Abstract
Recognition of microbial patterns by host pattern recognition receptors is a key step in immune activation in multicellular eukaryotes. Peptidoglycans (PGNs) are major components of bacterial cell walls that possess immunity-stimulating activities in metazoans and plants. Here we show that PGN sensing and immunity to bacterial infection in Arabidopsis thaliana requires three lysin-motif (LysM) domain proteins. LYM1 and LYM3 are plasma membrane proteins that physically interact with PGNs and mediate Arabidopsis sensitivity to structurally different PGNs from Gram-negative and Gram-positive bacteria. lym1 and lym3 mutants lack PGN-induced changes in transcriptome activity patterns, but respond to fungus-derived chitin, a pattern structurally related to PGNs, in a wild-type manner. Notably, lym1, lym3, and lym3 lym1 mutant genotypes exhibit supersusceptibility to infection with virulent Pseudomonas syringae pathovar tomato DC3000. Defects in basal immunity in lym3 lym1 double mutants resemble those observed in lym1 and lym3 single mutants, suggesting that both proteins are part of the same recognition system. We further show that deletion of CERK1, a LysM receptor kinase that had previously been implicated in chitin perception and immunity to fungal infection in Arabidopsis, phenocopies defects observed in lym1 and lym3 mutants, such as peptidoglycan insensitivity and enhanced susceptibility to bacterial infection. Altogether, our findings suggest that plants share with metazoans the ability to recognize bacterial PGNs. However, as Arabidopsis LysM domain proteins LYM1, LYM3, and CERK1 form a PGN recognition system that is unrelated to metazoan PGN receptors, we propose that lineage-specific PGN perception systems have arisen through convergent evolution.
Sensing microbial surface patterns via host-encoded pattern recognition receptors (PRRs) is a prerequisite for the activation of antimicrobial defenses in multicellular organisms (1–6). Microbial signatures triggering host innate immunity are collectively referred to as pathogen or microbe-associated molecular patterns (PAMPs/MAMPs) (7, 8). Several PAMPs, including bacterial lipopolysaccharides, flagellins, and fungal cell wall-derived glucan and chitin fragments, have been shown to possess immunogenic activities in metazoans and plants, thus suggesting evolutionary conservation of pattern recognition systems across lineage borders (1, 9). However, differences in the modular composition and ligand specificities of human (hTLR5) and plant (FLS2) flagellin receptors support the view that host sensors for microbial patterns have arisen independently through convergent evolution in different kingdoms (10). In addition, as flagellin is the only one of the aforementioned patterns for which both metazoan and plant receptors have been identified, sensible propositions on the evolutionary origin of eukaryotic innate immune systems require identification of additional host pattern recognition receptors.
Peptidoglycans (PGNs) constitute building blocks of the cell walls of Gram-positive and Gram-negative bacteria that are composed of alternating β(1,4)-linked N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) residues (11, 12). Polymeric heteroglycan chains are bridged by oligopeptides, thereby forming a crystal lattice structure that provides rigidity to the bacterial envelope. Lysine (Lys) and meso-diaminopimelic acid (DAP) residues in the peptide moiety specify bacterial PGNs as either Lys- or DAP-type. Monomeric and polymeric PGNs of both subtypes constitute bacterial PAMPs that trigger antibacterial defenses and promote innate immunity in mammalian hosts as well as in Drosophila melanogaster (13, 14). Recognition of PGNs in animal hosts is mediated through various PRRs, including scavenger receptors, nucleotide-binding oligomerization domain-containing proteins (NOD), peptidoglycan recognition proteins (PGRPs), PGN hydrolases, and TOLL-like receptor TLR2 (13–17).
Polymeric PGNs from Gram-positive and Gram-negative bacteria or a mixture of oligomeric muropeptides derived thereof act as PAMPs in the model plant Arabidopsis thaliana (18, 19). Our previous experiments suggested that the carbohydrate backbone of PGN is important for its immunogenic activity and that PGN is recognized in a receptor-mediated manner in Arabidopsis (19). Plant lysin-motif (LysM) domain proteins have been widely implicated in the recognition of GlcNAc-containing glycans. In legumes, establishment of symbiosis with soil-borne rhizobacteria requires LysM domain receptor proteins mediating the recognition of bacterial lipochitooligosaccharide nodulation (Nod) factors (20, 21). Likewise, recognition of the fungal PAMP chitin (an unbranched 1,4-linked GlcNAc homopolymer) and immune stimulation in Oryza sativa or Arabidopsis are dependent on LysM-type PRRs OsCEBiP/OsCERK1 or AtCERK1, respectively (22–25). Importantly, previous studies further suggested that Arabidopsis employs different perception systems for bacterial PGN and fungal chitin (19), despite the fact that the latter is structurally closely related to the carbohydrate moiety of bacterial PGN. Here we report the identification of a tripartite PGN recognition system in the plasma membrane of Arabidopsis with shared functions in PGN sensing and transmembrane signaling. This system comprises two LysM domain proteins implicated in PGN ligand binding (LYM1, LYM3) and a transmembrane LysM receptor kinase (CERK1) that is likely required for conveying the extracellular signal across the plasma membrane and for initiating intracellular signal transduction. Importantly, all three proteins were shown to be indispensable for PGN sensitivity and immunity to bacterial infection.
