Abstract
In the heterogeneous environment surrounding plant roots (the rhizosphere), microorganisms both compete and cooperate. Here, we show that two very different inhabitants of the rhizosphere, the nonmotile fungus Aspergillus fumigatus and the swarming bacterium Paenibacillus vortex, can facilitate each other's dispersal. A. fumigatus conidia (nonmotile asexual fungal spores) can be transported by P. vortex swarms over distances of at least 30 cm and at rates of up to 10.8 mm h−1. Moreover, conidia can be rescued and transported by P. vortex from niches of adverse growth conditions. Potential benefit to the bacteria may be in crossing otherwise impenetrable barriers in the soil: fungal mycelia seem to act as bridges to allow P. vortex to cross air gaps in agar plates. Transport of conidia was inhibited by proteolytic treatment of conidia or the addition of purified P. vortex flagella, suggesting specific contacts between flagella and proteins on the conidial surface. Conidia were transported by P. vortex into locations where antibiotics inhibited bacteria growth, and therefore, growth and sporulation of A. fumigatus were not limited by bacterial competition. Conidia from other fungi, similar in size to those fungi from A. fumigatus, were not transported as efficiently by P. vortex. Conidia from a range of fungi were not transported by another closely related rhizosphere bacterium, Paenibacillus polymyxa, or the more distantly related Proteus mirabilis, despite both being efficient swarmers.
Keywords: bacterial–fungal interaction, social motility, spore dispersal, microbial transport
A rich variety of microorganisms, mainly bacteria and fungi, is found in the high-nutrient soil environment surrounding plant roots—the rhizosphere. Life in this heterogeneous and highly competitive habitat involves both cooperative and antagonistic interactions between microorganisms. The rhizosphere contains both motile and nonmotile microorganisms with a wide range of dispersal strategies. Over recent years, motile microorganisms have been shown to move micrometer-scale objects such as beads or components of miniaturized devices (1–3). This finding raises questions that motivated this work. Can microbial transport phenomena also be relevant to the dispersal of nonmotile microorganisms? If so, what are the benefits to the parties involved, and how specific are such interactions?
Paenibacillus vortex is a Gram-positive bacterium capable of swarming and coordinated behavior that generates complex, spreading patterns on nutrient agars (4–6). The architecture of these intricate and dynamic macroscopic colonies is highly dependent on the environmental conditions. Bacteria successful in the rhizosphere often possess extensive signal transduction and regulatory networks (7). The 6.8-Mb P. vortex genome has been recently sequenced, and it reveals a microorganism capable of versatile signal transduction, with an extremely high number of two-component sensory transduction systems (8). Successful competition with other microorganisms seems to be, in part, reflected by the high capacity of the genome to encode antimicrobials, including putative antifungals. P. vortex is known to limit the growth of Verticillium dahliae, a fungal pathogen of plants (8). Members of the genus Paenibacillus have a variety of interactions with fungi, both antagonistic (9, 10) and cooperative; an example of the latter is stimulation of the growth of the fungus Glomus intraradices by P. validus (11).
Aspergillus fumigatus is another versatile inhabitant of the rhizosphere and many other environments; its 29.4-Mb genome has been fully sequenced (12). This saprophytic fungus has a mycelial mode of growth. Conidia (nonmotile asexual fungal spores) are generated in large numbers in specialized fruiting bodies (conidiophores) that develop from mycelia. These allergenic spores are widespread and readily inhaled. Systemic infections of A. fumigatus, particularly in immunocompromised individuals, can be fatal. A. fumigatus infections can be treated by triazole drugs, including voriconazole (13). Despite being a versatile and complex microorganism, A. fumigatus is not motile. Airborne dispersal of conidia is a long-ranged but passive process. Conidia have a hydrophobic surface, but during germination under favorable conditions, these spores become more hydrophilic and swell because of water uptake (14). Germ tubes typically emerge after ∼6–9 h and develop into mycelia. Under suitable conditions, sporulation occurs within a few days. Although reproduction is generally asexual, the genome sequence of A. fumigatus has revealed the potential for a sexual cycle (12).
In this work, we show that swarming P. vortex can transport conidia of A. fumigates, and we identify situations where this transportation is advantageous for the fungal cargo. Because fungal mycelia assist P. vortex in crossing otherwise impenetrable air gaps, there seems to be a mutual benefit to the two microorganisms in the association related to dispersal.
Results
Transport of Nonmotile A. fumigatus Conidia by P. vortex Swarms.
Imaging of P. vortex swarming.
