Abstract
Mammalian methionyl-tRNA synthetase (MRS) plays an essential role in initiating translation by transferring Met to initiator tRNA (tRNAiMet). MRS also provides a cytosolic anchoring site for aminoacyl-tRNA synthetase-interacting multifunctional protein-3 (AIMP3)/p18, a potent tumor suppressor that is translocated to the nucleus for DNA repair upon DNA damage. However, the mechanism by which this enzyme mediates these two seemingly unrelated functions is unknown. Here we demonstrate that AIMP3 is released from MRS by UV irradiation-induced stress. Dissociation was induced by phosphorylation of MRS at Ser662 by general control nonrepressed-2 (GCN2) following UV irradiation. Substitution of Ser662 to Asp (S662D) induced a conformational change in MRS and significantly reduced its interaction with AIMP3. This mutant possessed significantly reduced MRS catalytic activity because of loss of tRNAMet binding, resulting in down-regulation of global translation. According to the Met incorporation assay using stable HeLa cells expressing MRS S662A or eukaryotic initiation factor-2 subunit-α (eIF2α) S51A, inactivation of GCN2-induced phosphorylation at eIF2α or MRS augmented the role of the other, suggesting a cross-talk between MRS and eIF2α for efficient translational inhibition. This work reveals a unique mode of regulation of global translation as mediated by aminoacyl-tRNA synthetase, specifically MRS, which we herein identified as a previously unidentified GCN2 substrate. In addition, our research suggests a dual role for MRS: (i) as a coregulator with eIF2α for GCN2-mediated translational inhibition; and (ii) as a coupler of translational inhibition and DNA repair following DNA damage by releasing bound tumor suppressor AIMP3 for its nuclear translocation.
Translational regulation is a mechanism by which genetic expression can be modulated to cope with various biological conditions. In diseases such as cancer, dysregulation of protein synthesis is frequently observed; therefore, accurate translational control appears to be important for the maintenance of normal growth and proliferation (1, 2). Under stress conditions, global translational control mainly occurs at the point of translational initiation through modification of eukaryotic initiation factors (eIFs). A key regulatory mechanism of this process is phosphorylation of eIF2 subunit-α (eIF2α), which prevents formation of a ternary complex (TC) comprising eIF2, GTP, and Met-charged initiator tRNA (Met-tRNAiMet), thereby inhibiting further rounds of translation initiation (3).
Aminoacyl-tRNA synthetases (ARSs) are essential enzymes for protein synthesis, linking codons to their corresponding amino acids (4, 5). A key factor in translation initiation, methionyl-tRNA synthetase (MRS) produces Met-tRNAiMet, which is indispensable for TC formation. MRS has been also found in the nucleus, where it may play a role in the biogenesis of rRNA (6). Under oxidative stress, MRS charges Met to noncognate tRNAs at a high frequency, resulting in reduced translational fidelity (7). Stable overexpression of the MRS substrate tRNAiMet can cause oncogenic transformation (8). These reports indicate the unique significance of MRS in translational control; however, little is known regarding the regulation of its catalytic activity and noncanonical activities.
In mammalian systems, nine different ARSs including MRS form an intriguing macromolecular complex, the multisynthetase complex (MSC), which contains three nonenzymatic factors designated ARS-interacting multifunctional protein-1 (AIMP1)/p43, AIMP2/p38, and AIMP3/p18 (4, 5, 9). Within this complex, MRS forms a strong association with the tumor suppressor AIMP3 (10–12). Although AIMP3 is mainly anchored to MRS in the cytosol, it also activates ATM/ATR in the nucleus upon DNA damage or oncogenic stress (11, 13, 14). However, it is unclear whether nuclear AIMP3 is actually translocated from the cytosolic MSC. Reasoning that translational regulation and the DNA damage response should be closely coupled for the precise control of cell survival, we hypothesized cross-talk between nuclear translocation of AIMP3 and translational control following UV irradiation. This study was designed to address the roles of MRS in the regulation of translation and cellular localization of the tumor suppressor AIMP3 and its underlying molecular mechanism.
Results
AIMP3 and MRS Interact Through GST-Homology Domains.
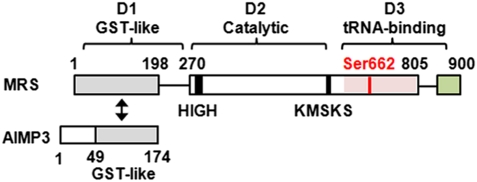
The structural organization of MRS varies significantly between species (15). Human MRS contains eukaryote-specific noncatalytic extensions, such as GST-homology and WHEP domains (16) (Fig. 1). WHEP domain is present in tryptophanyl-tRNA synthetase (WRS), histidyl-tRNA synthetase (HRS), and glutamyl-prolyl-tRNA synthetase (EPRS), for which the domain is named. AIMP3 also contains a C-terminal GST-homology domain. Although interaction between MRS and AIMP3 has been reported previously (10–12), it is unclear which domains mediate the interaction. To map the interaction between MRS and AIMP3, MRS deletion fragments, D1, D2, D3, and the N- and C-terminal domains of AIMP3 were attested for binding in a yeast two-hybrid assay. Among all of the pairs of LexA-AIMP3 and B42-MRS derivatives, AIMP3 strongly interacted with MRS D1 (Fig. S1A). We also tested the interaction of MRS with the AIMP3 C terminus in a pairwise test of LexA-MRS and B42-AIMP3 fragments (Fig. S1B). Pull-down assays revealed that AIMP3 also showed strong interaction with MRS D1 and MRS showed interaction with the AIMP3 C terminus (Fig. S1 C and D). Together, these results demonstrate that MRS and AIMP3 interact via their GST-homology domains (Fig. 1).
