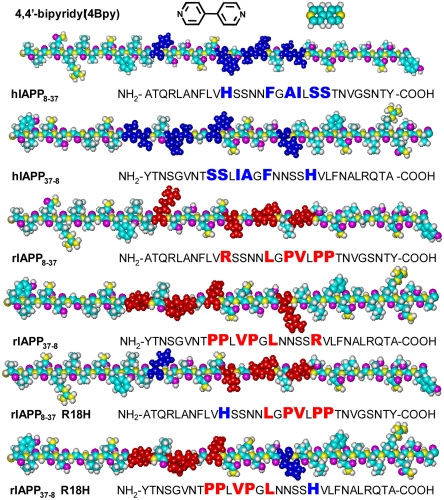
Abstract
We report here the identification of the key sites for the beta structure motifs of the islet amyloid polypeptide (IAPP) analogs by using scanning tunneling microscopy (STM). Duplex folding structures in human IAPP8–37 (hIAPP8–37) assembly were observed featuring a hairpin structure. The multiplicity in rIAPP assembly structures indicates the polydispersity of the rat IAPP8–37 (rIAPP8–37) beta-like motifs. The bimodal length distribution of beta structure motifs for rIAPP8–37 R18H indicates the multiple beta segments linked by turns. The IAPP8–37 analogs share common structure motifs of IAPP8–17 and IAPP26–37 with the most probable key sites at positions around Ser19/Ser20 and Gly24. These observations reveal the similar amyloid formation tendency in the C and N terminus segments because of the sequence similarity, while the differences in specific amino acids at each key site manifest the effect of sequence variations. The results could be beneficial for studying structural polymorphism of amyloidal peptides with multiple beta structure motifs.
Keywords: amylin, folding sites, self-assembly, amyloid
Human islet amyloid polypeptide (hIAPP, amylin) is a major component in pancreatic amyloid deposits, which have been found in a majority of patients with type 2 diabetes (1–3). Human IAPP with 37 amino acid residues is known to form soluble aggregates that further assemble into protofibrils and mature fibrils and eventually develop into plagues (4, 5), and many studies have been conducted to identify the core regions with amyloidogenic properties (6–8). By using coordinated fiber X-ray diffraction (XRD) (9, 10), electron diffraction, and cryoelectron microscopy (10), the fibrillar structures of hIAPP have been identified as a beta-sheet structure with beta-strands perpendicular to the fiber axis, and the separation between neighboring strands is 4.7 Å. The parallel beta-structures of hIAPP have been proposed by using electron paramagnetic resonance spectroscopy of spin-labeled derivatives of hIAPP (11), and the “serpentine” model has been derived based on electron microscopy and scanning transmission electron microscopy data (12). Recent XRD studies have revealed a structural motif of short segments derived from a variety of amyloidal proteins (13–15). The atomic level structural model for hIAPP21–27 revealed a pronounced bend facilitated by Gly24, and the model of hIAPP based on the crystalline structures of hIAPP21–27 and hIAPP28–33 segments indicated the beta-spine steric zipper structure for hIAPP23–37 (two beta-sheets with interdigitated side chains) (15). The beta-hairpin structure of hIAPP has also been obtained by solid-state nuclear magnetic resonance (NMR) (16), suggesting the parallel beta-sheet structures with 10 residues in the core domains. The fragment IAPP1–7 is believed to be non-beta-sheet structure (17) as the result of the disulfide bond between cysteine residues 2 and 7.
As an important analog of IAPP, rat islet amyloid polypeptide (rIAPP) is reported to form no fibrils in solutions (18, 19), while the single site mutant analog of rIAPP (rIAPP R18H) exhibits the ability of fibrillogenesis unexpectedly (20). Many efforts have been made in the aggregation behaviors of the mutant analogs of IAPP (2, 7, 12, 15, 21–24), while more evidence is still needed to gain insight into the relationship between primary sequences of the IAPP analogs and the conformational variants.
It is generally acknowledged that the noncrystalline nature of the fibril structures formed by amyloidal peptides makes it experimentally challenging for structural analyses at molecular level. The advances in analytical techniques to resolve the core regions of the beta-sheet and the folding configurations should be of genuine interest in this topical area. Scanning tunneling microscopy (STM) has been applied to study the molecular structures of peptides due to its high structural resolution and adaptability to various environments, especially for amyloid peptides lacking crystalline structures (25, 26).
Previous STM results revealed amyloid peptide assemblies at liquid–solid interfaces (25–28). An average of 11 amino acid residues have been identified for the core regions of hIAPP (26), which roughly agrees with the model proposed by Luca et al. (16). Furthermore, evidence of multiple folding structures of hIAPP hairpins has been obtained according to the statistical analysis of the core regions in STM observations (26). Evidence of similar polymorphism of folding structures for amyloid beta (Aβ1–42) has also been reported by STM observations (25). The above results demonstrated the feasibility of STM technique in studying the beta-structure motifs of the surface-bound peptide assemblies. Meanwhile, it should be noted that only a single beta motif has been observed in the previously reported observations (25–28); for peptides with multiple beta motifs (such as hairpin structures consisting of two beta-strands), new investigation strategies are keenly needed to identify multiple sites for beta-structure motifs.
