed by up-regulating Notch through the expression of a transgene carrying a wild-type copy of the receptor (Fig. 1 F). To probe the genetic circuitry capable of modulating the dx and krz synergy, we relied on the Dx and Krz coexpression wing-nicking phenotype to carry out a genetic screen for dominant modifiers using the Exelixis mutant collection (Artavanis-Tsakonas, 2004; Kankel et al., 2007).
Figure 1.
shrub modulates the synergy between dx and krz. (A) Wild-type adult wing. (B) Heterozygous Notch-null allele (N54l9/+) is associated with the typical wing notching. (C) Expression of full-length Notch driven by C96-Gal4 (C96-Gal4/UAS-NFL7) does not affect wing morphology under our experimental conditions. (D and F) Co-expression of Dx and Krz shows wing notching (UAS-Flag:Dx; C96-Gal4, UAS-HA:Krz/+) (D), consistent with Notch loss-of-function, which is rescued by expressing a transgene encoding wild-type Notch (UAS-Flag:Dx/+; C96-Gal4, UAS-HA:Krz/ UAS-NFL7 = 90%, n = 20) (F). (E) N54l9/+ enhances Dx- and Krz-mediated wing notching phenotype (N54l9/+; UAS-Flag:Dx/+; C96-Gal4, UAS-HA:Krz/+ = 100%, n = 17). (G) Wing notching phenotype associated with Dx and Krz is rescued by treatment with chloroquine (52%, n = 23). (H and I) Heterozygote shrub loss-of-function mutations, shrub4/+ (H), or overexpression of Shrub alone by C96-GAL4 in the developing wing (I) does not display wing notching. (J and K) The wing notching phenotype is suppressed by expressing ShrubRNAi (UAS-Flag:Dx/+; C96-Gal4, UAS-HA:Krz/UAS-ShrubRNAi = 76%, n = 33) (J), or by reducing shrub levels in heterozygous animals (shrub4/UAS-Flag:Dx; C96-Gal4, UAS-HA:Krz/+ = 91%, n = 11) (K). (L) The wing notching is enhanced by increasing shrub levels along with Dx and Krz (UAS-Flag:Dx/+; C96-Gal4, UAS-HA:Krz/UAS-Shrub = 83%, n = 23). Bars, 0.2 mm. Representative examples are shown.
The screen identified d02738, an insertion in shrub as a strong suppressor (see Materials and methods). shrub encodes a protein homologous to the yeast protein Snf7 and its mammalian orthologue vps32, a key core component of the ESCRT-III complex (Sweeney et al., 2006). The insertion in d02738 disrupted the shrub coding region, suggesting a loss-of-function mutation. Consistent with this, shrub4, an extant hypomorphic allele of shrub (Sweeney et al., 2006), failed to complement the lethality of d02738 (unpublished data). Moreover shrub4 as a heterozygote suppressed the wing-nicking phenotype caused by Dx and Krz (Fig. 1 K), as did the expression of an inverted repeat RNA corresponding to Shrub (ShrubRNAi; Fig. 1 J). Conversely, expression of Shrub enhanced the wing-nicking phenotype (Fig. 1 L).
shrub affects the subcellular localization of Notch
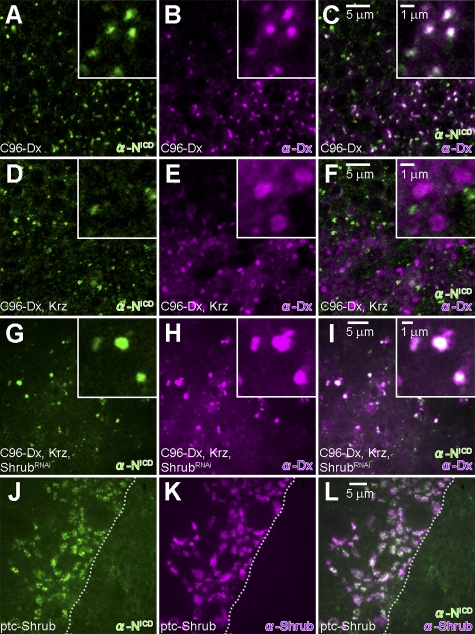
In wild-type epithelial cells, Notch is mostly concentrated in the apical lateral adhesion junctions (zonula adherens) with some protein detected more basally in intracellular vesicles (Fehon et al., 1991). When Dx, a molecule known to physically interact with Notch (Matsuno et al., 1995), is expressed in the wing margin, Notch is depleted from the adhesion junctions of the Dx-expressing cells (Hori et al., 2004). This cellular phenotype is paralleled by an enrichment of intracellular vesicles encompassing both Dx and Notch (Fig. 2, A–C) and accompanied by a slight up-regulation of Notch signals (Hori et al., 2004; Mukherjee et al., 2005; see also Fig. 3, G and H). Expression of Krz alone in the same cells did not alter the subcellular distribution of Notch (Mukherjee et al., 2005). However, consistent with the documented Dx- and Krz-dependent ubiquitinylation and degradation of Notch (Mukherjee et al., 2005), when both Dx and Krz were coexpressed, both proteins are localized in vesicles that are no longer positive for Notch (Fig. 2, D–F), a cellular phenotype associated with loss of Notch signaling and consequently a wing-nicking adult phenotype (Fig. 1 D; Mukherjee et al., 2005). Notably, when these flies are cultured in fly medium containing chloroquine, a reagent known to inhibit lysosomal degradation by raising intra-lysosomal pH (Chi et al., 2010), the wing-nicking phenotype is suppressed (Fig. 1 G), suggesting that Dx and Krz regulate the sorting and degradation of Notch protein via an endosomal/lysosomal pathway.
Figure 2.
Shrub regulates the synergistic effects of dx and krz on Notch by affecting trafficking. (A–C) In the wing disc expressing Dx driven by C96-Gal4 (UAS-Flag:Dx/+; C96-Gal4/+), Notch (green) is colocalized with Dx (purple) in enriched intracellular vesicles. (D–F) In the wing disc coexpressing Dx and Krz driven by C96-Gal4 (UAS-Flag:Dx/+; C96-Gal4, UAS-HA:Krz/+), Notch (green) does not colocalize with Dx (purple)-positive intracellular vesicles. (G–I) In wing discs expressing ShrubRNAi in addition to Dx and Krz by C96-GAL4 (UAS-Flag:Dx/+; C96-Gal4, UAS-HA:Krz/UAS-ShrubRNAi), Notch (green) colocalizes with Dx (purple). (J–L) Expression of Shrub by ptc-GAL4 driver (ptc-GAL4/+; UAS-Shrub/+) leads to accumulation of Notch (green) in Shrub (purple)-positive enlarged vesicles. Dashed lines indicate the boundary between cells induced to express Shrub versus wild-type cells.
The down-regulation of Shrub through the expression of ShrubRNAi at a cellular level prevents the depletion of Notch associated with overexpression of Dx with Krz (compare Fig. 2, G–I, with Fig. 2, D–F), suggesting that the above phenotypic interactions reflect a mechanistic relationship among the three proteins. We have not been able to document physical interaction between Dx and Shrub or Krz and Shrub, but our observations indicate that Shrub can be a rate-limiting factor in the Notch regulation mediated by Dx and Krz.
Consistent with this notion, we observe that Shrub expression influences the subcellular distribution of Notch. When Shrub alone is expressed along the anterior–posterior (AP) boundary of wing imaginal discs (using the ptc-GAL4 driver), the surface Notch staining in Shrub-expressing versus wild-type cells is not obviously altered (unpublished data). In contrast, there is a dramatic shift in the intracellular epitopes of Notch, with Notch accumulating in intracellular vesicles, which are also associated with Shrub (Fig. 2, J–L), and is accompanied by a clear, albeit low penetrance, wing-notching phenotype in the adult (ptc-GAL4/+; UAS-Shrub/+ = 8.4%, n = 166). These observations suggest that Shrub, a component of ESCRT-III, affects the membrane trafficking of Notch protein and the dx-krz–mediated Notch signal modulation.
Shrub is a negative regulator of Notch signaling
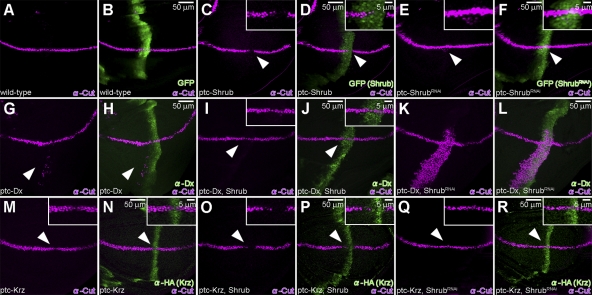
To confirm that Shrub actually affects the output of the Notch receptor, we examined the consequences of Shrub modulation on Cut expression, a downstream target of Notch signals in the dorsoventral (DV) boundary of the wing imaginal disc (de Celis et al., 1996; Neumann and Cohen, 1996), as an indicator of Notch activity. In the wild type, Cut is expressed in a narrow stripe along the DV wing boundary (Fig. 3, A and B), whereas endogenous Shrub is expressed ubiquitously throughout the third instar wing disc (Sweeney et al., 2006). Induction of Shrub expression along the AP boundary suppresses Cut expression at the AP–DV boundary intersection (Fig. 3, C and D). In contrast, the down-regulation of Shrub along the AP boundary, via the expression of ShrubRNAi, induced a weak but consistent ectopic Cut expression (Fig. 3, E and F) at 18°C, notably in the ventral part, a phenotype dramatically enhanced at higher temperatures (Fig. S1, A and B). We conclude that Shrub negatively regulates Notch signals during wing development.