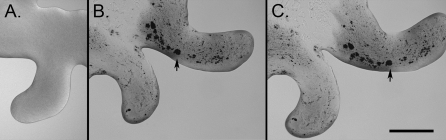
Time-lapse imaging by low-power microscopy was used to observe P. vortex swarming on reduced strength Mueller–Hinton agar (RMHA) plates maintained at 37 °C with a heated slide warmer (SI Materials and Methods). P. vortex, inoculated from swarming colonies, swarmed on fresh RMHA plates within 20 min to 1 h of inoculation (Fig. 1A). The lead bodies of the swarming bacteria, typically continuous snakelike masses on 0.5% wt/vol agar and tending to more discrete rotating colonies on higher percentage agars (up to 2% wt/vol), were capable of swarming at up to 11 mm h−1.
Fig. 1.
Direct imaging of conidial transport. (A) Swarming P. vortex without conidia. (B) Swarming mass of P. vortex transporting hundreds of ungerminated conidia (e.g., arrow indicates an aggregate of >20 conidia; conidia appear black) imaged after 4 h and 31 mm away from the coinoculation location. (C) Same as B but 1 min later. (Scale bar: 400 μm.)
Rates of conidia transport.
Direct transport of A. fumigatus conidia by swarming P. vortex was observed when the microorganisms were coinoculated in the center of an RMHA plate (Fig. 1 B and C, Figs. S1 and S2, and Movies S1, S2, and S3). Observation of the central inoculation point suggested that only a minority of conidia was transported from the inoculation point (<1%). Individual conidia and aggregates of conidia over 25 μm across could be observed in motion within masses of P. vortex several centimeters away from the origin of swarming within 1–4 h of inoculation. Typically, hundreds of conidia were visible within swarming colonies during the first few hours. The identity of these objects as conidia was confirmed by recovery with a toothpick followed by microscopy. Swarming P. vortex carried viable conidia, which was shown by recovery and selective fungal culture on Sabouraud agar. For most purposes, the intensely pigmented nature of the conidia was sufficient to permit their imaging within P. vortex colonies by transmission light microscopy. Transport was only seen in multilayered masses of cells. Conidia observed within monolayers of motile P. vortex were not in motion, and swarming multilayered aggregates of bacteria deposited an intermittent trail of conidia in their wake. Conidial transport rates of up to 180 μm min−1 (10.8 mm h−1) were observed (i.e., similar to the movement rates of P. vortex masses without conidia).
Conidia can be transported over long distances.
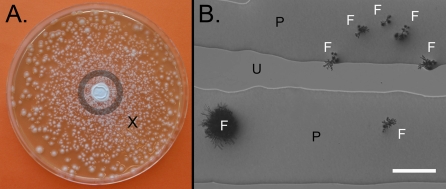
After they are loaded, the leading swarming microcolonies transporting conidia could be observed to reach the edge of 14-cm diameter RMHA plates within 7 h. To test conidial transport over longer distances and time periods, A. fumigatus conidia and swarming P. vortex bacteria were coinoculated in two kinds of test areas: at the center of a 14-cm round Petri dish (Fig. 2A) and at the corner of a 22-cm square Petri dish. In the round Petri dish, small growing colonies of A. fumigatus were observed within the P. vortex swarm at different locations from the center to the periphery (up to 7 cm from the inoculation point). In the larger, square RMHA plate, growing colonies of A. fumigatus were observed at distances of up to 30 cm (i.e., at the corner diagonally opposite the inoculation corner), with the distance limited by the size of the plate. The coinoculation experiments were repeated using RMHA containing 0.5 mM p-nitrophenyl glycerol (PNPG) to inhibit P. vortex swarming (5). PNPG at this concentration was not inhibitory to the germination or growth of A. fumigatus (judged from microcolony and visible colony diameter on RMHA plates with and without 0.5 mM PNPG). With PNPG, the spread of A. fumigatus was not facilitated by P. vortex but rather, was similar to the spread found when inoculating conidia alone (below).
Fig. 2.
Dispersal of A. fumigatus by swarming P. vortex. (A) A 14-cm diameter RMHA plate (1% wt/vol Eiken agar) was inoculated with both organisms and then cultured for 50 h. P. vortex have swarmed over the entire plate (not visible), and white fungal colonies are dispersed across the plate, with greater density closer to the center. The central ring shows the minimal and maximal radii of expansion of A. fumigatus colonies when inoculated in the absence of P. vortex (taken from control experiments after 50 h). (B) View by microscopy from a location on the same plate (marked by an X in A) after 22 h incubation. Microcolonies of A. fumigatus (F) are observed within the mass of P. vortex (P) but not within the regions of agar remaining uncolonized by the bacterium (U). (Scale bar: 100 μm.)
Fungal dispersal is facilitated by bacterial transport of conidia.