While the STM method has subangstrom resolution capability in lateral direction on surfaces, it is highly desired to overcome the inevitable fluctuations of peptide termini to obtain reliable peptide length measurement at liquid–solid interface. In the previous study, it has been demonstrated that the peptide C terminus could be immobilized to allow measurements of the peptide chain length by introducing coassembly species such as 4,4′-bipyridyl (4Bpy) (28, 29). Therefore, one will be able to determine the number of amino acids in the surface-bound peptide chains with beta-structure characteristics. This strategy could be readily extended to study the peptides that could develop into beta-structures consisting of multiple beta motifs.
In this work, we studied the molecular structures of hIAPP8–37, rIAPP8–37, and rIAPP8–37 R18H with both normal and reversed sequences (peptide sequences as shown in Fig. 1) by using STM. Coassemblies of peptides and chaperon-like molecules, 4Bpy (Fig. 1), show homogeneous beta-like structural characteristics. Duplex beta-like motifs of hIAPP and its derivatives could be identified according to the measured length distributions of C-terminal and N-terminal peptide strands.
Fig. 1.
Molecular structure of the chaperon-like molecule 4,4′-bipyridyl (4Bpy) and the sequences of hIAPP8–37, hIAPP37–8, rIAPP8–37, rIAPP37–8, rIAPP8–37 R18H, and rIAPP37–8 R18H. The residues in rIAPP8–37, rIAPP37–8, rIAPP8–37 R18H, and rIAPP37–8 R18H that differ from hIAPP8–37 and hIAPP37–8 are depicted in red, and the corresponding positions in hIAPP8–37 and hIAPP37–8 are depicted in blue. The residues in rIAPP8–37 R18H and rIAPP37–8 R18H depicted in blue are the mutation sites His18. Color code: cyan for C, gray for H, yellow for N, and pink for O.
Results and Discussion
hIAPP8–37 Folding Sites.
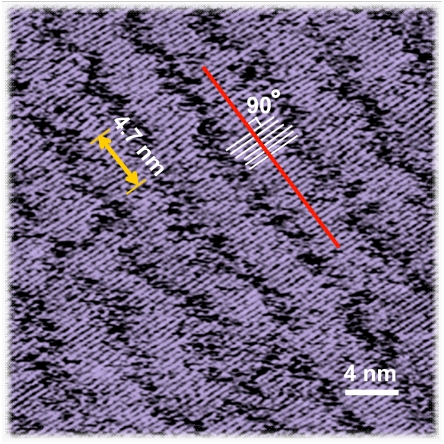
The assembly structures of hIAPP1–37 have been observed by STM with submolecular resolution in the previous work (26), which shows typical lamella characteristics. In the current study, fragment hIAPP1–7 is excluded from the peptide because hIAPP1–7 is non-beta-sheet structured and makes little contribution to the assembly characteristics due to the disulfide bond between cysteine residues 2 and 7. The assembly structures of hIAPP8–37 were also observed by STM showing lamella structures (Fig. 2). The bright stripes in Fig. 2 correspond to the core regions of hIAPP8–37 on the supporting surface. The molecular axes of hIAPP8–37 peptides (as highlighted by a group of short white lines) are observed to be approximately perpendicular to the long axes of the stripes (as highlighted by a red line). The averaged separation between two neighboring molecules is 4.7 ± 0.2 Å (indicated by yellow bars), which is consistent with the previous reports on hIAPP1–37 by STM (26) and X-ray fiber diffraction (9, 10).
Fig. 2.
High resolution STM image of hIAPP8–37 assembly. The length of the yellow bar, covering 10 molecules, is 4.7 nm. The molecular axes of hIAPP8–37 peptides indicated by a group of short white lines are perpendicular to the long axis of the stripe (highlighted by the red line). Tunneling conditions: I = 350.0 pA, V = 490.0 mV.
As an effort to gain further insight into the fine structural characteristics of the folding structures of hIAPP8–37, a chaperon-like molecule, 4Bpy, was introduced to coassemble with the hIAPP8–37 peptide. The STM image in Fig. 3A shows the coassembly structures of hIAPP8–37 and 4Bpy. The relatively high local electron density of states of the conjugated π-electron of 4Bpy moiety can be associated with the enhanced contrast in the linear bright arrays. The lamella structures with lower contrast between the 4Bpy arrays are associated with the peptide strands showing beta-like structure characteristics. The heterogeneous organic-peptide coassembly structure is stabilized by hydrogen bond between 4Bpy and the carboxyl terminus of hIAPP8–37 (28, 29). As the result, the C terminal strands of hIAPP8–37 peptides are stabilized by the coassembly molecule 4Bpy. Thus reliable length measurements of the peptide strands from C terminus could be achieved.
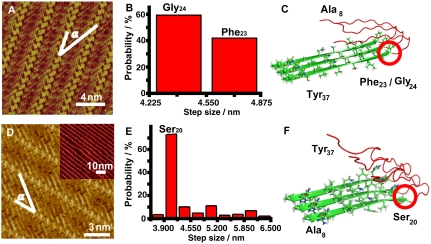
Fig. 3.
