Abstract
Deficiencies of subunits of the transcriptional regulatory complex Mediator generally result in embryonic lethality, precluding study of its physiological function. Here we describe a missense mutation in Med30 causing progressive cardiomyopathy in homozygous mice that, although viable during lactation, show precipitous lethality 2–3 wk after weaning. Expression profiling reveals pleiotropic changes in transcription of cardiac genes required for oxidative phosphorylation and mitochondrial integrity. Weaning mice to a ketogenic diet extends viability to 8.5 wk. Thus, we establish a mechanistic connection between Mediator and induction of a metabolic program for oxidative phosphorylation and fatty acid oxidation, in which lethal cardiomyopathy is mitigated by dietary intervention.
Keywords: heart, metabolism, peroxisome proliferator-activated receptor-γ coactivator-1α
The Mediator complex encompasses ∼30 proteins necessary for expression of RNA polymerase II-transcribed genes (1), binding simultaneously to Pol II and to gene-specific transcriptional activators and promoting preinitiation complex assembly (2, 3).
The fundamental requirement for Mediator in eukaryotic gene expression is demonstrated by the fact that all mutant alleles of murine Mediator subunits generated to date are homozygous lethal during embryonic development. Med1−/− mice do not develop further than embryonic day 11.5, and a Med1 hypomorphic mutant dies before embryonic day 13.5, displaying a plethora of developmental defects (4, 5). Mice deficient in Med24 die before embryonic day 9.5 with severe developmental abnormalities (6). Med21 is essential for survival past the blastocyst stage of mouse embryonic development (7). Other lethal effects have been reported during embryogenesis in mice and zebrafish, with truncations of Cdk8 and Med12, respectively (8, 9). So far, only Med1 and Med24 deficiencies permit survival to a sufficiently differentiated stage of development in which analysis of distinct tissues is possible. In humans, missense mutations of MED12 lead to neurological abnormalities observed in Opitz-Kaveggia syndrome (10) or Lujan-Fryns syndrome (11). Understanding the mechanisms and consequences of Mediator binding to various transcription factors in the context of cell or tissue type, developmental stage, and biological process has thus been hampered by the embryonic lethality of Mediator mutants.
In mammals, Mediator was first isolated by virtue of its direct association with nuclear hormone receptors in complexes, including TRAP (thyroid hormone receptor-associated proteins) and DRIP (vitamin D receptor-interacting proteins), and was shown to play a key role in hormone-dependent signaling (2, 3). However, the physiological relevance of individual complex components to metabolism or disease has only recently begun to emerge. Here we describe zeitgeist (zg), a recessive N-ethyl-N-nitrosurea (ENU)-induced phenotype attributed to a missense mutation in Med30. Initially isolated from the TRAP complex (1), MED30 is a component of the Mediator core thus far not associated with any phenotype in living organisms. In contrast to other models with Mediator deficiencies, homozygous zeitgeist mice are physically indistinguishable from littermates at the time of weaning, but develop progressive cardiomyopathy that is invariably fatal by 7 wk of age. Mechanistically, the Med30zg mutation causes a progressive and selective decline in the transcription of genes necessary for oxidative phosphorylation (OXPHOS) and mitochondrial integrity, eventually leading to cardiac failure. Remarkably, a ketogenic diet (KD) significantly extends the lifespan of homozygotes.
Results and Discussion
We serendipitously observed conspicuous and frequent premature death in a pedigree of C57BL/6J G3 mice homozygous for ENU-induced mutations. This phenotype was mapped to a 14.4-Mbp critical region on Chromosome 15 (Fig. 1A and Fig. S1) and ascribed to a single nonsynonymous A-to-T transversion in the first of the four exons of Med30 (Fig. 1B). The mutation resulted in an isoleucine to phenylalanine substitution at amino acid 44 of the 178-residue protein. An isoleucine at the corresponding position is conserved in MED30 of all vertebrate and invertebrate species examined (Fig. 1C). Complementation studies between a gene-trap allele of Med30, in which the last exon was deleted and the Med30zg allele, did not produce any compound heterozygotes, suggesting the zeitgeist mutation is hypomorphic and embryonic lethal when placed in trans with a null allele of the same gene (Table S1).
Fig. 1.