When preparations of purified conidia of A. fumigatus were inoculated onto RMHA in the same way as described above but without bacteria, a slowly expanding colony formed; the edge grew out at a rate of up to 6 mm/d (Fig. S3). After 54–70 h, conidiophores were produced. After 5 d or longer, small secondary fungal colonies were sometimes observed that resulted from the aerial spread of conidia from the central colony germinating within the uncolonized region of the agar plate. Typically, it took from 6 to 12 d for A. fumigatus to spread from a central inoculum to the edge of the 14-cm diameter plate. Therefore, the rapidity of dispersal (7 h until lead conidia reach the edge of the 14-cm diameter plate) in the presence of swarming P. vortex could not be explained by fungal growth, the germination of airborne conidia generated from the inoculation point, or other sources of airborne contamination.
Distribution and germination of transported conidia.
Examination of plates by microscopy (16–22 h after inoculation of P. vortex with conidia under swarming conditions) revealed microcolonies of A. fumigatus dispersed across the plate. In all cases (n = 100 for three replicates), the fungal microcolonies were growing within masses of P. vortex (Fig. 2B). This case was in a situation where, although lead elements of P. vortex had completely spread to the edge the 14-cm plate, 35–45% of the plate area remained uncolonized by the bacterium with a typical complex patterning colonial morphology. Transport of A. fumigatus by swarming P. vortex was possible on a variety of media (l-agar, Mueller–Hinton, and RMHA) using agars from different manufacturers (Difco and Eiken) as gelling agents. Quantification of the microcolony distribution after 16 h was by microscopy, starting outside the radius that fungal mycelia could reach by conventional growth from the inoculation point. Dispersal, as judged by this method, was optimal on RMHA (1% wt/vol Eiken agar), with a lower-percentage agar (0.5% wt/vol) being less effective (Fig. S4A).
Rescue of conidia from niches of adverse growth conditions.
Voriconazole (VOR), a triazole antimycotic effective against A. fumigatus (13), was used to create regions (a niche) on RMHA where conidial outgrowth and mycelial extension were inhibited. When P. vortex and A. fumigatus spores were coinoculated in a 10-μL aliquot of RMH medium containing 10 or 50 μg VOR, fungal growth was inhibited within a 2- to 3-cm radius of the inoculation point. However, conidia were transported by swarming P. vortex to regions outside the zone of inhibition of the VOR (Fig. S5A). This transport allowed successful conidial germination and outgrowth. It was also possible for P. vortex not loaded with conidia to swarm into an area seeded with conidia and VOR and in doing so, capture and rescue them from the antifungal agent (Fig. S5B). As with the other experiments, fungal dispersal was dependent on the presence of swarming P. vortex. This finding suggests that circumstances may exist in the soil where A. fumigatus conidia can be transported by P. vortex from niches where the fungus cannot thrive to more favorable locations. We note that the converse, movement of conidia to a less favorable niche, may also be possible, but this movement is likely to be less important from a fitness perspective. A small increase in transport in the presence of VOR (Fig. S4C, 40–49 mm from the inoculation point) was observed. Recovery of conidia transported from the VOR-containing region suggested that the antifungal agent restricted conidial germination and outgrowth. Recovery from exposure to VOR was possible under these circumstances: conidia retrieved by toothpick after 6 h (from the experiments using 10 μg VOR) showed >80% viability. This finding suggests that the restriction of microcolony development by VOR may have a benefit in increasing the transportability of A. fumigatus.
Transport of conidia into locations of antibiotic-limited bacterial growth.
P. vortex was even able to transport conidia into locations in which bacterial growth was inhibited by the presence of antibiotics. As detailed in SI Materials and Methods, Transport of Conidia into Locations Where Bacterial Growth Is Inhibited by Antibiotics, bacterial colonies inoculated outside a region of antibiotic inhibition (four different antibiotic agents were tested) can send pioneering swarmers (rotating bacterial groups or vortices) that are able to traverse several centimeters of antibiotic-impregnated agar to reach new areas favorable for growth (Figs. S6 and S7A). Swarming cells were shown to be temporarily refractory to the antibiotics tested, which has been observed previously for other swarming bacteria (15).
When inoculated in the center of the plate with conidia of A. fumigatus, pioneering groups of P. vortex were able to transport conidia into the regions of antibiotic action. In this situation, fungal outgrowth from the P. vortex colonies was rapid, and A. fumigatus quickly came to dominate the niche (Fig. S7 B and C). Furthermore, 70 h after inoculation, conidiophore development was observed only within the zone in which the antibiotics limited P. vortex growth (SI Materials and Methods, Transport of Conidia into Locations Where Bacterial Growth Is Inhibited by Antibiotics and Fig. S7D).
Conidia–Bacteria Interaction and the Transport Specificity.
Image processing and comparison with simulated virtual conidia.