Results
LysM Domain Proteins LYM1 and LYM3 Bind PGN, Mediate Plant Sensitivity to PGN, and Contribute to Immunity to Bacterial Infection.
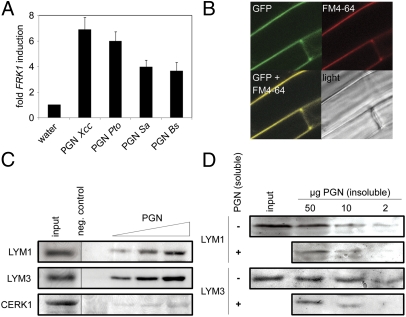
Infiltration into leaves of different PGN preparations results in the production of reactive oxygen intermediates and substantial reprogramming of the plant transcriptome, including enhanced expression of immune marker genes (18, 19). For example, flagellin-induced receptor kinase FRK1 gene expression is significantly enhanced upon treatment with PGNs from Gram-negative Xanthomonas campestris pv. campestris (Xcc) and Pseudomonas syringae pv. tomato (Pto) or from Gram-positive Bacillus subtilis (Bs) and Staphylococcus aureus (Sa) (Fig. 1A). As all PGNs tested were active triggers of FRK1 expression, the A. thaliana PGN perception system does not appear to discriminate between Lys-type (produced by Sa) or DAP-type PGNs (produced by Xcc, Pto, Bs). Notably, treatment with lysozyme, a muramidase that hydrolyzes O-glycosidic bonds between MurNAc and GlcNAc moieties within complex PGNs, strongly reduced the ability of Xcc PGN to trigger immune marker gene expression (SI Appendix, Fig. 1), suggesting that elicitor activities within the PGN preparations used were not due to contaminating bacterial cell wall components.
Fig. 1.
Arabidopsis LYM1 and LYM3 directly bind to PGN. (A) PGN induces FRK1 gene expression in Arabidopsis. Seedlings treated with 100 μg/mL PGN were subjected to RT-qPCR 6 h posttreatment. EF1a transcripts served normalization; corresponding water controls were set to 1. Data represent means ± SD of three independent experiments performed with 10–15 seedlings per genotype. (B) GFP-LYM3 is a plasma membrane protein. GFP fluorescence of lym3-1 plants stably expressing a p35S::GFP-LYM3 construct was analyzed by confocal laser-scanning microscopy. FM4-64 was used to stain the plasma membrane. (C and D) PGN binding assays. Recombinant, His6-tagged LYM1, LYM3, or CERK1 ectodomains (1 μg input) were incubated with 50 μg (or indicated amounts) PGN Sa, and bound protein was precipitated by centrifugation and analyzed by immunoblotting using anti-His6 antibodies. Negative controls did not contain PGN. Experiments were conducted at least three times. (C) Recombinant protein was precipitated by 10, 50, and 100 μg PGN (from left). (D) For competition experiments, LYM1-His6 or LYM3-His6 was incubated with PGN as indicated in the presence (+) or absence (−) of 100 μg soluble PGN.
Plant LysM domain proteins have been widely implicated in the recognition of GlcNAc-containing glycans, such as Nod factors (20, 21) and chitin (22–25). As our previous experiments had suggested receptor-mediated activation of PGN-induced defenses in Arabidopsis (19), we assumed that PGN recognition and subsequent immune activation might be controlled by a PRR with a LysM domain. The Arabidopsis genome harbors 14 genes encoding LysM domain proteins (SI Appendix, Fig. 2) comprising glycosylphosphatidylinositol-anchored LysM proteins (LYM1–3, LYP), LysM receptor kinases (CERK1, LYK2–5), and extracellular (LysMe) and intracellular nonsecretory LysM proteins (LysMn) (26). We focused on the functional characterization of the LYM3 protein as well as on the closely related protein LYM1 as putative PGN receptors (SI Appendix, Fig. 2). This choice was based on the presumed plasma membrane localization of LYM proteins (27) and of plant PRRs (1, 9), the structural similarity of LYM proteins to the rice chitin receptor CEBiP (22), and the suppression of LYM1 and LYM3 gene expression upon infection with virulent PtoDC3000 bacteria that was similar to that observed for the flagellin receptor FLS2 (SI Appendix, Fig. 3).