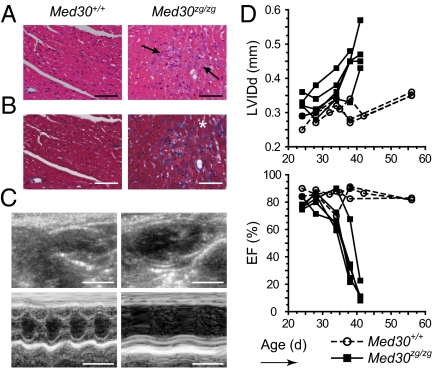
Zeitgeist phenotype; identification of the mutation and anatomical analysis of the heart. (A) Kaplan–Meier survival curve of C57BL/6J × C3H/HeN F1 hybrid zeitgeist mutants (n = 41; **P = 0.058). (B) Chromatogram showing an A195T transversion in exon 1 of Med30 (NM_027212). (C) The zeitgeist mutation causes an isoleucine to phenylalanine substitution at amino acid 44, a residue conserved in all species known to possess a Med30 gene. (D) The zeitgeist mutants displayed dilated and flaccid ventricles compared with control littermates (H&E staining). (Scale bars, 2 mm.) (F) MRI analysis of mutant and control hearts at the indicated ages shown in a coronal plane. Ao, aorta; LV, left ventricle, RV, right ventricle. (Scale bar, 5 mm.)
Other components of Mediator are critical for heart development and anatomy in mice (4–6). In humans, truncation or missense mutations in MED13L, a MED13 homolog, have been observed in some patients with transposition of the great arteries (12). In contrast, Med30zg/zg mice showed no developmental anomaly per se, only progressive heart failure postweaning. However, Med30zg/zg mice were born to heterozygous parents at a frequency beneath expectation, indicating that attrition of homozygotes occurs prenatally (Table S2). Genotyping of embryos at gestational day 15.5 suggested that death may occur after this time point (Table S2).
Dissection of moribund Med30zg/zg mice revealed a dilated cardiomyopathy (DCM), affecting all chambers of the heart (Fig. 1D), and ascites. In situ MRI analysis indicated progressive dilation of the ventricles, which became prominent at 32 d of age (Fig. 1E). Further histopathological investigations of Med30zg/zg mice showed fibrosis of the atrial wall (Fig. S2A), focal myocardial necrosis (Fig. 2A), myofibril loss, and diffuse interstitial fibrosis (Fig. 2B) of dilated hearts. Echocardiography of Med30zg/zg mutants shortly before death indicated ventricle and atrium dilation (Fig. 2C). Furthermore, the contractility of mutant hearts was very poor, with a fractional shortening of 13.9 ± 5.9% vs. 45.7 ± 4.0% for wild-type controls (P < 0.001) (Table S3). Dilation and functional impairment of mutant hearts were progressive and started after 4 wk of age (Fig. 2D and Fig. S2B). These data support the conclusion that Med30zg/zg mice die of heart failure stemming from myocardial necrosis, fibrosis, and consequent impaired myocardial contractility.
Fig. 2.
Zeitgeist homozygotes develop myocardial fibrosis and heart failure. Histological and functional analysis of mutant hearts shortly before death. (A) H&E staining of myocardium. (Scale bars, 100 μm.) (B) Masson's trichrome staining of the ventricular wall. Arrows show regions with cardiomyocyte necrosis and asterisks areas of prominent fibrosis. (Scale bars, 100 μm.) (C) Echocardiography of mutant and control hearts. (Scale bars, 0.5 cm.) (D) LV internal dimension in diastole (LVIDd) and ejection fraction (EF) as a function of age. n = 3 for wild-type; n = 5 for mutant mice.
The function of other vital organs, including the liver and kidneys, appeared intact in homozygotes, although moderate prerenal azotemia was apparent (Table S4). Blood lactate concentration did not differ between wild-type and Med30zg/zg mice, suggesting that the phenotypic anomaly is restricted to cardiac muscle (Fig. S3). Alternatively, the brief lifespan of mutants may not allow skeletal muscle or other organs to display a detectable phenotype.
