Abstract
The SnRK2 family members are plant-specific serine/threonine kinases involved in plant response to abiotic stresses and abscisic acid (ABA)-dependent plant development. SnRK2s have been classed into three groups; group 1 comprises kinases not activated by ABA, group 2 comprises kinases not activated or activated very weakly by ABA, and group 3 comprises kinases strongly activated by ABA. So far, the ABA-dependent kinases belonging to group 3 have been studied most thoroughly. They are considered major regulators of plant response to ABA. The regulation of the plant response to ABA via SnRK2s pathways occurs by direct phosphorylation of various downstream targets, for example, SLAC1, KAT1, AtRbohF, and transcription factors required for the expression of numerous stress response genes. Members of group 2 share some cellular functions with group 3 kinases; however, their contribution to ABA-related responses is not clear. There are strong indications that they are positive regulators of plant responses to water deficit. Most probably they complement the ABA-dependent kinases in plant defense against environmental stress. So far, data concerning the physiological role of ABA-independent SnRK2s are very limited; it is to be expected they will be studied extensively in the nearest future.
Introduction
In nature, plants are frequently exposed to harmful environmental conditions: abiotic and biotic stresses. In order to survive plants must be able to sense the changes of their environment and respond to these changes by various defense mechanisms. Stress signals are recognized and transmitted to different cellular compartments by specialized signaling pathways in which protein kinases and phosphatases are key components. Among the protein kinases involved in stress signal transduction in plants are those common to all eukaryotic organisms, for example, mitogen-activated protein kinases (MAPKs) (reviews: Cho et al., 2009; Mishra et al., 2006; Pitzschke et al., 2009; Rodriguez et al., 2010), Glycogen synthase kinase 3 (GSK3) (Jonak and Hirt, 2002; Koh et al., 2007), S6 kinase (S6K) (Mahfouz et al., 2006), as well as plant specific ones, for example, calcium-dependent protein kinases (CDPKs) (Das and Pandey, 2010; Hrabak et al., 2003; Wurzinger et al., 2011) and most of SNF1-related kinases (SnRKs).
Plant SnRKs are classified into the SNF1/AMPK family, which comprises also SNF1 (sucrose nonfermenting-1) kinase from the yeast Saccharomyces cerevisiae and mammalian AMPKs (AMP-activated protein kinases). AMPK was originally defined as a mammalian protein kinase allosterically activated by 5′-AMP, able to phosphorylate and inactivate enzymes of lipid and cholesterol synthesis (Carling et al., 1987; Hardie et al., 1989). In S. cerevisiae, the SNF1 gene was discovered by a screen for mutations that caused failure to grow on sucrose or on nonfermentable carbon sources such as glycerol and ethanol (Celenza and Carlson, 1986). It is well established now that AMPK in mammals and SNF1 in yeast are cellular energy sensors acting as central regulators of cellular energy homeostasis. They control energy balance by regulating metabolic processes and responses to different environmental and metabolic stresses. AMPK inhibits several anabolic processes (e.g., synthesis of lipids, glycogen, and proteins) and activates catabolic ones (fatty acid oxidation and glycolysis) by direct phosphorylation of enzymes controlling these processes (for review, see Hardie, 2007). Moreover, AMPK activates transcription of several genes involved in cellular adaptation to stress by modulating the activity of transcription regulators (for reviews, see Cantó and Auwerx, 2010; Hardie, 2007) and also through direct association with chromatin and phosphorylation of histone H2B (Bungard et al., 2010) and histone deacetylase (McGee et al., 2008). Disorders in AMPK functioning result in serious health problems (for reviews, see Luo et al. 2010; Poels et al., 2009; Rafalski and Brunet, 2011; Wang and Guan, 2009).
The SNF1 kinase exhibits very similar features to AMPK, including structural similarities (both are heterotrimeric), substrate specificity, mode of regulation (reversible phosphorylation of a conserved Thr in the kinase activation loop), and the role in regulation of cellular metabolism and response to environmental stresses. SNF1 activity rises dramatically during glucose deficit and also to some extent upon alkalization of yeast environment, exposure to high concentration of sodium ions, and oxidative stress (Hong and Carlson, 2007; Wilson et al., 1996). SNF1 is the main regulator of the shift from fermentation to respiratory metabolism, being a global regulator of transcription of genes involved in carbohydrate metabolism and respiration. Several excellent reviews on structural and biochemical features of AMPK and SNF1 as well as their functions have been published recently (Hardie, 2007, 2011; Hedbacker and Carlson 2008; Petranovic et al., 2010; Scott et al., 2009; Zhang et al., 2010a).
SnRKs are plant homologs of AMPK/SNF1 and similarly to them seem to be crucial in linking stress and metabolic responses (Coello et al., 2011; Halford and Hey, 2009; Hey et al., 2010; Polge and Thomas, 2006). They are divided into three subfamilies: SnRK1, SnRK2, and SnRK3 (for reviews, see Halford and Hardie, 1998; Harmon, 2003; Hrabak et al., 2003). This review is dedicated to the SnRK2 subfamily, whose role in regulation of plant response to abiotic stress was recently recognized
Brief Overview of SnRK1 and SnRK3 Subfamilies
SnRK1 subfamily
Enzymes belonging to the SnRK1 subfamily structurally and functionally resemble the yeast and animal SNF1/AMPK kinases. SnRK1s show approximately 62% amino acid sequence identity with SNF1 and AMPK in the catalytic domain and 48% overall (Halford et al., 2004). SnRK1 genes from various plants have been identified and characterized. SnRK1 kinases are involved in the regulation of global metabolism and energy status of the plant, for example, in response to low glucose/high sucrose level, dark period, hypoxia, salinity, pathogen or herbivore attack (for reviews, see Halford and Hey, 2009; Polge and Thomas, 2006). SnRK1s are postulated to control signaling pathways leading to modulation of nitrogen, sucrose, and lipid metabolism, organogenesis, and senescence. Like the AMPK/SNF1 kinases, SnRK1s regulate the above processes at the transcriptional and posttranslational levels (for reviews, see Baena-González and Sheen, 2008; Halford and Hardie, 1998; Halford et al., 2003) Several enzymes involved in basal metabolism (e.g., nitrate reductase, HMG-CoA reductase, sucrose phosphate synthase, trehalose phosphate synthase) are known to be directly phosphorylated and regulated by SnRK1s. Using various molecular biology techniques, a combination of systematic screens, as well as by analyzing public transcriptome databases the KIN10 and KIN11 protein kinases, members of the SnRK1 subfamily from Arabidopsis thaliana, have been shown to control convergent reprogramming of transcriptional response to darkness, changes of sugar level, and stress conditions (Baena-González et al., 2007). KIN10 promotes catabolism by regulating expression of a broad array of genes involved in a variety of major catabolic pathways that provide alternative sources of energy and metabolites. Conversely, a large set of genes involved in the energy-consuming ribosome biogenesis and anabolism were coordinately repressed by KIN10 (Baena-González et al., 2007). This functional analysis of Arabidopsis KIN10/11 provides evidence that SnRK1s, like yeast SNF1 and mammalian AMPK, have a central role in energy signaling. The results also reveal crucial and previously unrecognized regulatory roles of the plant SnRK1s in controlling a large number of genes encoding transcription factors, chromatin remodeling proteins, and a plethora of signal transduction components (Baena-González., 2010).
The SnRK2 and SnRK3 kinases have diverged further from SNF1/AMPK than members of the SnRK1 subfamily. These two subfamilies are specific to plants and are involved in plant development and environmental stress signaling (for reviews, see Boudsocq and Lauriere, 2005; Coello et al., 2011; Halford and Hey, 2009; Harmon, 2003; Hrabak et al., 2003).
SnRK3 Subfamily
The first identified serine/threonine protein kinase, which according to Halford and Hardie (1998) belongs to the SnRK3 subfamily, was WPK4 from wheat (Sano and Youssefian, 1994). WPK4 transcription was upregulated by light, nutrient deprivation, and cytokinins, and downregulated by sucrose (Ikeda et al., 1999; Sano and Youssefian, 1994). Results obtained by Ikeda et al. (2000) strongly suggest that WPK4 is responsible for controlling nitrogen metabolism and carbon assimilation during periods of low temperature, in which the kinase resembles the SnRK1 kinases. WPK4 orthologs are known in various plants (Chikano et al., 2001; Ohba et al., 2000); their expression is differentially and distinctly regulated by various external signals.
Today, it is considered that the SnRK3 subfamily comprises protein kinases interacting with calcineurin B-like (CBL) calcium-binding proteins. They were discovered independently by the research groups of Zhu, Kudla, and Luan. Therefore, different names are given to those enzymes in the literature: SOS2-like kinases or PKSs—protein kinases related to SOS2 (Gong et al., 2004; Guo et al., 2001) and CIPKs—CBL-interacting protein kinases (Shi et al., 1999). A similar situation occurs in the case of the calcium sensors interacting with them. In Arabidopsis, the calcineurin B-like proteins are alternatively designated as SOS3-like, SCaBPs, or CBLs (Gong et al., 2004; Halfter et al., 2000; Liu and Zhu, 1998; Shi et al., 1999). CBLs and CIPKs have been indentified in all higher plant species whose genomes have been sequenced. The Arabidopsis genome encodes 26 putative CIPK and 10 CBL proteins. CIPKs/SOS2-like kinases/PKSs directly interact with CBL/SCaBPs/SOS3-like proteins. Several studies have revealed that each CBL interacts with a subset of CIPKs and each CIPK interacts with one or more CBLs (for review, see Batistič and Kudla, 2009; Gong et al., 2004; Luan, 2009; Weinl and Kudla, 2009). An increase of Ca2+ concentration triggered by various environmental or cellular stimuli is detected by CBLs: their conformation changes upon Ca2+ binding, which in most cases results in activation of the CIPKs interacting with them. The CIPKs play an important role in the plant responses to drought, cold, ABA, sugar, salt, and pH changes (for reviews, see Coello et al., 2011; Gong et al., 2004; Hrabak et al., 2003; Luan, 2009). The best known signaling pathway in which CIPKs are involved is the SOS (salt overly sensitive) pathway regulating sensitivity to salinity in A. thaliana. The SOS3–SOS2 kinase complex, which is required for intracellular Na+ and K+ homeostasis and plant tolerance to high Na+ and low K+ environments, appears to control the expression of SOS1 and the activity of SOS1 protein, a plasma membrane Na+/H+ exchanger (antiporter) (Zhu, 2003). Moreover, SOS2 positively regulates activities of two vacuolar membrane transporters, the Na+/H+ antiporter AtNHX1 (Qiu et al., 2004) and the Ca2+/H+ antiporter CAX1 (Cheng et al., 2004), and negatively regulates the highly selective Na+ transporter AtHKT1 (Rus et al., 2001). The SOS pathway was found to control expression of several salt stress-specific tolerance determinant genes (Gong et al., 2001).
The data concerning the involvement of SnRK1 and SnRK3 kinases in stress response are combined in excellent reviews by Hrabak et al. (2003), Chinnusamy et al. (2004), Boudsocq and Lauriere (2005), Baena-González and Sheen (2008), Halford and Hey (2009), and Coello et al. (2011).