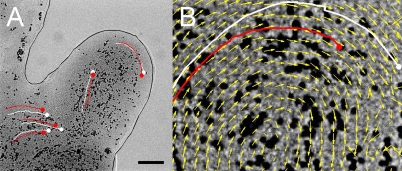
Movies of conidial transport by P. vortex were analyzed using a fast normalized cross-correlation method (SI Materials and Methods) to create a velocity field describing the motion of bacterial masses from frame to frame. Within this field, virtual conidia were placed, and the velocity field was used to predict their trajectory and velocity. The velocity (Table S1) of the virtual objects closely matched the velocity of the actual conidia. There was no significant difference in velocity between virtual and real conidia (Student t test, P > 0.1, n = 20), and the predicted path of the virtual conidia closely followed the actual trajectory of the closest conidia (Fig. 3). In the context of the model for virtual motion, this result suggested that the conidia were in some way connected to the bacteria and therefore, moving at the same speed (i.e., were tightly coupled).
Fig. 3.
Tracking and modeling conidial transport. (A) Still from a movie showing conidial transport (black spheres) in a background of moving P. vortex. Red trajectories are the tracks of individual conidia, and white trajectories are the tracks of virtual beads (calculation of the motion of bacteria aggregates that can carry beads was as explained in SI Materials and Methods). The continuous black line indicates the edge of the swarming mass. (B) Close-up view with the velocity field (yellow arrows) indicating the direction of local groups of microorganisms to overall describe a typical rotation (or vortex) motion. (Scale bar: A, 20 μm; B, 2 μm.)
EM of transport.
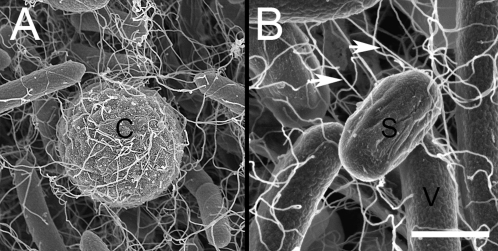
Imaging by scanning EM showed that the conidia were incorporated within the rafting mass of moving but interconnected bacteria, with the flagella of P. vortex both linking adjacent bacteria and entrapping the conidia (Fig. 4A), explaining the tight coupling suggested by the model (above). Each conidium was captured by from 6 to 30 flagella, typically derived from two to nine bacteria. P. vortex can also transport its own spores, which was shown when purified preparations of spores were coinoculated with swarming vegetative cells. In Fig. 4B, we show that, similar to the conidia, the bacterial spore is also apparently contacted by bacterial flagella and incorporated into the moving colony while transported.
Fig. 4.
Imaging of transport by scanning EM. (A) Conidium of A. fumigatus during transport by a P. vortex swarm imaged 3 h after inoculation at a location 24 cm from the inoculation point. C indicates the conidium. (B) Same as A but showing transport of a spore of P. vortex imaged 2 h after inoculation 2 cm away from the site of loading. Spore is marked by S. V indicates part of a P. vortex bacterium. Arrows indicate flagella. (Scale bar: A, 3 μm; B, 1 μm.)
Cargo specificity—dependency of the transport efficacy on the conidia type.
The transport of conidia from other fungi (Penicillium camemberti LCP66.584, P. citrinum S12, P. expansum BFE189, and Colletotrichum gloeosporoides f. sp. aeschynomene 3.1.3) by P. vortex was investigated. Transport was relatively inefficient compared with A. fumigatus conidia as determined by direct imaging of transported conidia (Fig. S8). Transport of P. camemberti and C. gloeosporoides f. sp. aeschynomene was particularly poor, with loading (conidia per colony) being only 2–13% of the loading achieved with A. fumigatus conidia.
Transporter specificity—comparison with other swarming bacteria.
Two other swarming bacteria, the closely related rhizosphere inhabitant P. polymyxa E681 and the Gram-negative bacterium Proteus mirabilis, were tested for their ability to transport conidia. Neither strain was able to transport conidia (from a range of fungi, which was noted in the previous section) tested under a wide range of conditions (SI Materials and Methods, Transporter Specificity—Comparison with Other Swarming Bacteria). Moreover, P. polymyxa did not even transport P. citrinum conidia, although this fungus can stimulate swarming of these bacteria by secretion of citrinin (16).
Surface properties of conidia are important for transport.
These experiments (described in the previous section) suggested a degree of specificity in transport. It seemed likely that the surface properties of conidia played a role. Indeed, when SDS or proteinase K was used to treat A. fumigatus conidia, subsequent transport by P. vortex was largely inhibited (Fig. S9). Addition of purified flagella from P. vortex (but not from P. mirabilis) inhibited the transport of A. fumigatus conidia by P. vortex. These data support the conclusions drawn from modeling (Fig. 3) and scanning EM (Fig. 4A) that P. vortex flagella are involved in capturing A. fumigatus conidia and likely to be contacting proteins on the surface of the fungal bodies.
