Abstract
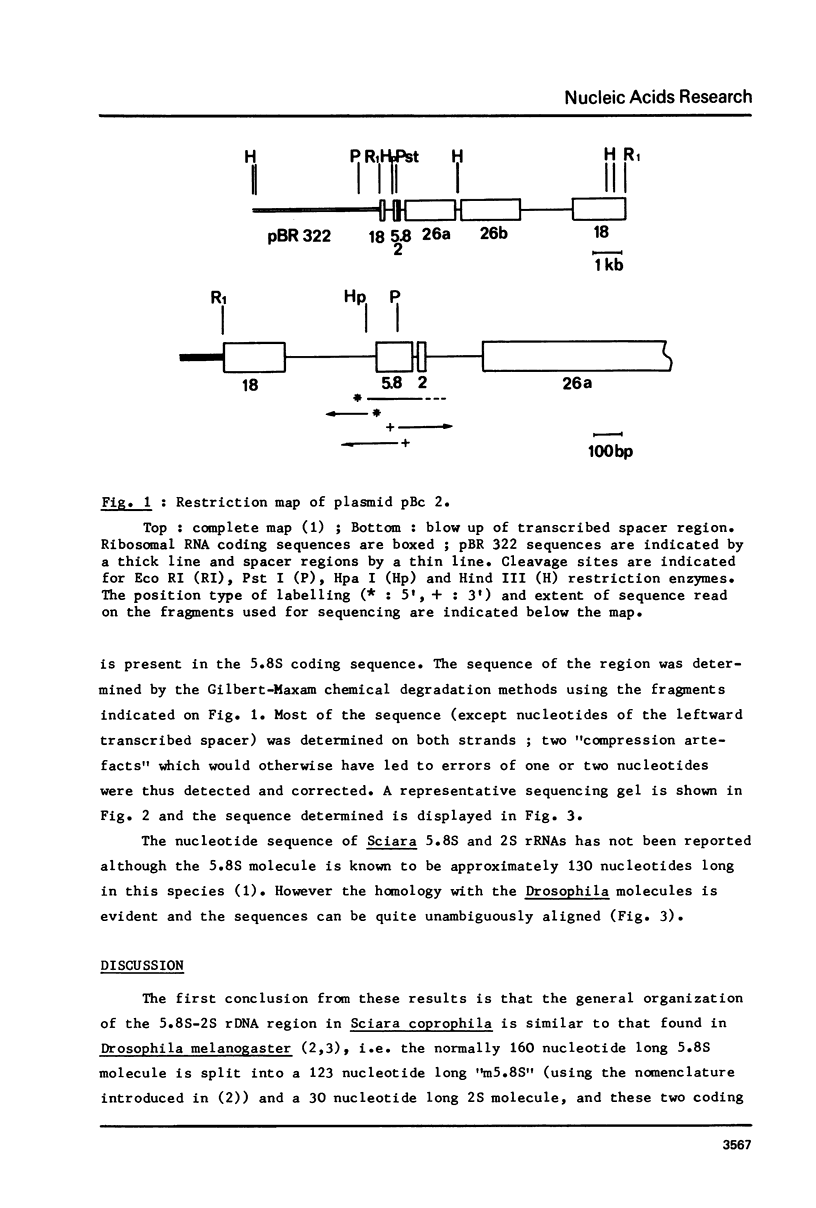
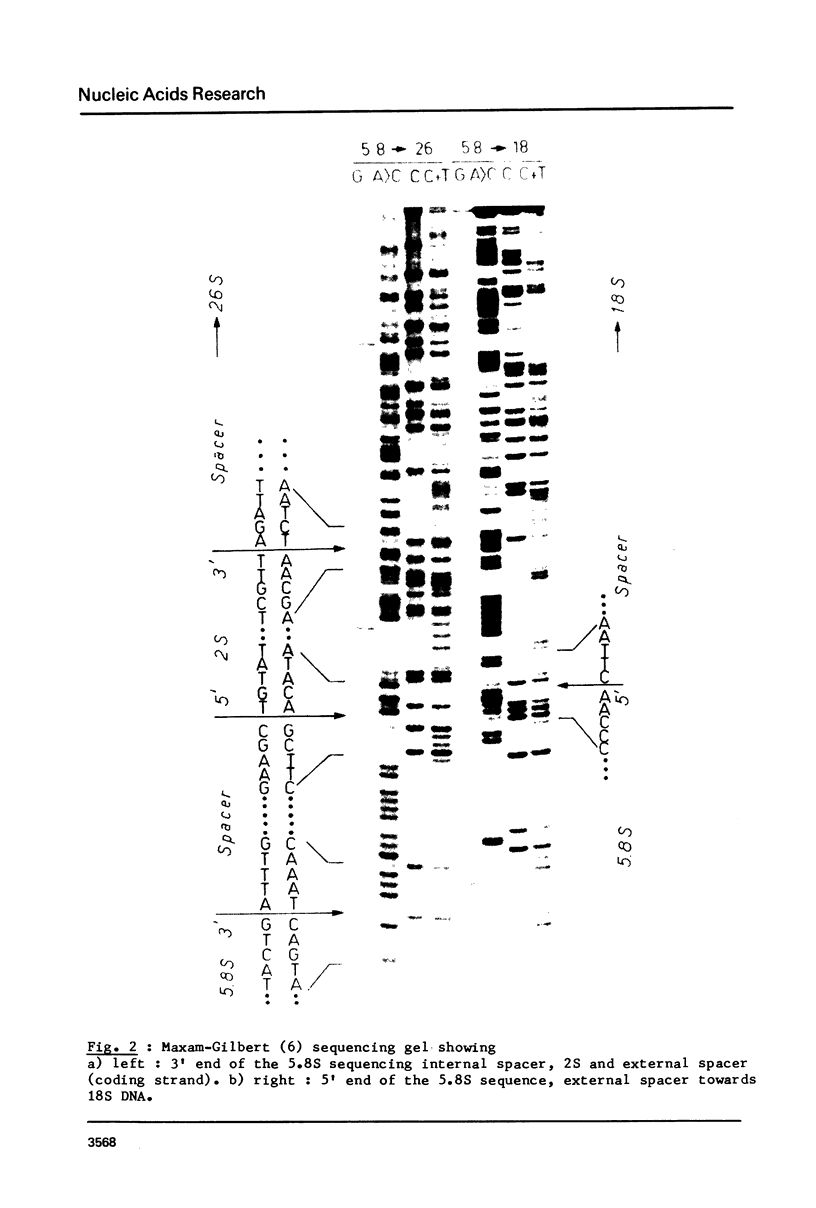
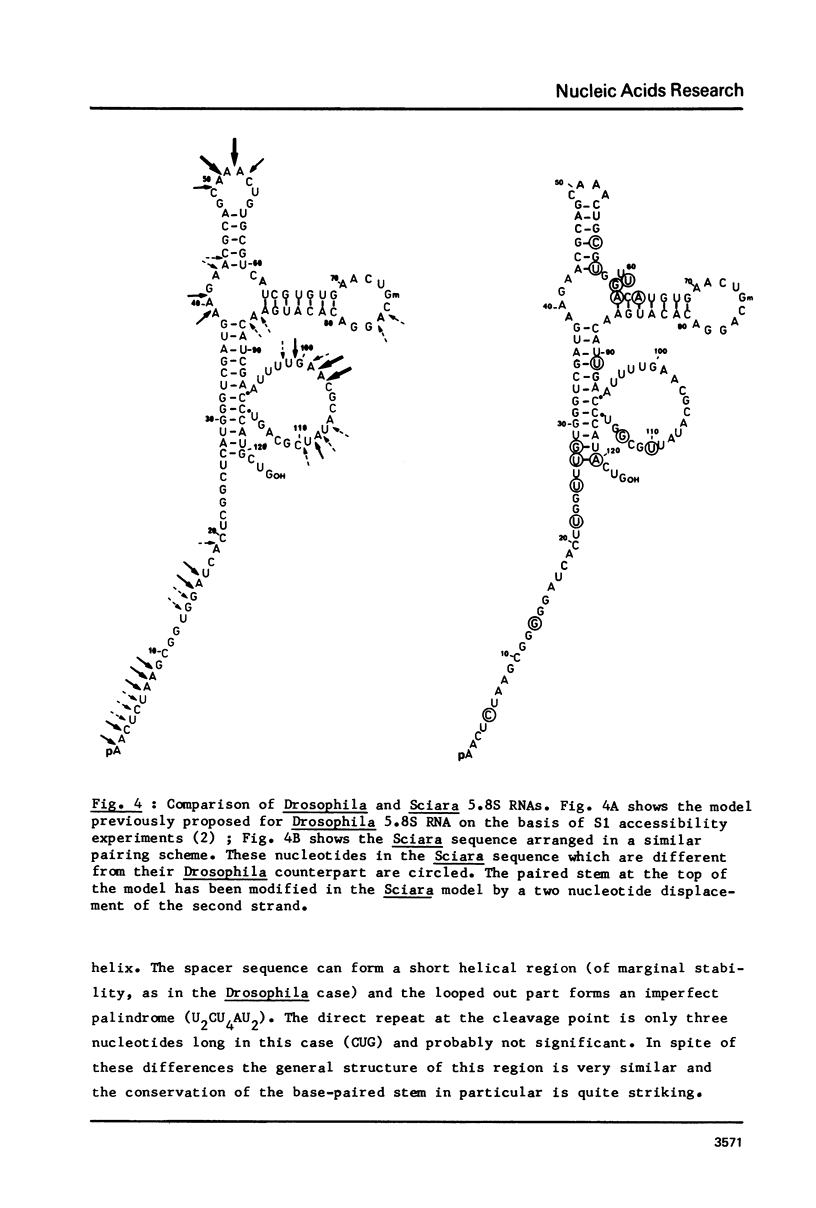
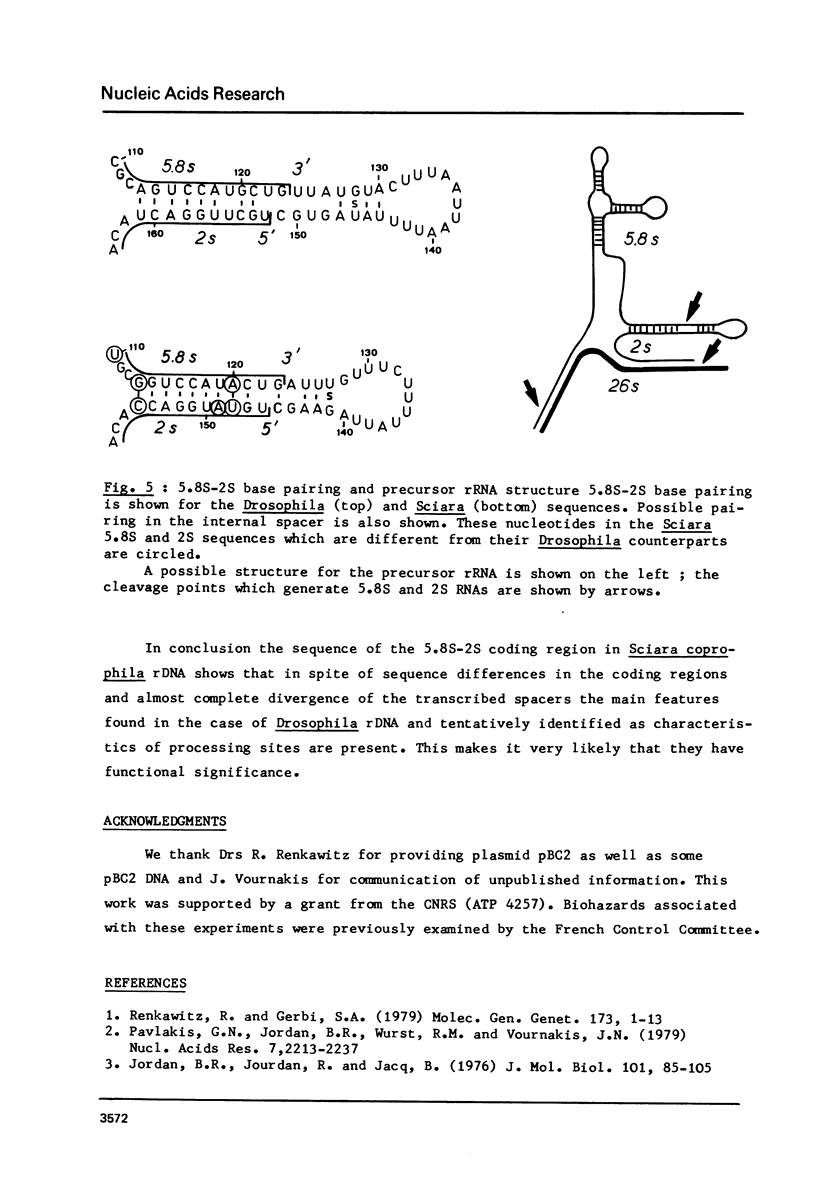
The sequence of 436 nucleotides around the region coding for 5.8S RNA in the Sciara coprophila rDNA transcription unit (1) has been determined. Regions coding for 5.8S and 2S RNAs have been identified; they are 80 - 90% homologous to the corresponding Drosophila sequences and are separated by a 22 nucleotide long spacer. This sequence as well as the two before the 5.8 and after the 2S coding region are very different from the corresponding Drosophila sequences. The main features reported in the Drosophila study (2) are however also found, i.e. all three spacers are very rich in A-T; the sequence of the internal spacer