Fig. 1.
Schematic representation of functional domain arrangements in human MRS and AIMP3. Human MRS contains an N-terminal extension that has homology to GST (gray). The central catalytic domain (white) contains class I signature motifs, such as HIGH and KMSKS. The C-terminal domain (pink for anticodon binding and green for WHEP domains) interacts with tRNAMet. The GST-homology domain of MRS specifically interacts with another GST-homology domain (gray) of AIMP3. Ser662 is phosphorylated by UV-activated GCN2. D1 (1–266 residues), D2 (267–597), and D3 (598–900) are the deletion fragments of MRS that were used for further assays.
AIMP3 Is Released from MRS and Translocated to Nucleus upon UV Stress.
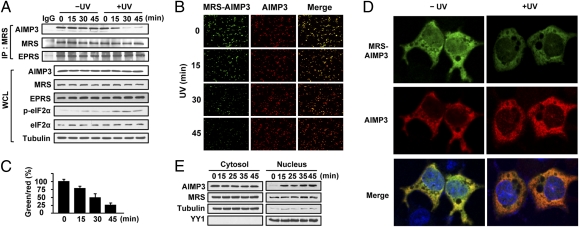
AIMP3 is localized to the nucleus following UV irradiation (14), prompting us to investigate whether UV irradiation affects the interaction between MRS and AIMP3. In immunoprecipitation (IP) assays using HeLa cells, we found that although the interaction between MRS and AIMP3 was maintained without UV irradiation, levels of MRS-bound AIMP3 gradually decreased after UV irradiation (Fig. 2A). MRS binding by EPRS, another component of MSC, was unaffected by UV, indicating that AIMP3 was specifically released from MSC (Fig. 2A). The dissociation of MRS and AIMP3 was coincident with induction of phosphorylated eIF2α (p-eIF2α), a known marker for UV-dependent translational inhibition (17).
Fig. 2.
AIMP3 dissociates from MRS and translocates to the nucleus following UV irradiation. (A) MRS-interacting proteins in UV-irradiated HeLa cells (60 J/m2) were determined by IP. Levels of proteins in whole cell lysates (WCL) were also detected by immunoblotting. (B) HCT116 cells cotransfected with Flag–AIMP3-VN and HA–MRS-VC were UV-irradiated. Reconstitution of Venus fluorescence by the interaction of MRS and AIMP3 is shown by green fluorescence. AIMP3 alone was observed by red fluorescence of the Alexa Fluor 555-conjugated anti-Flag Ab (200x). (C) The graph presents the relative ratios of green (MRS:AIMP3 complex) versus red (AIMP3) spot counts. Data are represented as mean ± SD (n = 3). (D) The dissociation of AIMP3 from MRS and nuclear localization was monitored at higher magnification (600x) using HeLa cells. The red fluorescence from AIMP3 merged with DAPI-stained nucleus, forming foci. (E) After UV irradiation, cytosolic and nuclear AIMP3 was analyzed by immunoblotting. YY1 and tubulin were used as controls.
To visualize the direct interaction between MRS and AIMP3, we used bimolecular fluorescence complement (BiFC) analysis by fusing the Venus N-terminal (VN) and C-terminal (VC) fragments to AIMP3 and MRS, respectively (18). VN and VC are nonfluorescent fragments; however, their close proximity, mediated by the interaction of the fused binding proteins, reconstituted Venus fluorescence. Using alternate constructs, we confirmed that cotransfection of Flag–AIMP3-VN and HA–MRS-VC produced green fluorescence, as mediated by MRS and AIMP3 interaction (Fig. S2A).
To test whether UV irradiation would induce dissociation of MRS and AIMP3, we transfected HCT116 cells with both fusion proteins and exposed the cells to UV. We found the number of green-fluorescent cells decreased, indicating UV-induced dissociation of MRS and AIMP3 (Fig. 2B). Red fluorescence from AIMP3 was unchanged, indicating that AIMP3 levels and cell viability were not affected (Fig. 2B). Counts of green versus red fluorescence indicate that dissociation of MRS and AIMP3 is a gradual process (Fig. 2C).
When BiFC was monitored at higher magnification, we observed UV-dependent nuclear AIMP3 foci formation (Fig. 2D), suggesting that AIMP3 dissociation from MRS is accompanied by nuclear localization. UV-induced nuclear foci formation was also observed when GFP-AIMP3 was overexpressed in HeLa cells (Fig. S2B). UV-induced nuclear translocation of AIMP3 was confirmed by cell fractionation and immunoblotting (Fig. 2E). Although AIMP3 did not appear to be significantly changed in the cytosol, an increase in AIMP3 in the nucleus following UV irradiation was clearly demonstrated. A portion of MRS was also found in the nuclear fraction, as previously reported (6); however, this localization occurred regardless of UV irradiation. We did not observe dissociation of MRS and AIMP3 by cycloheximide treatment (Fig. S2C), suggesting the dissociation is not induced by blockage of de novo protein synthesis.
Phosphorylation of MRS by UV-Activated General Control Nonrepressed-2 Is Responsible for Dissociation of AIMP3.