SNRK2 Subfamily
Until very recently not much attention was paid to this subfamily. Before the year 2000, there was basically no information concerning SnRK2 function, only a few data indicating that ABA and abiotic stresses induce the expression of some SnRK2 genes were published (Anderberg and Walker-Simmons, 1992; Holappa and Walker-Simons, 1995). In the year 2000, SnRK2s started to be recognized as enzymes involved in abiotic stress signal transduction in plants. That year two articles showing rapid activation of SnRK2 kinases in response to abiotic stresses were published; activation of Nicotiana tabacum osmotic stress-activated protein kinase (NtOSAK) in response to hyperosmotic stress in tobacco BY-2 cells (Mikołajczyk et al., 2000) and AAPK (ABA-activated protein kinase) by ABA in response to drought in guard cells of fava bean (Vicia faba) (Li et al., 2000) were presented. Li et al. (2000) showed that AAPK is involved in regulation of anion channels and stomatal closure. Studies during the following years extended our knowledge on SnRK2s—their structure, mechanism of activation, and function. The year 2009 was a very special year for understanding SnRK2s function. In 2009, independently, two laboratories obtained a triple snrk2.2/3/6 mutant, which allowed to establish the role of ABA-dependent SnRK2s in plant response to water deficit, seed maturation, and germination (Fujii and Zhu, 2009; Fujita et al., 2009; Nakashima et al., 2009). Moreover, the same year soluble ABA receptors were discovered, and this discovery led to understanding early events of ABA signaling pathway, in which SnRK2 kinases are involved. Now, after 10 years of studies, the SnRK2 subfamily members are considered as major players in plant response to osmotic stress and ABA-dependent plant development (Boudsocq and Lauriere, 2005; Fujii et al., 2007, 2011; Fujii and Zhu, 2009; Fujita et al., 2009; Nakashima et al., 2009). The SnRK2 kinases were identified in every plant studied. Ten members of the SnRK2 family are encoded by the Arabidopsis as well as rice and sorghum genomes (Boudsocq et al., 2004; Kobayashi et al., 2004; Li et al., 2010). Eleven SnRK2s have been identified in maize (Huai et al., 2008), and some SnRK2 representatives were described for tobacco (Kelner et al., 2004), wheat (Anderberg and Walker-Simmons, 1992; Holappa and Walker-Simons, 1995; Mao et al., 2010; Zhang et al., 2010b, 2011), soybean (Monks et al., 2001), fava bean (Li and Assmann, 1996; Li et al., 2000), and algae (Davies et al., 1999; Gonzalez-Ballester et al., 2008). Through transient expression in protoplasts it was shown that all Arabidopsis thaliana SnRK2s, except SnRK2.9, are rapidly activated by treatment with different osmolytes, such as sucrose, mannitol, sorbitol, or NaCl, and some of them also by abscisic acid (ABA), suggesting that these kinases are involved in a general response to osmotic stress (Boudsocq et al., 2004, 2005). In rice, all 10 members are activated by hyperosmotic stress and three of them also by ABA (SAPK8, SAPK9, and SAPK10). Rice SnRK2 kinases were designated SAPK1 through SAPK10, which stands for osmotic Stress/ABA-activated Protein Kinase (Kobayashi et al., 2004). SAPK3 is identical with the previously reported rice Ser/Thr protein kinase REK (Hotta et al., 1998). SAPK1 and SAPK2 are highly homologous to wheat (Triticum aestivum) PKABA1 (protein kinase ABA1) (Anderberg and Walker-Simmons, 1992; Holappa and Walker-Simmons, 1995), and barley (Hordeum vulgare) HvPKABA1 (Yamaguchi et al., 2002).
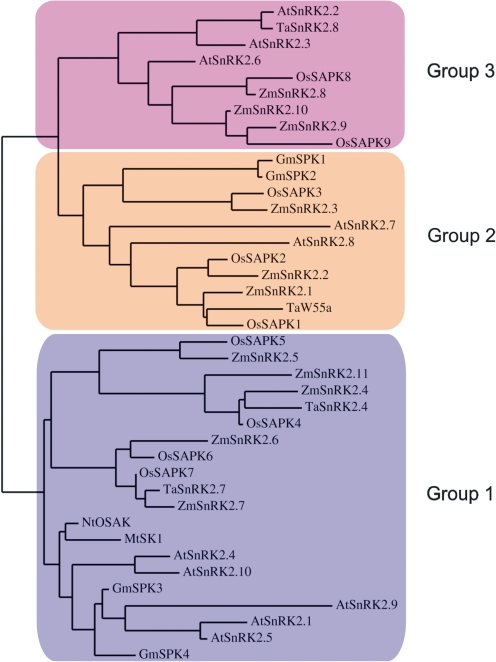
Based on a phylogenetic analysis, the SnRK2 family members have been divided into three groups (Fig. 1). Group 1 comprises kinases not activated by ABA, group 2—not activated or activated very weakly by ABA (depending on plant species), and group 3—strongly activated by ABA. Evolutional studies based on sequence data from algae (Chlamydomonas reinhardtii), mosses Physcomitrella patens and Selaginella moellendorffi, and several angiosperm genomes (Arabidopsis, poplar, rice) showed that all higher plants have all three groups of SnRK2, P. patens has only SnRK2s belonging to group 3, and S. moellendorffi kinases belonging to groups 2 and 3. Chlamydomonas reinhardtii SnRK2s are distinct from those of land plants (Mizoguguchi et al., 2010; Umezawa et al., 2009; and reviews by Hauser et al., 2011; Umezawa et al., 2010).These data indicate that group 3 is an ancient form of land plant SnRK2, whereas group 1 is the most recent. Another classification of SnRK2s can also be found in the literature (Halford and Hardie, 1998), which now has only historical meaning. In that classification the SnRK2 kinases were divided into two classes, SnRK2a and SnRK2b, based on the amino acid composition of their divergent C-terminal domains rich in acidic amino acids, either poly-Asp (SnRK2a) or poly-Glu (SnRK2b). Groups 2 and 3 of the phylogenic classification correspond to the SnRK2a subfamily, whereas group 1 to SnRK2b (Fig. 2).
FIG. 1.
Phylogenetic tree of selected higher plant SnRK2s. Group 1— ABA-independent kinases, group 2—kinases not dependent or weakly dependent on ABA, group 3—ABA-dependent kinases. The unrooted UPGMA (unweighted pair group method with arithmetic mean) tree was built using ClustalW2 and presented using TreeView.
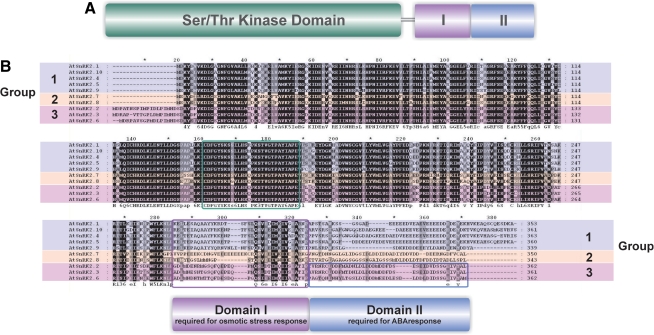
FIG. 2.
Structure of Arabidopsis thaliana SnRK2s. (A) Model of SnRK2 structure. Ser/Thr kinase domain—kinase catalytic domain, I—domain involved in ABA-independent activation in response to osmotic stress, characteristic for all SnRK2, II—domain needed for ABA-dependent activation of SnRK2s. (B) Alignment of Arabidopsis thaliana SnRK2s with characteristic domains marked: activation loop (green box), domain I (violet box), and domain II (blue box). Gaps introduced to maximize alignment are marked with dashes.
Structure of SnRK2
SnRK2s are monomeric serine/threonine protein kinases with a molecular weight around 40 kDa (Hardie, 1999; Mikołajczyk et al., 2000). Recently, crystals of mutated forms of SRK2E/OST1/SnRK2.6 unable to bind ATP were obtained (Yunta et al., 2011). Only preliminary X-ray analysis has been performed so far and the structure should be solved soon.
The amino acid sequences of all SnRK2s can be divided into two regions, the N-terminal highly conserved kinase domain, related to those of SNF1 and AMPK, and the regulatory C-terminal domain containing stretches of acidic amino acids, either Glu (group 1) or Asp (groups 2 and 3). Further, the C-terminal domain consists of two subdomains, Domain I and Domain II (Fig. 2). Domain I (about 30 amino acids starting from the kinase domain) is characteristic for all SnRK2 family members and is needed for activation by osmotic stress, independently of ABA, whereas Domain II (about 40 amino acids just after Domain I) is specific to the ABA-dependent SnRK2s only and is needed for the ABA response (Belin et al., 2006; Kobayashi et al., 2004; Yoshida et al., 2006). Yoshida et al. (2006) proposed a model in which Domain II of SRK2E/OST1/SnRK2.6 kinase was responsible for the kinase activation by ABA and interaction with the ABI1 phosphatase. Recently, several independent investigations have confirmed the interaction between ABA-dependent SnRK2s (from group 3) with members of the clade A PP2C phosphatases, among them ABI1 (Umezawa et al., 2009; Vlad et al., 2009), and have shown that these phosphatases are negative regulators of the ABA-dependent SnRK2s.
Regulation of SnRK2 activity
Results presented by several laboratories provide evidence that reversible phosphorylation in the kinase activation loop is responsible for regulation of SnRK2 activity (Belin et al., 2006; Burza et al., 2006; Boudsocq et al., 2007; Kobayashi et al., 2004; Mikołajczyk et al., 2000). In 2006, our group demonstrated that in response to osmotic stress the activity of NtOSAK (Nicotiana tabacum osmotic stress-activated protein kinase), a tobacco ABA-independent SnRK2 (member of group 1), is regulated by reversible phosphorylation of Ser154 and Ser158 within the kinase activation loop. Site-directed mutagenesis studies showed that phosphorylation of Ser-158 was crucial for the kinase activity, whereas phosphorylation of Ser-154 most probably facilitated the phosphorylation on Ser-158 (Burza et al., 2006). Several other laboratories also showed the crucial role of phosphorylation on a residue corresponding to Ser-158 of NtOSAK in activation of different members of the SnRK2 subfamily (Belin et al., 2006; Boudsocq et al., 2007; Chae et al., 2007). Using Pro-Q Diamond, a specific dye for phosphoproteins, Boudsocq et al. (2007) showed that all activated SnRK2s studied were phosphorylated in response to osmotic stress but the induced changes of phosphorylation were clearly different depending on the kinase. In addition, the increase of the global phosphorylation level induced by ABA application was lower than in the case of osmotic stress, suggesting that different phosphorylation mechanisms may be involved in SnRK2 activation by hyperosmolarity and ABA (Boudsocq et al., 2007). These differences have been recently confirmed (Vlad et al., 2010). These authors studied changes of phosphorylation of two SnRK2 kinases, SnRK2.6/OST1 (ABA-dependent kinase from group 3) and SnRK2.10 (ABA-independent kinase from group 1), in response to osmotic stress and ABA. They indentified two important phosphorylation sites within the activation loops of these kinases, one of them critical for the catalytic activity and both necessary for the full activation by hyperosmolarity or ABA of both SnRK2s studied. It was also shown that SnRK2.6 was most probably activated through independent phosphorylation of these two sites, whereas a sequential process occurred in SnRK2.10, phosphorylation of one serine being required for the phosphorylation of the other. The results, indicating that SnRK2.10 is activated by sequential phosphorylation of two serines in the kinase activation loop, are in agreement with our previous data concerning the mechanism of NtOSAK activation (Burza et al., 2006).