Plant PRRs are proposed to reside in the plant plasma membrane (1, 9). Confocal laser-scanning microscopy of Arabidopsis plants stably expressing a p35S::GFP-LYM3 construct revealed association of the LYM3 protein with the cell periphery, thus corroborating the proposed association of LYM proteins (27) with the plasma membrane (Fig. 1B). Immunoblotting and plasmolysis experiments further supported membrane localization of the protein (SI Appendix, Fig. 4). Likewise, LYM1 plasma membrane localization has been demonstrated previously (27). Next, we analyzed the ability of LYM1 and LYM3 to physically interact with bacterial PGNs using insoluble PGNs to precipitate soluble, recombinant His6-tagged LYM1 or LYM3, respectively. In addition, we tested the recombinant His6-tagged LysM domain of the chitin receptor CERK1 for PGN binding. As shown in Fig. 1C, PGN binding was demonstrated for both LYM1 and LYM3 but not for CERK1, thus reinforcing their roles as PGN-binding proteins. The PGN concentrations used (40–400 μg/mL) were very similar to those required to trigger FRK1 expression (Fig. 1A), thus demonstrating a quantitative correlation between concentrations required to bind LYM1 or LYM3 and to evoke a physiological response. Importantly, competition experiments conducted with excess soluble PGN fragments relative to insoluble PGNs revealed that the amount of precipitated LYM1 and LYM3 decreased in comparison to the control with insoluble PGN alone (Fig. 1D). These findings indicate specific interaction of the ligand with its cognitive binding proteins. More detailed kinetic analyses performed with recombinant LYM3 protein revealed a rapid and specific association with PGNs within 1 min upon incubation that increased over time (SI Appendix, Fig. 5A). Dissociation of preformed LYM3–PGN complexes was achieved by sudden dilution in excess buffer, demonstrating the reversible nature of this association event (SI Appendix, Fig. 5B). In experiments in which Nod factors or chitohexamers were used as competitors, no reduction in precipitated LYM3 was observed (SI Appendix, Fig. 5 C and D). Likewise, competition experiments performed with excess chitooctamers revealed that binding of PGNs to both recombinant LYM1 and LYM3 was unaltered in comparison with control experiments with insoluble PGN alone (SI Appendix, Fig. 5E). Altogether, our findings show that LYM1 and LYM3 bind PGNs in a ligand-specific manner, which is in agreement with the proposed pattern recognition receptor function of both proteins.
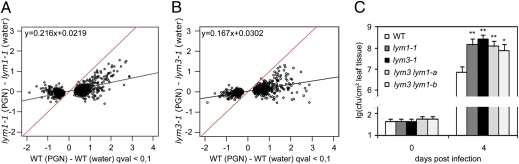
To examine the importance of LYM1 and LYM3 for the activation of PGN-inducible defenses in Arabidopsis, we isolated homozygous LYM1 or LYM3 T-DNA insertion mutants (lym1-1, lym1-2, lym3-1, lym3-2, ecotype Col-0) (SI Appendix, Fig. 6). When treated with PGN, all mutant alleles showed strongly reduced PGN-inducible marker gene expression (SI Appendix, Fig. 7). No reduction in FRK1 marker transcript levels was observed when lym1 and lym3 mutant genotypes were tested for chitin-inducible FRK1 expression (SI Appendix, Fig. 7), suggesting that LYM1 and LYM3 mediate a PGN-specific plant response. We further examined PGN-induced alterations in total gene expression in Col-0, lym1-1 and lym3-1 mutants. Seven hundred and fifty genes were differentially expressed in PGN-treated wild-type plants, 411 or 339 genes, respectively, of which expression was enhanced or reduced in PGN-treated Col-0 plants (Fig. 2 A and B; http://www.ncbi.nlm.nih.gov/geo, Gene Expression Omnibus accession no. GSE28004). Expression of virtually all PGN-responsive genes was altered in either lym1-1 or lym3-1 genotypes, suggesting that both LYM1 and LYM3 mediate activation of PGN responses. Deregulation in additional lym1-2 and lym3-2 alleles of PGN-responsive genes At1g51890, MLO12, PAD3, and CYP71A13 was confirmed by reverse transcription–quantitative PCR (RT-qPCR) (SI Appendix, Fig. 7). Importantly, lym1, lym3, as well as lym3 lym1 mutant alleles proved supersusceptible to infection with the virulent strain PtoDC3000 relative to Col-0 (Fig. 2C). Likewise, a lym3-1/p35S::GFP-LYM3 line used in LYM3 protein localization studies (Fig. 1B) or a genomic LYM3 complementation line exhibited wild-type levels of basal immunity (SI Appendix, Fig. 8 A and B). Increased growth on lym3 mutants was also found for nonpathogenic PtoDC3000 hrcC− and hypovirulent PtoDC3000 ΔAvrPto/PtoB strains (SI Appendix, Fig. 8 C and D). Altogether, we conclude that a lack of either LYM1 or LYM3 compromises PGN responsiveness. Further, a lack of LYM1, LYM3, or both proteins compromises immunity to bacterial infection. It is important to note here that supersusceptibility levels did not differ between lym single mutants and two double-mutant lines (Fig. 2C). This finding argues against functional redundancy in LYM1- and LYM3-dependent PGN-inducible immunity, and is in agreement with a cooperative role of both proteins in PGN sensing and immune activation.