Figure 3.
Shrub antagonizes Dx while it enhances Krz activity. (A and B) Wild-type Cut (purple) expression in larval wing disc, along the DV boundary in ptc-Gal4/UAS-GFP animals. (C and D) Suppression of Cut (purple, arrowhead) is seen when Shrub is expressed by ptc-GAL4 driver (ptc-Gal4/UAS-GFP; UAS-Shrub/+). (E and F) Ectopic Cut expression (purple, arrowhead) in the ventral region of the wing pouch is induced when shrub activity is inhibited through shrub RNAi expression driven by ptc-GAL4 (ptc-Gal4/UAS-GFP; UAS-ShrubRNAi/+) (see also Fig. S2). (A–F) Expression of GFP (green) marks ptc-Gal4 expression domain. (G and H) Expression of Dx (green) alone results in ectopic Cut (purple, arrowhead) expression (ptc-GAL4/UAS-Flag:Dx). (I and J) This effect is suppressed (purple, arrowhead) by the co-expression of Shrub with Dx (green) driven by ptc-Gal4 (ptc-Gal4/UAS-Flag:Dx; UAS-Shrub/+). (K and L) When ShrubRNAi is expressed along the AP boundary together with Dx (green) (ptc-Gal4/UAS-Flag:Dx; UAS-ShrubRNAi/+), a dramatic up-regulation of Cut (purple) is seen, albeit in the ventral part of the disc. (M and N) Expression of Krz (green) alone results in a slight but consistent suppression of Cut (purple, arrowhead) (ptc-Gal4/; UAS-HA:Krz/+). (O and P) Co-expression of Krz (green) and Shrub results in an obvious suppression of Cut (purple, arrowhead) (ptc-Gal4/+; UAS-HA:Krz/UAS-Shrub). (Q and R) Co-expression of Krz (green) with ShrubRNAi does not affect endogenous Cut (purple, arrowhead) levels (ptc-Gal4/+; UAS-HA:Krz/UAS-ShrubRANi). All crosses were performed at 18°C.
Shrub antagonizes Dx, whereas it enhances Krz activity
We examined the antagonistic role Shrub has on Notch signals and particularly how it affects the dx–krz synergy by probing how and if it can individually affect dx and krz activities.
As expected, expression of Dx alone in the AP axis results in weak ectopic expression of Cut (Fig. 3, G and H). This effect is not seen when Dx is coexpressed with Shrub (Fig. 3, I and J). Conversely, the down-regulation of Cut associated with Shrub expression (Fig. 3, C and D) is reversed by Dx (Fig. 3, I and J; n = 20). Thus, Dx and Shrub have opposing effects on Notch signaling. This is confirmed by the dramatic enhancement of Notch signaling when Dx and ShrubRNAi are coexpressed (Fig. 3, K and L).
We note again that this enhancement is context specific as it is detected only in the ventral region of the disc. In probing this behavior, our analysis showed that this context specificity is independent of Fringe (unpublished data), an obvious candidate for such regulation (Fleming et al., 1997). However, when we inhibit lysosomal functions with chloroquine, activation is triggered both ventrally and dorsally (Fig. S1 D), indicating that at least partially this tissue-specific regulation is regulated by lysosomal degradation.
Unlike Dx, expression of Krz alone down-regulates reproducible, albeit slight, Cut expression (Fig. 3, M and N), without resulting in an adult wing notching phenotype (0%, n = 40). When Shrub is coexpressed with Krz, this down-regulation is amplified (Fig. 3, O and P), resulting in a highly penetrant wing-notching adult phenotype (88.9%, n = 54). Consistently, when shrub expression was inhibited by RNAi, the down-regulation of Cut by Krz was no longer observed (Fig. 3, Q and R), defining Shrub as a positive regulator of krz activity. We can thus conclude that Shrub enhances the action Krz exerts on Notch, whereas it antagonizes the effects Dx has on the pathway.
Shrub and Dx modulate Notch signaling through MVBs/endosomal trafficking
To gain insight into the way Shrub affects the trafficking of Notch, we modulated Shrub activity and followed the internalization of the Notch molecules in live cells (Vaccari and Bilder, 2005). Notch was tagged on the surface of imaginal discs dissected from animals expressing different levels of Shrub. The discs were then fixed and stained, at different time points after incubation, to reveal the subcellular localization of the Notch extracellular antigens (Fig. S2, A–F). Remarkably, the same internalization kinetics of Notch are seen whether we down-regulate or up-regulate Shrub along the AP axis, an unexpected observation in view of the antagonistic relationship between Shrub and Notch (Fig. S2, C–F). Thus, even though in this assay low or high Shrub levels affect the internalization kinetics of the Notch receptor in a similar fashion, the effects on signaling are distinct and opposite between the two conditions.
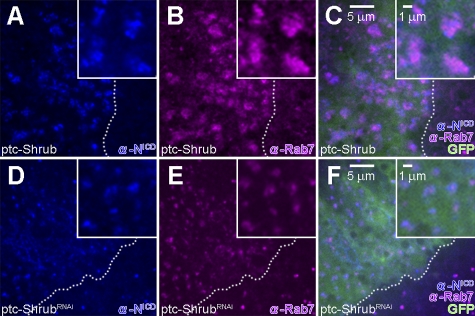
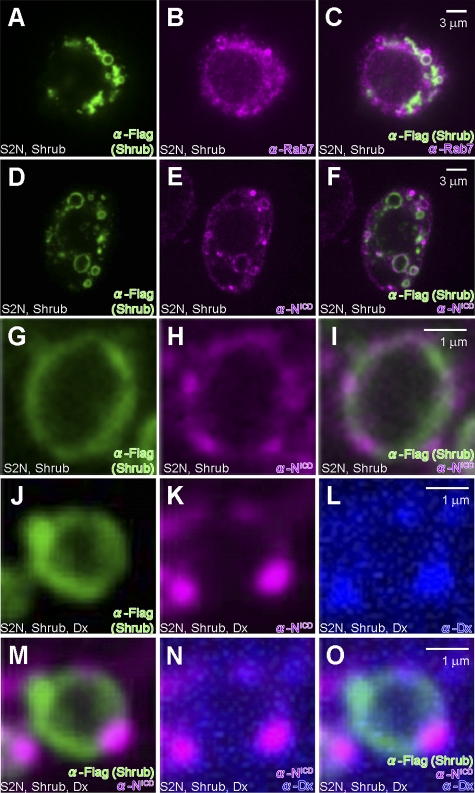
We further examined the subcellular localization of the receptor in the wing discs expressing Shrub (Fig. 4) as well as in S2-N cultured cells (Fig. 5), a cell line stably expressing full-length Notch (Rebay et al., 1991). As Shrub is a core element of the ESCRT-III complex, we expect it to localize on MVBs. Indeed, when Shrub is expressed in S2-N cells, we find that it localizes on the surface of large vesicular structures also positive for Rab7 and thus indicative of MVBs (Fig. 5, A–C). In wing discs overexpressing Shrub, Notch is mostly accumulated in the enlarged Rab7-positive MVBs (Fig. 4, A–C). Similarly, when we query where Notch is relative to Shrub in S2-N cells, we see both molecules coinciding on MVBs (Fig. 5, D–F). This is true whether we probe with antibodies specific for intracellular or extracellular Notch epitopes (Fig. S2, G–L). As can be seen in the higher magnification confocal images (Fig. 5, G–I), Shrub and Notch, while residing on the same vesicles, do not have identical distribution on the vesicle. These findings indicate that Shrub expression shifts the distribution of Notch to MVBs, eventually leading to the degradation of Notch, explaining the down-regulation of the Notch signal by Shrub.
Figure 4.
Shrub modulates the subcellular distribution of Notch. (A–C) In wing discs expressing Shrub by ptc-GAL4 (ptc-GAL4/UAS-GFP; UAS-Shrub/+), Notch (blue) is accumulated in Rab7 (purple)-positive large vesicles. (D–F) In the wing disc expressing ShrubRNAi by ptc-GAL4 (ptc-GAL4/UAS-GFP; UAS-ShrubRNA/+), Notch (blue) is localized in Rab7 positive vesicles (purple). Dashed lines indicate the boundary of the region (green) that is induced by ptc-GAL4 driver. All crosses were performed at 18°C.