(A and D) High resolution STM images of hIAPP8–37 (A) and hIAPP37–8 (D) coassembled with 4Bpy. The angles α between peptide molecular axes and the stripe directions are measured to be 34 ± 2° both in A and D. Tunneling conditions: (A) I = 314.3 pA, V = 605.2 mV; (D) I = 532.8 pA, V = 379.7 mV. Inset in D is large scale STM image of hIAPP37–8/4Bpy. (B and E) Length distribution histograms of beta-structure motifs for hIAPP8–37 and hIAPP37–8, respectively, measured from the STM images. The step size of the chart is 0.325 nm and is equivalent to the separation of two neighboring residues in the parallel beta-sheet structure. (C and F) Proposed models for the folding sites in hIAPP8–37 and hIAPP37–8 beta-structure motifs, respectively. Red rings highlight the folding sites in beta-structure motif. The green belts stand for the beta-structure motifs, and the flexible red strings for the undiscernible parts in STM observations. Color code: green for C, gray for H, blue for N, and red for O. The side-chain conformations are shown in schematic way. The crystalline structure of hIAPP21–27 and hIAPP28–33 are referred to for the proposal of this schematic illustration (15).
The separation between two neighboring peptide strands has been measured to be 5.4 ± 0.2 Å in the coassembly of hIAPP8–37 and 4Bpy molecules (Fig. 3A). This interstrand separation is slightly larger than 4.7 Å for pristine hIAPP8–37 assemblies (Fig. 2) and hIAPP1–37 assemblies (26) but still within the separation range of hydrogen bond formation (10). The difference in interstrand separation between pristine hIAPP self-assembly and the coassembly structures may be possibly due to the change of interchain hydrogen bond configuration associated with the assembling characteristics (30). The measured angle α between hIAPP8–37 molecular axes and peptide stripe orientation is measured to be 34 ± 2° in the coassembly system (Fig. 3A), different from the nearly 90° angle for pristine assembly (Fig. 2), which is also supportive to the hydrogen bond formation between C termini and 4Bpy (28, 31).
The measurement of peptide lengths has been defined in the previous study as the lengths of the dotted stripe-shaped features with relatively high contrast between the neighboring grooves with dark contrast (25) (as shown in Fig. S1A). The length is the full width between two positions with abrupt slope changes in the sectional profile (Fig. S1B). As for the lengths of the peptide strands labeled with 4Bpy, we define the length of single peptide as the stripe between two 4Bpy moieties with same criteria as shown above (Fig. S1 C and D). Due to the contrast variations in the peptide stand terminals, the uncertainty of the length measurements in the terminal regions is unavoidable, while the fidelity is based on the identical criteria in all the length measurements. The theoretical calculations and simulation of the fine peptide feature STM images should be keen in future studies to give rigorous identification of the peptide length.
The histogram of the length distribution of hIAPP8–37 molecules indicates that there are two discrete lengths of C-terminal strands (4.22 ∼ 4.55 nm, 4.55 ∼ 4.88 nm), as shown in Fig. 3B and Table S1 (statistical methods are shown in SI Text). In the parallel beta-sheet structures, the expected separation between two neighboring residues is 0.325 nm (32). The corresponding residue numbers for the above two C-terminal strands are therefore estimated at 14 and 15, respectively. As a result, the C-terminal beta-structure motifs of IAPP8–37 could preferentially begin from Phe23 or Gly24. Considering the reported hairpin structure of IAPP peptide (16) and the possibility of short peptide folding into beta-hairpin (33), we proposed the tentative model with possible folding sites at the residue Phe23 or Gly24, as shown in Fig. 3C. The green belts stand for the beta-structure motifs, which are the observable parts in STM images, and the flexible red strings for the undiscernible parts in STM observations. The side-chain conformations are shown in a schematic way and the crystalline structure of hIAPP21–27 and hIAPP28–33 are referred to for proposal of this schematic illustration (15). Peptide assembly with longer strand lengths (6.82 ∼ 8.77 nm, corresponding to 22 ∼ 27 residues) were also observed (Fig. S1 E and F and Table S2), which is attributed to the unfolded IAPP structures.
In order to obtain the key sites for all the beta-structure motifs in the hairpin structure, STM investigations on hIAPP37–8, which has the identical sequence of hIAPP8–37 but with reversed N and C termini, were performed for identification of the key site for N-terminal beta-structure motif. As shown in Fig. 3D, the STM images of the hIAPP37–8/4Bpy coassembly show similar results as that of hIAPP8–37/4Bpy. The histogram of the length distribution of hIAPP37–8 molecules indicates that the most probable length of the beta-structure motif is 3.90–4.22 nm, corresponding to 13 residues (Ala8-Ser20), as shown in Fig. 3E and Table S3. Again, considering the hairpin structure consists of two beta-structure motifs, residue Ser20 could be proposed as one of the folding sites of hIAPP37–8.