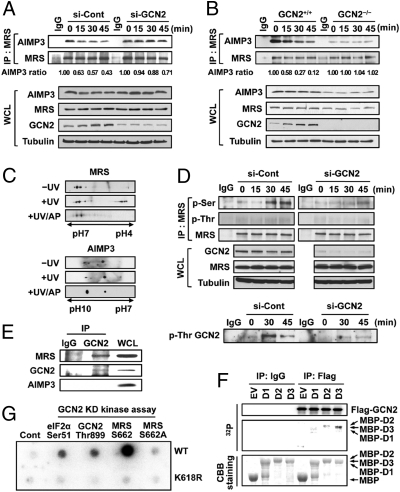
For UV-induced dissociation of MRS and AIMP3, we hypothesized the involvement of general control nonrepressed-2 (GCN2), as it is known to down-regulate translation via UV-dependent phosphorylation of eIF2α (17, 19). To test this hypothesis, we used GCN2-specific siRNA. Following UV irradiation, MRS and AIMP3 disassociated in HeLa cells transfected with control siRNA; however, they did not disassociate in cells with knockdown of GCN2 (Fig. 3A). Similar experiments were conducted using GCN2 mouse embryonic fibroblast (MEF) cells. Following UV irradiation, MRS and AIMP3 did not disassociate in GCN2−/− cells in contrast to in the WT cells (Fig. 3B). These data suggest that GCN2 is involved in controlling the interaction between MRS and AIMP3.
Fig. 3.
GCN2-induced phosphorylation is responsible for the dissociation of MRS and AIMP3. (A and B) HeLa cells treated with si-control or si-GCN2 for 72 h (A) or GCN2+/+ and GCN2−/− MEFs (B) were UV-irradiated. Interaction between MRS and AIMP3 was detected by IP and immunoblotting. The relative ratio of bound AIMP3 to MRS was quantified and presented to show the extent of AIMP3 dissociation. (C) Proteins from HeLa cells were separated by 2D-PAGE. Half of the UV-irradiated samples were treated with alkaline phosphatase (AP) for dephosphorylation. (D) The effect of GCN2 knockdown on MRS phosphorylation was detected (Upper). UV-dependent phosphorylation of GCN2 was validated using p-Thr Ab. (E) Cellular interaction of GCN2 with MRS was observed in UV-irradiated HeLa cells. (F) Kinase assay was done by incubation of Flag-GCN2, [γ-32P]ATP, and MBP-MRS D1, D2, and D3. After stained with Coomassie brilliant blue (CBB), gel was dried and exposed for autoradiography. (G) The immobilized GST-GCN2 kinase domain (KD) was mixed with biotinylated synthetic peptides containing known sequences of GCN2 substrates (GCN2 Thr899 and eIF2α Ser51) and [γ-32P]ATP to detect phosphorylation. An inactive form of GCN2 KD K618R mutant was used as a control.
To investigate posttranslational modifications (PTMs) of MRS and AIMP3 following UV irradiation, 2D-PAGE was used to compare the protein mobility. UV irradiation generated additional spots of MRS that shifted to the acidic side, which disappeared when lysates were treated with alkaline phosphatase (Fig. 3C). In contrast, no obvious UV-induced acidic protein shifts were observed for AIMP3.
To determine which amino acid residues were phosphorylated upon UV stress, MRS was immunoblotted with phospho-specific Abs in control and GCN2-knockdown cells. Phosphorylation of MRS at serine residues was observed in MRS extracted from the control cells and was significantly reduced in GCN2-knockdown cells (Fig. 3D, Upper). Under the same conditions, UV-dependent activation of GCN2 was confirmed by immunoblotting as previously reported (20) (Fig. 3D, Lower). Importantly, MRS, but not AIMP3, was coimmunoprecipitated with GCN2 following UV irradiation (Fig. 3E), suggesting that MRS is a direct substrate of GCN2.
We introduced both initiator and elongator tRNAMet (tRNAeMet) into HeLa cells and determined their effects on the interaction between MRS and AIMP3, as GCN2 is activated by uncharged tRNA (21). We confirmed the enhanced expression of the transfected tRNAs by Northern blotting (Fig. S3A). Increased expression of both tRNAs triggered the dissociation of AIMP3 from MRS and phosphorylation of MRS (Fig. S3B) and also enhanced p-GCN2 and p-eIF2α, as previously reported (22). A previous report shows that deprivation of amino acids also activates GCN2 by increasing free tRNAs (17), prompting us to test whether Met deprivation would also induce separation of AIMP3 from MRS; this was indeed the case, and the process was reversed by Met addition (Fig. S3C). As expected, this Met-sensitive separation was abolished when GCN2 was knocked (Fig. S3D). Taken together, these results suggest that GCN2 is responsible for MRS phosphorylation and causes release of AIMP3 from MRS.
To identify the phosphorylation sites induced by UV stress, two different MRS proteins were prepared. First, endogenous MRS was immunoprecipitated from UV-treated HeLa cells. Second, MRS was subjected to an in vitro GCN2 kinase assay. MRS bands extracted from gels after SDS/PAGE were digested and processed for nano-LC-MS/MS analysis. Based on the Mascot search results, phosphorylation at Ser229, Ser472, and Ser662 was detected in both cases (Fig. S4A). We then conducted the GCN2 kinase assay with maltose-binding protein (MBP)-MRS domains, D1, D2, and D3. Relatively strong radioactivity was detected in the D3 harboring Ser662 (Fig. 3F), and the signal was not observed in D3 WT incubated with catalytically inactive GCN2 or in the D3 S662A mutant (Fig. S4B).