Figure 5.
Shrub modulates endosomal trafficking and activation of Notch. (A–C) Shrub (green) localization partially overlaps with Rab7-positive vesicles (purple) in S2-N cells transfected with Shrub-Flag-HA. (D–F) Localization of Shrub (green) and Notch (purple) in subcellular compartments in S2-N cells transfected with Shrub-Flag-HA. (G–I) Relative localization of Notch (purple) and Shrub (green) on subcellular vesicles in S2-N cells transfected with Shrub-Flag-HA. (J–L) Relative Localization of Shrub (green), Notch (purple), and Dx (blue) on subcellular vesicles of S2-N cells transfected with Shrub-Flag-HA and Dx. M, N, and O are the merged images, respectively, of J and K, K and L, and J–L.
Given that Dx can counteract the negative effect Shrub has on Notch signaling (Fig. 3, I and J), we examined if Dx expression can affect Shrub dependent recruitment of Notch on MVBs. When S2-N cells were transfected with both Shrub and Dx, Shrub is localized on Rab7-positive MVBs and so is Notch (Fig. 5, J, K, and M). Dx is also detected on the same subcellular structures, and consistent with its ability to physically interact with Notch (Matsuno et al., 1995), the position of these two molecules coincides (Fig. 5, J–O).
In wing discs expressing ShrubRNAi, where Notch signal is up-regulated (Fig. 3, E and F), Notch is localized in Rab7-positive vesicles (Fig. 4, D–F). Because either the up- or down-regulation of Shrub expression is key in triggering the internalization and its localization on MVBs, tagged by Rab7, the functional outcome of the differential regulation is not the same. We propose that once Notch is trafficked on to MVBs, whether it will be degraded or not depends on the relative availability of Shrub and Dx. Down-regulating Shrub results in an up-regulation of the Notch signal (Fig. 3, E and F), while the up-regulation of Dx promotes the activation of the receptor, defining the Notch–Dx–Shrub–Krz circuitry as an important means for Notch signal regulation.
Shrub-Dx–regulated intracellular Notch signaling is ligand independent
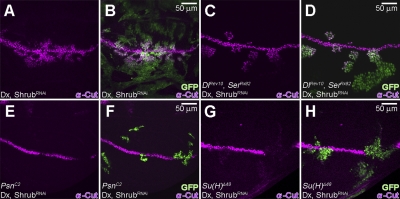
Given that our results indicate that Notch receptor can be induced to signal while not on the cell surface and potentially through a noncanonical signal mechanism, we wanted to examine if this signaling is ligand dependent. We generated somatic clones lacking both Notch ligands, Dl and Ser, and asked if the Notch signal modulated by Dx and ShrubRNAi (Fig. 3, K and L) is ligand dependent. Consistent with what we expect from the above analysis, cells in mosaic clones in the ventral part of the wing disc expressing Dx and ShrubRNAi (marked by GFP) up-regulated Notch signaling, as judged by the expression of Cut (Fig. 6, A and B). In cells lacking both DlREV10 and SerRX82 (Fig. 6, C and D) but overexpressing Dx and ShrubRNAi, Cut is still activated, confirming that the intracellular signaling we observed is independent of the ligand. Extending these observations, we also determined that this mode of Notch activation is not affected when Dl is down-regulated in vivo by DlRNAi (Fig. S3, A and B). Moreover, in a Notch-dependent reporter assay using S2 cells that express neither of the ligands, we find that the signal triggered by the expression of full-length Notch is modulated by Shrub (Fig. S2 M).
Figure 6.
Activation of Notch induced by Dx and ShrubRNAi is independent of Dl and Ser, but dependent on Psn and Su(H). (A and B) In the mosaic clones expressing Dx and ShrubRNAi (marked by GFP) (hs-FLP, tub-GAL4, UAS-GFP/+; UAS-Flag:Dx/UAS-ShrubRNAi; tub-GAL80, FRT82B/FRT82B), ectopic expression of Cut (purple) is induced in mostly the ventral part of the wing disc. (C and D) Co-expression of Dx and ShrubRNAi in DlREv10 and SerRX82 clones (marked by GFP) maintains the ectopic Cut expression (purple) (hs-FLP, tub-GAL4, UAS-GFP/+; UAS-Flag:Dx/UAS-ShrubRNAi; tub-GAL80, FRT82B/DlREV10, SerRX82, FRT82B). (E and F) In PsnC2 clones (marked by GFP), coexpression of Dx and ShrubRNAi does not induce the expression of Cut (purple) (hs-FLP, tub-GAL4, UAS-GFP/+; UAS-Flag:Dx/UAS-ShrubRNAi; tub-GAL80, FRT2A/PsnC2, FRT2A). (G and H) Su(H)Δ47 clones (marked by GFP), with coexpressing Dx and ShrubRNAi, fail to induce Cut expression (purple) (hs-FLP, tub-GAL4, UAS-GFP/+; tub-GAL80, FRT40A/Su(H)Δ47 FRT40A; UAS-Flag:Dx/UAS-ShrubRNAi).
Finally, we examined whether this noncanonical Shrub and Dx-dependent activation of Notch still depends on Presenilin (Psn), γ-secretase that is essential for the cleavage of intracellular Notch, and on the canonical effector Su(H). Through clonal analyses we found that cells lacking either Psn activity (PsnC2 clones marked by GFP; Fig. 6, E and F) or Su(H) activity (Su(H)Δ47 clones marked by GFP; Fig. 6, G and H), but coexpressing Dx and ShrubRNAi, cannot up-regulate Cut. Correspondingly, PsnRNAi or Su(H)RNAi inhibit the Notch receptor activation we describe (Fig. S3, C–F). Thus, the Shrub-Dx–regulated signaling, although ligand independent, still needs γ-secretase as well as Su(H).
Relating Dx activity to the ESCRT complexes
Given the association of Shrub with ESCRT-III and previous reports suggesting that loss-of-function mutations in other ESCRT elements display cellular interactions with Notch (Thompson et al., 2005; Vaccari and Bilder, 2005; Vaccari et al., 2008, 2009), we explored the relationship of the noncanonical Notch signaling we uncovered with the ESCRT family of complexes (Slagsvold et al., 2006; Hanson et al., 2009). We asked if the Notch noncanonical signaling, triggered by Dx and ShrubRNAi, can also be seen when the function of other members of the ESCRT family are disrupted.
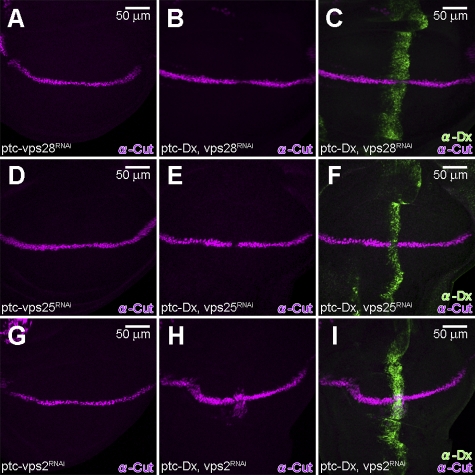
Inhibiting Vps23, Vps28 (ESCRT-I), Vps36, and Vps25 (ESCRT-II) by RNAi, in a Dx-expressing background, failed to show an up-regulation of Notch signaling (Fig. 7, B, C, E, and F; and Fig. S4, B, C, E, and F). We observed that Cut expression is slightly, but detectably, suppressed under these conditions. The suppression of Cut is a bit more prominent when we inhibited Vps28 or Vps25 by RNAi, using the Ay-GAL4 driver at 25°C (Fig. S5, A–D). Thus, the results suggest that these components are not involved in the dx-mediated activation of Notch signal in this context. This was corroborated by extant loss of function mutations in vps23 and vps36 (vps23e00381, vps23f00976, and vps36c04474), as heterozygotes failed to affect the wing-nicking phenotype associated with Dx and Krz coexpression (unpublished data), unlike loss-of-function shrub alleles.
Figure 7.
Notch signal induced by Dx and ShrubRNAi is dependent on ESCRT-III. (A, D, and G) Expression of double-strand RNA of vps28 (vps28RNAi) (ptc-GAL4/+; UAS-vps28RNAi/+) (A), vps25 (vps25RNAi) (ptc-GAL4/+; UAS-vps25RNAi/+) (D), or vps2 (vps2RNAi) (ptc-GAL4/+; UAS-vps2RNAi/+) (G), by ptc-GAL4 does not show significant effect on endogenous Cut levels (purple). (B, C, E, and F) Co-expression of Dx (green) with vps28RNAi (ptc-GAL4/UAS-Flag:Dx; UAS-vps28RNAi/+) (B and C) or vps25RNAi (ptc-GAL4/UAS-Flag:Dx; UAS-vps25RNAi/+) (E and F) results in the subtle but reproducible reduction of Cut (purple). (H and I) Co-expression of Dx (green) with vps2RNAi results in the ectopic activation of Cut (purple) (ptc-GAL4/UAS-Flag:Dx; UAS-vps2RNAi/+). All crosses were performed at 18°C.