Combining the above results for two constituent strands of the hIAPP hairpin structure, one may propose the duplex folding structures as shown in Fig. 3 C and F. This polymorphism in the peptide folding structures may be related to the side-chain registration associated with steric zipper interactions that are prevalent in amylome (14, 21, 34). The two beta motifs of hIAPP could be matched in two stacking ways with an amino acid mismatched by tight interdigitation of side chains between two neighboring beta-sheet strands, as manifested in reported polymorphic crystal structures of short peptide segments GNNQQNY, NNQQ, and MVGGVV (14).
The suggested hIAPP beta-structure motifs based on our STM results are in qualitative agreement with the model based on distance constraints from solid-state NMR (16) and the XRD results for key segments (NNFGAIL and SSTNVG, hIAPP21–27 and hIAPP28–33) (15). In the hairpin model proposed by Luca et al. (16), the core structures are formed by two beta-strands (hIAPP8–17 and hIAPP28–37) joined by a loop with residues 18–27. XRD results for the key segment of hIAPP, NNFGAIL (residues 21–27), revealed a pronounced bend in the peptide strand facilitated by Gly24 (15). The results in the current study suggesting the key site at Phe23 or Gly24 for hIAPP are in close agreement with the previous structural models (15, 16). However, in the present stage the identification of the side-chain conformations are still far out of reach of STM resolution capability at liquid–solid interface, considering the rich selection of possible conformations for peptides. Rigorous analysis of the fine features in STM images of peptide structure is deemed a very rewarding task both experimentally and theoretically (25).
rIAPP8–37 Folding Sites.
In the parallel studies, we also observed the rIAPP8–37/4Bpy coassemblies on HOPG surface. As shown in Fig. 4A, STM images of the rIAPP8–37/4Bpy coassembly present beta-sheet-like structures similar to that of hIAPP8–37. As suggested in the previous reports, solution conformation of rIAPP is random coils or alpha helix depending on the phospholipid membrane bounded or ion concentration, temperature, and pH (35–38). STM results indicated rIAPP8–37 may undergo structural transformation from random coils to beta-sheet-like structures possibly due to the surface-induced assembling effects (26, 27).
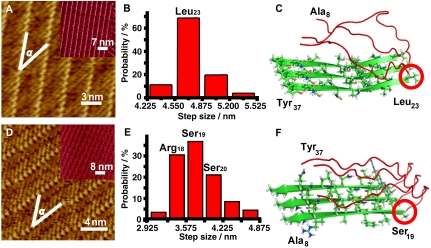
Fig. 4.
(A and D) High resolution STM images of rIAPP8–37 (A) and rIAPP37–8 (D) coassembled with 4Bpy. The angles α between peptide molecular axes and the stripe directions are measured to be 36 ± 2° (A) and 30 ± 2° (D). Tunneling conditions: (A) I = 550.1 pA, V = 322.8 mV; (D) I = 611.5 pA, V = 300.0 mV. Insets in A and D are large scale STM images. (B and E) Length distribution histograms of rIAPP8–37 (B) and rIAPP37–8 (E) beta-structure motifs measured from the STM images. The step sizes for the histograms are also 0.325 nm, which is the same as Fig. 3. (C and F) Proposed models for the folding sites in rIAPP8–37 and rIAPP37–8 beta-structure motifs respectively. The denotation for the colors and the shapes are the same as Fig. 3.
The histogram of the length distribution ranged from 4.22 to 5.52 nm for rIAPP8–37 C-terminal strands suggests the most probable number of 15 residues (Fig. 4B and Table S4). As a result, the C-terminal beta-structure motif of rIAPP8–37 could be proposed as preferentially starting from Leu23 as shown in Fig. 4C.
The structural information of rIAPP8–37 N -erminal strand can be obtained by similar experiments performed for the coassembly of rIAPP37–8 and 4Bpy (Fig. 4D). As shown in Fig. 4E and Table S5, the histogram of the length distribution ranges from 2.92 to 4.88 nm, and the most probable length covers 12 residues, equivalent to strands of Ala8-Ser19. Therefore, the key site for N-terminal beta-structure motif of rIAPP8–37 could be around residue Ser19 (possibly Arg18, Ser19, or Ser20) (Fig. 4F). The wide distribution different from that of hIAPP indicates the multiplicity of the rIAPP8–37 beta-structure motif as shown in Fig. 4E.
The beta-structure motifs of rIAPP8–37 observed in STM experiments may be composed of native beta motifs in physiological environment or beta-sheet-like structures induced by substrate surface (7, 24). Enhanced propensity for forming beta-like-structures for rIAPP on hydrophobic surfaces has been suggested by Rivera et al. (38) by using a combination of Monte Carlo and molecular dynamics simulations. The solution NMR spectroscopy results about rIAPP structures (39) could be a corroborative evidence for the current STM experiments on the key sites of rIAPP beta-like motif. In the model described by Nanga et al. (39), the structure of rIAPP is composed of an ordered helix (rIAPP5–17), a more disordered helix (rIAPP20–23), and an unstructured segment (rIAPP24–37), which is qualitatively consistent with the two possible sites around Ser19, and Leu23 obtained from the STM experiments. The polydispersity of the rIAPP8–37 beta-like motif could be associated with its less fibrillogenic characteristics.