Germination, Outgrowth, and Sporulation of Transported Conidia.
Transport effect on germination and outgrowth.
The effect of transport and growth of A. fumigatus within P. vortex colonies was studied. To investigate a possible effect on germination, transported conidia were recovered from within swarming P. vortex colonies using a toothpick and imaged by microscopy. The diameters of these recovered conidia were compared with the diameters of conidia incubated on RMHA in the absence of bacteria. In both situations, after 6 h, >90% of conidia were swollen, suggesting that P. vortex did not inhibit the early stages of germination of transported conidia. In other experiments detailed in SI Materials and Methods, Effect of Transport on Germination and Outgrowth, we found that the P. vortex bacteria did not inhibit the outgrowth of the fungal microcolonies after germination.
Inhibition of conidia and conidiophore formation.
For A. fumigatus conidia inoculated on RMHA plates in the absence of P. vortex, new conidiophores developed after 48–56 h. By contrast, for A. fumigatus growing on RMHA within P. vortex, conidia and conidiophore formation was not observed even after 80 h, despite the appearance of aerial mycelia within 26 h, suggesting that fungal sporulation was inhibited by P. vortex.
Effect of germination on the efficiency of conidia transport.
The transport of conidia was tested after pregermination in liquid RMH medium. We found that transport of conidia after 5–9 h germination, as the conidia developed into fungal microcolonies, was significantly decreased in efficiency compared with ungerminated conidia (Fig. S4B). This finding was confirmed by transferring (by toothpick) conidia and microcolonies at different stages in development into swarming P. vortex. Microcolonies above 10–20 μm in diameter were not transported when directly loaded into swarming P. vortex aggregates. The results indicate that P. vortex can distinguish between ungerminated conidia and germinating conidia.
Potential Benefits of Conidial Transport for P. vortex.
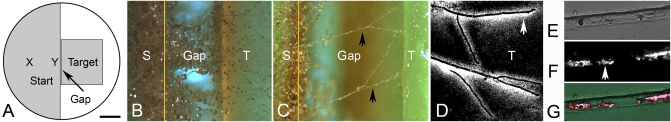
The transport of conidia by P. vortex poses the question as to what, if any, is the benefit to the bacterium. One notable feature of dispersal of microorganisms in the soil is that fungal mycelia are known to grow across air-filled regions between soil particles. Such air gaps are a more significant barrier to many species of bacteria (17, 18). An experimental system was devised where the bacterium was tested for its ability to cross a 0.5-mm-wide air gap in an RMHA plate (Fig. 5 A and B). In initial experiments, a mixture of P. vortex and A. fumigatus conidia (or each individually) was inoculated adjacent to the gap. Swarming P. vortex (alone) was unable to cross an air gap in all of 30 trials with incubation for 5 d at 37 °C. Swarming masses of P. vortex generally ceased swarming when reaching the air gap, with limited numbers of bacteria moving down the vertical wall on the inoculation side. However, the latter groups of bacteria were unable to cross the plastic of the Petri dish at the base of the air gap. No bacteria were detected on the far side when the fungus was inoculated alone. When the swarming bacteria were loaded with A. fumigatus conidia, P. vortex crossed the air gap in 16 of 30 trials. Microscopy of the air gap indicated that some transported conidia had germinated near the gap and grown across (Fig. 5B). When the air gap was increased to 0.8-mm wide, a smaller number of mycelia was able to bridge the gap (Fig. 5C), but these few mycelia were insufficient to facilitate P. vortex reaching the far side (0/30). When 0.5 mm PNPG was added to the RMHA and P. vortex was inoculated with conidia, the bacterium was successful in crossing 0.5-mm air gaps less frequently (i.e., in only 6 of 30 trials). This finding suggested a role for swarming and/or flagella in the process but not an absolute dependency. P. vortex was found to be associated with these bridging mycelia in large numbers on the far side of the air gap (Fig. 5D) and smaller numbers on some mycelia over the air gap (Fig. 5E). Examination of the mycelia from over the 0.5-mm air gaps by fluorescence microscopy indicated that hexidium iodide-stained P. vortex could be detected on 5–10% of mycelia (Fig. 5 E–G). In a second series of experiments, the two organisms were inoculated together or independently 3.5 cm from a 0.5-mm-wide air gap. In this situation, neither the fungus nor the bacterium inoculated alone could cross the air gap within 5 d. Coinoculation resulted in success in 9 of 30 trials. This test required P. vortex to transport conidia to the air gap and A. fumigatus to then facilitate the bacterium in crossing. These data suggest that there is a degree of mutualism between P. vortex and A. fumigates in that each one can assist the dispersal of the other in the right situation.
Fig. 5.