In contrast, the disruption of ESCRT-III members other than Shrub emphasizes the particular role ESCRT-III displays in modulating the Notch signaling mode we describe here. Inhibiting Vps2 (through Vps2RNAi expression) in a Dx-expressing background using two independent drivers results, like Shrub, in the up-regulation of Notch signals (Fig. 7, H and I; and Fig. S5, E and F). The disruption of Vps4 activity (through Vps4RNAi expression) in a Dx-expressing background shows a severe developmental defect of the wing disc (unpublished data).
Examining the subcellular localization of Notch in the wing discs expressing Dx when representative members of ESCRT-I (Vps28), -II (Vps25), and -III (Vps2) are inhibited by RNAi corroborated the distinct role of ESCRT-III. Vps2 inhibition resulted in the up-regulation of the Notch signal (Fig. 7, H and I; and Fig. S5, E and F) and the colocalization of Notch and Dx (Fig. S5, M–O). Alternatively, when Vps28RNAi or Vps25RNAi is expressed along with Dx, we observed that Notch accumulates in vesicles, but Notch is not colocalized with Dx (Fig. S5, G–L) and clearly signaling is not activated (Fig. 7, B, C, E, and F; and Fig. S5, A–D). These results also highlight that colocalization of Notch with Dx is necessary for the endosomal activation of Notch signaling we document with loss of ESCRT-III components. We conclude that the Dx-dependent, ligand-independent Notch activation by ESCRT-III components is distinct from the previously reported ESCRT-I– and ESCRT-II–dependent Notch activation in eye disc (Vaccari and Bilder, 2005; Vaccari et al., 2009).
Shrub and Dx regulate the extent of Notch ubiquitinylation
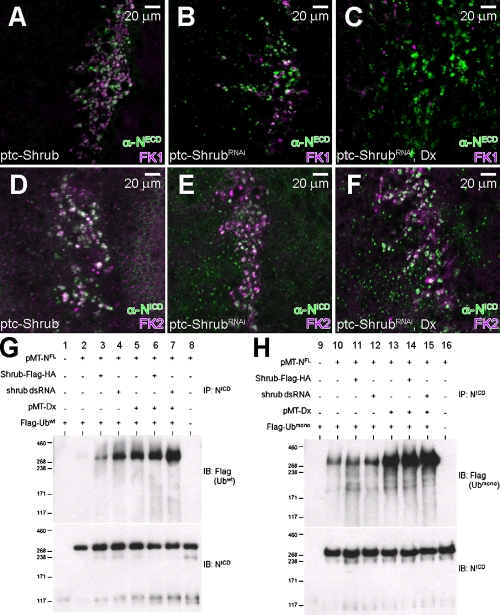
Given that the ESCRT complexes are associated with the trafficking of ubiquitinylated proteins (Slagsvold et al., 2006; Saj et al., 2010) and that Dx function is also associated with ubiquitinylation (Mukherjee et al., 2005; Wilkin et al., 2008), we examined the ubiquitinylation status of Notch under conditions where Dx and Shrub levels are modulated. When Shrub is expressed with the ptc-GAL4 driver, 95% of Notch-bearing vesicles in Shrub-expressing cells are also positive for FK1, an antibody that recognizes poly-ubiquitinated proteins (Fig. 8 A; n = 100). On the other hand, upon inhibiting Shrub through ShrubRNAi, where Notch signal is slightly up-regulated (Fig. 3, E and F), the FK1 antigens coincide with the Notch antigens less frequently (Fig. 8 B; 60%, n = 100). Because up-regulation of Notch by ShrubRNAi is robustly enhanced by expression of Dx (Fig. 3, K and L), we examined if Dx can alter this ubiquitinylation pattern. When both Dx and ShrubRNAi are expressed, the vast majority of the Notch-positive vesicles do not coincide with those positive for poly-ubiquitinated proteins (Fig. 8 C; 5%, n = 80). Using an antibody that recognizes both mono- and poly-ubiquitinated proteins (FK2), the expression pattern of Notch in cells expressing Shrub, ShrubRNAi, or ShrubRNAi and Dx does not display significant differences (Fig. 8 D–F). Thus, Dx can influence the ubiquitinylation state of Notch, a phenotype paralleled by a dramatic up-regulation of Notch signaling (Fig. 3, K and L).
Figure 8.
Shrub and Dx regulate the ubiquitinylation status of Notch. (A) Expression of Shrub by ptc-GAL4 results in the accumulation of Notch (green) in endosomal vesicles marked by FK1 (purple), an antibody that recognizes poly-ubiquitinated proteins (ptc-Gal4/+; UAS-Shrub/+). (B) Expression of ShrubRNAi by ptc-GAL4 results in a significant increase of Notch-containing vesicles (green) that are not marked by FK1 (purple) (ptc-Gal4/+; UAS-ShrubRNAi/+). (C) Co-expression of ShrubRNAi and Dx results in Notch accumulation (green) in subcellular vesicles that are negative for FK1 (purple) (ptc-Gal4/UAS-Flag:Dx; UAS-ShrubRNAi/+). (D–F) Expression of Shrub (D), ShrubRNAi (E), or ShrubRNAi and Dx (F), driven by ptc-GAL4, increases the number of Notch-containing vesicles (green) that are marked by FK2 (purple). (G and H) Ubiquitinylation assay in S2R+ cells transfected with pMT-NFL, pMK33-Shrub-Flag-HA, pMT-Dx, pMT-Flag-Ubwt (G), or pMT-Flag-Ubmono (H) shows a significant increase in Notch ubiquitinylation in the presence of Dx, suggesting that Dx drives Notch to a mono-ubiquitinated form. The cells were treated with shrub dsRNA to deplete shrub levels.
To directly assess the relative roles of Dx and Shrub in the ubiquitinylation of Notch we relied on the S2R+ cultured cells that do not express Notch endogenously. Cells were transfected with full-length Notch and a flag-tagged wild-type Ubiquitin (Flag-Ubwt) and 24 h later treated with shrub dsRNA (ShrubRNAi) for another 24 h (Fig. 8, G and H). When Notch alone is expressed in S2R+ cells together with Flag-Ubwt we detect low levels of ubiquitinylated Notch species. In contrast, the down-regulation of Shrub increases the levels of ubiquitinylated Notch species. Analogous experiments involving Dx and Notch also show that Dx modulation affects the ubiquitinylation status of Notch, as previously determined (Mukherjee et al., 2005). If Dx is expressed in the presence of ShrubRNAi, Notch ubiquitinylation is further enhanced. When instead of Flag-Ubwt we use Flag-Ubmono as the ubiquitinylation substrate, which can only participate in mono-ubiquitinylation events, we observe an almost identical pattern of Notch ubiquitinylation, arguing that the phenomenon we monitor in this cellular context is probably associated with mono-ubiquitinylation events.
In summary, the in vivo immunocytochemical evidence indicates that the down-regulation of Shrub in the presence of Dx results in shifting Notch away from vesicles positive for a poly-ubiquitin marker. The biochemical analysis indicates that this shift is associated with high levels of mono-ubiquitinylation on the receptor. The functional consequence of these events is a Dx-associated promotion of Notch signaling that reverses the negative regulation Shrub exerts on Notch.
Discussion
The extraordinary sensitivity of normal development to the dosage of the Notch receptor is manifested through the haploinsufficient and triplomutant behavior of the Notch locus (Artavanis-Tsakonas et al., 1999). Dosage sensitivity is consistent with the fact that the Notch signaling mechanism relies on stoichiometric interactions rather than enzymatic amplification steps to bring the signal from the surface to the nucleus. This also provides a rationale for the observation that cellular events involved in trafficking/turnover are emerging as major Notch signal–controlling mechanisms (Artavanis-Tsakonas et al., 1999; Fortini, 2009; Tien et al., 2009; Yamamoto et al., 2010). The canonical pathway relies on the activation of the receptor triggered by its interaction with membrane-bound ligands on an apposing cell but the possibility that the receptor can also be activated intracellularly, in a ligand-independent fashion, as several studies, including the present one, suggest, has important implications for the biology and pathobiology of Notch. The rules governing how and where a receptor, trafficking through the endocytic compartments, can be activated, in the presence or absence of the ligand, are still not completely defined. Moreover, we do not understand how such events are integrated into the genetic circuitry that affects the regulation of endosomal compartment assembly and function.