To validate the phosphorylation site mapping, we synthesized a biotinylated MRS peptide spanning Ser662 and the same peptide with a S662A substitution. Although GCN2-induced phosphorylation was apparent in the WT MRS peptide, it was not observed in the S662A peptide (Fig. 3G). Results of two positive control peptides derived from GCN2 and eIF2α (19, 20, 23) and a negative control, the GCN2 K618R inactive mutant, validated the GCN2 kinase assay. We next detected MRS phosphorylation using p-Ser662–specific Ab and found that UV-dependent p-Ser662 signal was specific to GCN2 and Ser662 residue (Fig. S4 C and D). This result suggests that under conditions of UV stress, GCN2 mediates phosphorylation of MRS at Ser662.
Phosphorylation of MRS at Ser662 Induces a Conformational Change to Release AIMP3.
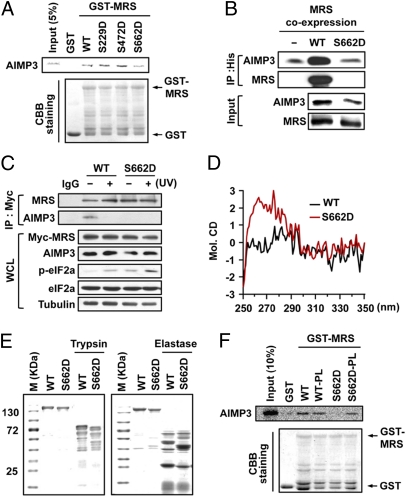
To determine the effect of GCN2-induced phosphorylation on the interaction between MRS and AIMP3, we introduced S229D, S472D, and S662D mutations into MRS, and examined how these mutations would affect the interaction of MRS with AIMP3. Using a GST pull-down assay, we found the binding of MRS S662D to AIMP3 was reduced in comparison with the WT and other mutants (Fig. 4A). We then prepared DNA constructs whereby His-AIMP3 is coexpressed with WT MRS or the S662D mutant. When His-AIMP3 was purified from cells coexpressing the constructs, the bound S662D mutant was significantly reduced compared with WT MRS (Fig. 4B). Although the dissociation of MRS S662D and AIMP3 was confirmed by IP assay using HeLa cells stably expressing MRS WT or the S662D (Fig. 4C), the MRS S662A mutant interacted stably with AIMP3 regardless of UV stress (Fig. S5A). These results suggest that phosphorylation of MRS at Ser662 may decrease the binding affinity of MRS to AIMP3.
Fig. 4.
GCN2-induced phosphorylation may trigger a conformational change in MRS and induce AIMP3 dissociation. (A) Radioactively labeled AIMP3 was incubated with immobilized GST-MRS WT and MRS mutants. (B) MRS WT or the S662D mutant was coexpressed with His-AIMP3 using a dual protein expression system, and the bound protein to AIMP3 was detected by immunoblotting. (C) The interaction of MRS WT and the S662D mutant with AIMP3 by UV was determined by IP using HeLa cells stably expressing Myc-tagged MRS proteins. (D) CD spectra of the purified MBP-MRS WT and S662D mutant were obtained in the near UV. (E) Purified MBP-MRS proteins were digested with trypsin or elastase, and the cleaved fragments were visualized by CBB staining. (F) Peptide linker was inserted between the residues 233 and 234 of MRS WT and of the S662D mutant. The native and peptide linker-inserted MRS proteins were subjected to an in vitro pull-down assay with radioactively labeled AIMP3.
Because Ser662 is distal to the N-terminal extension of MRS (Fig. 1), phosphorylation at Ser662 is unlikely to directly affect the interaction with AIMP3; instead, it may induce a conformational change that leads to the dissociation of AIMP3. We tested this hypothesis by comparing circular dichroism (CD) spectra of MRS WT and the S662D mutant. The MRS S662D mutant had a different CD spectrum in the near UV range (Fig. 4D), suggesting a difference in its tertiary structure. The two proteins were also subjected to trypsin and elastase digestion. With both enzymes, the MRS S662D mutant showed a slightly different digestion pattern (Fig. 4E), further suggesting a conformational difference. Next, we inserted a Gly-Gly-Gly-Gly-Ser peptide linker into the region between the N-terminal AIMP3 binding and catalytic domains of MRS. We expected the insertion of this flexible peptide to loosen the conformational linkage between the AIMP3-binding region and Ser662-harboring domain and thus provide conformational flexibility. We did not expect the selected position for the peptide-linker insertion to alter MRS activity or protein–protein interactions (12, 24). Indeed, we found that in the WT background, the aminoacylation activity of MRS was not influenced by peptide-linker insertion (Fig. S5B), and AIMP3 bound to the normal and peptide linker-inserted MRS with similar affinity (Fig. 4F). In contrast, the insertion of the peptide linker into MRS S662D restored its ability to bind AIMP3 (Fig. 4F). These data suggest that Ser662 phosphorylation may induce a conformational change that is propagated to the N-terminal extension, thereby releasing the bound AIMP3.
MRS Phosphorylation at S662 Reduced Catalytic Activity and Global Translation.
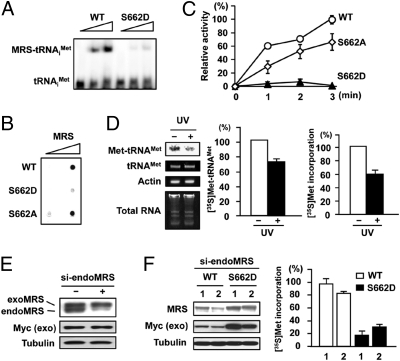
Ser662 is located within the anticodon-binding motif (25, 26); therefore, its mutation may disrupt tRNA binding and aminoacylation activity. We tested this theory using in vitro tRNA binding and aminoacylation assays. When a tRNAiMet probe was reacted with MRS WT and the S662D mutant, MRS WT generated the tRNA-bound complex in a dose-dependent manner, but the MRS S662D mutant did not bind tRNA (Fig. 5A). Substitution of Ser662 to Ala did not disturb the complex formation with tRNAiMet (Fig. 5B). If the MRS S662D mutant cannot bind tRNA, it cannot produce Met-tRNAMet. As expected, the MRS S662D mutant showed negligible aminoacylation activity (Fig. 5C). The reduced activity was not caused by nonspecific amino acid substitution, as the MRS S662A mutant remained active.