Effect of Hot Spots in Peptide Sequence and Folding Sites.
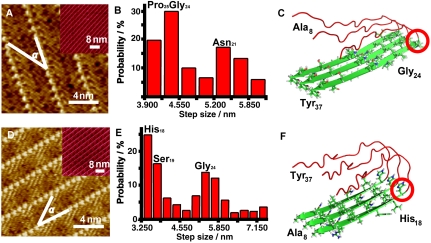
With the aim of understanding the relationship between “hot spots” (40, 41) in peptide primary sequence and beta-like-structure stability, STM experiments were carried out on the coassembly structures of peptide rIAPP8–37 R18H and 4Bpy (Fig. 5A). Coexistence of peptide rows with different width can be observed in STM images, and the length of peptides is nearly constant within a row (as shown in Fig. S2A). The histogram of peptide strand lengths exhibits bimodal distribution with the two most probable lengths of the beta-structure motifs, 13 or 14 residues for the predominant distribution and 17 residues for the minor distribution, respectively (Fig. 5E and Table S6). It could be plausible to propose that the preferential site for beta-like motifs in rIAPP8–37 R18H is located at around Gly24 (possibly Gly24 or Pro25) as shown in Fig. 5C. The minor distribution with strand length of 17 residues corresponds to the beta-structure motif rIAPP21–37 R18H, in which segment rIAPP21–23 (or segment rIAPP21–24) could be ascribed to the surface-induced meta-stable beta-turn-like structures, which is attached to the primary beta-structure motif rIAPP24–37 R18H or rIAPP25–37 R18H. This result differs apparently from that of rIAPP in which residue Leu23 is attributed as the end site for the N-terminal beta-structure motif.
Fig. 5.
(A and D) High resolution STM images of rIAPP8–37 R18H (A) and rIAPP37–8 R18H (D) coassembled with 4Bpy. The angles (α) between peptide molecular axes and the stripe directions are measured to be 34 ± 2° (A) and 36 ± 2° (D). Tunneling conditions: (A) I = 612.9 pA, V = 300.4 mV; (D) I = 575.9 pA, V = 334.4 mV. Insets in A and D are large scale STM images. (B and E) Length distribution histograms of rIAPP8–37 R18H (B) and hIAPP37–8 R18H (E) beta-structure motifs measured from the STM images. The step sizes for the histograms are also 0.325 nm, which is the same as Fig. 3. (C and F) Proposed models for the folding sites in rIAPP8–37 R18H and rIAPP37–8 R18H beta-structure motifs respectively. The denotation for the colors and the shapes are the same as Fig. 3.
Similar experiments were performed for the coassembly of rIAPP37–8 R18H and 4Bpy (Fig. 5D). As shown in Fig. 5E and Table S7, there are two distribution regions of peptide strand lengths, 3.25 to 4.88 nm (11–15 residues, predominant distribution), 4.88 to 7.48 nm (16–23 residues, minor distribution). The coexistence of parallel peptide rows of different width is shown in Fig. S2B. Therefore, the most probable key site for beta-structure motif of rIAPP37–8 R18H could be identified around His18 (possibly His18 or Ser19). Segment rIAPP19–24 R18H (or rIAPP20–24 R18H) could be attributed to a surface-induced meta-stable beta-sheet-like structure motif attached to the primer beta-structure motif rIAPP8–18 R18H (or rIAPP8–19 R18H). This difference in the key sites of beta-structure motifs between rIAPP8–37 and rIAPP8–37 R18H may be considered as evidence of possible collateral effect as the result of mutation of His18 with Arg18.
The results of R18H/F23L replacements (42, 43) suggested cooperativity between hIAPP residues 18 and 23, which agrees well with the suggested collateral effect in rIAPP8–37 R18H. The proposed preferential sites for rIAPP R18H motifs located at around His18 (possibly His18 or Ser19) and Gly24 (possibly Gly24 or Pro25) based on STM results are different from that of rIAPP. This observation coincides with the recognition of residue His18 to play a long-range effect in modulating peptide stacking interactions (12, 43, 44). Moreover, Pro25 is believed to contribute to rIAPP8–37 R18H fibrillogenesis due to the well-known favoring of the beta-turns structure and intermolecular assembly (45–47), while the residues Pro28-Pro29 are a part of peptide in the typical study of the polyproline II helix conformation (48). Two proline residues adopt (φ, ψ) backbone dihedral angles of roughly (-75°, 150°) and have trans-isomers of their peptide bonds, endowing rigidity for C terminus structure motif (48) in rIAPP and rIAPP R18H and facilitating the STM observations of the nearly straight strands in both of the assemblies.