Here, we provide insight into these questions by showing that the interplay between the Notch signal modulator Dx, the nonvisual β-arrestin orthologue Krz, and a critical component of the ESCRT-III complex, Shrub, directs Notch either into a degradation or into a ligand-independent activation path, which is paralleled by distinct ubiquitinylation states of Notch. The ESCRT pathway, recently described as a “cargo-recognition and membrane-sculpting machine,” defines a complex, multipurpose cellular machinery with cellular roles and molecular mechanisms that are not fully elucidated (Henne et al., 2011). ESCRT is crucial in mediating the various steps leading to the sorting of membrane proteins into MVBs on their way to lysosomal degradation. The implication of Shrub in Notch signaling–related processes was revealed through an unbiased genetic screen for isogenic modifiers of the dx-krz–dependent phenotype, which is based on the endosomal/lysosomal degradation of the Notch receptor (Mukherjee et al., 2005). Our study is not the first to provide a general link between Notch signaling with the ESCRT machinery (Moberg et al., 2005; Thompson et al., 2005; Vaccari and Bilder, 2005; Vaccari et al., 2008, 2009; Herz et al., 2009), but both our genetic screen as well as our subsequent analysis points to the differential and major role of the ESCRT-III complex in the dx-krz–dependent, ligand-independent mode of Notch signaling we describe here.
Several studies established that as the Notch receptor enters an endocytic path, it can be activated inside the cell in both a ligand-dependent as well as ligand-independent fashion (Hori et al., 2004; Sakata et al., 2004; Wilkin et al., 2004, 2008; Mukherjee et al., 2005; Childress et al., 2006). Consequently, several elements of the endosomal machinery including elements of the ESCRT complexes were shown to influence the intracellular accumulation and activation of the Notch receptor (Moberg et al., 2005; Thompson et al., 2005; Vaccari and Bilder, 2005; Vaccari et al., 2008, 2009; Herz et al., 2009). Our data are compatible with these studies and indeed extend and complement them. These studies are not directly comparable, not only because of differing genetic backgrounds, a crucial element in evaluating genetic interactions, but also because we are analyzing the impact of ESCRT function on the modulation of Notch signaling via the synergistic action of Dx and Krz, which may well define a different but specific path. Considering the entire body of work related to various aspects of Notch receptor trafficking it seems that there may be several, distinct ways the receptor can be activated after entering the endocytic path. Some studies link early endosomes with the activation of the receptor (Vaccari and Bilder, 2005; Childress et al., 2006; Vaccari et al., 2009), whereas others (Wilkin et al., 2008) implicate late endosomal compartments with ligand-independent activation of Notch. Particularly relevant to the activation mode we document here are the genetic studies of Wilkin et al. (2008), which associated Notch activation with the HOPS (homotypic fusion and vacuole protein sorting) and AP-3 (adaptor protein-3) complexes, demonstrating the existence of a Notch activation path that is dependent on late endosomal compartments.
We found that the expression of Shrub triggers a dramatic subcellular shift of the Notch receptor to MVBs, consistent with the fact that ESCRT-III mediates the cargo de-ubiquitination, budding, and scission of intraluminal vesicles (Wollert et al., 2009, Henne et al., 2011), which control the delivery of the cargo to the lysosomes. The down-regulation of Notch signals by Shrub is apparently associated with the recruitment of Notch in intraluminal vesicles and its eventual degradation. On the other hand, disrupting the cellular equilibrium between Dx and Shrub by down-regulating Shrub and/or up-regulating Dx activates the receptor in a ligand-independent manner. It is also clear that the mode of Notch activation we document is independent of the ligands and is linked to the ubiquitinylation status of the Notch receptor, which is in turn is modulated by Dx and Krz. Krz was shown to modulate Notch activity through its ability to regulate the levels of the Notch protein (Mukherjee et al., 2005). We note with interest that although β-arrestins have been implicated as adaptors during clathrin-dependent endocytosis, Ram8, an arrestin homologous protein in yeast, has also been associated with the recruitment of the ESCRT machinery to MVBs loaded with a G-coupled receptor cargo (Herrador et al., 2010). If Krz has a similar relationship with the ESCRT machinery, it may be involved in sorting Notch on MVBs and hence the eventual recruitment of Notch in intraluminal vesicles for degradation, a notion compatible with the observation that Krz enhances the Shrub-dependent down-regulation of Notch.
In order for the receptor to enter intraluminal vesicles, a de-ubiquitinylation of the cargo must take place (Slagsvold et al., 2006; Ma et al., 2007; Henne et al., 2011). It is noteworthy that Snf7 (the Shrub orthologue in yeast) recruits the de-ubiquitinating enzyme Doa4 necessary for such cargo de-ubiquitination (Odorizzi et al., 2003; Luhtala and Odorizzi, 2004). In our cell culture studies, where we could clearly follow the relative subcellular localization of Notch, Shrub, and Dx, we observed localization of Notch, Shrub, and Dx with MVB membranes. Because this subcellular phenotype is paralleled by a dramatic ligand-independent activation of the receptor and a shift from poly- to a mono-ubiquitination status of the receptor, this leads us to suggest that Dx, which physically interacts with Notch, interferes with processes that are essential for loading the receptor on intraluminal vesicles.
It is clear that a more detailed analysis of subcellular dynamics in vivo is necessary to address many of the questions raised by the present study. On the basis of the data we present here we propose a model for the activation mode we uncovered (Fig. 9). We suggest that the ESCRT-III component Shrub can regulate receptor cycling, diverting it to a signaling path, a fate modulated by Dx and Krz. Thus, Notch signaling can be attenuated inside the cell in a ligand-independent fashion. It remains to be determined how such intracellular signaling serves the developmental logic of Notch which couples the fate of one cell to that of the next door cellular neighbor. It is possible for such mode of Notch action to be useful to modulate the fate of a cell that, for example, circulates, as was elegantly demonstrated by Mukherjee et al. (2011), and is thus not necessarily in contact with a ligand-expressing neighbor. Irrespective of the potential role ligand-independent activation may play in normal development, activating the receptor can have profound pathological consequences. Therefore, understanding pathways capable of modulating Notch activity in an intracellular, ligand-independent manner is of great importance.
Figure 9.
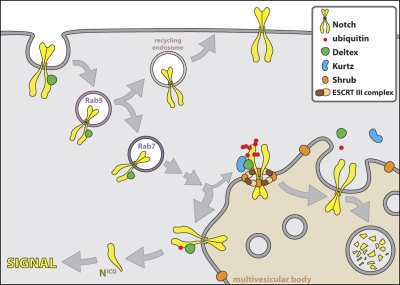
Shrub-Dx-Krz–dependent modulation of Notch signaling. The ubiquitinylation state of the Notch receptor regulates its activation fate as it enters the endocytic path. Although some steps in this path have been characterized, some simply define working hypothesis. Our studies indicate that Dx in synergy with Krz promotes the poly-ubiquitinated state of the receptor, which leads to the degradation of Notch, through the MVBs, a step regulated by Shrub, a core component of the ESCRT-III complex. The close proximity of the Shrub–ESCRT-III complex with Notch in the cartoon is not meant to imply a direct association of Shrub with Notch, given that we could not find evidence favoring such interaction. Our evidence is consistent with the notion that Shrub “surrounds” the ubiquitinylated receptor, a role compatible with the previously suggested role of the yeast homologue Snf7 (Wollert et al., 2009). The expression of Dx, which physically interacts with Notch, favors a mono-ubiquitinated state of the receptor, which leads to a ligand-independent intracellular activation of Notch (NICD: the cleaved, activated form of Notch).
Materials and methods
Genetic strains
We used the following mutant alleles: N54l9 (Lindsley and Zimm, 1992), krz1 (Roman et al., 2000), shrub4 (Sweeney et al., 2006), PsnC2 (Lukinova et al., 1999), Su(H)Δ47 (Morel and Schweisguth, 2000), and DlRevF10 and SerRX82 double mutant (Micchelli et al., 1997). The UAS lines used were UAS-Flag:Dx (Mukherjee et al., 2005), UAS-HA:Krz (Mukherjee et al., 2005), UAS-Shrub (Sweeney et al., 2006), UAS-ShrubRNAi (Sweeney et al., 2006), UAS-GFP (Bloomington Drosophila Stock Center, Bloomington, IN), UAS-vps23RNAi (Vienna Drosophila RNAi Center, Vienna, Austria), UAS-vps28RNAi (Vienna Drosophila RNAi Center), UAS-vps36RNAi (Vienna Drosophila RNAi Center), UAS-vps25RNAi (Vienna Drosophila RNAi Center), UAS-vps20RNAi (Vienna Drosophila RNAi Center), UAS-vps24RNAi (Vienna Drosophila RNAi Center), UAS-vps4RNAi (Vienna Drosophila RNAi Center), UAS-vps2RNAi (Vienna Drosophila RNAi Center), UAS-DlRNAi (Vienna Drosophila RNAi Center), UAS-PsnRNAi (Vienna Drosophila RNAi Center), and UAS-Su(H)RNAi (Vienna Drosophila RNAi Center). The UAS constructs were driven by C96-Gal4 (Gustafson and Boulianne, 1996), ptc-Gal4 (Speicher et al., 1994), tub-GAL4 (Wang and Struhl, 2005), or Ay-GAL4 (Ito et al., 1997) as indicated in the figure legends. All crosses were performed at 25°C unless otherwise stated.