Fig. 2.
LYM1 and LYM3 mediate PGN sensitivity and immunity to bacterial infection. (A and B) Analysis of global gene expression induced by PGN treatment in wild type and lym1-1 (A) and lym3-1 (B) mutants. Leaves of adult plants were treated with water or 100 μg/mL of PGN for 6 h, and extracted RNA was used for microarray analyses. Significantly induced or suppressed genes by either treatment were selected (750 genes; Storey's q value < 0.1), and the log2 ratios of transcript levels in PGN-treated plants versus water controls were plotted. Linear regression lines indicate gene expression levels in Col-0 (red) or mutants (black). (C) lym3 lym1 double mutants are as supersusceptible to bacterial infection as lym1-1 and lym3-1 single mutants. Growth of Pto DC3000 in lym1-1 and lym3-1 plants and two independent lym3 lym1 double-mutant lines (a, b) after infiltration of 104 colony-forming units/mL. Data represent means ± SD of six replicate measurements per genotype per data point. Statistical significance compared with wild type is indicated by asterisks (*P ≤ 0.1, **P ≤ 0.05, Student's t test). One of three independent experiments is shown.
LysM Receptor Kinase CERK1 Is Required for Sensitivity to PGNs and Immunity to Bacterial Infection.
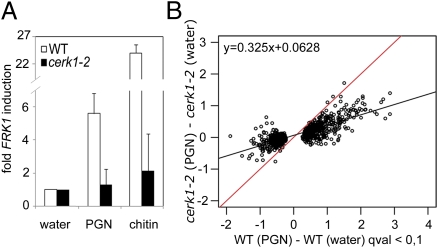
PGN-mediated immune stimulation requires initiation of an intracellular signal transduction cascade culminating in stimulus-dependent reprogramming of the transcriptome. As LYM1 and LYM3 lack cytoplasmic domains, transmembrane signaling likely requires an additional protein(s). Recently, an Arabidopsis mutant lacking the PRR CERK1 [CERK1 binds fungal chitin and mediates basal resistance to fungal infection (23, 24, 28, 29)] was shown to be compromised in immunity to bacterial infection (30). Moreover, chitin recognition and immunity to fungal infection in rice require the LysM domain proteins OsCEBiP and OsCERK1 (22, 25). We thus assumed that CERK1, in addition to LYM1 and LYM3, might be involved in PGN sensitivity, resulting in immunity to bacterial infection. Notably, cerk1-2 mutants exhibited neither fungal chitin- nor bacterial PGN-inducible FRK1 gene expression (Fig. 3A). Likewise, microarray experiments revealed that PGN-induced transcriptome changes were strongly reduced in the cerk1-2 mutant (Fig. 3B), which was also confirmed for selected genes in the same as well as in a second cerk1 allele, cerk1-3 (SI Appendix, Fig. 9 A and B). Further, we confirmed reported findings (30) that cerk1-2 mutants are supersusceptible to PtoDC3000 infection (SI Appendix, Fig. 9C). Thus, we provide evidence for the involvement of CERK1 in both PGN sensitivity and immunity to bacterial infection.
Fig. 3.
The LysM receptor kinase CERK1 mediates PGN sensitivity. (A) PGN-induced FRK1 transcript accumulation in cerk1-2 mutant Arabidopsis plants. FRK1 gene expression in seedlings treated with 100 μg/mL PGN or chitin were analyzed as described in Fig. 1A. The experiment was repeated five times. (B) Microarray analysis of the cerk1-2 mutant compared with Col-0 wild type performed as described in Fig. 2 A and B indicates that PGN-induced changes in the transcriptome are abolished in the cerk1-2 mutant.