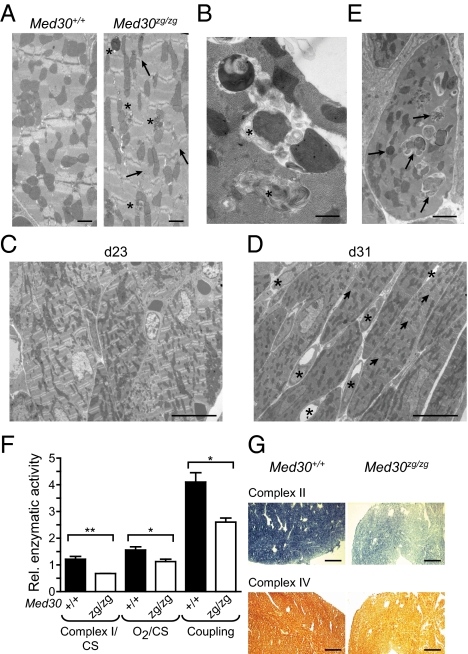
Mutations in cytoskeletal or sarcomeric proteins are commonly responsible for inherited DCM (13), but mitochondrial dysfunction can also instigate cardiomyopathy (14). We therefore examined cardiomyocytes in the left ventricle (LV) of homozygous Med30zg/zg hearts by electron microscopy. We observed disorganization of the Z-band pattern and shortening of I bands (Fig. 3A). We also observed abnormalities in mitochondrial morphology characterized by cristolysis and the formation of membranous swirls within individual mitochondria (Fig. 3B), features that became apparent after 4 wk of age (Fig. 3 C and D). The degree and extent of mitochondrial alterations were irregular, both among neighboring cells and within single cardiomyocytes (Fig. 3 B and E). In the most severe cases, such changes were associated with disappearance of myofibrillar structure (Fig. 3E), augmentation of the interstitial space between the cardiomyocytes (Fig. 3D), and increased numbers of intercalated fibroblasts (Figs. 2B and 3D). Similar mitochondrial anomalies result from deficiencies affecting OXPHOS (15, 16). Therefore, we assessed the enzymatic activity of complexes of the electron transport chain (ETC) in mutant cardiac mitochondria. Analysis of total cardiac mitochondria and in situ analysis revealed a significant decline in the function of several ETC complexes, in the rate of oxygen consumption, and in coupling efficiency (Fig. 3 F and G). We observed no differences in the number or volume of mitochondria relative to those of wild-type mice, nor accumulation of lipid droplets (Fig. 3A and Fig. S4), as previously reported for other models of cardiomypathy (15, 17, 18). We conclude that in Med30zg/zg mice, impaired OXPHOS leads to mitochondrial deterioration and dysfunction.
Fig. 3.
Altered ultrastructure and defective function of Med30zg/zg mitochondria. (A) Electron micrographs of LV sections from 34-d-old wild-type and mutant mice. Arrows indicate disarrayed Z bands and asterisks aberrant mitochondria. (Scale bars, 1 μm.) (B) High magnification view of aberrant mitochondria. Asterisks indicate membranous swirls. (Scale bar, 500 nm.) (C and D) Cardiomyocytes of 3-wk-old mutant mice display normal myofibrillar structure; ultrastructural and histological alterations are visible in mutant mice ≥ 4 wk old. Electron micrographs of LV sections of Med30zg/zg hearts on day 23 (C) and day 31 (D). (Scale bars, 10 μm.) Asterisks indicate intercalated fibroblasts and arrows disarrayed Z bands. (E) An isolated cardiomyocyte from a 31-d-old Med30zg/zg LV. Arrows show examples of defective mitochondria. (Scale bar, 2 μm.) (F) Measurements of respiratory chain function in cardiac mitochondria relative to citrate synthase (CS) activity in 32-d-old hearts. Complex I activity (**P = 0.0076), oxygen consumption (O2) (*P = 0.0434), and coupling efficiency (*P = 0.0173) were measured. n = 3. Values are represented as mean ± SEM. (G) In situ activity of Complex II and Complex IV in day 39 LV as measured by staining for succinate dehydrogenase and cytochrome c oxidase, respectively. (Scale bars, 100 μm.)
Mediator functions as a transcriptional regulator. To gain insight into the transcriptional defect imparted by Med30zg/zg, we used a candidate approach and analyzed the expression level of selected genes involved in OXPHOS, in the generation of substrates for electron transport, and in the regulation of mitochondrial biogenesis. Examination at 32–39 d of age of the transcriptional profile of the LV of zeitgeist homozygotes with DCM compared with that of wild-type mice revealed 81% and 73% reductions, respectively, in transcripts of Ppargc1a and Esrra, which encode PGC-1α (peroxisome proliferator-activated receptor-γ coactivator-1α) and estrogen-related receptor (ERR)α, key transcriptional regulators of mitochondrial biogenesis, OXPHOS, and fatty acid oxidation (FAO) (19–21). PGC-1α acts as a coactivator that binds to and enhances the transcriptional activity of the transcription factor ERRα. There was also significant down-modulation of the expression of the genes encoding citrate synthase (Cs) and several subunits of complex I, II, and IV of the ETC (Table 1). Many OXPHOS genes, including Cox5b, Cox6a2, Ndufs2, Sdha, and Sdhb, are direct targets for PGC-1α-ERR–mediated coactivation (22). Thus, reduced Ppargc1a expression in homozygous zeitgeist hearts may prevent adequate expression of OXPHOS genes (Table 1), resulting in a corresponding diminution in the activity of ETC complexes (Fig. 3 F and G).
Table 1.