Isolation of shrub mutant
The Exelixis collection is composed of 16,000 transposon-induced gene disruptions, resulting in mutations in ∼53% of the Drosophila genome (https://drosophila.med.harvard.edu; Artavanis-Tsakonas, 2004; Kankel et al., 2007). The collection was screened for genes that dominantly modify the wing-nicking phenotype associated with simultaneous C96-Gal4–directed expression of Dx and Krz using the UAS-Flag:Dx/+; C96-Gal4, UAS-HA:Krz/+ strain (C96-Dx+Krz). In the primary screen, C96-Dx+Krz virgin females were crossed with males carrying autosomal or viable X-linked insertions. C96-Dx+Krz males were crossed with virgin females carrying lethal insertions on X chromosome and the F1 progeny was scored for phenotypic modifications. Modifying transposons were categorized as enhancers or suppressors of weak, moderate, or strong intensity. Out of 805 primary screen modifiers, 388 were retested with C96-Dx+Krz to confirm modification. A secondary test was performed by crossing confirmed modifiers to flies carrying the C96-Gal4 alone to eliminate the false modifiers that affect wing development. Positive secondary tests were performed to examine the interaction with dx or krz, using wing phenotypes that result from expression of Dx (UAS-Flag:Dx/+; C96-Gal4/+), expression of Krz (C96-Gal4, UAS-HA:Krz/+), dx mutant (dx152;; C96-Gal4/+), or krz mutant (krz1, C96-Gal4/+). As a result, we recovered 127 modifiers, including a single transposon insertion in shrub (d02738).
Generation of mosaics
Mitotic clones were generated by Flp-mediated mitotic recombination (Xu and Rubin, 1993). Recombination was induced in the second instar larvae by a 60-min heat shock at 37°C. To generate the clones expressing Dx with ShrubRNAi, hs-FLP, tub-GAL4, UAS-GFP/Y; UAS-Flag:Dx/+; tub-GAL80, FRT82B/+ males were crossed with UAS-ShrubRNAi/CyO,GFP; FRT82B/TM6B virgin females. To generate the double-mutant clones of DlREV10 and SerRX82, expressing Dx with ShrubRNAi, hs-FLP, tub-GAL4, UAS-GFP/Y; UAS-Flag:Dx/+; tub-GAL80, FRT82B/+ males were crossed with UAS-ShrubRNAi/CyO,GFP; DlREV10, SerRX82, FRT82B/TM6B virgin females. To generate the mutant clones of PsnC2 expressing Dx and ShrubRNAi, hs-FLP, tub-GAL4, UAS-GFP/Y; UAS-Flag:Dx/+; tub-GAL80, FRT2A/+ males were crossed with UAS-ShrubRNAi/CyO,GFP; PsnC2, FRT2A/TM6B virgin females. To generate the Su(H)Δ47 expressing Dx and ShrubRNAi, hs-FLP, tub-GAL4, UAS-GFP/Y; tub-GAL80, FRT40A/CyO, UAS-Flag:Dx/+ males were crossed with Su(H)Δ47 FRT40A/CyO; UAS-ShrubRNAi/TM6B.
Immunohistochemistry
Wing discs from third instar larvae were dissected in PBS and fixed in PLP (2% paraformaldehyde, 0.01 M NaIO4, 0.075 M lysine, and 0.037 M sodium phosphate, pH 7.2; Matsuno et al., 2002). Discs were washed in PBS-DT (0.3% sodium deoxycholate and 0.3% Triton X-100 in PBS) and incubated with the following antibodies: mouse anti-NICD (9C6, 1:500; Fehon et al., 1990), mouse anti-NECD (2H, 1:500; Fehon et al., 1990), mouse anti-Cut (2B10, 1:100; Developmental Studies Hybridoma Bank, Iowa City, IA), rabbit anti-Flag (1:1,000; Sigma-Aldrich), rabbit anti-HA (1:1,000; Sigma-Aldrich), rat anti-Dx (17A, 1:50; Busseau et al., 1994), mouse anti-polyubiquitinated protein (FK1, 1:100; Enzo Life Sciences), mouse anti-mono/polyubiquitinated protein (FK2, 1;1000; Enzo Life Sciences), rabbit anti-Rab7 (1:1,000; Chinchore et al., 2009), and rabbit anti-Shrub (1:1,000; Sweeney et al., 2006). After several washes in PBS-DT, the discs were incubated with Alexa 488–, 594–, or 647–conjugated secondary (1:300; Invitrogen) antibodies, followed by washing in PBS-T (0.1% Triton X-100 in PBS). The samples were mounted in FluoroGuard Antifade Reagent (Bio-Rad Laboratories) at room temperature. For the endocytosis assay, the wing discs were incubated in S2 medium containing rat anti-NECD (1:500; Fehon et al., 1990) or mouse anti-Dl (9B) (1:500; Fehon et al., 1990) for 40 min at room temperature. After washing with S2 medium for 10 min three times, the wing discs were fixed in PLP for 40 min on ice. For immunostaining of cultured cells, pMK33-Shrub-Flag-HA was transfected into S2-N cells using Effectene (QIAGEN). Cells were induced with 0.35 mM CuSO4 overnight, and fixed in PLP. Subsequent washes, antibody incubations, and visualization were performed as described above. For chloroquine feeding assays, the flies were cultured in vials with fly medium containing 1 mg/ml chloroquine (Chi et al., 2010).
Microscopy
Fluorescent images of imaginal discs were collected with either a Nikon C1Si spectral point scanning confocal microscope or a Nikon A1R confocal microscope. Nikon C1 is equipped with a Nikon TE2000 inverted microscope with DIC, phase, and epifluorescence optics (with 40x Plan Fluor NA 1.4 and 60x Plan Apo NA 1.4 objective lens), Melles Griot solid state diode lasers (405 nm, 488 nm [10 mW], and 561 [10 mW]), and a 100-mW mercury arc lamp illumination for viewing fluorescence by eye. Nikon A1R confocal is quipped with a Nikon Ti-E motorized inverted microscope with Perfect Focus System (with 60x Plan Apo NA 1.4 lens), three solid state lasers (405, 561, and 640 nm), argon-krypton laser (457, 488, and 514 nm), and Nikon intensilight fluorescence light source for viewing by eye. The cultured cells were scanned using a Yokogawa spinning disk confocal microscope on a Zeiss 200M inverted fluorescence microscope (with 63x Plan Apo PH NA 1.4), equipped with a computer-controlled Spherical Aberration Correction unit (SAC, Intelligent Imaging Innovations; Saffarian et al., 2009), environmental chamber containing heated sample holder (20/20 Technology) to maintain air temperature, Stage controller (Piezo ‘Z’ PZ-2000, X&Y Stage MS-2000; Applied Scientific Instrumentation), Xenon lamp (Stutter Instruments), and lasers (488, 561, and 640 nm; Cobalt and Coherent; 40–50 mW). The image acquisition software used was Nikon Ez-C1, Nikon Elements, or Slidebook 4 (Intelligent Imaging Innovations). Adobe Photoshop CS5 was used to pseudocolor images.
Ligand-independent luciferase assay
Drosophila S2R+ cells were cultured in standard M3 medium (Invitrogen) supplemented with 10% fetal bovine serum. dsRNA was generated by in vitro transcription using the T7 RNA polymerase (Invitrogen). RNAi experiments were performed as described previously (Saleh et al., 2006). In brief, cells were plated at 106 cells/ml in serum free media, and incubated with 10 µg dsRNA at RT for 30 min. 2 d after adding media with serum, the cells were transfected with E(spl)m3-luc (Mukherjee et al., 2005), pRL (a Renilla luciferase in pIZ vector for transfection control), pMT-NFL, or pMK33-Flag-HA-Shrub (Guruharsha et al., 2011) using Effectene (QIAGEN). Equal amounts of plasmid were transfected, and the total amount of DNA was kept constant by adding empty vector or GFP dsRNA. Cells were induced with 0.35 mM CuSO4 overnight. The luciferase activity was measured using the Dual-Glo Luciferase Assay System (Promega). The ratio of E(spl)m3-luc/pRL was calculated for each sample as an activity of the Notch signal. Experiments were performed in triplicate.