Testing the ability of P. vortex to cross air gaps with the assistance of A. fumigatus. (A) Experimental setup. A 90-mm diameter agar plate was filled with RMHA. A sterile razor blade was used to remove the RMHA from the right-hand one-half of the plate, leaving the inoculation region (Start). A second RMHA block (Target) was positioned to create an air gap (Gap) 0.5 or 0.8 mm wide. P. vortex with A. fumigatus conidia (or as a control, the bacteria alone) were inoculated at either position X (3.5 cm from air gap) or position Y (adjacent to air gap) in the inoculation region. This setup was incubated in a humidity chamber at 37 °C for 5 d. (B) Mycelial growth across a 0.5-mm air gap (Gap) from the starting region (S) to the target region (T). The left-hand edge of the air gap is obscured by fungal growth and is indicated by a vertical orange line. (C) Same as B but a smaller number of mycelia (black arrows) crosses a 0.8-mm air gap. (D) Fluorescence microscopy of target region (T) showing mycelia growing from the air gap (from the left) with associated P. vortex 3 d after inoculation. The bacteria are labeled with hexidium iodide, and the fungal mycelia are not stained but are visible as a negative image because of the surrounding bacteria (white). (E) Section of a mycelium crossing an air gap imaged by transmission light microscopy. (F) Same section of mycelia imaged by fluorescence microscopy revealing clusters of P. vortex associated with the fungus (labeled with hexidium iodide; e.g., white arrow). (G) Merged image combining E and F. (Scale bar: A, 17 mm; B and C, 0.3 mm; D, 80 μm; E–G, 25 μm.)
Discussion
Fungi use a diverse and fascinating range of methods to spread their conidia (19). Wind and water currents are common mechanisms. Additionally, a variety of ballistic methods (such as the impact of water droplets on the conidiophore) are used for local spread or to get conidia airborne. Transport by larger organisms, including insects, birds, and mammals (20–22), enables long-range dispersal. The work presented here shows that one microorganism can act as a vector for the dispersal of conidia from another microorganism. It is interesting that a swarm of smaller microorganisms (i.e., bacteria) can effectively cooperate to move a larger one.
One apparent benefit is rapid local dispersal. Also, it is likely that not all environmental niches are accessible to airborne conidia, and therefore, there may be gains in reaching new territory. Conidia present in an environment where they cannot germinate can be transported to regions where germination and outgrowth are possible, which was indicated by our finding that P. vortex swarms are able to rescue A. fumigatus spores from an antifungal agent (VOR). It is possible that agents that reversibly inhibit germination and outgrowth may increase dispersal by limiting the size of the cargo organism, and therefore, it remains transportable. We note a minor reversible effect of VOR (Fig. S4C), an antifungal compound that restricts fungal growth with limited exposure (23).
Treatments that damaged the conidial surface of A. fumigatus, particularly by the action of a protease but also by a strong detergent, inhibited transport. In addition, modeling, scanning EM, and inhibiting transport by the addition of purified P. vortex flagella (Fig. S9) all support a role for flagella in making specific contacts with the surface of the A. fumigatus conidia. Flagella are known to mediate attachment of bacteria to fungal mycelia (24), and this work suggests a role in conidial binding.
The soil is a heterogeneous environment, and P. vortex is capable of swarming in pioneering groups that can traverse unfavorable niches into distant, better ones. We found that the conidia can ride the pioneer groups into regions where antibiotics limited bacterial growth. Because conidial germination and outgrowth in these regions were favorable, the fungus might exploit the bacterium to enter niches where it can outgrow the transporting swarm.
In coculture with A. fumigatus, P. vortex limits sporulation of the fungus and reduces the size of microcolonies growing within areas of bacterial growth. The relationship between P. vortex and A. fumigatus seems competitive in some instances. However, the fungus can still effectively form aerial mycelia, and these aerial mycelia can grow out from bacterial masses and bridge air gaps that P. vortex cannot cross unaided. Because PNPG reduced the efficiency of this process, it is likely that swarming and/or flagella production (involved in motility or attachment to mycelia) play a part (24). Other workers have observed a phenomenon that may be related: fungal mycelia facilitate the spread of motile bacteria in the soil, acting as highways for motile bacteria (17, 18). In this work, there is an interesting mutualism with respect to dispersal: P. vortex can carry A. fumigatus conidia rapidly and rescue them from unfavorable niches, whereas the fungal mycelia allow the bacterium to cross a type of barrier, an air gap, common in many soils. However, any reproductive benefit of such mutualism in the soil remains to be shown.
Swarming P. vortex is able to transport its own spores when added as a cargo to vegetative cells (Fig. 4B). Given that sporulation takes time and resources and the rapidity by which P. vortex can swarm into unfavorable environments (Fig. S6), it may be a survival advantage to carry preformed spores rather than generate new ones in a crisis. Spore transport may act as a bet hedging strategy against the failure of adventurous swarms to find a favorable niche.