Gene expression in Med30zg/zg (n = 3) relative to Med30+/+ LV (n = 2) from 32- to 39-d-old mice
| Function | Gene | Med30zg/zg/Med30+/+ | P |
| Transcription | |||
| Energy metabolism | Ppargc1a | 0.19 | * |
| Energy metabolism | Ppara | 0.21 | ns |
| Energy metabolism | Esrra | 0.27 | * |
| Energy metabolism | Ppard | 0.55 | ns |
| Mediator complex | Med23 | 0.62 | ns |
| Energy metabolism | Pparg | 0.69 | ns |
| Mediator complex | Med1 | 0.69 | ns |
| Mediator complex | Med17 | 0.71 | ns |
| Mediator complex | Med12 | 0.76 | ns |
| Mediator complex | Med30 | 1.31 | ns |
| Mediator complex | Med15 | 1.41 | ns |
| Mediator complex | Med14 | 1.56 | ns |
| OXPHOS | |||
| Complex II | Sdhb | 0.21 | ** |
| Complex I | Ndufs7 | 0.22 | * |
| Complex IV | Cox10 | 0.24 | * |
| Complex II | Sdhd | 0.25 | ** |
| Complex IV | Cox6a2 | 0.28 | * |
| Complex II | Sdha | 0.28 | ** |
| Complex I | Ndufs2 | 0.30 | * |
| Complex I | Ndufb7 | 0.35 | ** |
| Complex II | Sdhc | 0.35 | ** |
| Complex I | Ndufa10 | 0.43 | * |
| Complex IV | Cox5b | 0.44 | ** |
| Complex I | Ndufv1 | 0.55 | * |
| Complex I | Ndufb3 | 0.58 | * |
| Complex I | Ndufa1 | 0.67 | ns |
| Complex IV | Surf1 | 0.79 | ns |
| Metabolism | |||
| Fatty acid metabolism | Lpl | 0.12 | ** |
| Krebs cycle | Cs | 0.24 | ** |
| Glucose metabolism | Pdk4 | 0.37 | ns |
| Fatty acid metabolism | Fabp4 | 0.65 | ns |
| Fatty acid metabolism | Cpt1b | 0.67 | ns |
| Fatty acid metabolism | Angptl4 | 1.23 | ns |
| Glucose metabolism | Hk1 | 1.39 | ns |
| ROS detoxification | |||
| Superoxide dismutase | Sod2 | 0.33 | ** |
| Catalase | Cat | 0.58 | ns |
| Superoxide dismutase | Sod1 | 0.85 | ns |
| Mitochondrial | |||
| Transcription factor | Tfam | 0.39 | ns |
| Complex IV | mt-Co1 | 0.44 | * |
| ATP synthase | mt-Atp6 | 0.58 | ns |
| Proton carrier | Ucp2 | 0.84 | ns |
ns, nonsignificant; ROS, reactive oxygen species;
*P < 0.05;
**P ≤ 0.01. Values of quantitative results represent gene expression level in Med30zg/zg LV over that in Med30+/+ LV.
When examined at weaning age (23 d, when mice appear healthy) and at 30 d of age (just before the precipitous decline in heart function), selected transcriptional regulator and OXPHOS transcripts showed a progressive decline in expression relative to levels in wild-type controls (Fig. 4A). In particular, the levels of Ppargc1a and Esrra transcripts were similar to those in wild-type mice on day 23, but were reduced 50% relative to wild-type levels on day 30 before heart failure. These data suggest that the progressive decline in expression of genes required for electron transport between postnatal days 23 and 30 leads to mitochondrial dysfunction and subsequent heart failure in Med30zg/zg mice. Notably, the expression of Med30 itself did not differ significantly between mutants and wild-type mice at 23 or 30 d of age (Fig. S5A). PGC-1α is also a regulator of enzymes important for detoxification of reactive oxygen species (23), consistent with the 67% down-modulation of Sod2 transcript expression in mutant hearts (Table 1).
Fig. 4.
Partial rescue of zeitgeist phenotype by KD. (A) Relative expression of selected genes at 23, 30, and 45 d of age as measured by quantitative PCR. Mice were fed either chow (CD; solid lines) or ketogenic (KD; dotted lines) diet beginning on day 23, when they were weaned. Gene expression in Med30zg/zg over that in Med30+/+ whole hearts is represented. For each biological sample, quantitative PCR reactions were performed in duplicate and expression was normalized to Rpl32 expression. For each indicated time point and diet, P values were determined by comparing the relative expression of the indicated gene in Med30zg/zg vs. Med30+/+ hearts using an unpaired Student's two-tailed t test. Statistically significant P values are indicated. For Med30zg/zg hearts, n = 3 for day 23 CD; n = 8 for day 30 CD; n = 5 for day 30 KD; n = 5 for day 45 KD. For Med30+/+ hearts, n = 2 for day 23 CD; n = 3 for day 30 CD; n = 5 for day 30 KD; n = 4 for day 45 KD. (B) Kaplan–Meier survival curve of Med30zg/zg mice (n = 3; ***P = 0.0002) on a KD compared with Med30zg/zg mice on a regular diet (n = 71). Significance was determined by log-rank test. ns, not significant.
