Abstract
The process of lipid droplet (LD) formation is an evolutionarily conserved process among all eukaryotes and plays an important role in both cellular physiology and disease. Recently, fat storage-inducing transmembrane proteins 1 and 2 (FIT1/FITM1 and FIT2/FITM2) were discovered as an evolutionarily conserved family of proteins involved in fat storage. In mammals, FIT1 is expressed primarily in skeletal muscle and FIT2 is expressed primarily in adipose, raising the possibility that FIT1 and FIT2 have unique functions. These proteins are exclusively localized to the endoplasmic reticulum (ER) and mediate triglyceride-rich LD accumulation when overexpressed in cells, mouse liver, or muscle. Unlike the ER-resident diacylglycerol O-acyltransferase family of triglyceride-synthesizing enzymes, FITs do not synthesize triglyceride, but rather partition triglyceride into LDs. The mechanism by which FIT proteins mediate this process has not been determined. A simple hypothesis was tested that FIT proteins bind to triglyceride to mediate LD formation. Here, it is shown that FIT proteins purified in detergent micelles directly bind triolein with specificity and saturation-binding kinetics. A FIT2 gain-of-function mutant that formed larger LDs, FLL(157–9)AAA, showed increased binding to triolein relative to wild-type FIT2, whereas FIT1 and a FIT2 partial loss-of-function mutant, N80A, had significantly lower triolein binding and produced smaller LDs. In summary, FIT proteins are transmembrane domain-containing proteins shown to bind triglyceride. These findings indicate that FITs have a unique biochemical mechanism in mediating LD formation and implicates triglyceride binding as important for FIT-mediated LD formation.
Keywords: lipid droplets, diabetes, obesity, adipocytes
Lipid droplets are cytosolic structures found in cells of all eukaryotes and are composed of a monolayer of phospholipids surrounding a core of uncharged lipids such as triglyceride (TAG) and sterol esters. These structures are considered organelles largely based on the findings that they contain a unique proteome and are dynamic in nature (1). Lipid droplet (LD) formation involves the partitioning of neutral lipids from their site of synthesis at the endoplasmic reticulum (ER) to the cytosol and is considered to be a rapid process (2). The proteins that mediate the partitioning of triglyceride into LDs have not been previously identified; thus, the mechanism of LD formation is not known. Several forward genetic screens were recently conducted in model organisms and cells to identify proteins important in LD biology. These screens have surprisingly revealed that more than 1% of the genes in eukaryotic genomes are involved in LD biology (3–5), underscoring the importance of the LD in normal cellular physiology. Several members of the perilipin family, namely plin2, plin3, and plin4, have been implicated as having roles in LD formation (6–8). However, most evidence supports a role of the plin family in the regulated lipolysis of LDs (9). Interestingly, the ER-resident membrane protein seipin, which is responsible for the most severe known form of generalized lipodystrophy, has been suggested to play a role in either LD formation or regulation of droplet morphology based on loss-of-function studies (5, 10). By unknown mechanisms, it has also been suggested that seipin has tissue-specific effects in regulating lipogenesis and also plays a role in adipogenesis (11–13).
We recently identified an evolutionarily conserved family of ER-resident transmembrane proteins that are important in triglyceride LD formation and named them fat storage-inducing transmembrane protein 1 and 2 (FITM1/FIT1 and FITM2/FIT2) (14). FIT1 and FIT2 are 292- and 262-aa long, respectively, and have six transmembrane domains with both N and C termini facing the cytosol (14, 15). FIT1 and FIT2 share a similarity of 50% at the amino acid level, but do not share homology to known protein domains or other protein families. FIT2 is the ancient orthologue of the FIT family and has orthologues found in Saccharomyces cerevisiae. Orthologues of FIT1 are found as early as bony fish. In mammals, FIT1 is primarily expressed in skeletal muscle, whereas FIT2 is ubiquitously expressed at low levels in mammalian tissues, having the highest expression in adipose tissue (14). These findings suggest that FIT1 and FIT2 have distinct physiological roles in mammals. Expression of FIT1 or FIT2 in cell culture resulted in triglyceride droplet formation. FIT proteins exhibit a unique function in LD biology in that they mediate the partitioning of de novo synthesized triglyceride into the LD without mediating triglyceride biosynthesis (14). In line with these findings, a recent report (16) demonstrated that mouse embryonic fibroblasts deficient in both diacylglycerol O-acyltransferases 1 and 2 (DGAT1 and DGAT2)—enzymes that catalyze the rate-limiting step in the formation of triglyceride—are incapable of forming triglyceride LDs, providing strong genetic evidence that no other mammalian proteins can synthesize triglyceride. Importantly, FIT2 deficiency in adipocytes resulted in reduced size and number of LDs per cell, indicating an important role of FIT2 in LD formation (14). Given these combined findings we hypothesize and demonstrate here that FIT proteins bind directly and specifically to triglyceride and that triglyceride binding is important for FIT-mediated LD formation.