So far, an upstream kinase(s) phosphorylating and activating SnRK2s has not been identified. Some authors suggest that SnRK2s can be activated by autophosphorylation (Fujii et al., 2009); however, this issue remains controversial.
Even though the kinases upstream of SnRK2s are still not known, several negative regulators of SnRK2s' activity have been identified. All SnRK2s are inactivated by dephosphorylation; therefore, obviously specific phosphatases should act as their negative regulators. Recently, it has been shown that members of clade A PP2C-type protein phosphatases interact with ABA-dependent SnRK2s and inactivate them efficiently via dephosphorylation of Ser and Thr residues in the kinase activation loop (Umezawa et al., 2009; Vlad et al., 2009). This discovery was reported at the same time as the identification of ABA receptors—RCAR/PYR/PYL (RCAR, regulatory component of ABA Receptor/PYR1, pyrabactin resistance 1/PYL, PYR1-like) (for reviews, see Cutler et al., 2010; Hubbard et al., 2010; Klingler et al., 2010; Melcher et al., 2010; Umezawa et al., 2010; Weiner et al., 2010). The identification of the ABA receptors was a turning point in studies on ABA signaling and on the regulation of SnRK2s activity. The phosphatases from group A of the PP2C family form a complex with RCAR/PYR/PYL proteins, sometimes they are even called ABA coreceptors. In the absence of ABA, phosphatases belonging to the clade A PP2C interact with and dephosphorylate ABA-activated SnRK2 kinases (from group 3), keeping them in an inactive form. Upon ABA binding the conformation of the RCAR/PYR/PYL-PP2C complex changes resulting in inactivation of the phosphatase and its release from the SnRK2 kinase active site. This allows activation of the kinase (Hirayama and Umezawa, 2010; Joshi-Saha et al., 2011; Ma et al., 2009; Melcher et al., 2009; Nishimura et al., 2009, 2010; Park et al., 2009; Santiago et al., 2009; Umezawa et al., 2009, 2010; Vlad et al., 2009). It should be stressed that the above-mentioned mechanism was described only for the ABA-dependent SnRK2s belonging to group 3. Kinases from group 2 (Arabidopsis SnRK2.7 and SnRK2.8) interact with PP2C phosphatases as well (Umezawa et al., 2009), but so far their inactivation by PP2C in vivo has not been shown. The ABA-independent SnRK2s (group 1) do not interact with PP2C phosphatases in the yeast two hybrid system (Umezawa et al., 2009; Vlad et al., 2010, our unpublished results).
Recently, another negative regulator of SnRK2, SCS (SnRK2-interacting Calcium Sensor), has been identified in our laboratory (Bucholc et al., 2010). SCS is a plant-specific calcium-binding protein that interacts with all SnRK2s studied by us, both ABA-dependent and ABA-independent, and inhibits their activity. These results present not only a new mechanism of regulation of SnRK2s activity but also a link between SnRK2 and Ca2+ signaling.
Besides phosphorylation, also other posttranslational modifications can possibly regulate SnRK2 activity in plant cells. It has been demonstrated that nitric oxide (NO) induces the activity of NtOSAK in tobacco cells and can be a key component in NtOSAK activation in response to hyperosmotic and salinity stresses (Lamotte et al., 2006; Wawer et al., 2010). This suggested that presumably S-nitrosylation of NtOSAK could be involved in the regulation of its activity. However, S-nitrosylation studies have indicated that cysteine residues of NtOSAK do not undergo direct chemical modifications; it is more likely that some of the S-nitrosylated cellular partners of the kinase, for example, GAPDH (glyceraldehyde-3-phosphate dehydrogenase) are responsible for the regulation of the activity of NtOSAK by NO (Wawer et al., 2010) by an as-yet unknown mechanism. So far, it is not known if NO is involved in the regulation of the ABA-dependent SnRK2s; however, it is probable because NO is a key element of ABA-dependent stomatal closure (for review, see Neill et al., 2008).
It should also be mentioned that an ABA-independent kinase, SnRK2.10 has been identified among proteins interacting with phosphatidic acid (Testerink et al., 2004); therefore, it cannot be ruled out that this phospholipid, whose role in stress signal transduction is well documented, can be a regulator of at least some of the SnRK2 subfamily members.
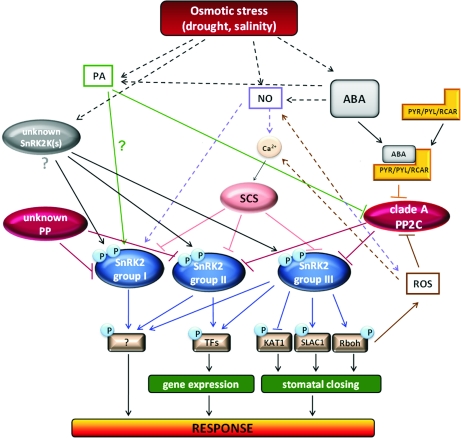
Possible posttranslational modifications involved (or most probably involved) in regulation of SnRK2 kinases activity are presented in Figure 3.
FIG. 3.
Simplified model of SnRK2s pathways involved in plant response to osmotic stress. Model was done based on published and unpublished data described in the main text. ABA, abscisic acid; NO, nitric oxide; PA, phosphatidic acid; ROS, reactive oxygen species; unknown SnRK2K(s), an SnRK2 upstream kinase(s)— still unknown; unknown PP, an unknown phosphoprotein phosphatase dephosphorylating and inactivating ABA-independent SnRK2s; clade A PP2C, clade A PP2C phosphoprotein phosphatase; SCS, SnRK2-interacting calcium sensor; PYR/PYL/RCAR, soluble ABA receptor; Rboh, plasma membrane NADPH oxidase; SLAC1, Slow Anion Channel 1; KAT1, inward-rectifying potassium channel; TFs, transcription factors. Connections represent positive (arrow) and negative (block) regulation. Bold line—direct regulation/interaction, dashed line—indirect regulation. Question marks indicate currently unknown or not confirmed regulation.
Regulation of SnRK2s expression
SnRK2s are primarily regulated at the posttranslational level, by phosphorylation. However, expression of genes encoding these kinases is also regulated in response to changes of the plant environment. Kabayashi et al. (2004) showed that expression of various Oryza sativa SnRK2s is differently regulated by ABA, mannitol, and NaCl in different plant organs (roots, blades, and sheaths); some of them are upregulated, whereas others, on the contrary, are downregulated. In rice, high salt significantly induces SAPK4 expression (Diédhiou et al., 2008). Triticum aestivum PKABA1 (protein kinase induced by abscisic acid 1), the first cloned SnRK2 from wheat, as well as other wheat SnRK2 genes: TaSnRK2.4, TaSnRK2.7, and TaSnRK2.8 are upregulated in response to plant treatment with different stressors. Their expression is induced by polyethylene glycol, NaCl, and cold, and additionally expression of TaSnRK2.4 (from group 1), TaSnRK2.8 (from group 3), and PKABA1 (from group 2) is upregulated by ABA (Anderberg and Walker-Simmons, 1992; Holappa and Walker-Simons, 1995; Mao et al., 2010; Zhang et al., 2010b, 2011). A recent report by Xu et al. (2009) concerned expression of the W55a gene, encoding a protein kinase belonging to the SnRK2 family from wheat, whose amino acid sequence is the most homologous to rice SAPK1 (group 2 of SnRK2s). Expression of W55a was upregulated not only by drought, salt, and exogenously applied ABA, but also by salicylic acid, ethylene, and methyl jasmonate, whereas it was not responsive to cold stress. In maize, expression of SnRK2s genes was enhanced in response to NaCl (ZmSnRK2.3, ZmSnRK2.6, and ZmSnRK2.4), ABA (ZmSnRK2.2, ZmSnRK2.4, ZmSnRK2.5, ZmSnRK2.7, and ZmSnRK2.10), and cold (ZmSnRK2.3, ZmSnRK2.7), whereas expression of some of them (ZmSnRK2.5, ZmSnRK2.6 and ZmSnRK2.9) was suppressed by heat (Huai et al., 2008; Zou et al., 2006). Hyperosmolarity (dehydration and high salinity) induces the expression of four soybean (Glycine max) SnRK2 genes, SPK1, SPK2, SPK3, and SPK4 (serine/threonine protein kinase 1, 2, 3, and 4) (Monks et al., 2001; Yoon et al., 1997).
It should also be mentioned that expression of at least some SnRK2s can be regulated by the light–dark cycle, for example, SnRK2.8 (Shin et al., 2007), SnRK2.4 and SnRK2.1 (Park et al., 1993).