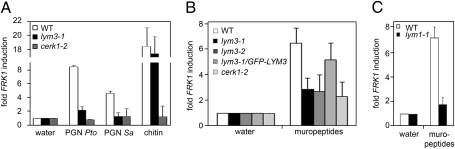
Gram-negative bacteria produce DAP-type PGNs, whereas the majority of Gram-positive bacteria produce Lys-type PGNs. We have shown that plants mount immunity-associated defenses upon treatment with either subtype of PGN (Fig. 1A). This finding raised the question of whether plants possess specific receptors for PGN subtypes or whether plants harbor PGN receptors mediating recognition of PGNs in general. Precedence for both is found, as mammals use nonspecific PGN receptors NOD1/NOD2 for PGN-mediated immune activation, whereas D. melanogaster has evolved PGN subtype-specific receptors (PGRP-LC recognizes DAP-type PGNs; PGRP-SA recognizes Lys-type PGNs) (13–17). We therefore tested DAP-type PGNs from Pto (in addition to Xcc PGN; Figs. 1 A–C and 3) and Lys-type PGN from Sa for FRK1 induction in lym3-1 and cerk1-2 mutants. As shown in Fig. 4A, FRK1 marker gene expression was virtually abolished in both genotypes, suggesting that the Arabidopsis PGN perception system apparently does not discriminate between different PGN subtypes. Moreover, mixtures of soluble oligomeric PGN fragments have previously been shown to stimulate plant immune responses in Arabidopsis (18, 19), suggesting that partially hydrolyzed ligands could potentially serve PRR-mediated immune activation. We now show that Arabidopsis senses soluble PGN fragments in a LYM1 LYM3 CERK1-dependent manner (Fig. 4 B and C), and conclude that this plant employs the same sensor system to recognize soluble PGN fragments and complex, insoluble PGNs.
Fig. 4.
Recognition of DAP-type PGN, Lys-type PGN, and muropeptides by the LYM1 LYM3 CERK1 system. FRK1 transcript accumulation in indicated mutant genotypes. Col-0, lym1-1, lym3-1, lym3-2, cerk1-2, and lym3-1/p35S::GFP-LYM3 (lym3-1/GFP-LYM3) seedlings were treated with 100 μg/mL PGN (Pto or Sa), chitin (A), or muropeptides (Xcc) (B and C), and FRK1 gene expression was analyzed via RT-qPCR as described in Fig. 1A.
Discussion
By using biochemical and reverse-genetics technologies, we have shown that the Arabidopsis PGN perception system comprises three LysM domain proteins for bacterial sensing and activation of host immunity. LYM1 and LYM3 are plasma membrane proteins that lack cytoplasmic signaling domains and that physically bind PGNs, whereas the LysM receptor kinase CERK1 is a transmembrane protein that itself does not bind PGNs. We further demonstrate a genetic and functional link between LYM1, LYM3, and CERK1, as all three proteins are required for the activation of PGN-inducible defenses and immunity to bacterial infection. Importantly, supersusceptibility to PtoDC3000 infection in lym1, lym3, and two independent lym3 lym1 mutants were virtually indistinguishable (Fig. 2C), strongly suggesting that neither protein operates in additive, functionally redundant pathways, but might cooperate in PGN sensing and immune activation. We therefore hypothesize that LYM1 and LYM3 proteins form a heteromeric PGN-binding module that in conjunction with the transmembrane receptor kinase CERK1 builds a receptor complex that is required for ligand binding and initiation of an intracellular signaling cascade. Ligand-induced receptor activation eventually culminates in the activation of basal defenses against host-adapted bacterial pathogens or immunity to infection by host nonadapted microbes.
The Arabidopsis LYM1 LYM3 CERK1 perception system is partially similar to the OsCEBiP OsCERK1 complex that mediates chitin perception and immunity to fungal infection in rice (25). In both cases, pattern perception and transmembrane signaling require LysM protein and LysM receptor kinase activity. Our genetic evidence now suggests that PGN perception in Arabidopsis requires the concerted activity of two LysM domain proteins (LYM1, LYM3). Whether other members of the OsCEBiP protein family are also implicated in chitin perception in rice remains to be investigated.
The growing body of structural and functional information on plant PRRs reveals that there are mainly two types of receptors mediating sensing of structurally different classes of microbial patterns. Leucine-rich repeat (LRR) proteins and LRR receptor kinases might be implicated in host immune activation preferentially to peptide patterns (1, 31), whereas LysM domain proteins and LysM receptor kinases might mediate microbial sensing and host immunity preferentially through recognition of glycan patterns (22–25). A wider implication of our findings is that LysM domain proteins appear to constitute a class of plant receptors that preferentially bind N-acetylglucosamine-containing ligands. This view is further corroborated by LysM-type receptors from Lotus japonicus and Medicago truncatula that are implicated in bacterial lipochitooligosaccharide Nod factor perception (20, 21). However, unlike Arabidopsis LYM1 and LYM3 and rice OsCEBiP, plant Nod factor receptors have not been shown yet to bind bacterial GlcNAc-containing ligands.