Previous studies on microbial transport have either covalently attached nonliving cargo objects to individual motile algae (3) or used layers of bacteria confined in microfabricated chambers or other arrays of immobile organisms with active flagella to move beads (2). Here, we have shown that none of these methods are necessary. P. vortex can pick up and retain cargo objects without covalent attachment, and they move them over tens of centimeters. Within the microengineering field, there is interest in microbial transport in patterning or as part of microfabricated devices. The sophisticated pattern-forming capability of P. vortex combined with its formidable transport capability may make it a good candidate.
Although swarming is a common phenomenon among soil microorganisms, nonmotile microorganisms must also find new niches. This fact raises the fascinating question as to whether there is a whole set of transporter–cargo interactions between the motile and sessile microbiota. This idea is supported by a recent report that the social amoeba Dictostylium discoideum can transport bacteria (25). To date, with rare exceptions (17), swarming has been studied in monoculture, a process in which even different strains of the same species may be incompatible (26). The situation may be a great deal more interesting in a complex and heterogeneous environment such as the soil.
Materials and Methods
Bacteria (P. vortex, P. polymyxa, or P. mirabilis) swarming at 37 °C on RMHA (14 g Oxoid MH broth/L) were harvested and resuspended to 109 cfu mL−1 in RMH broth. Fungal conidia or P. vortex spores for transport assays (cargo) were concentrated by centrifugation and resuspended to 108 cfu mL−1. Inoculations of bacteria alone, putative cargo objects alone, or both together were made by pipetting 10 μL droplets into the center of an RMHA plate (up to 107 bacteria and/or 106 cargo objects per inoculation). Incubation was at 37 °C for 4 h up to several days. Treatment of specific areas of plates with antibacterial or antifungal compounds as selective agents used NeoSensi Tabs as recommended by the manufacturer (Rosco Diagnostics).
The progress and behaviors of the swarming bacteria and cargo objects were monitored by microscopy. Real-time imaging was done using an Olympus BX-41 microscope equipped with ×4 and ×10 Fluorotar objective lenses (5). Agar plates were maintained at 37 °C on the microscope stage using a heated slide warmer and imaged by transmission light microscopy or fluorescence microscopy. Images were captured by a Kappa CCD camera. Individual frames were compiled into movies and analyzed using the ImageJ software suite, version 1.47 (http://rsb.info.nih.gov/ij/) (Movies S1, S2, and S3).
Recovery of microorganisms from specific regions of agar plates followed by selective viable counting were used to determine the degree of colonization by bacteria and fungi. Excision of specific regions from swarm plates followed by rapid fixation and scanning EM were as previously described (5). Swarm assays, imaging, and viable counting methods are described in more detail in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Joëlle Dupont, Rolf Geisen, Peter Schneeberger, and Amir Sharon for strains of fungi and advice on fungal culture and Adriaan van Aelst for help with EM. This research has been supported by the Maguy–Glass Chair in Physics of Complex Systems and a grant from the Tauber Family Foundation at Tel Aviv University.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102097108/-/DCSupplemental.
References
- 1.Darnton N, Turner L, Breuer K, Berg HC. Moving fluid with bacterial carpets. Biophys J. 2004;86:1863–1870. doi: 10.1016/S0006-3495(04)74253-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hiratsuka Y, Miyata M, Tada T, Uyeda TQP. A microrotary motor powered by bacteria. Proc Natl Acad Sci USA. 2006;103:13618–13623. doi: 10.1073/pnas.0604122103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weibel DB, et al. Microoxen: Microorganisms to move microscale loads. Proc Natl Acad Sci USA. 2005;102:11963–11967. doi: 10.1073/pnas.0505481102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Jacob E, et al. Generic modelling of cooperative growth patterns in bacterial colonies. Nature. 1994;368:46–49. doi: 10.1038/368046a0. [DOI] [PubMed] [Google Scholar]
- 5.Ingham CJ, Ben Jacob E. Swarming and complex pattern formation in Paenibacillus vortex studied by imaging and tracking cells. BMC Microbiol. 2008;8:36. doi: 10.1186/1471-2180-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Jacob E, et al. Bacterial cooperative organization under antibiotic stress. Physica A. 2000;282:247–282. [Google Scholar]