The physiological role of SnRK2 subfamily members; SnRK2 downstream targets
Ample data indicate that SnRK2s are positive regulators of plant response to drought. AAPK (ABA-activated protein kinase) is activated by ABA in guard cells of fava bean (Vicia faba) in response to drought and is involved in the regulation of anion channels and stomatal closure (Li and Assmann, 1996; Li et al., 2000). The closest homolog of AAPK in A. thaliana is the SRK2E/OST1/SnRK2.6 protein kinase. So far, this kinase is the most broadly studied kinase in respect to SnRK2 role in plant response to environmental stresses. The srk2e/ost1/snrk2.6 mutant cannot cope with a rapid decrease in humidity and has a wilty phenotype (Yoshida et al., 2002). Loss of the kinase function causes an ABA-insensitive phenotype in stomatal closure. It has been shown that OST1 mediates the regulation of stomatal aperture by ABA (Mustilli et al., 2002; Yoshida et al., 2002) and acts upstream of reactive oxygen species (ROS) production (Mustilli et al., 2002). ROS are essential intermediates in ABA signaling in guard cells. They are needed for regulation of Ca2+ influx by activation of Ca2+ channels (Kwak et al., 2003; Murata et al., 2001). It is well established that Ca2+ influx is required for stomatal closure. Two partially redundant guard cell NADPH oxidases (AtrbohD and AtrbohF) function in ROS-dependent ABA signaling in Arabidopsis (Kwak et al., 2003). Therefore, it is very likely that ABA-dependent regulation of ROS production by OST1 is achieved through phosphorylation of the plasma membrane NADPH oxidases. In the case of AtrbohD, it was shown that phosphorylation and Ca2+ binding is needed for its activation (Ogasawara et al., 2008). Sirichandra et al. (2009) showed that the N-terminal part of the recombinant AtrbohF produced in a bacterial system is phosphorylated by OST1 in vitro. Moreover, OST1 interacts with AtrbohF in bimolecular fluorescence complementation (BIFC) assay, suggesting that NADPH oxidase could be an SnRK2.6/OST1 substrate in vivo. However, there are no direct data so far presenting phosphorylation and regulation of Atrbohs activity by SnRK2 in planta. Specific ion channels of the plasma membrane of guard cells, activated in response to ABA are responsible for the ABA-dependent stomatal closure. Recently, a guard cell slow-anion channel 1 (SLAC1), functioning in stomatal response to high CO2 and ABA, was identified and characterized at the molecular level (Negi et al., 2008; Vahisalu et al., 2008). It appeared that SLAC1 is a critical channel of guard cells, responsible for stomatal movements. It was shown that the activity of this channel was regulated by reversible phosphorylation, the phosphorylated form being the active one. It was established that OST1 together with CDK21 and possibly CDPK23 were responsible for the phosphorylation and activation of SLAC1 (Lee et al., 2009; Geiger et al., 2009, 2010). Another ion channel in Arabidopsis guard cells mediating the aperture of stomata pores is an inward-rectifying potassium channel KAT1 (Pilot et al., 2001). The activity of this channel is also regulated by phosphorylation catalyzed by SnRK2.6/OST1 and most probably by CDPK (Sato et al., 2009). However, in the case of KAT1, the phosphorylation has a negative effect on its activity. It should be stressed, however, that in contrast to SLAC1, downregulation of KAT1 activity is needed for stomatal closure. The above results indicate that SnRK2.6/OST1 phosphorylates both anion (SLAC1) and kation (KAT1) channels that are crucial for stomatal movements, and these phosphorylations are required for the ABA-dependent stomatal closing in response to water deficit. Very recently two other reports concerning OST1, SLAC1 and stomatal closing were published. It was shown that OST1 phosphorylates and activates SLAC1 not only in response to ABA, but also in response to elevated level of intercellular CO2 (Xue et al. 2011) or ozone (Vahisalu et al., 2010). Moreover, OST1 is responsible for stomata closing in response to pathogens (Melotto, 2006). The regulation of plant response to water stress by SnRK2.6/OST1 is not restricted to the regulation of stomata movement. Already in 2002, Yoshida et al. presented results indicating that this kinase is responsible for the positive regulation of expression of several stress response genes, for example, RD29B and RD22. Now, our knowledge on the regulation of gene expression by SnRK2s is significantly broader.
Some functional redundancy between SnRK2.6, SnRK2.2, and SnRK2.3 has been postulated. All of them are members of group 3 (ABA-dependent SnRK2s) of the SnRK2 family and are key regulators of plant response to ABA. The Arabidopsis triple mutant snrk2.2/snrk2.3/snrk2.6, also known as srk2d/i/e, disrupted in three ABA-dependent SnRK2s (SnRK2.2/SRK2D, SnRK2.3/SRK2I, and SnRK2.6/SRK2E), is extremely insensitive to ABA and exhibits greatly reduced tolerance to drought, not only by the disruption of stomatal closing but also as a result of downregulation of ABA- and water stress-induced genes (Fujii and Zhu, 2009; Fujita et al., 2009). Studies involving this mutant indicate that these kinases are major regulators of ABA signaling in response to water stress. The intensity of the triple mutant phenotype is far stronger than that of any of the single or double mutants. Previous studies using snrk2.2, snrk2.3, and double snrk2.2/snrk2.3 T-DNA knockout mutants indicated that SnRK2.2 and SnRK2.3 kinases were involved in the regulation of seed dormancy and germination (Fujii et al., 2007). Double mutant snrk2.2snrk2.3 was insensitive to ABA in terms of seed dormancy, germination, and seedling growth, in contrast to the snrk2.6 mutant (Fujii et al., 2007; Yoshida et al., 2002). Moreover, snrk2.2snrk2.3 was barely affected in leaf water loss, suggesting that SnRK2.2 and SnRK2.3 have only minor roles in the control of stomatal movement in drought response, which further distinguished their function from that of SnRK2.6 (Fujii et al., 2007). However, the phenotype of the triple mutant clearly showed that all three kinases are partially redundant and all of them are important for ABA responses, both in terms of water stress response, as well as ABA-dependent developmental processes (seed development and dormancy) (Fujii and Zhu, 2009; Fujita et al., 2009; Nakashima et al., 2009).
As mentioned above, OST1/SnRK2.6, as well as other ABA-dependent SnRK2s, are involved in the regulation of expression of stress–response genes. The Arabidopsis triple mutant snrk2.2/snrk2.3/snrk2.6 showed strong impairment of ABA- and water stress-dependent gene expression under water deficit conditions (Fujita et al., 2009). On the other hand, jasmonic acid-dependent genes and those involved in regulation of flowering were upregulated. Those authors showed that expression of almost all genes encoding dehydration-responsive late embryogenesis abundant (LEA) proteins as well as those encoding group A PP2C (negative regulators of the SnRKs activity) were strongly downregulated in the snrk2.2/snrk2.3/snrk2.6 mutant under water stress (NaCl or ABA treatment). Similar results were also obtained when the mutant was analyzed in terms of regulation of gene expression during seed development and dormancy. Stress-related genes were downregulated in this mutant in comparison to the wild type, whereas those related to photosynthesis and tetrapyrrole synthesis were upregulated (Nakashima et al., 2009). It was concluded that ABA-dependent SnRK2s (SnRK2.2, SnRK2.3, and SnRK2.6) are essential for the control of seed development and dormancy through an extensive control of gene expression.
ABA-regulated genes have conserved ABA-responsive elements (ABRE) in their promoters, usually in several copies. The transcription factors which bind to ABRE and trigger transcription of the ABA-regulated genes, known as ABRE-binding proteins (AREBs) or ABRE-binding factors (ABFs), are regulated by phosphorylation. Furihata et al. (2006) showed that Arabidopsis ABA-dependent SnRK2s from group 3 (SnRK2.2, SnRK2.3, and SnRK2.6) phosphorylate ABFs in vitro at sites crucial for their activity. That result was confirmed by Fujii et al. (2007). Later on it was shown that these kinases play an essential role in phosphorylation of ABFs, including ABI5 during seed maturation and germination (Nakashima et al., 2009), and also in response to environmental stress. There are several examples of phosphorylation of ABFs by SnRK2s also in plants other than Arabidopsis (Johnson et al. 2002; Kobayashi et al., 2005). A rice ABF, TRAB1, has been shown to be activated via ABA-dependent phosphorylation. SAPK8, SAPK9, and SAPK10 (orthologs of SnRK2.2, SnRK2.3, and SnRK2.6) directly phosphorylate TRAB1 in response to ABA (Kobayashi et al., 2005). Not only genes with an ABRE in their promoters are likely to be regulated by SnRK2s. For example, in both the snrk2.6 and obviously in the snrk2.2/snrk2.3/snrk2.6 mutant an impairment of RD22 expression was observed (Fujii and Zhu, 2009; Yoshida et al., 2002). Expression of RD22 is controlled by Myc and Myb transcription factors; therefore, it can be speculated that also the activity of these transcription factors is modulated by SnRK2 phosphorylation. More information on the ABA-mediated transcription regulation in response to osmotic stress in plants, including regulation by SnRK2, can be found in a very recent review by Fujita et al. (2011).
There are indications that SnRK2s from group 3 are involved in the regulation of plant metabolism. Recently, Zheng et al. (2010) showed that SnRK2.6 participates in the regulation of oil synthesis in Arabidopsis seeds, sucrose synthesis and fatty acid desaturation in leaves.
Much less is known on the physiological role of SnRK2s from group 2 (SnRK2.7 and SnRK2.8 in Arabidopsis). Umezawa et al. (2004) showed that SKR2C/SnRK2.8 plays a significant role in drought tolerance. Arabidopsis knockout mutants srk2c/snrk2.8 were hypersensitive to drought, whereas Arabidopsis transgenic plants overexpressing SRK2C/SnRK2.8 were significantly more tolerant to drought than wild-type plants. The kinase functions mainly in root tips and apparently does not affect stomatal movement, in contrast to SRK2E/SnRK2.6/OST1, because the water-loss level of the mutant was similar to that of wild-type plants. On the other hand, root elongation and lateral root number were significantly lower in the srk2c/snrk2.8 mutant than in wild-type plants. DNA microarray analysis of 35S::SRKC/SnRK2.8-GFP and control plants revealed upregulation of many stress-responsive genes, for example, RD29A, COR15A, DREB1A in transgenic plants overexpressing SRKC/SnRK2.8, indicating that, most probably, the regulation of water stress tolerance by SnRK2.8 is due to the control of stress-responsive gene expression. Recently, an analysis of the expression profiles of snrk2.7/snrk2.8 double mutant and wild-type seedlings exposed to drought stress confirmed previous data indicating that kinases from group 2 of the SnRK2 subfamily play an important role in drought response (Mizoguchi et al., 2010). The results indicated that SnRK2.7 and SnRK.8 upregulate the expression of some AREB/ABF target genes, albeit to an extent lower than the SnRK2.2/2.3/2.6 kinases. Those authors showed that SRK2C/SnRK2.8 interacts with an ABA-related bZIP protein, EEL. SRK2C/SnRK2.8 and EEL are coexpressed in root tips and SRK2C/SnRK2.8 phosphorylates EEL as well as another bZIP protein (ABF1) in vitro (Mizoguchi et al., 2010). These data indicate that EEL and maybe some other bZIP proteins could be cellular substrates of SnRK2.8. Johnson et al. (2002) have identified TaABF, a wheat member of the ABF family, that is specifically bound and phosphorylated by wheat kinase PKABA1 (from group 2 of the SnRK2 subfamily). The data suggested that TaABF may serve as a physiological substrate for PKABA1 in the ABA signal transduction pathway during grain maturation, dormancy, and could lead to suppression of germination-associated gene expression and germination itself.
Recent analyses have addressed the question about the role of group 2 and 3 SnRK2s from wheat. cDNAs encoding wheat kinases were expressed in Arabidopsis and stress tolerance analysis of the obtained transgenic plants was performed. Expression of TaSnRK2.8 (from group 3) or TaSnRK2.4 (from group 2) in Arabidopsis enhanced the plant tolerance to drought, salinity, and low temperature (Mao et al., 2010; Zhang et al., 2010b). Expression of TaSnRK2.4 resulted in delayed seedling establishment, longer primary roots, and higher yield in normal growth condition. The authors also suggest that TaSnRK2.4 and TaSnRK2.8 are involved in the regulation of carbon metabolism, which is closely connected with stress response. Independently, Xu et al. (2009) showed that transgenic Arabidopsis plants in which W55a (a wheat SnRK2 belonging to group 2) has been expressed exhibited higher tolerance to drought. Even though recent studies indicate that some SnRK2s belonging to group 2, for example, Arabidopsis SnRK2.7 and SnRK2.8, are weakly activated by ABA treatment and upregulate expression of some AREB/ABF target genes in response to stress (drought, NaCl), they do not regulate gene expression in response to direct plant treatment with ABA; therefore, their role in ABA signaling is still unknown.