The LYM1 LYM3 CERK1 PGN perception system does not discriminate between complex and soluble PGN structures or between different subtypes of PGN originating from Gram-negative (DAP-type) or Gram-positive (Lys-type) bacteria, respectively. These conclusions are based on the findings that Arabidopsis did recognize different PGN preparations, and that sensitivity in lym1, lym3, and cerk1 genotypes to complex PGNs and soluble, fragmented PGNs was compromised. An emerging question concerns the minimum ligand structures required for receptor binding and host immune activation. Recognition by plants of both PGN subtypes through the same recognition system strongly suggests that the carbohydrate backbone is crucial to LYM1 LYM3 CERK1-mediated plant immune activation, as PGN subtypes differ in the composition of peptide bridges. This conclusion is further supported by the fact that mutanolysin-mediated cleavage of O-glycosidic bonds in the PGN carbohydrate backbone, but not lysostaphin-mediated cleavage of glycylglycine bonds in the peptide moiety of Sa PGN, eliminated PGN elicitor activity (19). Likewise, muramyldipeptides, which are known triggers of NOD2-mediated immune responses in mammalian systems, failed to stimulate immunity-associated defenses in Arabidopsis (19). Thus, PGN-mediated bacterial sensing and immunity to bacterial infection is most likely not mediated through recognition of a small peptidoglycan motif. Instead, the polymer chain length or 3D structures of bacterial PGNs might determine its PAMP activity in Arabidopsis. These ligand structure requirements resemble those of Drosophila PGRP-LC and PGRP-SA that are PGN receptors mediating the recognition of PGNs derived from Gram-negative or Gram-positive bacteria through Imd and Toll pathways, respectively (32). Likewise, PGN-induced antibacterial immune activation in Bombyx mori requires at least two repeating GlcNAc–MurNAc units with peptide chains (33). There is also precedence that plant LysM receptors recognize oligomeric GlcNAc-containing ligands, because the rice LysM domain protein OsCEBiP was reported to bind chitin oligomers with a degree of polymerization >6 only (34, 35). Thus, oligomerized GlcNAc–MurNAc disaccharide fragments may constitute the most bioactive ligands for the LYM1 LYM3 CERK1 perception system.
Peptidoglycans from Gram-negative and Gram-positive bacteria trigger host immune responses in mammals, insects, and plants. Peptidoglycans are thus not only common microbial patterns but also constitute immunogenic ligands of which host perception systems are extraordinarily widely distributed among multicellular eukaryotes. Our current report on a plant PGN receptor complex now closes a knowledge gap, as it adds plant LysM proteins to eukaryotic proteins implicated in sensing bacterial PGNs and host immunity to bacterial infection. A lack in primary sequence conservation among plant LysM proteins and mammalian PGN receptors NOD1, NOD2, TLR2, or PGLYRP1–4 as well as members of the Drosophila PGRP family strongly suggests, however, that these immune sensors have arisen independently through convergent evolution in different phylae. This view is further supported by the fact that PGN receptors from different lineages exhibit different ligand structure requirements for mediating host immunity. In sum, we propose that functionality of PGN receptor systems, but not their evolutionary origins, are conserved across lineage borders (13–17).
Methods
Plant Material and Pathogen Infection.
lym1-1, lym1-2, lym3-1, lym3-2, and cerk1-2 mutants are in the Columbia (Col-0) background, whereas cerk1-3 was isolated in the Wassilewskija (Ws-4) background. Origins of mutant lines and plant growth conditions are described inSI Appendix, Methods. Infection assays on Arabidopsis using P. syringae pv. tomato or P. syringae pv. phaseolicola were performed as described previously (36) on leaves of the same leaf stage of 4- to 5-wk-old plants. Data were analyzed with a two-tailed, unpaired Student's t test. P values of less than 0.05 were considered significant.
PGN Preparations and Other Carbohydrates.
The purification of X. campestris pv. campestris PGN and muropeptides derived thereof was performed as described (18). PGNs from S. aureus and P. syringae pv. tomato were prepared as described in SI Appendix, Methods. B. subtilis PGN and soluble PGN from Escherichia coli were purchased from Invivogen. All described PGNs, chitohexamer, chitooctamer (Seikagaku), and chitin (Sigma) were dissolved in water at a concentration of 1 or 10 mg/mL. Nod factors [LCO-IV(S, C16:2)] were described previously (37).
DNA Constructs and Transgenic Plants.
For a detailed description of the generation of p35S::GFP-LYM3 and pLYM3::LYM3 transgenic plants and the generation of His6-tagged fusion proteins, see SI Appendix, Methods.
Carbohydrate Binding Assay.