Fig. 5.
The effect of MRS phosphorylation on tRNA binding and global translation. (A and B) The tRNAiMet probe was incubated with His-MRS WT and mutant proteins and separated by nondenaturing PAGE (A) or filtered through a nitrocellulose membrane (B). (C) Relative catalytic activities of His-MRS WT and mutants were assessed by aminoacylation assay. Data are represented as mean ± SD (n = 3). (D) HeLa cells were incubated with [35S]Met with or without UV irradiation. Total RNA was extracted and charged tRNAMet was detected by autoradiography (Left). The relative Met-tRNAMet amount was quantified by digital imaging (Center). The values are represented as mean ± SD (n = 2). Protein synthesis under the same conditions was monitored by the [35S]Met incorporation assay (Right). Data are represented as mean ± SD (n = 3). (E) Endogenous MRS in stable HeLa cells expressing Myc-tagged MRS WT were knocked down by siRNA. (F) Endogenous MRS was down-regulated in MRS WT and S662D stable HeLa cells (Left) and stable cell clones were assayed for global protein synthesis. We chose stable cells highly expressing the S662D mutant to see its overexpressional effect on translation. Data are represented as mean ± SD (n = 3) (Right).
Given that UV-dependent MRS phosphorylation blocks the charging ability of MRS, the level of Met-tRNAMet should be reduced under UV. Indeed, decreased levels of Met-tRNAMet was detected in the UV-treated cells (Fig. 5D, Left and Center), and decreased global protein synthesis was also observed under the same conditions (Fig. 5D, Right). The UV-dependent reduction of Met-tRNAiMet was also confirmed in GCN2 MEFs and HeLa cells by Northern blotting (Fig. S6 A and B). To test whether phosphorylation of MRS at Ser662 would affect global translation, we selected two independent HeLa stable cell lines expressing MRS WT or the S662D mutant. To remove any contribution from endogenous MRS, we designed siRNA targeting the 3′UTR sequence of the endogenous MRS transcript but not affecting exogenous MRS (Fig. 5E). Cells expressing the MRS S662D mutant revealed significant reduction of protein synthesis compared with that in MRS WT cells (Fig. 5F), and showed little amounts of Met-tRNAiMet with knockdown of endogenous MRS (Fig. S6C). In the same context, portions of charged tRNAiMet were reduced by UV stress in stable cells where ectopic MRS WT was substituted for endogenous MRS; however, it was not observed in stable cells expressing the MRS S662A mutant (Fig. S6D). These data suggest that phosphorylation at Ser662 in MRS not only triggers the release of AIMP3 for nuclear translocation but also down-regulates global translation by blocking binding of tRNAMet to MRS.
MRS and eIF2α Cooperate for Translational Inhibition in Response to DNA Damage.
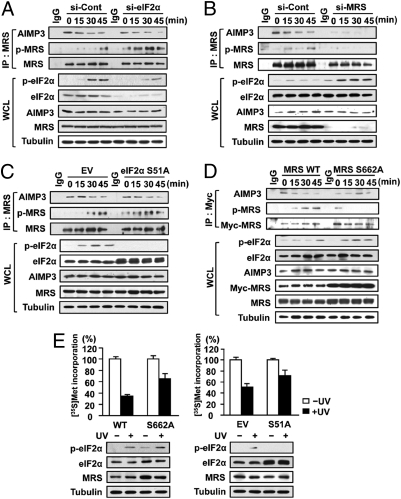
GCN2 phosphorylates eIF2α to inhibit global translation (19). Because MRS is also a GCN2 substrate and can inhibit translation under UV stress, we investigated the potential for cross-talk between the two pathways. We observed that the level of UV-induced phosphorylated MRS (p-MRS) was elevated when eIF2α level was reduced (Fig. 6A). Conversely, knockdown of MRS enhanced the level of p-eIF2α induced by UV irradiation (Fig. 6B). Likewise, stable overexpression of the dominant-negative eIF2α S51A mutant adversely affected endogenous p-eIF2α production and enhanced UV-induced p-MRS (Fig. 6C). HeLa stable cell lines expressing the MRS S662A mutant also showed increased levels of p-eIF2α, compared with stable cells expressing MRS WT (Fig. 6D). Dissociation of AIMP3 from MRS following UV irradiation was observed under all experimental conditions, except when the MRS S662A mutant was immunoprecipitated for the analysis of bound AIMP3. This result indicates that phosphorylation of MRS at Ser662 is the prerequisite step for AIMP3 release. Knockdown or overexpression of MRS or eIF2α did not affect the total protein level of its counterpart. These data suggest that the levels of p-eIF2α and p-MRS can be controlled in a complementary fashion.
Fig. 6.