In our attempts to understand the progressive course of the Med30zg/zg phenotype, we considered environmental changes that precede disease onset. In the developing heart, the early postnatal period is characterized by a metabolic shift from fetal glucose and lactate oxidation to mitochondrial FAO, a program depending on the PGC-1 and ERR isoforms (21, 24). Because the health of homozygous zeitgeist mice deteriorates within a few days of separation from their mothers, we hypothesized a role for MED30 in accommodation to the dietary change associated with weaning, which entails a reduction in fat intake. Mouse milk is exceptionally rich in fat, containing 42% lipids in the C57BL/6J strain (25). Regular chow has a mere 5–9% fat content. Strikingly, newly weaned mutants fed a KD had a significantly increased lifespan, with all mice surviving at least 10 d after 100% lethality was observed in mutants weaned to chow (Fig. 4B). Moreover, in 30-d-old KD-fed Med30zg/zg mice, cardiac expression of Ppargc1a, Esrra, Sod2, and several OXPHOS genes was increased compared with chow-fed Med30zg/zg controls, albeit to levels below those measured in wild-type hearts (Fig. 4A). These data are consistent with reports that a KD induces up-regulation of Ppargc1a expression (26) and reduces oxidative stress (27). We infer that the KD counterbalances the detrimental effects of the zeitgeist mutation by stimulating, through unknown mechanisms, the expression of Ppargc1a, Esrra, and other OXPHOS genes. Rescue is impermanent, however, and by 45 d of age, gene expression has declined in mice maintained on a KD (Fig. 4A).
Although the nature of the association between MED30, PGC-1α, and ERRα has not been explored, PGC-1α can directly engage Mediator through MED1 (28), for which a role has been demonstrated in FAO and glucose metabolism (29, 30). MED30zg, as part of Mediator, might disturb this interaction, and in turn the interaction of the PGC-1α–Mediator complex with target genes. However, immunoprecipitation of the Mediator complex from nuclear extracts from primary cells using either MED4 or MED30 antibodies did not reveal any major perturbation of Mediator complex integrity by endogenous MED30zg (Fig. S5B). Of note, in patients with heart failure, including DCM and ischemic heart disease, FAO is reduced but glucose metabolism is favored, recapitulating the fetal metabolic program. These changes are associated with diminution of Ppargc1a and Esrra expression (31, 32). The signals leading to their down-regulation during heart failure are still elusive (33), although MYC has been proposed as a potential upstream repressor of Ppargc1a transcription (34).
Our findings establish a connection between Mediator function and the induction of a metabolic program for mitochondrial OXPHOS and FAO, with a specific impact on cardiac function. Mitochondrial cardiomyopathy has been associated with numerous mutations in both mitochondrial and nuclear DNA-encoded genes (14, 35–38). Our data indicate that genetic lesions affecting Mediator complex subunits should also be considered as a possible cause of primary cardiomyopathies in humans, particularly in pediatric cases given the importance of MED30 to early cardiac function during the period of weaning in mice. Such diseases may not be associated with anatomic abnormalities, such as shunts. Our findings also illustrate that, at least in some instances, morbidity and mortality may be mitigated by dietary modifications.
Materials and Methods
Mice.
All mouse studies were performed in accordance with institutional regulations governing animal care and use and were approved by the Scripps Research Institute Institutional Animal Care and Use Committee. C57BL/6J mice were bred locally at The Scripps Research Institute and treated with ENU, as previously described (39). The zeitgeist strain is described at http://mutagenetix.utsouthwestern.edu. The gene trap line was reconstituted from the SIGTR ES cell line CH0776 obtained from the Mutant Mouse Regional Resource Center. Bio-Serv F3666 diet was used for the KD.
Genetic Mapping and Mutation Identification.
Death was used as a phenotype to assign the mutation to a region on chromosome 15 (Fig. S1). Genetic mapping was performed as described (http://mutagenetix.utsouthwestern.edu/protocol/protocol_rec.cfm?pid=14) using a genome-wide panel of 128 simple sequence-length polymorphisms distinguishing C57BL/6J from the mapping strain, C3H/HeN. Six additional markers were used for meiotic mapping as indicated in Fig. S1A. Only mice with phenotype were used for mapping. All coding sequences within 71 annotated genes in the critical region were subjected to bidirectional capillary sequencing and 71.2% coverage to a phred score of 30 or greater was achieved on at least one strand.