Carbohydrate binding was assayed at 4 °C with 1 μg Ni2+-nitrilotriacetic acid (NTA) batch-purified protein. Purified protein was mixed with 50 μg or the given amount of PGN from S. aureus in 250 μL of 100 mM phosphate buffer (pH 7.0) for 10 min at 4 °C if not stated otherwise. Bound protein retained in the PGN pellet after centrifuging the incubation mixture at 13,000 × g for 10 min at 4 °C was washed twice with 100 mM phosphate buffer (pH 7.0) and then dissolved in SDS sample buffer. Following SDS/polyacrylamide gel separation, samples were analyzed by immunoblot with an anti-His antibody (Sigma). For a detailed description of association, dissociation, and competition studies, see SI Appendix, Methods.
RT-qPCR and Microarray Analysis.
For RT-qPCR analyses, Arabidopsis seeds were surface-sterilized and germinated on half-strength Murashige–Skoog (MS) medium supplemented with 0.8% (wt/vol) agarose. Seedlings were grown for 8 d at long-day conditions (16-h photoperiod, 22 °C, 40–60% humidity) and used for elicitor treatment (23). The seedlings were preincubated with fresh half-strength MS medium supplemented with 1% (wt/vol) sucrose in a 48-well microtiter plate overnight and then treated with 100 μg/mL Xcc PGN (if not stated otherwise), muropeptides, or chitin for 6 h. RNA isolation and RT-qPCR analysis were performed as described previously (19). All quantifications were made in duplicate on RNA samples obtained from three independent experiments, each performed with a pool of two leaves or 10–15 seedlings. EF1a transcripts served normalization; corresponding water controls were set to 1. The sequences of the primers used for PCR amplifications are indicated in SI Appendix, Table 1.
Microarray experiments were performed on A. thaliana Col-0 plants and T-DNA insertion lines lym1-1, lym3-1, and cerk1-2. Leaves of 4- to 5-wk-old plants were treated for 6 h with PGNs or water as a control. Total RNA was profiled using the NimbleGen DNA microarray (A. thaliana Gene Expression 12 × 135K Array TAIR 9.0) following the manufacturer's protocol (Roche). Three independent experiments (biological replicates) were performed. For a detailed description of microarray analysis, see SI Appendix, Methods.
Supplementary Material
Acknowledgments
We thank Marcelo Desimone, Claudia Oecking, Frédéric Brunner, Hugues Driguez, Eric Samain, Martin Ohsten Rasmussen, Naoto Shibuya, and Marc Ongena for providing materials, David Nittner for technical support, and Wolfgang Knogge and Jürg Felix for critical discussions. This work was supported by the Deutsche Forschungsgemeinschaft (SFB766 to A.A.G. and T.N.; SPP1212 to T.N.), the National Science Foundation (MCB-0918908 to F.K.), the Danish Council for Independent Research, Technology and Production Sciences (G.E. and M.-A.N.), and the European Commission Marie Curie Research Training Network (MRTN-CT-2006-035546 “NODPERCEPTION”), which supported the stay of R.W. at the Laboratory of Plant-Microbe Interactions, Toulouse, France.
Footnotes
The authors declare no conflict of interest.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE28004).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112862108/-/DCSupplemental.
References
- 1.Boller T, Felix G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 2.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Ishii KJ, Koyama S, Nakagawa A, Coban C, Akira S. Host innate immune receptors and beyond: Making sense of microbial infections. Cell Host Microbe. 2008;3:352–363. doi: 10.1016/j.chom.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 5.Segonzac C, Zipfel C. Activation of plant pattern-recognition receptors by bacteria. Curr Opin Microbiol. 2011;14(1):54–61. doi: 10.1016/j.mib.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 6.Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: Discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6(1):10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 8.Thomma BP, Nürnberger T, Joosten MH. Of PAMPs and effectors: The blurred PTI-ETI dichotomy. Plant Cell. 2011;23(1):4–15. doi: 10.1105/tpc.110.082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nürnberger T, Brunner F, Kemmerling B, Piater L. Innate immunity in plants and animals: Striking similarities and obvious differences. Immunol Rev. 2004;198:249–266. doi: 10.1111/j.0105-2896.2004.0119.x. [DOI] [PubMed] [Google Scholar]
- 10.Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nat Immunol. 2005;6:973–979. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- 11.Glauner B, Höltje JV, Schwarz U. The composition of the murein of Escherichia coli. J Biol Chem. 1988;263:10088–10095. [PubMed] [Google Scholar]
- 12.Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dziarski R, Gupta D. Review: Mammalian peptidoglycan recognition proteins (PGRPs) in innate immunity. Innate Immun. 2010;16(3):168–174. doi: 10.1177/1753425910366059. [DOI] [PubMed] [Google Scholar]