MRS coordinates with eIF2α to regulate translation upon UV irradiation. (A and B) HeLa cells treated with si-eIF2α or si-MRS were UV-irradiated and p-MRS and p-eIF2α was monitored. (C and D) UV-induced phosphorylation of MRS and eIF2α was monitored in stable cell lines expressing the eIF2α S51A or MRS S662A mutant. Empty vector (EV)-transfected cells were used as a control (C). The Myc-tagged eIF2α S51A mutant migrated slowly and appeared in the upper band of endogenous eIF2α. (E) Protein synthesis after UV irradiation was measured in stable cells expressing MRS WT and the S662A mutant (Left) and EV and the eIF2α S51A mutant (Right). Endogenous MRS in cells expressing MRS WT and the S662A mutant was knocked down by siRNA treatment.
To observe how both proteins contribute to global translational regulation, we performed a [35S]Met-incorporation assay in those stable HeLa cells. In MRS WT cells protein synthesis was reduced to about 33% by UV stress, whereas in cells expressing the MRS S662A mutant, protein synthesis was decreased to 65% (Fig. 6E, Left). Similar results were obtained in stable cells expressing the eIF2α S51A mutant (Fig. 6E, Right). MRS and eIF2α, therefore, seem to cooperate for efficient control of translation under UV stress. Phosphorylation of each protein may be augmented when regulation cannot be conducted by its counterpart. In conclusion, GCN2-induced phosphorylation of MRS links translational regulation and the DNA damage response via the release of AIMP3, which is translocated to the nucleus where it mediates the p53-dependent DNA damage response (Fig. 7).
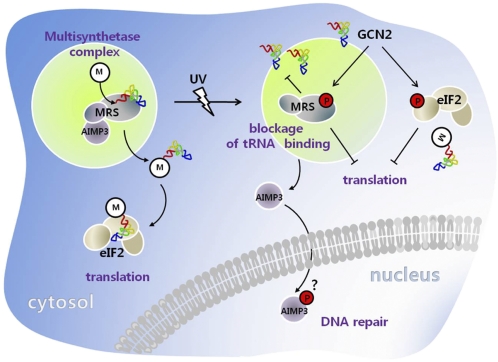
Fig. 7.
Schematic representation for the dual role of MRS. Under normal condition, MRS catalyzes aminoacylation of tRNAMet for translational initiation. AIMP3 is mainly bound to the N-terminal extension of MRS. Upon UV irradiation, MRS is phosphorylated at Ser662 by GCN2, and binding of tRNAMet was blocked. Phosphorylation of MRS induces a conformational change, causing the dissociation of AIMP3 from MRS. Released AIMP3 is translocated to the nucleus to mediate DNA damage repair. GCN2 also phosphorylates eIF2α and p-MRS and p-eIF2α work together to control global translation.
Discussion
UV-activated GCN2 phosphorylates eIF2α, causing a reduction in global translation by inhibition of TC formation (19). Here, we provide evidence that MRS is an additional GCN2 substrate that works coordinately with eIF2α for translational regulation under UV stress. Phosphorylation of MRS can block new Met-tRNAMet production but p-eIF2α inhibits TC formation with preexisting pools of Met-tRNAiMet. Considering that only partial translation inhibition was observed when one of them was made nonphosphorylatable, dual blockage of sequential steps by p-MRS and p-eIF2α seems to be required for the precise control of translation.
Interestingly, the influence of MRS phosphorylation after UV irradiation was not restricted to translational control. MRS also linked translational inhibition to the DNA damage response by regulating its molecular interaction with the tumor suppressor AIMP3. The dissociation between MRS and AIMP3 was striking, as observed in BiFC analysis (Fig. 2B). Decrease of AIMP3 in the cytosol, however, was not so apparent, as increased AIMP3 in the nucleus was clearly detected (Fig. 2E). This finding suggests that only a portion of AIMP3 released from MRS is translocated to the nucleus. Perhaps, the released AIMP3 may have to undergo another processing event for its nuclear import to occur. To test the possibility of AIMP3 modification, we fractionated Met-starved and tRNAiMet-overexpressing HeLa cells. In these cells, MRS is phosphorylated without DNA damage, which augments the levels of free AIMP3. If further AIMP3 modification is not necessary for translocation, nuclear AIMP3 levels in these cells after UV irradiation should remain unchanged, and similarly, elevated nuclear AIMP3 levels should occur in the absence of UV irradiation. However, UV-dependent nuclear translocation of AIMP3 was observed and basal nuclear AIMP3 was not elevated (Fig. S7 A and B), suggesting the possibility of PTM of free AIMP3. We found that phosphorylation is a possible modification, based on the 2D-PAGE analysis of nuclear AIMP3 (Fig. S7C).
AIMP3 is functionally versatile. In addition to its tumor-suppressive activity, increased cellular AIMP3 levels were recently shown to enhance cellular senescence through negative regulation of lamin A (27). Considering its diverse roles, it is plausible that multiple PTMs may guide AIMP3 for its functional destination. The additional role of AIMP3 in the cytosol and its PTMs will be the subject of further investigation.
GCN2 also appears to be involved in control of AIMP3 and MRS expression. In GCN2−/− MEFs, bound AIMP3 to MRS was basally low in the absence of UV (Fig. 3B). This effect appears related to reduced expression of MRS and AIMP3, specifically AIMP3, in the GCN2−/− MEFs. Interestingly, among the translation-related proteins tested in this study, only the levels of MRS and AIMP3 were reduced by the absence of GCN2 (Fig. S8). In contrast, when GCN2 was transiently suppressed by siRNA, MRS and AIMP3 levels were little affected (Fig. 3A). Perhaps, in GCN2−/− cells constitutively lacking the GCN2 activity, a risk of uncontrolled translation may be compensated by reducing cellular levels of MRS and AIMP3, although the precise role of GCN2 in this connection is currently unknown.