allows base pairing; 5.8S and 2S RNAs can pair through their 3' and 5' terminal regions respectively. The features previously proposed as processing sites in the Drosophila case are thus all found in Sciara in spite of very different spacer sequences.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beckingham K., White R. The ribosomal DNA of Calliphora erythrocephala; and analysis of hybrid plasmids containing ribosomal DNA. J Mol Biol. 1980 Mar 15;137(4):349–373. doi: 10.1016/0022-2836(80)90162-x. [DOI] [PubMed] [Google Scholar]
- Jordan B. R., Jourdan R., Jacq B. Late steps in the maturation of Drosophila 26 S ribosomal RNA: generation of 5-8 S and 2 S RNAs by cleavages occurring in the cytoplasm. J Mol Biol. 1976 Feb 15;101(1):85–105. doi: 10.1016/0022-2836(76)90067-x. [DOI] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Alternative pathways in the processing of ribosomal RNA precursor in Drosophila melanogaster. J Mol Biol. 1980 Apr 25;138(4):873–878. doi: 10.1016/0022-2836(80)90070-4. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlakis G. N., Jordan B. R., Wurst R. M., Vournakis J. N. Sequence and secondary structure of Drosophila melanogaster 5.8S and 2S rRNAs and of the processing site between them. Nucleic Acids Res. 1979 Dec 20;7(8):2213–2238. doi: 10.1093/nar/7.8.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkawitz R., Gerbi S. A., Glätzer K. H. Ribosomal DNA of fly Sciara coprophila has a very small and homogeneous repeat unit. Mol Gen Genet. 1979 May 23;173(1):1–13. doi: 10.1007/BF00267685. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]