The minor distribution in rIAPP8–37 R18H assembly indicated one more possible key site at residue Asn21, which suggested the possible extension of the beta-structure motif even with Pro25 in the middle of the strand. The minor length distribution in rIAPP37–8 R18H assembly also suggested one more possible key site at residue Gly24, indicating the possible extension of the beta-structure motif rIAPP8–18 R18H or rIAPP8–19 R18H. The overlapping possible beta-structure motif rIAPP20–23 R18H could be ascribed to the three beta segments in rIAPP8–37 R18H linked by turns at His18-Ser19 and Gly24-Pro25, which may contribute to the enhanced tendency of beta-structure and fibrillogenesis effect (20) with site mutation R18H in rIAPP.
The observations of the above analogs of IAPP revealed both similarity and differences of the key sites. For example, the IAPP8–37 analogs share common structure motifs of IAPP8–17 and IAPP26–37 with the most probable key sites at positions around Ser19/Ser20 and Gly24. This observation illustrates the similar amyloid formation tendency in the C and N terminus segments because of the sequence similarity, while the differences in specific amino acids at each key site manifest the effect of sequence variations. The identification of the side groups of the amino acid residues are still far out of reach of this study. The detailed models with atomic specificity for the peptide assembly are still challenging. The influence of the sample preparation conditions on the peptide self-assembly process is also a crucial issue need to be understood at molecular level, which will be the focus in our future studies.
Conclusion
We observed human IAPP8–37, rat IAPP8–37, and rat IAPP8–37 R18H assemblies with beta-like-structures and identified the motif end sites on the basis of the STM experiments. The observed multiplicity in key sites for beta-structure motifs may be associated with the polymorphism in hairpin structures or fibrillogenesis characteristics. Substitutions in amyloidal peptide sequence play a vital role in motif formation. Overall, it is a feasible and beneficial method to obtain key sites in peptide multiple beta-structure motifs by studying peptide assembly polymorphisms with STM.
Materials and Methods
Sample Preparation.
Synthetic hIAPP8–37 was obtained from American Peptide Co. Synthetic human IAPP with reverse sequence IAPP37–8, wild-type rIAPP8–37 and rIAPP37–8, and variant peptides rIAPP8–37 R18H and rIAPP37–8 R18H were purchased from Shanghai Science Peptide Biological Technology Co., Ltd. The 4,4′-bipyridyl was purchased from Sigma-Aldrich Co., Ltd. All the materials were purchased and used without further purification.
Lyophilized powders of hIAPP8–37 were dispersed in hexafluoroisopropanol (HFIP) first in eppendorf tube. After the solvent HFIP was totally evaporated, the peptides were dissolved in Milli-Q water into a concentration of 1 mg/mL. The hIAPP8–37 solution (15 μL) was first deposited on the surface of freshly cleaved highly oriented pyrolytic graphite (HOPG) for approximately 1 min incubation, and then the excess solution was blown away from the HOPG surface and the surface are blew dry by using high purity nitrogen gas prior to STM experiments.
For the coassembly experiments, hIAPP8–37, hIAPP37–8, rIAPP8–37, rIAPP37–8, rIAPP8–37 R18H, or rIAPP37–8 R18H was dissolved in chromatographic grade acetonitrile (Sigma), respectively. The solid powder of 4Bpy was dissolved in Milli-Q water. For each coassembly experiment, the peptide solution was mixed with 4Bpy solution to reach the final peptide concentration ca. 1 mg/mL with peptide/4Bpy molar ratio 1∶1. A drop of the mixed solution of peptide and 4Bpy (1 μL) was deposited directly onto the surface of freshly cleaved HOPG followed by the STM experiments after solvents were completely evaporated.
STM Measurements.
STM experiments were performed in constant-current mode under ambient conditions (Nanoscope IIIa SPM system, Veeco). The STM tips were mechanically formed by Pt/Ir wire (80/20). The tunneling conditions are described in the corresponding figure captions. Experiments were repeated independently using different tips for reproducibility.
Supplementary Material
Acknowledgments.
The authors thank Prof. Yanmei Li and Dr. Jed Wiltzius for their helpful discussions. This work was supported by the National Basic Research Program of China (2009CB930100, 2011CB932800) and the Chinese Academy of Sciences (CAS) (KJCX2-YW-M15). Financial support from the National Natural Science Foundation of China (20911130229) and CAS Key Laboratory for Biological Effects of Nanomaterials and Nanosafety are also gratefully acknowledged.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102971108/-/DCSupplemental.