Previous reports provide several examples of how the dynamic relationship of MSC components is controlled by phosphorylation. For example, in IFN-γ–activated U937 cells, EPRS dissociates and forms new complexes called gamma-interferon activated inhibitor of translation (GAIT) with the 3′UTR of target transcripts, which mediate translational gene silencing (28). In addition, in mast cells, lysyl-tRNA synthetase is phosphorylated and translocates to the nucleus, where it controls the activity of the microphthalmia transcription factor (29). Among the three nonenzymatic components of MSC, AIMP2 is phosphorylated and translocated to the nucleus, where it binds p53 upon DNA damage (30). The effects of p-MRS can be distinguished from these earlier reported modes of regulation. Specifically, p-MRS is not disassociated from MSC, and in its associated state releases bound AIMP3. Moreover, phosphorylation of MSC components has not previously been shown to affect global translation. Here, we found that phosphorylation significantly suppresses the catalytic activity of MRS, leading to the down-regulation of global translation. This finding provides evidence for a unique role of MRS as a coordinator of cytosolic translation and nuclear DNA repair.
MRS phosphorylation links UV stress to the DNA damage response through AIMP3 translocation, whereas eIF2α phosphorylation increases the NF-κB response upon UV irradiation (23). Although AIMP3 release from MRS and nuclear translocation were observed within 1 h after UV irradiation, NF-κB activation by p-eIF2α is only observed starting 1 h after UV irradiation. Thus, GCN2 may use MRS-AIMP3 as an early response to UV-induced DNA damage and eIF2α for the transcriptional control of NF-κB–dependent genes in the later stages. GCN2-mediated delay of cellular entry into the S phase by UV stress has been reported; however, the underlying mechanism and cross-talk between GCN2 and ATM/ATR in cell-cycle regulation and DNA repair are not clearly understood (31). Our work suggests that the activation of GCN2 is functionally linked to the activation of ATM/ATR via AIMP3 released from p-MRS, although the actual relationship between ATM/ATR, NF-κB, p-eIF2α, and AIMP3 needs further investigation. In conclusion, we have demonstrated a dual role for human MRS in the regulation of cytosolic translation and molecular interaction with AIMP3, thus elucidating a mechanism for coupling the control of protein synthesis to the DNA damage response. This work is also significant in that MRS represents a previously undetected gate for translational control, suggesting the possibility that other ARSs may play unique roles in translational control and cell signaling.
Materials and Methods
Protocols for well established procedures (yeast two-hybrid assay, pull-down assay, BiFC, cell fractionation, 2D-PAGE, kinase assay, CD spectrum, aminoacylation assay, gel shift assay, Northern blotting, and Met incorporation assay) can be found in SI Materials and Methods.
Cell Culture.
GCN2 MEFs and HeLa were grown in DMEM supplemented with 10% (vol/vol) FBS and 1% (vol/vol) penicillin/streptomycin in 5% (vol/vol) CO2 at 37 °C. For the selection of stable cell line, plasmids were transfected with FuGENE HD (Roche) and transfected cells were selected and maintained with 1 mg/mL geneticin (G418, Duchefa Biochemie). For IP or BiFC, cells were UV-irradiated (60 J/m2), recovered with serum-free medium, and harvested at the indicated time points.
IP Assay.
Cells were lysed with IP buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.5% Triton X-100, 5 mM EDTA, and 10% (vol/vol) glycerol) containing protease inhibitor and phosphatase inhibitor. Cell lysates were centrifuged and incubated overnight with IgG or specific Abs and then with Protein A/G agarose. IP proteins were separated by SDS/PAGE and detected by immunoblotting.
Supplementary Material
Acknowledgments
We thank Dr. D. Ron for providing GCN2 mouse embryonic fibroblast cells and pCDNA3-GCN2, and Dr. C.-D. Hu for providing pBiFC-VN173 and pBiFC-VC155. This research was supported by the World Class University Project (R31-2008-000-10103-0) of Korea Science and Engineering Foundation, Global Frontier Research Grant NRF-M1AXA002-2010-0029785, and the Basic Science Research Program (2010-0023292) of the National Research Foundation funded by the Ministry of Education, Science and Technology of Korea.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103922108/-/DCSupplemental.