Results
Solubilization and Purification of FIT2.
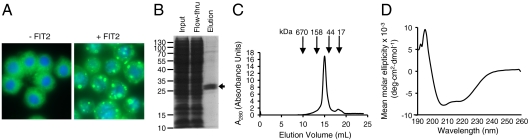
To purify FIT2, we used a baculovirus expression system in Hi5 insect cells to express the protein linked to C-terminal tandem His6 and StrepII (Trp-Ser-His-Pro-Gln-Phe-Glu-Lys) tags. The resulting recombinant protein is designated FIT2-His6-StrepII. To prove that FIT2-His6-StrepII was functional we examined insect cells expressing FIT2-His6-StrepII for the presence of LDs. Insect cells transduced with recombinant FIT2 baculovirus showed massive accumulation of LDs compared to cells transduced with control baculovirus (Fig. 1A). This effect of FIT2 on LD accumulation was consistent with previous work (14, 15). Based on the use of Fos-choline 13 (a zwitterionic phospholipid-like detergent) to purify the sterol-sensing domain of sterol response element-binding protein cleavage activating protein (SCAP) and the ER-resident membrane-spanning protein Insig-2 (17, 18), we purified FIT2-His6-StrepII using this detergent. Similar to SCAP and Insig-2, FIT2-His6-StrepII was only stable in the Fos-choline series of detergents (see Materials and Methods). Following solubilization in Fos-choline 13 detergent, the recombinant protein was purified to homogeneity using a tetrameric Strep-tactin column (Fig. 1B, Coomassie blue-stained gel). FIT2-His6-StrepII ran as an apparent doublet when expressed in Hi5 cells. The upper band of this doublet was consistent with the form of the protein that retains its signal sequence, a common finding with overexpression of membrane proteins in insect cells in which the signal peptide peptidase is saturated with recombinant substrate (19).
Fig. 1.
Purification and characterization of FIT2-His6-StrepII. (A) Immunofluorescence microscopy of Sf9 cells stained for LDs (BODIPY 493/503, green) and nuclei (Hoechst 33342, blue) 36 h posttransduction with pIEX-Bac1-FIT2-His6-StrepII baculovirus revealed that FIT2 overexpression in insect cells resulted in the accumulation of LDs. Images are representative of three independent experiments. (B) Recombinant FIT2-His6-StrepII protein was purified as described in Materials and Methods, subjected to 15% SDS-PAGE, and visualized by Coomassie blue staining. Input designates total cellular lysate; flow-through designates protein not bound to Strep-tactin resin; elution designates protein bound specifically to column. Arrow indicates purified FIT2-His6-StrepII. (C) Gel filtration chromatography of purified FIT2-His6-StrepII. Molecular mass is indicated with arrows. The apparent molecular mass of FIT2-His6-StrepII is 60 kDa. FPLC traces are representative of three independent experiments. (D) CD spectroscopy of FIT2-His6-StrepII. The average of six spectra is shown.
Gel filtration of FIT2-His6-StrepII showed that the recombinant protein is monodisperse and eluted as a ≈60 kDa species in this detergent. Because the calculated molecular mass of FIT2-His6-StrepII monomer is 33 kDa and that of a Fos-choline 13 micelle is ≈25 kDa, the estimated molecular weight was consistent with FIT2-His6-StrepII as a monomeric protein (Fig. 1C). Far UV CD of FIT2-His6-StrepII in Fos-choline 13 micelles indicated a highly helical protein with characteristic minima at approximately 206 and 222 nm (Fig. 1D). This result supports previous findings showing that the majority of FIT2 is comprised of transmembrane domains (15) and suggests that the transmembrane domains are α-helical in nature.
FIT2 Specifically Binds Triglyceride.