- 14.Kurata S. Extracellular and intracellular pathogen recognition by Drosophila PGRP-LE and PGRP-LC. Int Immunol. 2010;22(3):143–148. doi: 10.1093/intimm/dxp128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Müller-Anstett MA, et al. Staphylococcal peptidoglycan co-localizes with Nod2 and TLR2 and activates innate immune response via both receptors in primary murine keratinocytes. PLoS One. 2010;5:e13153. doi: 10.1371/journal.pone.0013153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Royet J, Dziarski R. Peptidoglycan recognition proteins: Pleiotropic sensors and effectors of antimicrobial defences. Nat Rev Microbiol. 2007;5:264–277. doi: 10.1038/nrmicro1620. [DOI] [PubMed] [Google Scholar]
- 17.Strober W, Murray PJ, Kitani A, Watanabe T. Signalling pathways and molecular interactions of NOD1 and NOD2. Nat Rev Immunol. 2006;6(1):9–20. doi: 10.1038/nri1747. [DOI] [PubMed] [Google Scholar]
- 18.Erbs G, et al. Peptidoglycan and muropeptides from pathogens Agrobacterium and Xanthomonas elicit plant innate immunity: Structure and activity. Chem Biol. 2008;15:438–448. doi: 10.1016/j.chembiol.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 19.Gust AA, et al. Bacteria-derived peptidoglycans constitute pathogen-associated molecular patterns triggering innate immunity in Arabidopsis. J Biol Chem. 2007;282:32338–32348. doi: 10.1074/jbc.M704886200. [DOI] [PubMed] [Google Scholar]
- 20.Limpens E, et al. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science. 2003;302:630–633. doi: 10.1126/science.1090074. [DOI] [PubMed] [Google Scholar]
- 21.Radutoiu S, et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–592. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- 22.Kaku H, et al. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc Natl Acad Sci USA. 2006;103:11086–11091. doi: 10.1073/pnas.0508882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miya A, et al. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:19613–19618. doi: 10.1073/pnas.0705147104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wan J, et al. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell. 2008;20:471–481. doi: 10.1105/tpc.107.056754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimizu T, et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010;64:204–214. doi: 10.1111/j.1365-313X.2010.04324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang XC, Cannon SB, Stacey G. Evolutionary genomics of LysM genes in land plants. BMC Evol Biol. 2009;9:183. doi: 10.1186/1471-2148-9-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borner GH, Lilley KS, Stevens TJ, Dupree P. Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A proteomic and genomic analysis. Plant Physiol. 2003;132:568–577. doi: 10.1104/pp.103.021170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iizasa E, Mitsutomi M, Nagano Y. Direct binding of a plant LysM receptor-like kinase, LysM RLK1/CERK1, to chitin in vitro. J Biol Chem. 2010;285:2996–3004. doi: 10.1074/jbc.M109.027540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petutschnig EK, Jones AM, Serazetdinova L, Lipka U, Lipka V. The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J Biol Chem. 2010;285:28902–28911. doi: 10.1074/jbc.M110.116657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gimenez-Ibanez S, et al. AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol. 2009;19:423–429. doi: 10.1016/j.cub.2009.01.054. [DOI] [PubMed] [Google Scholar]
- 31.Zipfel C. Pattern-recognition receptors in plant innate immunity. Curr Opin Immunol. 2008;20(1):10–16. doi: 10.1016/j.coi.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 32.Leulier F, et al. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat Immunol. 2003;4:478–484. doi: 10.1038/ni922. [DOI] [PubMed] [Google Scholar]
- 33.Iketani M, Nishimura H, Akayama K, Yamano Y, Morishima I. Minimum structure of peptidoglycan required for induction of antibacterial protein synthesis in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 1999;29(1):19–24. doi: 10.1016/s0965-1748(98)00099-x. [DOI] [PubMed] [Google Scholar]
- 34.Ito Y, Kaku H, Shibuya N. Identification of a high-affinity binding protein for N-acetylchitooligosaccharide elicitor in the plasma membrane of suspension-cultured rice cells by affinity labeling. Plant J. 1997;12:347–356. doi: 10.1046/j.1365-313x.1997.12020347.x. [DOI] [PubMed] [Google Scholar]
- 35.Okada M, Matsumura M, Ito Y, Shibuya N. High-affinity binding proteins for N-acetylchitooligosaccharide elicitor in the plasma membranes from wheat, barley and carrot cells: Conserved presence and correlation with the responsiveness to the elicitor. Plant Cell Physiol. 2002;43:505–512. doi: 10.1093/pcp/pcf060. [DOI] [PubMed] [Google Scholar]
- 36.Kemmerling B, et al. The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr Biol. 2007;17:1116–1122. doi: 10.1016/j.cub.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 37.Ohsten Rasmussen M, Hogg B, Bono JJ, Samain E, Driguez H. New access to lipo-chitooligosaccharide nodulation factors. Org Biomol Chem. 2004;2:1908–1910. doi: 10.1039/b403575e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.