There are several indications that SnRK2s from group 2 are involved in the regulation of plant metabolism. SnRK2.8 is a positive regulator of plant growth, most probably by phosphorylation and regulation of the activity of several enzymes involved in basic metabolism (Shin et al., 2007). There is evidence that expression of SnRK2.8 is downregulated by N, P, or K deprivation, and this downregulation is accompanied by poor plant growth, whereas overexpression of cDNA encoding this kinase results in improved growth of Arabidopsis under normal and nutrient-limited conditions (Shin et al., 2007). Several target proteins have been identified as being phosphorylated by SnRK2.8. Certain targets, such as 14-3-3 proteins, regulate as yet unidentified proteins, whereas other targets, such as glyoxalase I and ribose 5-phosphate isomerase, detoxify byproducts of glycolysis and catalyze one of the final steps in carbon fixation, respectively. Also, adenosine kinase and a 60S ribosomal protein were confirmed as targets of SnRK2.8.
So far, information on the role of ABA-independent SnRK2 family members (group 1) is scant. Diédhiou et al. (2008) have revealed that overexpression of SAPK4 (a rice SnRK2 from group 1) increases rice tolerance to salinity and oxidative stress. The authors studied seed germination as well as development of rice seedlings and growth of mature plants under salt stress. In plants overexpressing SAPK4 transcript levels of the vacuolar H+-ATPase and catalase were higher in comparison to wild-type plants, whereas those encoding the vacular Na+/H+ antiporter OsNHX1 and the Cl− channel OsCLC1 were lower. Most likely that was why the transgenic plants exhibited lower oxidative stress and accumulation of Na+ and Cl− ions and overall enhanced salt tolerance (Diédhiou et al., 2008). Zhang et al. (2011) reported that TaSnRK2.7 encoding an ABA-independent kinase expressed in Arabidopsis enhanced plant drought, cold, and salinity tolerance. It was also shown that two soybean (Glycine max) ABA-independent SnRK2s, SPK1, and SPK2, were able to phosphorylate soybean phosphoinositide binding protein Ssh1p, that may function as a component of the hyperosmotic stress response pathway (Kearns et al., 1998; Monks et al., 2001). These examples suggest that ABA-independent SnRK2s act as positive regulators of plant response to abiotic stresses. However, they may have other cellular functions as well. Several years ago it was found that MtSK1, a gene encoding an SnRK2 kinase from group 1 in Medicago truncatula, was involved in the induction of somatic embryogenesis (Nolan et al., 2006).
Very recently, an extremely difficult large scale experiment leading to establishing the role of SnRK2s, including the ABA-independent members, was undertaken by Fujii et al. (2011). They constructed and characterized Arabidopsis decuple mutants carrying mutations in all 10 SnRK2 genes, mutants defective in SnRK2.1/4/5/7/8/9/10 and also quadruple snrk2.1/4/5/10 and quintuple snrk2.1/4/5/9/10 mutants. First of all, the results suggest that all SnRK2s are important in plant response to osmotic stress. The features of those mutants (expression level of selected stress-response genes, ABA, and proline production in response to osmotic stress and ABA treatment) have revealed some differences between the mutants studied. The analysis of proline level, for example, indicated that ABA-independent SnRK2s (SnRK2.1/4/5/9/10) could act as negative regulators of ABA-induced proline accumulation. This feature is exactly opposite to that of the ABA-dependent kinases from group 3. However, because the data concerning functions of ABA-independent members of the SnRK2 subfamily is scarce, it is very difficult to make any general statements in terms of their function; a lot of work remains to be done to establish the role of the ABA-independent SnRK2s.
In algae, several SnRK2s involved in response to abiotic stress have been identified. Two Chlamydomonas reinhardtii SnRK2 kinases, SNRK2.1 and SNRK2.2 (previous name Sac3) are involved in response to sulfur limitation (Davies et al., 1999; Gonzalez-Ballester et al., 2008). SNRK2.1 positively regulates sulfur-starvation responsive genes, for example, SAC-like Transporter 1 (SLT1), SAC-like Transporter 2 (SLT2), Sulfate Transporter 2 (SULTR2) needed for maintaining cell viability during S-deprivation (Gonzalez-Ballester et al., 2008; Pootakham et al., 2010). SNRK2.2, on the other hand, is responsible for repression of S-inducible genes when cells are depleted of S (Davies et al., 1999; Gonzalez-Ballester et al., 2008). It seems likely that also in higher plants at least some SnRK2s are involved in sulfur metabolism as well. Kimura et al. (2006) showed that mRNA levels of four Arabidopsis kinases, both ABA-dependent and ABA-independent, SnRK2.1, SnRK2.3, SnRK2.4, and SnRK2.6, increased in response to sulfur limitation. Additionally, it was shown that SnRK2.3 controls the production of O-acetyl-L-serine, a putative signaling compound of the sulfur starvation response, and expression of the sulfate transporter SULTR2-2 gene under limited sulfur supply (Kimura et al., 2006). Taking into consideration that in lower plants and most probably also in higher plants SnRK2s are involved in sulfur metabolism, which is closely connected with regulation of heavy metal tolerance, one can expect that SnRK2s might be involved not only in the plant response to water stress and nutrient deprivation but also in the response to heavy metal stress.
Conclusions
All presented findings indicate a key role of SnRK2 kinases in the regulation of plant response to abiotic stresses and nutrient limitation in an ABA-dependent and independent manner. Similarly to SnRK1, they link ABA and stress signaling with plant metabolism. The ABA-dependent kinases belonging to group 3 of the SnRK2 subfamily are major regulators of plant response to ABA, both in environmental stress conditions as well as during plant development, seed maturation, and germination. They are a central hub in ABA signaling. Regulation of the response to ABA via SnRK2s occurs by direct phosphorylation of various effector proteins, such as ion channels (SLAC1, KAT1), and by regulation of expression of stress response genes, most probably by phosphorylation and activation of ABFs and other specific transcription factors. Even though ABA-dependent kinases in comparison to other SnRK2s have been studied the most deeply still only a few cellular substrates of members of this group were identified. It is well established that most SnRK2s are localized both in the nucleus and cytoplasm, but nothing is known on the mechanism of their nuclear–cytoplasmic translocation; their structure lacks a typical nuclear localization signals.
SnRK2s of group 2 are also positive regulators of plant responses to water deficit, most probably by regulation of stress response genes. However, this regulation is not as strong as in the case of the ABA-dependent SnRK2s from group 3.
The least is known on the physiological role of ABA-independent SnRK2s. We therefore envision that this group will be widely studied in the nearest feature. So far, there are mostly question marks concerning their role, mechanism(s) of regulation of their activity and localization (localization only of some of them was studied). Very intriguing is the fact that group 1 kinases, whose activity is not dependent on ABA, have an influence on ABA-dependent proline accumulation (Fujii et al., 2011), as well as other ABA-dependent processes (our unpublished results). Moreover, phosphoproteomic data obtained last year by Kline et al. (2010) showed a strong increase of SnRK2.4 phosphorylation in the kinase activation loop in response to ABA treatment. The increase of SnRK2.4 phosphorylation was similar to that observed for ABA-dependent kinases—SnRK2.2, SnRK2.3, and SnRK2.6. Why then is this kinase not activated by ABA? Are there some other mechanisms, beside phosphorylation, controlling SnRK2s activity? What is the role of PA and NO in SnRK2s function? Which phosphatases are responsible for inactivation of ABA-independent SnRK2? Is SnRK2 activation dependent on phosphorylation by an so far unknown upstream kinase(s)? In which stress signaling pathways, beside osmotic stress, are SnRK2s involved? Still much work needs to be done in order to identify the SnRK2s' cellular substrates and to establish the connections, or crosstalk, of SnRK2s with other signaling elements and various hormonal signaling pathways. All these issues should be addressed to learn more about the role of SnRK2 in plant response to abiotic stresses. Drought and salinity severely affect plant growth and productivity. In order to obtain plants with enhanced tolerance to environmental stresses the knowledge on plant stress signal transduction pathways and plant defense against given stressors is required. Therefore, studies on SnRK2s are of high priority, as they seem to be perfect candidates for improvement of crop tolerance to environmental stresses and water use efficiency.
Acknowledgments
Financial support from the Ministry of Science and Higher Education (Grant 500/N-COST/2009/0 to GD), EU (COST FA0605), and FNP (Foundation for Polish Science) (Grant TEAM/2008-2/4) fellowship to I.W. is gratefully acknowledged.
Author Disclosure Statement
No competing financial interests exist.