References
- 1.Opie EL. The relation of diabetes mellitus to lesions of the pancreas: hyaline degradation of the islands of Langerhans. J Exp Med. 1901;5:527–540. doi: 10.1084/jem.5.5.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Westermark P, et al. Amyloid fibrils in human insulinoma and islets of Langerhans of the diabetic cat are derived from a neuropeptide-like protein also present in normal islet cells. Proc Natl Acad Sci USA. 1987;84:3881–3885. doi: 10.1073/pnas.84.11.3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooper GJS, Willis AC, Leighton B. Amylin hormone. Nature. 1989;340:272–272. doi: 10.1038/340272b0. [DOI] [PubMed] [Google Scholar]
- 4.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 5.Kayed R, et al. Conformational transitions of islet amyloid poly-peptide (IAPP) in amyloid formation in vitro. J Mol Biol. 1999;287:781–796. doi: 10.1006/jmbi.1999.2646. [DOI] [PubMed] [Google Scholar]
- 6.Glenner GG, Eanes D, Wiley C. Amyloid fbrils formed from a segment of the pancreatic islet amyloid protein. Biochem Biophys Res Commun. 1988;155:608–614. doi: 10.1016/s0006-291x(88)80538-2. [DOI] [PubMed] [Google Scholar]
- 7.Moriarty DF, Raleigh DP. Effects of sequential proline substitutions on amyloid formation by human amylin(20–29) Biochemistry. 1999;38:1811–1818. doi: 10.1021/bi981658g. [DOI] [PubMed] [Google Scholar]
- 8.Tenidis K, et al. Identification of a penta- and hexapeptide of islet amyloid polypeptide (IAPP) with amyloidogenic and cytotoxic properties. J Mol Biol. 2000;295:1055–1071. doi: 10.1006/jmbi.1999.3422. [DOI] [PubMed] [Google Scholar]
- 9.Jaikaran ETAS, et al. Identification of a novel human islet amyloid polypeptide beta-sheet domain and factors influencing fibrillogenesis. J Mol Biol. 2001;308:515–525. doi: 10.1006/jmbi.2001.4593. [DOI] [PubMed] [Google Scholar]
- 10.Makin OS, Serpell LC. Structural characterisation of islet amyloid polypeptide fibrils. J Mol Biol. 2004;335:1279–1288. doi: 10.1016/j.jmb.2003.11.048. [DOI] [PubMed] [Google Scholar]
- 11.Jayasinghe SA, Langen R. Identifying structural features of fibrillar islet amyloid polypeptide using site-directed spin labeling. J Biol Chem. 2004;279:48420–48425. doi: 10.1074/jbc.M406853200. [DOI] [PubMed] [Google Scholar]
- 12.Kajava AV, Aebi U, Steven AC. The parallel superpleated beta-structure as a model for amyloid fibrils of human amylin. J Mol Biol. 2005;348:247–252. doi: 10.1016/j.jmb.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 13.Nelson R, et al. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawaya MR, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 15.Wiltzius JJW, et al. Atomic structure of the cross-beta spine of islet amyloid polypeptide (amylin) Protein Sci. 2008;17:1467–1474. doi: 10.1110/ps.036509.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luca S, Yau WM, Leapman R, Tycko R. Peptide conformation and supramolecular organization in amylin fibrils: Constraints from solid-state NMR. Biochemistry. 2007;46:13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaikaran ET, Clark A. Islet amyloid and type 2 diabetes: From molecular misfolding to islet pathophysiology. Biochim Biophys Acta. 2001;1537:179–203. doi: 10.1016/s0925-4439(01)00078-3. [DOI] [PubMed] [Google Scholar]
- 18.Yano BL, Hayden DW, Johnson KH. Feline insular amyloid: Association with diabetes mellitus. Vet Pathol. 1981;18:621–627. doi: 10.1177/030098588101800507. [DOI] [PubMed] [Google Scholar]
- 19.Betsholtz C, et al. Sequence divergence in a specific region of islet amyloid polypeptide (IAPP) explains differences in islet amyloid formation between species. FEBS Lett. 1989;251:261–264. doi: 10.1016/0014-5793(89)81467-x. [DOI] [PubMed] [Google Scholar]
- 20.Green J, et al. Full-length rat amylin forms fibrils following substitution of single residues from human amylin. J Mol Biol. 2003;326:1147–1156. doi: 10.1016/s0022-2836(02)01377-3. [DOI] [PubMed] [Google Scholar]
- 21.Wiltzius JJW, et al. Molecular mechanisms for protein-encoded inheritance. Nat Struct Mol Biol. 2009;16:973–978. doi: 10.1038/nsmb.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilead S, Gazit E. The role of the 14–20 domain of the islet amyloid polypeptide in amyloid formation. Exp Diabetes Res. 2008;2008:256954. doi: 10.1155/2008/256954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang P, Wei L, Pervushin K, Mu Y. pH-dependent interactions of human islet amyloid polypeptide segments with insulin studied by replica exchange molecular dynamics simulations. J Phys Chem B. 2010;114:10176–10183. doi: 10.1021/jp101811u. [DOI] [PubMed] [Google Scholar]
- 24.Abedini A, Raleigh DP. Destabilization of human IAPP amyloid fibrils by proline mutations outside of the putative amyloidogenic domain: Is there a critical amyloidogenic domain in human IAPP? J Mol Biol. 2006;355:274–281. doi: 10.1016/j.jmb.2005.10.052. [DOI] [PubMed] [Google Scholar]