Histological Studies.
Paraffin-embedded sections were stained with H&E or with Masson's trichrome.
LV from paraformaldehyde- and glutaraldehyde-fixed heart tissues were used for electron microscopy and examined with a Philips CM100 electron microscope. Images were documented with a Megaview III CCD camera (Olympus).
MR Imaging.
For MRI, mice were anesthetized and hearts were arrested in diastole phase by injecting a KCl solution. Whole mice were fixed in formalin before in situ imaging. Hearts were scanned using a 7T micromagnetic resonance imaging system (Biospin 7T/30; Bruker). MRI scanning was carried out using a 3D T1-weighted image protocol (RARE T1, TE = 12.3 ms, TR = 1,300 ms; Bruker Paravision). Field of view was set as 40 × 40 × 40 mm (512 × 512 × 256 voxels). MRI scan was taken for ∼10 h. Acquired 3D-stack MRI images were then converted to standard DICOM images and multiplane image reconstruction was performed using OsiriX software (OsiriX v3.8.1) and a computer (Mac Pro, Apple Inc.).
Echocardiography Measurements.
A Philips IE33 ultrasound unit with a 17-MHz linear transducer was used for echocardiography of anesthetized animals. Long- and short-axis M-mode at the level of the papillary muscle was used for measurement of diastolic and systolic diameter of LV, as well as wall thickness. Pulsed-wave Doppler was performed at the LV outflow tract, right ventricular outflow tract, mitral, and tricuspid valves for recording the flow pattern and velocity.
Mitochondrial Isolation and OXPHOS Enzyme Analysis.
Mitochondria were isolated from mouse hearts by homogenization and differential centrifugation, as previously described (40). Briefly, mouse hearts were collected, washed with isolation buffer, minced, digested with collagenase, and homogenized using a Dounce homogenizer. The homogenate was subjected to low-speed centrifugation to remove nuclei and cell debris and high-speed centrifugation to collect mitochondria. Respiration and enzymatic assays were performed using standard protocols (40).
Mitochondrial Histochemical Staining.
Mouse hearts were collected and frozen freshly in OCT compound with isopentane supercooled by liquid nitrogen. Six-micrometer sections were cut with a cryostat and stained for succinate dehydrogenase (complex II) or cytochrome c oxidase (complex IV) activities, as described previously (41).
Quantitative RT-PCR of Heart Tissue.
Mutant and wild-type heart were perfused with ice-cold PBS and LV were isolated and homogenized in TRIzol reagent (Invitrogen). Total RNA was purified according to the manufacturer's recommendations and subjected to first-strand cDNA synthesis using a RT super mix kit (BioPioneer). RNA levels were analyzed by quantitative RT-PCR with a SYBR Green-based kit (qPCR supermix; BioPioneer). For each biological sample, quantitative PCR reactions were performed in duplicate, and expression was normalized to Rpl32 expression, which has been reported to be a stable reference in mouse failing myocardium (42). P values were determined by comparing the relative expression of indicated gene in Med30zg/zg vs. Med30+/+ LV.
Statistical Analyses.
For Kaplan–Meier survival curve, significance of difference was determined by log-rank test. For differences between observed and expected numbers of progeny, significance was determined by either χ2 test or the Freeman–Halton extension of the Fisher exact probability test. For all of the other tests, the statistical significance of differences was determined by unpaired Student's two-tailed t test. All tests were calculated using GraphPad Prism version 4.00 software. For all figures: ns, nonsignificant; unless a specific P value is indicated, *P < 0.05; **P ≤ 0.01; ***P ≤ 0.001. Values of quantitative results are expressed as mean ± SEM.
Supplementary Material
Acknowledgments
We thank Margaret Chadwell for technical support and Dr. Nissi Varki for help with the initial evaluation of the histology. This work was supported by National Institutes of Health (NIH) Grant 5P01AI070167 and Contract HHSN272200700038C (to B.B.); NIH Grants DK062434 and HL105278 and the Helmsley Charitable Trust (to R.M.E.); fellowships from the European Molecular Biology Organization and the Swiss National Science Foundation (P.K.); and a fellowship from the Salk Center for Nutritional Genomics (to W.F.). R.M.E. is an Investigator of the Howard Hughes Medical Institute at the Salk Institute and March of Dimes Chair in Molecular and Developmental Biology.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117835108/-/DCSupplemental.