References
- Anderberg R.J. Walker-Simmons M.K. Isolation of a wheat cDNA clone for an abscisic acid-inducible transcript with homology to protein kinase. Proc Natl Acad Sci USA. 1992;89:10183–10187. doi: 10.1073/pnas.89.21.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E. Energy signaling in the regulation of gene expression during stress. Mol Plant. 2010;3:300–313. doi: 10.1093/mp/ssp113. [DOI] [PubMed] [Google Scholar]
- Baena-González E. Sheen J. Convergent energy and stress signaling. Trends Plant Sci. 2008;13:474–482. doi: 10.1016/j.tplants.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-Gonález E. Rolland F. Thevelein J. Sheen J. A central integrator of transcription networks in plant stress and energy signaling. Nature. 2007;443:938–943. doi: 10.1038/nature06069. [DOI] [PubMed] [Google Scholar]
- Batistic O. Kudla J. Plant calcineurin B-like proteins and their interacting protein kinases. Biochim Biophys Acta. 2009;1793:985–992. doi: 10.1016/j.bbamcr.2008.10.006. [DOI] [PubMed] [Google Scholar]
- Belin C. De Franco P.-O. Bourbousse C. Chaignepain S. Schmitter J.-M. Vavasseur A., et al. Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant Physiol. 2006;141:1316–1327. doi: 10.1104/pp.106.079327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M. Lauriere C. Osmotic signaling in plants. Multiple pathways mediated by emerging kinase families. Plant Physiol. 2005;138:1185–1194. doi: 10.1104/pp.105.061275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M. Barbier-Brygoo H. Lauriere C. Identification of nine SNF1-related protein kinase 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J Biol Chem. 2004;279:41758–41766. doi: 10.1074/jbc.M405259200. [DOI] [PubMed] [Google Scholar]
- Boudsocq M. Droillard M.J. Barbier-Brygoo H. Lauriere C. Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Mol Biol. 2007;63:491–503. doi: 10.1007/s11103-006-9103-1. [DOI] [PubMed] [Google Scholar]
- Bucholc M. Ciesielski A. Goch G. Anielska-Mazur A. Kulik A. Krzywińska E., et al. SNF1-related protein kinases 2 are negatively regulated by a plant-specific calcium sensor. J Biol Chem. 2011;286:3429–3441. doi: 10.1074/jbc.M110.115535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bungard D. Fuerth B.J. Zeng P.Y. Faubert B. Maas N.L. Viollet B., et al. Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science. 2010;329:1201–1205. doi: 10.1126/science.1191241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burza A.M. Pękala I. Sikora J. Siedlecki P. Małagocki P. Bucholc M., et al. Nicotiana tabacum osmotic stress-activated kinase is regulated by phosphorylation on Ser-154 and Ser-158 in the Kinase activation loop. J Biol Chem. 2006;281:34299–34311. doi: 10.1074/jbc.M601977200. [DOI] [PubMed] [Google Scholar]
- Cantó C. Auwerx J. AMP-activated protein kinase and its downstream transcriptional pathways. Cell Mol Life Sci. 2010;67:3407–3423. doi: 10.1007/s00018-010-0454-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carling D. Zammit V.A. Hardie D.G. A common bicycle protein kinase cascade inactivates the regulatory enzymes of fatty acid and cholesterol biosynthesis. FEBS Lett. 1987;223:217–222. doi: 10.1016/0014-5793(87)80292-2. [DOI] [PubMed] [Google Scholar]
- Celenza J.L. Carlson M. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science. 1986;233:1175–1180. doi: 10.1126/science.3526554. [DOI] [PubMed] [Google Scholar]
- Chae M.J. Lee J.S. Nam M.H. Cho K. Hong J.Y. Yi S.A., et al. A rice dehydration-inducible SNF1-related protein kinase 2 phosphorylates an abscisic acid responsive element-binding factor and associates with ABA signaling. Plant Mol Biol. 2007;63:151–169. doi: 10.1007/s11103-006-9079-x. [DOI] [PubMed] [Google Scholar]
- Cheng N.H. Pittman J.K. Zhu J.K. Hirschi K.D. The protein kinase SOS2 activates the Arabidopsis H(+)/Ca(2+) antiporter CAX1 to integrate calcium transport and salt tolerance. J Biol Chem. 2004;279:2922–2926. doi: 10.1074/jbc.M309084200. [DOI] [PubMed] [Google Scholar]
- Chikano K. Ogawa M. Ikeda Y. Koizumi N. Kusano T. Sano H. Two novel genes encoding SNF1-related protein kinases from Arabidopsis thaliana: differential accumulation of AtSR1 and AtSR2 transcripts in response to cytokinins and sugars, and phosphorylation of sucrose synthase by AtSR2. Mol Gen Genet. 2001;264:674–681. doi: 10.1007/s004380000354. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V. Schumaker K. Zhu J.K. Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J Exp Botany. 2004;55:225–236. doi: 10.1093/jxb/erh005. [DOI] [PubMed] [Google Scholar]
- Cho K. Agrawal G.K. Jwa N.S. Kubo A. Rakwal R. Rice OsSIPK and its orthologs: a “central master switch” for stress responses. Biochem Biophys Res Commun. 2009;379:649–653. doi: 10.1016/j.bbrc.2008.12.107. [DOI] [PubMed] [Google Scholar]
- Coello P. Hey S.J. Halford N.G. The sucrose non-fermenting-1-related (SnRK) family of protein kinases: potential for manipulation to improve stress tolerance and increase yield. J Exp Botany. 2011;62:883–893. doi: 10.1093/jxb/erq331. [DOI] [PubMed] [Google Scholar]
- Cutler S.R. Rodriguez P.L. Finkelstein R.R. Abrams S.R. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–669. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- Das R. Pandey G.K. Expressional analysis and role of calcium regulated kinases in abiotic stress signaling. Curr Genomics. 2010;11:2–13. doi: 10.2174/138920210790217981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J.P. Yildiz F.H. Grossman A.R. Sac3, an Snf1-like serine/threonine kinase that positively and negatively regulates the responses of Chlamydomonas to sulfur limitation. Plant Cell. 1999;11:1179–1190. doi: 10.1105/tpc.11.6.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diédhiou C.J. Popova O.V. Dietz K.-J. Golldack D. The SNF1-type serine-threonine protein kinase SAPK4 regulates stress-responsive gene expression in rice. BMC Plant Biol. 2008;8:49. doi: 10.1186/1471-2229-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H. Zhu J.-K. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci USA. 2009;106:8380–8385. doi: 10.1073/pnas.0903144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H.M. Verslues P.E. Zhu J.-K. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth and gene expression in Arabidopsis. Plant Cell. 2007;19:485–494. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H. Chinnusamy V. Rodrigues A. Rubio S. Antoni R. Park S.-Y., et al. In vitro reconstruction of an abscisic acid signaling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H. Verslues P.E. Zhu J.-K. Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc Natl Acad Sci USA. 2011;108:1717–1722. doi: 10.1073/pnas.1018367108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y. Nakashima K. Yoshida T. Katagiri T. Kidokoro S. Kanamori N., et al. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 2009;50:2123–2132. doi: 10.1093/pcp/pcp147. [DOI] [PubMed] [Google Scholar]
- Fujita Y. Fujita M. Shinozaki K. Yamaguchi-Shinozaki K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res. 2011;124:509–525. doi: 10.1007/s10265-011-0412-3. [DOI] [PubMed] [Google Scholar]
- Furihata T. Maruyama K. Fujita Y. Umezawa T. Yoshida R. Shinozaki K., et al. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci USA. 2006;103:1988–1993. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D. Scherzer S. Mumm P. Stange A. Marten I. Bauer H., et al. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci USA. 2009;106:21425–21430. doi: 10.1073/pnas.0912021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D. Scherzer S. Mumm P. Marten I. Ache P., et al. Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc Natl Acad Sci USA. 2010;107:8023–8028. doi: 10.1073/pnas.0912030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z. Koiwa H. Cushman M.A. Ray A. Bufford D. Kore-Eda S., et al. Genes that are uniquely stress regulated in salt overly sensitive (sos) mutants. Plant Physiol. 2001;126:363–375. doi: 10.1104/pp.126.1.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong D. Guo Y. Schumaker K.S. Zhu J.-K. The SOS3 family of calcium sensor and SOS2 family of protein kinases in Arabidopsis. Plant Physiol. 2004;134:919–926. doi: 10.1104/pp.103.037440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Ballaster D. Pollock S.V. Pootakham W. Grossman A.R. The central role of a SnRK2 kinase in sulfur deprivation responses. Plant Physiol. 2008;147:216–227. doi: 10.1104/pp.108.116137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y. Halfter U. Ishitani M. Zhu J.-K. Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell. 2001;13:1383–1399. doi: 10.1105/tpc.13.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford N.G. Hardie D.G. SNF1-related protein kinases: global regulators of carbon metabolism in plants? Plant Mol Biol. 1998;37:735–748. doi: 10.1023/a:1006024231305. [DOI] [PubMed] [Google Scholar]
- Halford N.G. Hey S.J. Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochem J. 2009;419:247–259. doi: 10.1042/BJ20082408. [DOI] [PubMed] [Google Scholar]
- Halford N.G. Hey S. Jhurreea D. Laurie S. McKibbin R.S, et al. Metabolic signaling and carbon portioning: role of Snf1-related (SnRK1) protein kinase. J Exp Botany. 2003;54:467–475. doi: 10.1093/jxb/erg038. [DOI] [PubMed] [Google Scholar]
- Halford N.G. Hey S. Jhurreea D. Laurie S. McKibbin R.S. Zhang Y., et al. Highly conserved protein kinases involved in the regulation of carbon and amino acid metabolism. J Exp Botany. 2004;55:25–42. doi: 10.1093/jxb/erh019. [DOI] [PubMed] [Google Scholar]
- Halfter U. Ishitani M. Zhu J.-K. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA. 2000;97:3735–3740. doi: 10.1073/pnas.040577697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie D.G. Plant protein serine/threonine kinases: classification and functions. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:97–131. doi: 10.1146/annurev.arplant.50.1.97. [DOI] [PubMed] [Google Scholar]
- Hardie D.G. AMP-activate /SNF1 protein kinases: conserved guardians of cellular energy. Nature. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- Hardie D.G. Sensing of energy and nutrients by AMP-activated protein kinase. Am J Clin Nutr. 2011;93:891S–896S. doi: 10.3945/ajcn.110.001925. [DOI] [PubMed] [Google Scholar]
- Hardie D.G. Carling D. Sim A.T.R. The AMP-activated protein kinase—a multisubstrate regulator of lipid metabolism. Trends Biochem Sci. 1989;14:20–23. [Google Scholar]
- Harmon A.C. Calcium-regulated protein kinases of plants. Gravitat Space Biol Bull. 2003;16:83–90. [PubMed] [Google Scholar]
- Hauser F. Waadt R. Schroeder J.I. Evolution of abscisic acid synthesis and signaling mechanisms. Curr Biol. 2011;21:R346–R355. doi: 10.1016/j.cub.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedbacker K. Carlson M. SNF1/AMPK pathways in yeast. Front Biosci. 2008;13:2408–2420. doi: 10.2741/2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey S.J. Byrne E. Halford N.G. The interface between metabolic and stress signaling. Ann Botany. 2010;105:197–203. doi: 10.1093/aob/mcp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T. Umezawa T. The PP2C-SnRK2 complex. The central regulator of an abscisic acid signaling pathway. Plant Signal Behav. 2010;5:160–163. doi: 10.4161/psb.5.2.10460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holappa L.D. Walker-Simmons M.K. The wheat abscisic acid-responsive protein kinase mRNA, PKABA1, is up-regulated by dehydration, cold temperature, and osmotic stress. Plant Physiol. 1995;108:1203–1210. doi: 10.1104/pp.108.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.P. Carlson M. Regulation of snf1 protein kinase in response to environmental stress. J Biol Chem. 2007;282:16838–16845. doi: 10.1074/jbc.M700146200. [DOI] [PubMed] [Google Scholar]