References
- 1.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pavon-Eternod M, et al. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 2009;37:7268–7280. doi: 10.1093/nar/gkp787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee SW, Cho BH, Park SG, Kim S. Aminoacyl-tRNA synthetase complexes: Beyond translation. J Cell Sci. 2004;117:3725–3734. doi: 10.1242/jcs.01342. [DOI] [PubMed] [Google Scholar]
- 5.Park SG, Ewalt KL, Kim S. Functional expansion of aminoacyl-tRNA synthetases and their interacting factors: new perspectives on housekeepers. Trends Biochem Sci. 2005;30:569–574. doi: 10.1016/j.tibs.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Ko YG, Kang YS, Kim EK, Park SG, Kim S. Nucleolar localization of human methionyl-tRNA synthetase and its role in ribosomal RNA synthesis. J Cell Biol. 2000;149:567–574. doi: 10.1083/jcb.149.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Netzer N, et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature. 2009;462:522–526. doi: 10.1038/nature08576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marshall L, Kenneth NS, White RJ. Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell. 2008;133:78–89. doi: 10.1016/j.cell.2008.02.035. [DOI] [PubMed] [Google Scholar]
- 9.Park SG, Choi EC, Kim S. Aminoacyl-tRNA synthetase-interacting multifunctional proteins (AIMPs): A triad for cellular homeostasis. IUBMB Life. 2010;62:296–302. doi: 10.1002/iub.324. [DOI] [PubMed] [Google Scholar]
- 10.Han JM, et al. Hierarchical network between the components of the multi-tRNA synthetase complex: Implications for complex formation. J Biol Chem. 2006;281:38663–38667. doi: 10.1074/jbc.M605211200. [DOI] [PubMed] [Google Scholar]
- 11.Park BJ, et al. The haploinsufficient tumor suppressor p18 upregulates p53 via interactions with ATM/ATR. Cell. 2005;120:209–221. doi: 10.1016/j.cell.2004.11.054. [DOI] [PubMed] [Google Scholar]
- 12.Rho SB, et al. Genetic dissection of protein-protein interactions in multi-tRNA synthetase complex. Proc Natl Acad Sci USA. 1999;96:4488–4493. doi: 10.1073/pnas.96.8.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim KJ, et al. Determination of three-dimensional structure and residues of the novel tumor suppressor AIMP3/p18 required for the interaction with ATM. J Biol Chem. 2008;283:14032–14040. doi: 10.1074/jbc.M800859200. [DOI] [PubMed] [Google Scholar]
- 14.Park BJ, et al. AIMP3 haploinsufficiency disrupts oncogene-induced p53 activation and genomic stability. Cancer Res. 2006;66:6913–6918. doi: 10.1158/0008-5472.CAN-05-3740. [DOI] [PubMed] [Google Scholar]
- 15.Kaminska M, Deniziak M, Kerjan P, Barciszewski J, Mirande M. A recurrent general RNA binding domain appended to plant methionyl-tRNA synthetase acts as a cis-acting cofactor for aminoacylation. EMBO J. 2000;19:6908–6917. doi: 10.1093/emboj/19.24.6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo M, Yang XL, Schimmel P. New functions of aminoacyl-tRNA synthetases beyond translation. Nat Rev Mol Cell Biol. 2010;11:668–674. doi: 10.1038/nrm2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 18.Shyu YJ, Suarez CD, Hu C-D. Visualization of ternary complexes in living cells by using a BiFC-based FRET assay. Nat Protoc. 2008;3:1693–1702. doi: 10.1038/nprot.2008.157. [DOI] [PubMed] [Google Scholar]
- 19.Deng J, et al. Activation of GCN2 in UV-irradiated cells inhibits translation. Curr Biol. 2002;12:1279–1286. doi: 10.1016/s0960-9822(02)01037-0. [DOI] [PubMed] [Google Scholar]
- 20.Berlanga JJ, Santoyo J, De Haro C. Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2alpha kinase. Eur J Biochem. 1999;265:754–762. doi: 10.1046/j.1432-1327.1999.00780.x. [DOI] [PubMed] [Google Scholar]
- 21.Dong J, Qiu H, Garcia-Barrio M, Anderson J, Hinnebusch AG. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol Cell. 2000;6:269–279. doi: 10.1016/s1097-2765(00)00028-9. [DOI] [PubMed] [Google Scholar]
- 22.Wek SA, Zhu S, Wek RC. The histidyl-tRNA synthetase-related sequence in the eIF-2 alpha protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol Cell Biol. 1995;15:4497–4506. doi: 10.1128/mcb.15.8.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang HY, Wek RC. GCN2 phosphorylation of eIF2alpha activates NF-kappaB in response to UV irradiation. Biochem J. 2005;385:371–380. doi: 10.1042/BJ20041164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He R, Zu LD, Yao P, Chen X, Wang ED. Two non-redundant fragments in the N-terminal peptide of human cytosolic methionyl-tRNA synthetase were indispensable for the multi-synthetase complex incorporation and enzyme activity. Biochim Biophys Acta. 2009;1794:347–354. doi: 10.1016/j.bbapap.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Nakanishi K, Ogiso Y, Nakama T, Fukai S, Nureki O. Structural basis for anticodon recognition by methionyl-tRNA synthetase. Nat Struct Mol Biol. 2005;12:931–932. doi: 10.1038/nsmb988. [DOI] [PubMed] [Google Scholar]
- 26.Ghosh A, Vishveshwara S. A study of communication pathways in methionyl- tRNA synthetase by molecular dynamics simulations and structure network analysis. Proc Natl Acad Sci USA. 2007;104:15711–15716. doi: 10.1073/pnas.0704459104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh YS, et al. Downregulation of lamin A by tumor suppressor AIMP3/p18 leads to a progeroid phenotype in mice. Aging Cell. 2010;9:810–822. doi: 10.1111/j.1474-9726.2010.00614.x. [DOI] [PubMed] [Google Scholar]
- 28.Arif A, et al. Two-site phosphorylation of EPRS coordinates multimodal regulation of noncanonical translational control activity. Mol Cell. 2009;35:164–180. doi: 10.1016/j.molcel.2009.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee YN, Nechushtan H, Figov N, Razin E. The function of lysyl-tRNA synthetase and Ap4A as signaling regulators of MITF activity in FcepsilonRI-activated mast cells. Immunity. 2004;20:145–151. doi: 10.1016/s1074-7613(04)00020-2. [DOI] [PubMed] [Google Scholar]
- 30.Han JM, et al. AIMP2/p38, the scaffold for the multi-tRNA synthetase complex, responds to genotoxic stresses via p53. Proc Natl Acad Sci USA. 2008;105:11206–11211. doi: 10.1073/pnas.0800297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grallert B, Boye E. The Gcn2 kinase as a cell cycle regulator. Cell Cycle. 2007;6:2768–2772. doi: 10.4161/cc.6.22.4933. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.