Ubiquitinylation assay
pMT-NFL was cotransfected into S2R+ cells with pMT-Flag-Ubwt (Mukherjee et al., 2005), pMT-Flag-Ubmono (Mukherjee et al., 2005), pMK33-Shrub-Flag-HA, pMT-Dx, shrub dsRNA, as indicated in Fig. 6, D and E. Equal amounts of plasmid were transfected, and the total amount of DNA was kept constant by adding empty vector or GFP dsRNA. Plasmid expression was induced with 0.35 mM CuSO4 overnight. Cells were treated for 4 h at 25°C A with the proteasome inhibitor MG132 (50 mM). Subsequently, cells were lysed in buffer containing 50 mM Tris, pH 7.5, 125 mM NaCl, 1.5 mM MgCl2, 5% glycerol, 0.2% NP-40, 1 mM DTT, 25 mM NaF, 1 mM Na3VO4, and complete protease inhibitor (Roche), and were frozen in dry ice/ethanol. Lysates were cleared by centrifugation at 14,000 g for 10 min, and incubated with anti-NICD (9C6, 1:500) for 3 h, followed by an overnight incubation with Protein G–Agarose (Roche). Precipitates were repeatedly washed with lysis buffer, protein complexes were eluted with LDS sample buffer (Invitrogen), separated on 3–7% Tris-Acetate gel, transferred onto PVDF membrane (Invitrogen). Membranes were incubated with rabbit anti-Flag (1:1,000; Sigma-Aldrich), followed by goat anti–mouse or –rabbit HRP-conjugated antibody (1:5,000; GE Healthcare), and developed using the ECL+ chemiluminescence detection system (GE Healthcare).
Online supplemental material
Fig. S1 shows that down-regulation of Shrub results in the activation Notch. Fig. S2 describes an endocytosis assay to monitor Notch trafficking in live wing disc cells and S2-N cells. Fig. S3 shows that Notch signaling induced by Dx and ShrubRNAi is independent of Dl and Ser, but dependent on Psn and Su(H). Fig. S4 shows that Notch signal induced by Dx and ShrubRNAi is dependent on ESCRT-III. Fig. S5 shows that ESCRT-III is primarily responsible for dx-mediated Notch signal. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201104146/DC1.
Acknowledgments
We would like to thank F.-B. Gao, K. Matsuno, the Bloomington Drosophila Stock Center and Vienna Drosophila RNAi Center for Drosophila fly stocks, as well as F.-B. Gao, D. Patrick, and the Developmental Studies Hybridoma Bank for antibodies. We are grateful to K.G. Guruharsha for critically reading the manuscript and J. Iwasa for her creative advice. We acknowledge the Nikon Imaging Center at Harvard Medical School for help with imaging.
This work was supported by National Institutes of Health grants NS26084 and CA98402 (S. Artavanis-Tsakonas), grant 07572 (T. Kirchhausen), a JSPS Postdoctoral Fellowship for Research Abroad (K. Hori), and a Postdoctoral Fellowship from the FSMA (A. Sen).
Footnotes
Abbreviations used in this paper:
- AP
- anterior–posterior
- Dl
- Delta
- DV
- dorsoventral
- Dx
- Deltex
- ESCRT
- endosomal sorting complex required for transport
- Krz
- Kurtz
- MVB
- multivesicular body
- NECD
- extracellular domain of Notch
- NICD
- intracellular domain of Notch
- Psn
- Presenilin
- Ser
- Serrate
- Su(H)
- Suppressor of Hairless
- UAS
- upstream activating sequence
References
- Artavanis-Tsakonas S. 2004. Accessing the Exelixis collection. Nat. Genet. 36:207 10.1038/ng1316 [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S., Rand M.D., Lake R.J. 1999. Notch signaling: cell fate control and signal integration in development. Science. 284:770–776 10.1126/science.284.5415.770 [DOI] [PubMed] [Google Scholar]
- Bray S.J. 2006. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7:678–689 10.1038/nrm2009 [DOI] [PubMed] [Google Scholar]
- Busseau I., Diederich R.J., Xu T., Artavanis-Tsakonas S. 1994. A member of the Notch group of interacting loci, deltex encodes a cytoplasmic basic protein. Genetics. 136:585–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi C., Zhu H., Han M., Zhuang Y., Wu X., Xu T. 2010. Disruption of lysosome function promotes tumor growth and metastasis in Drosophila. J. Biol. Chem. 285:21817–21823 10.1074/jbc.M110.131714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childress J.L., Acar M., Tao C., Halder G. 2006. Lethal giant discs, a novel C2-domain protein, restricts notch activation during endocytosis. Curr. Biol. 16:2228–2233 10.1016/j.cub.2006.09.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchore Y., Mitra A., Dolph P.J. 2009. Accumulation of rhodopsin in late endosomes triggers photoreceptor cell degeneration. PLoS Genet. 5:e1000377 10.1371/journal.pgen.1000377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coumailleau F., Fürthauer M., Knoblich J.A., González-Gaitán M. 2009. Directional Delta and Notch trafficking in Sara endosomes during asymmetric cell division. Nature. 458:1051–1055 10.1038/nature07854 [DOI] [PubMed] [Google Scholar]
- de Celis J.F., Garcia-Bellido A., Bray S.J. 1996. Activation and function of Notch at the dorsal-ventral boundary of the wing imaginal disc. Development. 122:359–369 [DOI] [PubMed] [Google Scholar]
- Fehon R.G., Kooh P.J., Rebay I., Regan C.L., Xu T., Muskavitch M.A., Artavanis-Tsakonas S. 1990. Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell. 61:523–534 10.1016/0092-8674(90)90534-L [DOI] [PubMed] [Google Scholar]
- Fehon R.G., Johansen K., Rebay I., Artavanis-Tsakonas S. 1991. Complex cellular and subcellular regulation of notch expression during embryonic and imaginal development of Drosophila: implications for notch function. J. Cell Biol. 113:657–669 10.1083/jcb.113.3.657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming R.J., Gu Y., Hukriede N.A. 1997. Serrate-mediated activation of Notch is specifically blocked by the product of the gene fringe in the dorsal compartment of the Drosophila wing imaginal disc. Development. 124:2973–2981 [DOI] [PubMed] [Google Scholar]
- Fortini M.E. 2009. Notch signaling: the core pathway and its posttranslational regulation. Dev. Cell. 16:633–647 10.1016/j.devcel.2009.03.010 [DOI] [PubMed] [Google Scholar]
- Gridley T. 2003. Notch signaling and inherited disease syndromes. Hum. Mol. Genet. 12(Spec No 1):R9–R13 10.1093/hmg/ddg052 [DOI] [PubMed] [Google Scholar]
- Guruharsha K.G., Rual J.F., Zhai B., Mintseris J., Vaidya P., Vaidya N., Beekman C., Wong C., Rhee D.Y., Cenaj O., et al. 2011. A protein complex network of Drosophila melanogaster. Cell. 147:690–703 10.1016/j.cell.2011.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson K., Boulianne G.L. 1996. Distinct expression patterns detected within individual tissues by the GAL4 enhancer trap technique. Genome. 39:174–182 10.1139/g96-023 [DOI] [PubMed] [Google Scholar]
- Hanson P.I., Shim S., Merrill S.A. 2009. Cell biology of the ESCRT machinery. Curr. Opin. Cell Biol. 21:568–574 10.1016/j.ceb.2009.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne W.M., Buchkovich N.J., Emr S.D. 2011. The ESCRT pathway. Dev. Cell. 21:77–91 10.1016/j.devcel.2011.05.015 [DOI] [PubMed] [Google Scholar]
- Herrador A., Herranz S., Lara D., Vincent O. 2010. Recruitment of the ESCRT machinery to a putative seven-transmembrane-domain receptor is mediated by an arrestin-related protein. Mol. Cell. Biol. 30:897–907 10.1128/MCB.00132-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz H.M., Woodfield S.E., Chen Z., Bolduc C., Bergmann A. 2009. Common and distinct genetic properties of ESCRT-II components in Drosophila. PLoS ONE. 4:e4165 10.1371/journal.pone.0004165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K., Fostier M., Ito M., Fuwa T.J., Go M.J., Okano H., Baron M., Matsuno K. 2004. Drosophila deltex mediates suppressor of Hairless-independent and late-endosomal activation of Notch signaling. Development. 131:5527–5537 10.1242/dev.01448 [DOI] [PubMed] [Google Scholar]
- Ito K., Awano W., Suzuki K., Hiromi Y., Yamamoto D. 1997. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 124:761–771 [DOI] [PubMed] [Google Scholar]
- Kankel M.W., Hurlbut G.D., Upadhyay G., Yajnik V., Yedvobnick B., Artavanis-Tsakonas S. 2007. Investigating the genetic circuitry of mastermind in Drosophila, a notch signal effector. Genetics. 177:2493–2505 10.1534/genetics.107.080994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R., Ilagan M.X. 2009. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 137:216–233 10.1016/j.cell.2009.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley, D.L., and G.G. Zimm. 1992. The Genome of Drosophila melanogaster. Academic Press, San Diego, CA. 1133 pp