- Hotta H. Aoki N. Matsuda T. Adachi T. Molecular analysis of a novel protein kinase in maturing rice seed. Gene. 1998;213:47–54. doi: 10.1016/s0378-1119(98)00207-8. [DOI] [PubMed] [Google Scholar]
- Hrabak E.M. Chan C.W.M. Gribskov M. Harper J.F. Choi J.H. Halford N., et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003;132:666–680. doi: 10.1104/pp.102.011999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huai J. Wang M. He J. Zheng J. Dong Z. Lv H., et al. Cloning and characterization of the SnRK2 gene family from Zea mays. Plant Cell Rep. 2008;12:1861–1868. doi: 10.1007/s00299-008-0608-8. [DOI] [PubMed] [Google Scholar]
- Hubbard K.E. Nishimura N. Hitomi K. Getzoff E.D. Schroeder J.I. Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev. 2010;24:1695–1708. doi: 10.1101/gad.1953910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y. Koizumi N. Kusano T. Sano H. Sucrose and cytokinin modulation of WPK4, a gene encoding a SNF1-related protein kinase from wheat. Plant Physiol. 1999;121:813–820. doi: 10.1104/pp.121.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y. Koizumi N. Kusano T. Sano H. Specific binding of a 14-3-3 protein to autophosphorylated WPK4, an SNF1-related wheat protein kinase, and to WPK4-phosphorylated nitrate reductase. J Biol Chem. 2000;275:31695–31700. doi: 10.1074/jbc.M004892200. [DOI] [PubMed] [Google Scholar]
- Johnson R. Wagner R. Verhey S.D. Walker-Simmons M.K. The ABA-responsive kinase PKABA1 interacts with a seed-specific ABA response element binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol. 2002;130:837–846. doi: 10.1104/pp.001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonak C. Hirt H. Glycogen synthase kinase 3/SHAGGY-like kinases in plants: an emerging family with novel functions. Trends Plant Sci. 2002;7:457–461. doi: 10.1016/s1360-1385(02)02331-2. [DOI] [PubMed] [Google Scholar]
- Joshi-Saha A. Valon C. Leung J. Abscisic acid signal off the STARting block. Mol Plant. 2011;4:562–580. doi: 10.1093/mp/ssr055. [DOI] [PubMed] [Google Scholar]
- Kearns M.A. Monks D.E. Fang M. Rivas M.P. Courtney P.D. Chen J., et al. Novel developmentally regulated phosphoinositide binding proteins from soybean whose expression bypasses the requirement for an essential phosphatidylinositol transfer protein in yeast. EMBO J. 1998;17:4004–4017. doi: 10.1093/emboj/17.14.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelner A. Pękala I. Kaczanowski S. Muszyńska G. Hardie D.G. Dobrowolska G. Biochemical characterization of the Nicotiana tabacum 42-kD protein kinase activated by osmotic stress. Plant Physiol. 2004;136:3255–3264. doi: 10.1104/pp.104.046151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T. Shibagaki N. Ohkama-Ohtsu N. Hayashi H. Yoneyama T. Davies J.P., et al. Arabidopsis SNRK2.3 protein kinase is involved in the regulation of sulfur-responsive gene expression and O-acetyl-L-serine accumulation under limited sulfur supply. Soil Sci Plant Nutr. 2006;52:211–220. [Google Scholar]
- Kline K.G. Barrett-Wilt G.A. Sussman M.R. In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc Natl Acad Sci USA. 2010;107:15986–15991. doi: 10.1073/pnas.1007879107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingler J.P. Batelli G. Zhu J.-K. ABA receptors: the START of a new paradigm in phytohormone signalling. J Exp Botany. 2010;61:3199–3210. doi: 10.1093/jxb/erq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y. Yamamoto S. Minami H. Kagaya Y. Hattori T. Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell. 2004;16:1163–1177. doi: 10.1105/tpc.019943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y. Murata M. Minami H. Yamamoto S. Kagaya Y. Hobo T., et al. Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 2005;44:939–949. doi: 10.1111/j.1365-313X.2005.02583.x. [DOI] [PubMed] [Google Scholar]
- Koh S. Lee S.C. Kim M.K. Koh J.H. Lee S. An G., et al. T-DNA tagged knockout mutation of rice OsGSK1, an orthologue of Arabidopsis BIN2, with enhanced tolerance to various abiotic stresses. Plant Mol Biol. 2007;65:453–466. doi: 10.1007/s11103-007-9213-4. [DOI] [PubMed] [Google Scholar]
- Kwak J.M. Mori I.C. Pei Z.M. Leonhardt N. Torres M.A. Dangl J.L., et al. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 2003;22:2623–2633. doi: 10.1093/emboj/cdg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamotte O. Courtois C. Dobrowolska G. Besson A. Pugin A. Wendehenne D. Mechanisms of nitric-oxide-induced increase of free cytosolic Ca2+ concentration in Nicotiana plumbaginifolia cells. Free Radic Biol Med. 2006;40:1369–1376. doi: 10.1016/j.freeradbiomed.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Lee S.C. Lan W. Buchanan B.B. Luan S. A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc Natl Acad Sci USA. 2009;106:21419–21424. doi: 10.1073/pnas.0910601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Assmann S.M. An abscisic acid-activated and calcium-independent protein kinase from guard cells of fava bean. Plant Cell. 1996;8:2359–2368. doi: 10.1105/tpc.8.12.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Wang X.Q. Watson M.B. Assmann S.M. Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science. 2000;287:300–303. doi: 10.1126/science.287.5451.300. [DOI] [PubMed] [Google Scholar]
- Li L.-B. Zhang Y.-R. Liu K.-C. Ni Z.-F. Fang Z.-J. Sun Q.-X., et al. Identification and bioinformatics analysis of SnRK2 and CIPK family genes in sorghum. Agric Sci China. 2010;9:19–30. [Google Scholar]
- Liu J. Zhu J.K. A calcium sensor homolog required for plant salt tolerance. Science. 1998;280:1943–1945. doi: 10.1126/science.280.5371.1943. [DOI] [PubMed] [Google Scholar]
- Luan S. The CBL–CIPK network in plant calcium signaling. Trends Plant Sci. 2009;14:37–42. doi: 10.1016/j.tplants.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Luo Z. Zang M. Guo W. AMPK as a metabolic tumor suppressor: control of metabolism and cell growth. Fut Oncol. 2010;6:457–470. doi: 10.2217/fon.09.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y. Szostkiewicz I. Korte A. Moes D. Yang Y. Christmann A., et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- Mahfouz M.M. Kim S. Delauney A.J. Verma D.P. Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell. 2006;18:477–490. doi: 10.1105/tpc.105.035931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X. Zhang H. Tian S. Chang X. Jing R. TaSnRK2.4, an SNF1-type serine/threonine protein kinase of wheat (Tricitum aestivum L.), confers enhanced multistress tolerance in Arabidopsis. J Exp Botany. 2010;61:683–696. doi: 10.1093/jxb/erp331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee S.L. Van Denderen B.J. Howlett K.F. Mollica J. Schertzer J.D. Kemp B.E., et al. AMP-activated protein kinase regulates GLUT4 transcription by phosphorylating histone deacetylase 5. Diabetes. 2008;57:860–867. doi: 10.2337/db07-0843. [DOI] [PubMed] [Google Scholar]
- Melcher K. Ng L.-M. Zhou X.E. Soon F.-F. Xu Y. Suino-Powell K.M., et al. A gate-latch-lock mechanism for hormone signaling by abscisic acid receptors. Nature. 2009;462:602–608. doi: 10.1038/nature08613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher K. Zhou X.E. Xu H.E. Thirsty plants and beyond: structural mechanisms of abscisic acid perception and signaling. Curr Opin Struct Biol. 2010;20:722–729. doi: 10.1016/j.sbi.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M. Underwood W. Koczan J. Nomura K. He S.Y. Plant stomata function in innate immunity against bacterial invasion. Cell. 2006;126:969–980. doi: 10.1016/j.cell.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Mikołajczyk M. Awotunde O.S. Muszyńska G. Klessig D.F. DOBROWOLSKA G. Osmotic stress induces rapid activation of a salicylic acid-induced protein kinase and a homolog of protein kinase ASK1 in tobacco cells. Plant Cell. 2000;12:165–178. [PMC free article] [PubMed] [Google Scholar]
- Mishra N.S. Tuteja R. Tuteja N. Signaling through MAP kinase networks in plants. Arch Biochem Biophys. 2006;452:55–68. doi: 10.1016/j.abb.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Mizoguchi M. Umezawa T. Nakashima K. Kidokoro S. Takasaki H. Fujita Y., et al. Two closely related subclass II SnRK2 protein kinases cooperatively regulate drought-inducible gene expression. Plant Cell Physiol. 2010;51:842–847. doi: 10.1093/pcp/pcq041. [DOI] [PubMed] [Google Scholar]
- Monks D.E. Aghoram K. Courtney P.D. Dewald D.B. Dewey R.E. Hyperosmotic stress induces the rapid phosphorylation of a soybean phosphatidylinositol transfer protein homolog through activation of the protein kinases SPK1 and SPK2. Plant Cell. 2001;13:1205–1219. doi: 10.1105/tpc.13.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y. Pei Z.M. Mori I.C. Schroeder J. Abscisic acid activation of plasma membrane Ca2+ channels in guard cells requires cytosolic NAD(P)H and is differentially disrupted upstream and downstream of reactive oxygen species production in abi1-1 and abi2-1 protein phosphatase 2C mutants. Plant Cell. 2001;13:2513–2523. doi: 10.1105/tpc.010210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli A.-C. Merlot S. Vavasseur A. Frenzi F. Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstrean of reactive oxygen species production. Plant Cell. 2002;14:3089–3099. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K. Fujita Y. Kanamori N. Katagiri T. Umezawa T. Kidokoro S., et al. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009;50:1345–1363. doi: 10.1093/pcp/pcp083. [DOI] [PubMed] [Google Scholar]
- Negi J. Matsuda O. Nagasawa T. Oba Y. Takahashi H. Kawai-Yamada M., et al. CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature. 2008;452:483–486. doi: 10.1038/nature06720. [DOI] [PubMed] [Google Scholar]
- Neill S. Barros R. Bright J. Desikan R. Hancock J. Harrison, et al. Nitric oxide, stomatal closure, and abiotic stress. J Exp Botany. 2008;59:165–176. doi: 10.1093/jxb/erm293. [DOI] [PubMed] [Google Scholar]
- Nishimura N. Hitomi K. Arvai A.S. Rambo R.P. Hitomi C. Cutler S.R., et al. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science. 2009;326:1373–1379. doi: 10.1126/science.1181829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N. Sarkeshi K A. Nito K. Park S.Y. Wang A. Carvalho P.C., et al. PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 2010;61:290–299. doi: 10.1111/j.1365-313X.2009.04054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan K.E. Saeed N.A. Rose R.J. The stress kinase gene MtSK1 in Medicago truncatula with particular reference to somatic embryogenesis. Plant Cell Rep. 2006;25:711–722. doi: 10.1007/s00299-006-0135-4. [DOI] [PubMed] [Google Scholar]
- Ogasawara Y. Kaya H. Hiraoka G. Yumoto F. Kimura S. Kadota Y., et al. Synergistic activation of the Arabidopsis NADPH oxidase AtrbohD by Ca2+ and phosphorylation. J Biol Chem. 2008;283:8885–8892. doi: 10.1074/jbc.M708106200. [DOI] [PubMed] [Google Scholar]