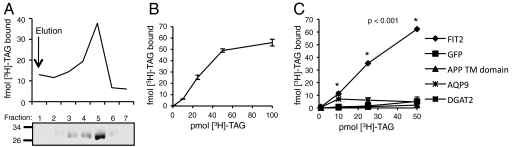
Based on assays developed for SCAP and Insig-2 (17, 18), we probed the direct binding of [9,10-3H(N)]-triolein ([3H]-TAG) to FIT2-His6-StrepII bound to Strep-tactin resin. FIT2-His6-StrepII was incubated with [3H]-TAG followed by elution of FIT2-His6-StrepII from Strep-tactin beads. Upon elution of FIT2-His6-StrepII from the Strep-tactin column, it was observed that both FIT2-His6-StrepII and [3H]-TAG coeluted, indicating that recombinant FIT2 bound [3H]-TAG in this assay (Fig. 2A). Equilibrium binding was achieved after approximately 3 h (Fig. S1). As an indication of specific binding to triglyceride, we performed a dose-response assay with increasing concentrations of [3H]-TAG at a constant FIT2-His6-StrepII concentration. FIT2-His6-StrepII bound [3H]-TAG in Fos-choline 13 micelles with saturation binding kinetics, reaching half-maximal binding at ≈100–250 nM as judged from multiple experiments (Fig. 2B). Maximal binding of [3H]-TAG was substoichiometric. At saturation, approximately one molecule of triglyceride was bound to approximately 150–160 molecules of FIT2-His6-StrepII. This substoichiometric binding was likely caused by the dilution effects of detergent micelles, as previously observed for the substoichiometric binding of cholesterol to SCAP (17).
Fig. 2.
Binding of [9,10-3H(N)]-triolein to FIT2-His6-StrepII is saturable and specific. (A) Binding and elution assay demonstrates that FIT2-His6-StrepII and [9,10-3H(N)]-triolein ([3H]-TAG) coelute from an affinity column (see Materials and Methods). (Top) Shows 3H-radioactivity eluted from affinity column. (Bottom) Anti-His6 immunoblot for FIT2-His6-StrepII. This experiment was reproduced five times with similar results. (B) Dose-response curve of FIT2-His6-StrepII binding to [9,10-3H(N)]-triolein ([3H]-TAG) shows saturable binding kinetics. Data are represented as mean ± SD. (C) Saturable binding of [3H]-triolein ([3H]-TAG) to FIT2-His6-StrepII is specific. Dose-response curves as in B were generated for GFP-His6-StrepII (GFP), APP TM Domain-His6-StrepII (APP TM Domain), aquaporin 9-His6 (AQP9), and DGAT2-His6-StrepII (DGAT2) alongside FIT2-His6-StrepII. Data are represented as mean ± SD. This experiment was replicated three times with similar results.
As a further test of specificity, we examined the ability of other recombinant proteins to bind [3H]-TAG. Purified recombinant GFP-His6-StrepII and amyloid-β precursor protein transmembrane domain-His6-StrepII failed to bind to [3H]-TAG (Fig. 2C, Fig. S2). Purified aquaporin 9-His6, like FIT2 has six transmembrane domains and is of similar molecular weight, also failed to bind to [3H]-TAG (Fig. 2C, Fig. S2). As a further indication of specificity the purified ER-resident membrane protein, DGAT2-His6-StrepII, failed to bind to [3H]-TAG (Fig. 2C, Fig. S2). Taken together, these experiments rule out the possibility that [3H]-TAG was binding to the affinity tags used to purify FIT2 and indicate that FIT2 binds to TAG with specificity.
Recombinant FIT2 Binds a Specific Set of Uncharged Glycerolipids.