- Luhtala N., Odorizzi G. 2004. Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. J. Cell Biol. 166:717–729 10.1083/jcb.200403139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukinova N.I., Roussakova V.V., Fortini M.E. 1999. Genetic characterization of cytological region 77A-D harboring the presenilin gene of Drosophila melanogaster. Genetics. 153:1789–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y.M., Boucrot E., Villén J., Affar B., Gygi S.P., Göttlinger H.G., Kirchhausen T. 2007. Targeting of AMSH to endosomes is required for epidermal growth factor receptor degradation. J. Biol. Chem. 282:9805–9812 10.1074/jbc.M611635200 [DOI] [PubMed] [Google Scholar]
- Matsuno K., Diederich R.J., Go M.J., Blaumueller C.M., Artavanis-Tsakonas S. 1995. Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development. 121:2633–2644 [DOI] [PubMed] [Google Scholar]
- Matsuno K., Ito M., Hori K., Miyashita F., Suzuki S., Kishi N., Artavanis-Tsakonas S., Okano H. 2002. Involvement of a proline-rich motif and RING-H2 finger of Deltex in the regulation of Notch signaling. Development. 129:1049–1059 [DOI] [PubMed] [Google Scholar]
- Micchelli C.A., Rulifson E.J., Blair S.S. 1997. The function and regulation of cut expression on the wing margin of Drosophila: Notch, Wingless and a dominant negative role for Delta and Serrate. Development. 124:1485–1495 [DOI] [PubMed] [Google Scholar]
- Moberg K.H., Schelble S., Burdick S.K., Hariharan I.K. 2005. Mutations in erupted, the Drosophila ortholog of mammalian tumor susceptibility gene 101, elicit non-cell-autonomous overgrowth. Dev. Cell. 9:699–710 10.1016/j.devcel.2005.09.018 [DOI] [PubMed] [Google Scholar]
- Morel V., Schweisguth F. 2000. Repression by suppressor of hairless and activation by Notch are required to define a single row of single-minded expressing cells in the Drosophila embryo. Genes Dev. 14:377–388 [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A., Veraksa A., Bauer A., Rosse C., Camonis J., Artavanis-Tsakonas S. 2005. Regulation of Notch signalling by non-visual β-arrestin. Nat. Cell Biol. 7:1191–1201 10.1038/ncb1327 [DOI] [PubMed] [Google Scholar]
- Mukherjee T., Kim W.S., Mandal L., Banerjee U. 2011. Interaction between Notch and Hif-alpha in development and survival of Drosophila blood cells. Science. 332:1210–1213 10.1126/science.1199643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann C.J., Cohen S.M. 1996. A hierarchy of cross-regulation involving Notch, wingless, vestigial and cut organizes the dorsal/ventral axis of the Drosophila wing. Development. 122:3477–3485 [DOI] [PubMed] [Google Scholar]
- Odorizzi G., Katzmann D.J., Babst M., Audhya A., Emr S.D. 2003. Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. J. Cell Sci. 116:1893–1903 10.1242/jcs.00395 [DOI] [PubMed] [Google Scholar]
- Rebay I., Fleming R.J., Fehon R.G., Cherbas L., Cherbas P., Artavanis-Tsakonas S. 1991. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell. 67:687–699 10.1016/0092-8674(91)90064-6 [DOI] [PubMed] [Google Scholar]
- Roman G., He J., Davis R.L. 2000. kurtz, a novel nonvisual arrestin, is an essential neural gene in Drosophila. Genetics. 155:1281–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffarian S., Cocucci E., Kirchhausen T. 2009. Distinct dynamics of endocytic clathrin-coated pits and coated plaques. PLoS Biol. 7:e1000191 10.1371/journal.pbio.1000191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saj A., Arziman Z., Stempfle D., van Belle W., Sauder U., Horn T., Dürrenberger M., Paro R., Boutros M., Merdes G. 2010. A combined ex vivo and in vivo RNAi screen for notch regulators in Drosophila reveals an extensive notch interaction network. Dev. Cell. 18:862–876 10.1016/j.devcel.2010.03.013 [DOI] [PubMed] [Google Scholar]
- Sakata T., Sakaguchi H., Tsuda L., Higashitani A., Aigaki T., Matsuno K., Hayashi S. 2004. Drosophila Nedd4 regulates endocytosis of notch and suppresses its ligand-independent activation. Curr. Biol. 14:2228–2236 10.1016/j.cub.2004.12.028 [DOI] [PubMed] [Google Scholar]
- Saleh M.C., van Rij R.P., Hekele A., Gillis A., Foley E., O’Farrell P.H., Andino R. 2006. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat. Cell Biol. 8:793–802 10.1038/ncb1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweisguth F. 2004. Regulation of notch signaling activity. Curr. Biol. 14:R129–R138 [PubMed] [Google Scholar]
- Slagsvold T., Pattni K., Malerød L., Stenmark H. 2006. Endosomal and non-endosomal functions of ESCRT proteins. Trends Cell Biol. 16:317–326 10.1016/j.tcb.2006.04.004 [DOI] [PubMed] [Google Scholar]
- Speicher S.A., Thomas U., Hinz U., Knust E. 1994. The Serrate locus of Drosophila and its role in morphogenesis of the wing imaginal discs: control of cell proliferation. Development. 120:535–544 [DOI] [PubMed] [Google Scholar]
- Sweeney N.T., Brenman J.E., Jan Y.N., Gao F.B. 2006. The coiled-coil protein shrub controls neuronal morphogenesis in Drosophila. Curr. Biol. 16:1006–1011 10.1016/j.cub.2006.03.067 [DOI] [PubMed] [Google Scholar]
- Thompson B.J., Mathieu J., Sung H.H., Loeser E., Rørth P., Cohen S.M. 2005. Tumor suppressor properties of the ESCRT-II complex component Vps25 in Drosophila. Dev. Cell. 9:711–720 10.1016/j.devcel.2005.09.020 [DOI] [PubMed] [Google Scholar]
- Tien A.C., Rajan A., Bellen H.J. 2009. A Notch updated. J. Cell Biol. 184:621–629 10.1083/jcb.200811141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccari T., Bilder D. 2005. The Drosophila tumor suppressor vps25 prevents nonautonomous overproliferation by regulating notch trafficking. Dev. Cell. 9:687–698 10.1016/j.devcel.2005.09.019 [DOI] [PubMed] [Google Scholar]
- Vaccari T., Lu H., Kanwar R., Fortini M.E., Bilder D. 2008. Endosomal entry regulates Notch receptor activation in Drosophila melanogaster. J. Cell Biol. 180:755–762 10.1083/jcb.200708127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccari T., Rusten T.E., Menut L., Nezis I.P., Brech A., Stenmark H., Bilder D. 2009. Comparative analysis of ESCRT-I, ESCRT-II and ESCRT-III function in Drosophila by efficient isolation of ESCRT mutants. J. Cell Sci. 122:2413–2423 10.1242/jcs.046391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Struhl G. 2005. Distinct roles for Mind bomb, Neuralized and Epsin in mediating DSL endocytosis and signaling in Drosophila. Development. 132:2883–2894 10.1242/dev.01860 [DOI] [PubMed] [Google Scholar]
- Wharton K.A., Johansen K.M., Xu T., Artavanis-Tsakonas S. 1985. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 43:567–581 10.1016/0092-8674(85)90229-6 [DOI] [PubMed] [Google Scholar]
- Wilkin M.B., Baron M. 2005. Endocytic regulation of Notch activation and down-regulation (review). Mol. Membr. Biol. 22:279–289 (review) 10.1080/09687860500129778 [DOI] [PubMed] [Google Scholar]
- Wilkin M.B., Carbery A.M., Fostier M., Aslam H., Mazaleyrat S.L., Higgs J., Myat A., Evans D.A., Cornell M., Baron M. 2004. Regulation of notch endosomal sorting and signaling by Drosophila Nedd4 family proteins. Curr. Biol. 14:2237–2244 10.1016/j.cub.2004.11.030 [DOI] [PubMed] [Google Scholar]
- Wilkin M., Tongngok P., Gensch N., Clemence S., Motoki M., Yamada K., Hori K., Taniguchi-Kanai M., Franklin E., Matsuno K., Baron M. 2008. Drosophila HOPS and AP-3 complex genes are required for a Deltex-regulated activation of notch in the endosomal trafficking pathway. Dev. Cell. 15:762–772 10.1016/j.devcel.2008.09.002 [DOI] [PubMed] [Google Scholar]
- Wollert T., Wunder C., Lippincott-Schwartz J., Hurley J.H. 2009. Membrane scission by the ESCRT-III complex. Nature. 458:172–177 10.1038/nature07836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Rubin G.M. 1993. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 117:1223–1237 [DOI] [PubMed] [Google Scholar]
- Yamamoto S., Charng W.L., Bellen H.J. 2010. Endocytosis and intracellular trafficking of Notch and its ligands. Curr. Top. Dev. Biol. 92:165–200 10.1016/S0070-2153(10)92005-X [DOI] [PMC free article] [PubMed] [Google Scholar]