- 25.Ma XJ, et al. Amyloid beta (1–42) folding multiplicity and single-molecule binding behavior studied with STM. J Mol Biol. 2009;388:894–901. doi: 10.1016/j.jmb.2009.03.054. [DOI] [PubMed] [Google Scholar]
- 26.Mao XB, et al. Structural characteristics of the beta-sheet-like human and rat islet amyloid polypeptides as determined by scanning tunneling microscopy. J Struct Biol. 2009;167:209–215. doi: 10.1016/j.jsb.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 27.Mao XB, et al. Molecular-level evidence of the surface-induced transformation of peptide structures revealed by scanning tunneling microscopy. Langmuir. 2009;25:8849–8853. doi: 10.1021/la901342r. [DOI] [PubMed] [Google Scholar]
- 28.Liu L, et al. Chaperon-mediated single molecular approach toward modulating Aβ peptide aggregation. Nano Lett. 2009;9:4066–4072. doi: 10.1021/nl902256b. [DOI] [PubMed] [Google Scholar]
- 29.Zhang L, Yang YL, Wang C. Regulation of aggregation and cytotoxicity of β-amyloid by molecular modulators. Acta Biophys Sin. 2010;26:649–661. [Google Scholar]
- 30.Yin SX, et al. Studies of the effects of hydrogen bonding on monolayer structures of C18H37X (X = OH, SH) on HOPG. Chem Phys Lett. 2001;348:321–328. [Google Scholar]
- 31.Xu B, et al. Identification of hydrogen bond characterization of isomeric 4Bpy and 2Bpy by STM. Surf Interface Anal. 2001;32:245–247. [Google Scholar]
- 32.Garrett RH, Grisham CM. Biochemistry. 2nd ed. London: Saunders College Publishing; 1999. pp. 168–169. [Google Scholar]
- 33.Blanco FJ, Rivas G, Serrano L. A short linear peptide that folds into a native stable β-hairpin in aqueous solution. Nat Struct Biol. 1994;1:584–590. doi: 10.1038/nsb0994-584. [DOI] [PubMed] [Google Scholar]
- 34.Goldschmidt L, Teng PK, Riek R, Eisenberg D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc Natl Acad Sci USA. 2010;107:3487–3492. doi: 10.1073/pnas.0915166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reddy AS, et al. Solution structures of rat amylin peptide: Simulation, theory, and experiment. Biophys J. 2010;98:443–451. doi: 10.1016/j.bpj.2009.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pappalardo G, et al. Environmental factors differently affect human and rat IAPP: Conformational preferences and membrane interactions of IAPP17-29 peptide derivatives. Eur Chem J. 2007;13:10204–10215. doi: 10.1002/chem.200700576. [DOI] [PubMed] [Google Scholar]
- 37.Engel MFM, et al. Islet amyloid polypeptide inserts into phospholipid monolayers as monomer. J Mol Biol. 2006;356:783–789. doi: 10.1016/j.jmb.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 38.Rivera E, Straub J, Thirumalai D. Sequence and crowding effects in the aggregation of a 10-residue fragment derived from islet amyloid polypeptide. Biophys J. 2009;96:4552–4560. doi: 10.1016/j.bpj.2009.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nanga RPR, et al. Three-dimensional structure and orientation of rat islet amyloid polypeptide protein in a membrane environment by solution NMR spectroscopy. J Am Chem Soc. 2009;131:8252–8261. doi: 10.1021/ja9010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ventura S, et al. Short amino acid stretches can mediate amyloid formation in globular proteins: The Src homology 3 (SH3) case. Proc Natl Acad Sci USA. 2004;101:7258–7263. doi: 10.1073/pnas.0308249101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ivanova MI, Sawaya MR, Gingery M, Attinger A, Eisenberg D. An amyloid-forming segment of beta 2-microglobulin suggests a molecular model for the fibril. Proc Natl Acad Sci USA. 2004;101:10584–10589. doi: 10.1073/pnas.0403756101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gazit E. A possible role for pi-stacking in the self-assembly of amyloid fibrils. FASEB J. 2002;16:77–83. doi: 10.1096/fj.01-0442hyp. [DOI] [PubMed] [Google Scholar]
- 43.Tracz SM, Abedini A, Driscoll M, Raleigh DP. Role of aromatic interactions in amyloid formation by peptides derived from human amylin. Biochemistry. 2004;43:15901–15908. doi: 10.1021/bi048812l. [DOI] [PubMed] [Google Scholar]
- 44.Brender JR, Hartman K, Reid KR, Kennedy RT, Ramamoorthy A. A single mutation in the nonamyloidogenic region of islet amyloid polypeptide greatly reduces toxicity. Biochemistry. 2008;47:12680–12688. doi: 10.1021/bi801427c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hutchinson EG, Thornton JM. A revised set of potentials for beta-turn formation in proteins. Protein Sci. 1994;3:2207–2216. doi: 10.1002/pro.5560031206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richardson JS. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- 47.Rose GD, Gierasch LM, Smith JA. Turns in peptides and proteins. Adv Protein Chem. 1985;37:1–109. doi: 10.1016/s0065-3233(08)60063-7. [DOI] [PubMed] [Google Scholar]
- 48.Adzhubei AA, Sternberg MJE. Left-handed polyproline II helices commonly occur in globular proteins. J Mol Biol. 1993;229:472–493. doi: 10.1006/jmbi.1993.1047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.