- 7.Galperin MY. A census of membrane-bound and intracellular signal transduction proteins in bacteria: Bacterial IQ, extroverts and introverts. BMC Microbiol. 2005;5:35–54. doi: 10.1186/1471-2180-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sirota-Madi A, et al. Genome sequence of the pattern forming Paenibacillus vortex bacterium reveals potential for thriving in complex environments. BMC Genomics. 2010;11:710. doi: 10.1186/1471-2164-11-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dijksterhuis J, Sanders M, Gorris LG, Smid EJ. Antibiosis plays a role in the context of direct interaction during antagonism of Paenibacillus polymyxa towards Fusarium oxysporum. J Appl Microbiol. 1999;86:13–21. doi: 10.1046/j.1365-2672.1999.t01-1-00600.x. [DOI] [PubMed] [Google Scholar]
- 10.Budi SW, van Tuinen D, Martinotti G, Gianinazzi S. Isolation from the Sorghum bicolor mycorrhizosphere of a bacterium compatible with arbuscular mycorrhiza development and antagonistic towards soilborne fungal pathogens. Appl Environ Microbiol. 1999;65:5148–5150. doi: 10.1128/aem.65.11.5148-5150.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hildebrandt U, Ouziad F, Marner FJ, Bothe H. The bacterium Paenibacillus validus stimulates growth of the arbuscular mycorrhizal fungus Glomus intraradices up to the formation of fertile spores. FEMS Microbiol Lett. 2006;254:258–267. doi: 10.1111/j.1574-6968.2005.00027.x. [DOI] [PubMed] [Google Scholar]
- 12.Nierman WC, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 13.Verweij PE, Mellado E, Melchers WJ. Multiple-triazole-resistant aspergillosis. N Engl J Med. 2007;356:1481–1483. doi: 10.1056/NEJMc061720. [DOI] [PubMed] [Google Scholar]
- 14.Campbell CK. Fine structure and physiology of conidial germination in Aspergillus fumigatus. Trans Br Mycol Soc. 1971;57:393–402. [Google Scholar]
- 15.Butler MT, Wang Q, Harshey RM. Cell density and mobility protect swarming bacteria against antibiotics. Proc Natl Acad Sci USA. 2010;107:3776–3781. doi: 10.1073/pnas.0910934107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park S-Y, et al. Citrinin, a mycotoxin from Penicillium citrinum, plays a role in inducing motility of Paenibacillus polymyxa. FEMS Microbiol Ecol. 2008;65:229–237. doi: 10.1111/j.1574-6941.2008.00492.x. [DOI] [PubMed] [Google Scholar]
- 17.Warmink JA, van Elsas JD. Migratory response of soil bacteria to Lyophyllum sp. strain Karsten in soil microcosms. Appl Environ Microbiol. 2009;75:2820–2830. doi: 10.1128/AEM.02110-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nazir R, Warmink JA, Boersma H, van Elsas JD. Mechanisms that promote bacterial fitness in fungal-affected soil microhabitats. FEMS Microbiol Ecol. 2010;71:169–185. doi: 10.1111/j.1574-6941.2009.00807.x. [DOI] [PubMed] [Google Scholar]
- 19.Van Leeuwen MR, Van Doorn TM, Golovina EA, Stark J, Dijksterhuis J. Water- and air-distributed conidia differ in sterol content and cytoplasmic microviscosity. Appl Environ Microbiol. 2010;76:366–369. doi: 10.1128/AEM.01632-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raguso RA, Roy BA. ‘Floral’ scent production by Puccinia rust fungi that mimic flowers. Mol Ecol. 1998;7:1127–1136. doi: 10.1046/j.1365-294x.1998.00426.x. [DOI] [PubMed] [Google Scholar]
- 21.Nagarajan S, Singh DV. Long-distance dispersion of rust pathogens. Annu Rev Phytopathol. 1990;28:139–153. doi: 10.1146/annurev.py.28.090190.001035. [DOI] [PubMed] [Google Scholar]
- 22.Frank JL, et al. Rodent dispersal of fungal spores promotes seedling establishment away from mycorrhizal networks on Quercus garryana. Botany. 2009;87:821–829. [Google Scholar]
- 23.Chryssanthou E, Loebig A, Sjölin J. Post-antifungal effect of amphotericin B and voriconazole against germinated Aspergillus fumigatus conidia. J Antimicrob Chemother. 2008;61:1309–1311. doi: 10.1093/jac/dkn129. [DOI] [PubMed] [Google Scholar]
- 24.Sen R, Nurmiaho-Lassila EL, Haahtela K, Korhonen T. Mycorrhizas in integrated systems from genes to plant development. In: Azcon-Aguilar C, Barea JM, editors. Proceedings of the 4th European Symposium on Mycorrhizae. Granada, Spain: European Commission; 1996. pp. 661–664. [Google Scholar]
- 25.Brock DA, Douglas TE, Queller DC, Strassmann JE. Primitive agriculture in a social amoeba. Nature. 2011;469:393–396. doi: 10.1038/nature09668. [DOI] [PubMed] [Google Scholar]
- 26.Gibbs KA, Urbanowski ML, Greenberg EP. Genetic determinants of self identity and social recognition in bacteria. Science. 2008;321:256–259. doi: 10.1126/science.1160033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.