- Ohba H. Steward N. Kawasaki S. Berberich T. Ikeda Y. Koizum I.N., et al. Diverse response of rice and maize genes encoding homologs of WPK4, an SNF1-related protein kinase from wheat, to light, nutrients, low temperature and cytokinins. Mol Gen Genet. 2000;263:359–366. doi: 10.1007/s004380051179. [DOI] [PubMed] [Google Scholar]
- Park S.-Y. Fung P. Nishimura N. Jensen D.R. Hiroaki F. Zhao Y., et al. Abscisic acid inhibits PP2Cs via the PYR/PYL family of ABA binding START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.S. Hong S.W. Oh S.A. Kwak J.M. Lee H.H. Nam H.G. Two putative protein kinase from Arabidopsis thaliana contain highly acidic domains. Plant Mol Biol. 1993;22:615–624. doi: 10.1007/BF00047402. [DOI] [PubMed] [Google Scholar]
- Petranovic D. Tyo K. Vemuri G.N. Nielsen J. Prospects of yeast systems biology for human health: integrating lipid, protein and energy metabolism. FEMS Yeast Res. 2010;10:1046–1059. doi: 10.1111/j.1567-1364.2010.00689.x. [DOI] [PubMed] [Google Scholar]
- Pilot G. Lacombe B. Gaymard F. Chérel I. Boucherez J. Thibaud J.-B., et al. Guard cell inward K+ channel activity in Arabidopsis involves expression of the twin channel subunits KAT1 and KAT2. J Biol Chem. 2001;276:3215–3221. doi: 10.1074/jbc.M007303200. [DOI] [PubMed] [Google Scholar]
- Pitzschke A. Schikora A. Hirt H. MAPK cascade signalling networks in plant defence. Curr Opin Plant Biol. 2009;12:421–426. doi: 10.1016/j.pbi.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Poels J. Spasic M.R. Callaerts P. Norga K.K. Expanding roles for AMP-acticated protein kinases in neuronal survival and autophagy. BioEssays. 2009;31:944–952. doi: 10.1002/bies.200900003. [DOI] [PubMed] [Google Scholar]
- Polge C. Thomas M. SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci. 2006;12:20–28. doi: 10.1016/j.tplants.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Pootakham W. Gonzalez-Ballester D. Grossman A.R. Identification and regulation of plasma membrane sulfate transporters in Chlamydomonas. Plant Physiol. 2010;153:1653–1668. doi: 10.1104/pp.110.157875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Q.S. Guo Y. Quintero F.J. Pardo J.M. Schumaker K.S. Zhu J.-K. Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway. J Biol Chem. 2004;279:207–215. doi: 10.1074/jbc.M307982200. [DOI] [PubMed] [Google Scholar]
- Rafalski V.A. Brunet A. Energy metabolism in adult neural stem cell fate. Prog Neurobiol. 2011;93:182–203. doi: 10.1016/j.pneurobio.2010.10.007. [DOI] [PubMed] [Google Scholar]
- Rodriguez M.C. Petersen M. Mundy J. Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol. 2010;61:621–649. doi: 10.1146/annurev-arplant-042809-112252. [DOI] [PubMed] [Google Scholar]
- Rus A. Yokoi S. Sharkhuu A. Reddy M. Lee B.-H. Matsumoto T.K., et al. AtHKT1 is a salt tolerance determinant that controls Na+ entry into plant roots. Proc Natl Acad Sci USA. 2001;98:14150–14155. doi: 10.1073/pnas.241501798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H. Youssefian S. Light and nutritional regulation of transcripts encoding a wheat protein kinase homolog is mediated by cytokinins. Proc Natl Acad Sci USA. 1994;91:2582–2586. doi: 10.1073/pnas.91.7.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J. Dupeux F. Round A. Antoni R. Park S.Y. Jamin M., et al. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature. 2009;462:665–668. doi: 10.1038/nature08591. [DOI] [PubMed] [Google Scholar]
- Sato A. Sato Y. Fukao Y. Fujiwara M. Umezawa T. Shinozaki K., et al. Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem J. 2009;424:439–448. doi: 10.1042/BJ20091221. [DOI] [PubMed] [Google Scholar]
- Scott J.W. Oakhill J.S. Van Denderen B.J. AMPK/SNF1 structure: a menage a trois of energy-sensing. Front Biosci. 2009;14:596–610. doi: 10.2741/3266. [DOI] [PubMed] [Google Scholar]
- Shi J. Kim K.-N. Ritz O. Albrecht V. Gupta R. Harter K., et al. Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell. 1999;11:2393–2405. doi: 10.1105/tpc.11.12.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R. Alvarez S. Burch A.Y. Jez J.M. Schachtman D.P. Phosphoproteomic identification of targets of the Arabidopsis sucrose nonfermenting-like kinase SnRK2.8 reveals a connection to metabolic processes. Proc Natl Acad Sci USA. 2007;104:6460–6465. doi: 10.1073/pnas.0610208104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirichandra C. Gu D. Hu H.-C. Davanture M. Lee S. Djaoui M., et al. Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett. 2009;583:2982–2986. doi: 10.1016/j.febslet.2009.08.033. [DOI] [PubMed] [Google Scholar]
- Testerink C. Dekker H.L. Lim Z.Y. Johns M.K. Holmes A.B. Koster C.G., et al. Isolation and identification of phosphatidic acid targets from plants. Plant J. 2004;39:527–536. doi: 10.1111/j.1365-313X.2004.02152.x. [DOI] [PubMed] [Google Scholar]
- Umezawa T. Yoshida R. Maruyama K. Yamaguchi-Shinozaki K. Shinozaki K. SnRK2C, a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2004;101:17306–17311. doi: 10.1073/pnas.0407758101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T. Sugiyama N. Mizoguchi M. Hayashi S. Myouga F. Yamaguchi-Shinozaki K., et al. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:17588–17593. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T. Nakashima K. Miyakawa T. Kuromori T. Tanokura M. Shinozaki K., et al. Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol. 2010;51:1821–1839. doi: 10.1093/pcp/pcq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahisalu T. Kollist H. Wang Y.F. Nishimura N. Chan W.Y. Valerio G., et al. SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature. 2008;452:487–491. doi: 10.1038/nature06608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahisalu T. Puzõrjova I. Brosché M. Valk E. Lepiku M. Moldau H., et al. Ozone-triggered rapid stomatal response involves the production of reactive oxygen species, and is controlled by SLAC1 and OST1. Plant J. 2010;62:442–453. doi: 10.1111/j.1365-313X.2010.04159.x. [DOI] [PubMed] [Google Scholar]
- Vlad F. Rubio S. Rodrigues A. Sirichandra C. Belin C. Robert N., et al. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell. 2009;21:3170–3184. doi: 10.1105/tpc.109.069179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F. Droillard M.J. Valot B. Khafif M. Rodrigues A. Brault M., et al. Phospho-site mapping, genetic and in planta activation studies reveal key aspects of the different phosphorylation mechanisms involved in activation of SnRK2s. Plant J. 2010;63:778–790. doi: 10.1111/j.1365-313X.2010.04281.x. [DOI] [PubMed] [Google Scholar]
- Wang W. Guan K.-L. AMP-activated protein kinase and cancer. Acta Physiol. 2009;196:55–63. doi: 10.1111/j.1748-1716.2009.01980.x. [DOI] [PubMed] [Google Scholar]
- Wawer I. Bucholc M. Astier J. Anielska-Mazur A. Dahan J. Kulik A., et al. Regulation of Nicotiana tabacum osmotic stress-activated protein kinase and its cellular partner GAPDH by nitric oxide in response to salinity. Biochem J. 2010;429:73–83. doi: 10.1042/BJ20100492. [DOI] [PubMed] [Google Scholar]
- Weiner J.J. Peterson F.C. Volkman B.F. Cutler S.R. Structural and functional insights into core ABA signaling. Curr Opin Plant Biol. 2010;13:495–502. doi: 10.1016/j.pbi.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinl S. Kudla J. The CBL-CIPK Ca(2+)-decoding signaling network: function and perspectives. New Phytol. 2009;184:517–528. doi: 10.1111/j.1469-8137.2009.02938.x. [DOI] [PubMed] [Google Scholar]
- Wilson W.A. Hawley S.A. Hardie D.G. The mechanism of glucose repression/derepression in yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr Biol. 1996;6:1426–1434. doi: 10.1016/s0960-9822(96)00747-6. [DOI] [PubMed] [Google Scholar]
- Wurzinger B. Mair A. Pfister B. Teige M. Cross-talk of calcium-dependent protein kinase and MAP kinase signaling. Plant Signal Behav. 2011;6:8–12. doi: 10.4161/psb.6.1.14012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z.S. Liu L. Ni Z.Y. Liu P. Chen M. Li L.C., et al. W55a encodes a novel protein kinase that is involved in multiple stress responses. J Integrat Plant Biol. 2009;51:58–66. doi: 10.1111/j.1744-7909.2008.00776.x. [DOI] [PubMed] [Google Scholar]
- Xue S. Hu H. Ries A. Merilo E. Kollist H. Schroeder J.I. Central functions of bicarbonate in S-type anion channel activation and OST1 protein kinase in CO(2) signal transduction in guard cell. EMBO J. 2011;30:1645–1658. doi: 10.1038/emboj.2011.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi D. Zentella R. Ho T.D. Molecular analysis of the barley (Hordeum vulgare) gene encoding the protein kinase PKABA1 capable of suppressing gibberellin action in aleurone layers. Planta. 2002;215:319–326. doi: 10.1007/s00425-002-0740-6. [DOI] [PubMed] [Google Scholar]
- Yoon H.W. Kim M.C. Shin P.G. Kim J.S. Kim C.Y. Lee S.Y., et al. Differential expression of two functional serine/threonine protein kinases from soybean that have an unusual acidic domain at the carboxy terminus. Mol Gen Genet. 1997;255:359–371. doi: 10.1007/s004380050507. [DOI] [PubMed] [Google Scholar]
- Yoshida R. Hobo T. Ichimura K. Mizoguchi T. Takahashi F. Aronso J., et al. ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 2002;43:1473–1483. doi: 10.1093/pcp/pcf188. [DOI] [PubMed] [Google Scholar]
- Yoshida R. Umezawa T. Mizoguchi T. Takahashi S. Takahashi F. Schinozaki K. The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates ABA and osmotic stress signals controlling stomatal closure in Arabidopsis. J Biol Chem. 2006;281:5310–5318. doi: 10.1074/jbc.M509820200. [DOI] [PubMed] [Google Scholar]
- Yunta C. Martinez-Ripoll M. Albert A. SnRK2.6/OST1 from Arabidopsis thaliana: cloning, expression, purification, crystallization and preliminary X-ray analysis of K50N and D160A mutants. Acta Crystallogr F Struct Biol Crystallizat Commun. 2011;67:364–368. doi: 10.1107/S1744309110053807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. Mao X. Jing R. Chang X. Xie H. Characterization of a common wheat (Triticum aestivum L.) TaSnRK2.7 gene involvement in abiotic stress responses. J Exp Botany. 2011;62:975–988. doi: 10.1093/jxb/erq328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. Mao X. Wang C. Jing R. Overexpression of a common wheat gene TaSnRK2.8 enhances tolerance to drought, salt and low temperature in Arabidopsis. PLoS One. 2010b;5:e16041. doi: 10.1371/journal.pone.0016041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Vemuri G. Nielsen J. Systems biology of energy homeostasis in yeast. Curr Opin Microbiol. 2010a;13:382–388. doi: 10.1016/j.mib.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Zheng Z. Xu X. Crosley R.A. Greenwalt S.A. Sun Y. Blakeslee B., et al. The protein kinase SnRK2.6 mediates the regulation of sucrose metabolism and plant growth in Arabidopsis. Plant Physiol. 2010;153:99–113. doi: 10.1104/pp.109.150789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.-K. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.-K. Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol. 2003;6:441–445. doi: 10.1016/s1369-5266(03)00085-2. [DOI] [PubMed] [Google Scholar]
- Zou H. Zhang X. Zhao J.R. Yang Q. Wu Z. Wang F, et al. Cloning and characterization of maize ZmSPK1, a homologue to nonfermenting1-related protein kinase2. Afr J Biotechnol. 2006;5:490–496. [Google Scholar]