References
- 1.Baek HJ, Malik S, Qin J, Roeder RG. Requirement of TRAP/mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAF(II)s. Mol Cell Biol. 2002;22:2842–2852. doi: 10.1128/MCB.22.8.2842-2852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 4.Ito M, Yuan CX, Okano HJ, Darnell RB, Roeder RG. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol Cell. 2000;5:683–693. doi: 10.1016/s1097-2765(00)80247-6. [DOI] [PubMed] [Google Scholar]
- 5.Landles C, et al. The thyroid hormone receptor-associated protein TRAP220 is required at distinct embryonic stages in placental, cardiac, and hepatic development. Mol Endocrinol. 2003;17:2418–2435. doi: 10.1210/me.2003-0097. [DOI] [PubMed] [Google Scholar]
- 6.Ito M, Okano HJ, Darnell RB, Roeder RG. The TRAP100 component of the TRAP/Mediator complex is essential in broad transcriptional events and development. EMBO J. 2002;21:3464–3475. doi: 10.1093/emboj/cdf348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tudor M, Murray PJ, Onufryk C, Jaenisch R, Young RA. Ubiquitous expression and embryonic requirement for RNA polymerase II coactivator subunit Srb7 in mice. Genes Dev. 1999;13:2365–2368. doi: 10.1101/gad.13.18.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong SK, et al. The zebrafish kohtalo/trap230 gene is required for the development of the brain, neural crest, and pronephric kidney. Proc Natl Acad Sci USA. 2005;102:18473–18478. doi: 10.1073/pnas.0509457102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Westerling T, Kuuluvainen E, Mäkelä TP. Cdk8 is essential for preimplantation mouse development. Mol Cell Biol. 2007;27:6177–6182. doi: 10.1128/MCB.01302-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Risheg H, et al. A recurrent mutation in MED12 leading to R961W causes Opitz-Kaveggia syndrome. Nat Genet. 2007;39:451–453. doi: 10.1038/ng1992. [DOI] [PubMed] [Google Scholar]
- 11.Schwartz CE, et al. The original Lujan syndrome family has a novel missense mutation (p.N1007S) in the MED12 gene. J Med Genet. 2007;44:472–477. doi: 10.1136/jmg.2006.048637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muncke N, et al. Missense mutations and gene interruption in PROSIT240, a novel TRAP240-like gene, in patients with congenital heart defect (transposition of the great arteries) Circulation. 2003;108:2843–2850. doi: 10.1161/01.CIR.0000103684.77636.CD. [DOI] [PubMed] [Google Scholar]
- 13.Jefferies JL, Towbin JA. Dilated cardiomyopathy. Lancet. 2010;375:752–762. doi: 10.1016/S0140-6736(09)62023-7. [DOI] [PubMed] [Google Scholar]
- 14.Fosslien E. Review: Mitochondrial medicine—Cardiomyopathy caused by defective oxidative phosphorylation. Ann Clin Lab Sci. 2003;33:371–395. [PubMed] [Google Scholar]
- 15.Bushdid PB, Osinska H, Waclaw RR, Molkentin JD, Yutzey KE. NFATc3 and NFATc4 are required for cardiac development and mitochondrial function. Circ Res. 2003;92:1305–1313. doi: 10.1161/01.RES.0000077045.84609.9F. [DOI] [PubMed] [Google Scholar]
- 16.Walker DW, Benzer S. Mitochondrial “swirls” induced by oxygen stress and in the Drosophila mutant hyperswirl. Proc Natl Acad Sci USA. 2004;101:10290–10295. doi: 10.1073/pnas.0403767101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehman JJ, et al. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng L, et al. Cardiomyocyte-restricted peroxisome proliferator-activated receptor-delta deletion perturbs myocardial fatty acid oxidation and leads to cardiomyopathy. Nat Med. 2004;10:1245–1250. doi: 10.1038/nm1116. [DOI] [PubMed] [Google Scholar]
- 19.Giguère V, Yang N, Segui P, Evans RM. Identification of a new class of steroid hormone receptors. Nature. 1988;331:91–94. doi: 10.1038/331091a0. [DOI] [PubMed] [Google Scholar]
- 20.Puigserver P, et al. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 21.Dufour CR, et al. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRalpha and gamma. Cell Metab. 2007;5:345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 23.St-Pierre J, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 24.Lai L, et al. Transcriptional coactivators PGC-1alpha and PGC-lbeta control overlapping programs required for perinatal maturation of the heart. Genes Dev. 2008;22:1948–1961. doi: 10.1101/gad.1661708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meier H, Hoag WG, McBurney JJ. Chemical characterization of inbred-strain mouse milk. I. Gross composition and amino acid analysis. J Nutr. 1965;85:305–308. doi: 10.1093/jn/85.3.305. [DOI] [PubMed] [Google Scholar]
- 26.Jornayvaz FR, et al. A high-fat, ketogenic diet causes hepatic insulin resistance in mice, despite increasing energy expenditure and preventing weight gain. Am J Physiol Endocrinol Metab. 2010;299:E808–E815. doi: 10.1152/ajpendo.00361.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarrett SG, Milder JB, Liang LP, Patel M. The ketogenic diet increases mitochondrial glutathione levels. J Neurochem. 2008;106:1044–1051. doi: 10.1111/j.1471-4159.2008.05460.x. [DOI] [PubMed] [Google Scholar]
- 28.Wallberg AE, Yamamura S, Malik S, Spiegelman BM, Roeder RG. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Mol Cell. 2003;12:1137–1149. doi: 10.1016/s1097-2765(03)00391-5. [DOI] [PubMed] [Google Scholar]
- 29.Jia Y, et al. Transcription coactivator PBP, the peroxisome proliferator-activated receptor (PPAR)-binding protein, is required for PPARalpha-regulated gene expression in liver. J Biol Chem. 2004;279:24427–24434. doi: 10.1074/jbc.M402391200. [DOI] [PubMed] [Google Scholar]
- 30.Chen W, Zhang X, Birsoy K, Roeder RG. A muscle-specific knockout implicates nuclear receptor coactivator MED1 in the regulation of glucose and energy metabolism. Proc Natl Acad Sci USA. 2010;107:10196–10201. doi: 10.1073/pnas.1005626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dávila-Román VG, et al. Altered myocardial fatty acid and glucose metabolism in idiopathic dilated cardiomyopathy. J Am Coll Cardiol. 2002;40:271–277. doi: 10.1016/s0735-1097(02)01967-8. [DOI] [PubMed] [Google Scholar]
- 32.Sebastiani M, et al. Induction of mitochondrial biogenesis is a maladaptive mechanism in mitochondrial cardiomyopathies. J Am Coll Cardiol. 2007;50:1362–1369. doi: 10.1016/j.jacc.2007.06.035. [DOI] [PubMed] [Google Scholar]
- 33.Schilling J, Kelly DP. The PGC-1 cascade as a therapeutic target for heart failure. J Mol Cell Cardiol. 2010;51:578–583. doi: 10.1016/j.yjmcc.2010.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahuja P, et al. Myc controls transcriptional regulation of cardiac metabolism and mitochondrial biogenesis in response to pathological stress in mice. J Clin Invest. 2010;120:1494–1505. doi: 10.1172/JCI38331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lazarou M, Thorburn DR, Ryan MT, McKenzie M. Assembly of mitochondrial complex I and defects in disease. Biochim Biophys Acta. 2009;1793:78–88. doi: 10.1016/j.bbamcr.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 36.Lazarou M, Smith SM, Thorburn DR, Ryan MT, McKenzie M. Assembly of nuclear DNA-encoded subunits into mitochondrial complex IV, and their preferential integration into supercomplex forms in patient mitochondria. FEBS J. 2009;276:6701–6713. doi: 10.1111/j.1742-4658.2009.07384.x. [DOI] [PubMed] [Google Scholar]
- 37.Galmiche L, et al. Exome sequencing identifies MRPL3 mutation in mitochondrial cardiomyopathy. Hum Mutat. 2011;32:1225–1231. doi: 10.1002/humu.21562. [DOI] [PubMed] [Google Scholar]
- 38.Zaragoza MV, Brandon MC, Diegoli M, Arbustini E, Wallace DC. Mitochondrial cardiomyopathies: How to identify candidate pathogenic mutations by mitochondrial DNA sequencing, MITOMASTER and phylogeny. Eur J Hum Genet. 2011;19:200–207. doi: 10.1038/ejhg.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoebe K, et al. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature. 2003;424:743–748. doi: 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
- 40.Trounce IA, Kim YL, Jun AS, Wallace DC. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods Enzymol. 1996;264:484–509. doi: 10.1016/s0076-6879(96)64044-0. [DOI] [PubMed] [Google Scholar]
- 41.Graham BH, et al. A mouse model for mitochondrial myopathy and cardiomyopathy resulting from a deficiency in the heart/muscle isoform of the adenine nucleotide translocator. Nat Genet. 1997;16:226–234. doi: 10.1038/ng0797-226. [DOI] [PubMed] [Google Scholar]
- 42.Brattelid T, et al. Reference gene alternatives to Gapdh in rodent and human heart failure gene expression studies. BMC Mol Biol. 2010;11:22. doi: 10.1186/1471-2199-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.