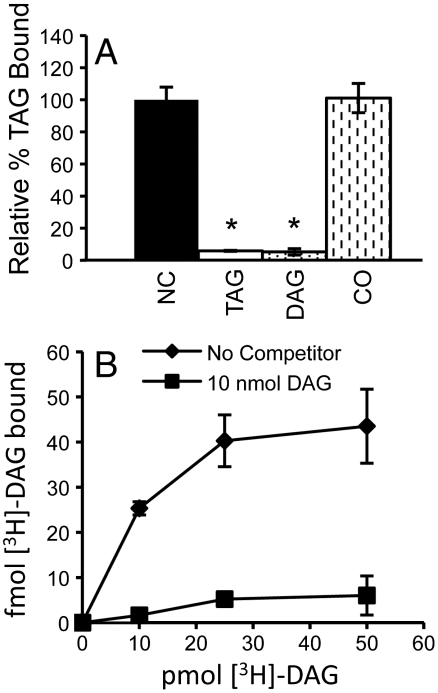
Because LDs contain other noncharged lipids in addition to TAG, namely diacylglycerol (DAG) and steryl-esters, we tested the possibility that FIT2 binds to these lipids. A competitive binding assay was developed to probe lipid specificity by mixing [3H]-TAG with FIT2-His6-StrepII with or without saturating amounts of unlabeled competitor lipid. A concentration of [3H]-TAG that resulted in half-maximal binding at a fixed concentration of FIT2-His6-StrepII was chosen for these experiments. Competition experiments showed that excess unlabeled triolein and 1-palmitoyl-2-oleoyl-sn-glycerol [diacylglycerol (DAG)] competed for the binding of [3H]-TAG to FIT2-His6-StrepII (Fig. 3A). To further probe the validity of this competition assay, direct binding of FIT2-His6-StrepII to [1,2,3-3H]-dioleoyl-rac-glycerol ([3H]-DAG) was also measured. FIT2 bound [3H]-DAG with saturation binding kinetics that could be competed with excess unlabeled DAG (Fig. 3B). Similar to [3H]-TAG binding by FIT2, half-maximal binding to [3H]-DAG was achieved at ≈100–250 nM (Fig. 3B). These experiments indicated that FIT2 specifically bound [3H]-TAG and [3H]-DAG.
Fig. 3.
FIT2 exhibits lipid binding specificity. (A) Competition binding assay. FIT2-His6-StrepII was incubated with [9,10-3H(N)]-triolein in the absence or presence of the indicated excess unlabeled lipid. Data are represented as mean ± SD. These results are representative of five experiments. (NC, no competitor; TAG, triolein; DAG, 1-palmitoyl-2-oleoyl-sn-glycerol; CO, cholesteryl oleate). (B) Direct binding of FIT2-His6-StrepII to [1,2,3-3H]-dioleoyl-rac-glycerol ([3H]-DAG) and competition by unlabeled 1-palmitoyl-2-oleoyl-sn-glycerol. Data are represented as mean ± SD. This experiment was replicated twice with similar results.
Given that competitive binding existed between these neutral glycerolipids, we tested if another neutral lipid found in LDs of mammalian cells, cholesteryl oleate (CO) was able to bind FIT2. CO did not compete for [3H]-TAG binding to purified FIT2-His6-StrepII (Fig. 3A). These experiments indicate that FIT2 binds specifically to TAG and DAG, and further demonstrates the specificity of FIT2 as a triglyceride-binding protein.
FIT Triglyceride-Binding Affinity Correlates with Lipid Droplet Size.
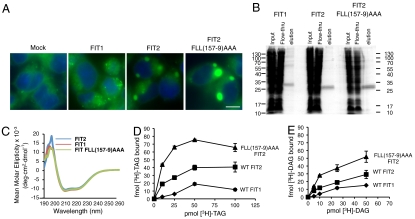
FIT1 is primarily expressed in skeletal muscle, which characteristically produces small LDs, whereas FIT2 is highly expressed in adipose tissue, which is characterized by large LDs. Indeed, expression of FIT2 resulted in larger LDs than those produced by FIT1 when expressed in HEK293 cells (Fig. 4A). Quantification of LD number and size in FIT1- versus FIT2-expressing cells using three-dimensional reconstructions of z-stacked confocal microscopy images indicated that the LDs produced by FIT2 were ≈80% larger than FIT1; however, there was no significant difference in the number of LDs produced by either FIT1 or FIT2 (Fig. S3). Previously, we identified a gain-of-function FIT2 mutant, FLL(157–9)AAA, that formed LDs that were significantly larger than wild-type FIT2 in HEK293 cells (Fig. 4A) (15). In these experiments, FIT1-V5, FIT2-V5, and FIT2-FLL(157–9)AAA-V5 expression levels were similar, and all were localized to the endoplasmic reticulum (Fig. S4, Fig. S5). Importantly, FIT2-FLL(157–9)AAA had an altered conformation relative to wild-type FIT2 as determined by limited proteolysis, suggesting that FIT2 undergoes an activating conformational change (15). We hypothesized that if TAG binding is important for function, then the FIT2-FLL(157–9)AAA gain-of-function mutant will have increased binding to [3H]-TAG. If this hypothesis is true, then it would be expected that triglyceride-binding affinity by FIT1, FIT2, and FIT2-FLL(157–9)AAA should be correlated with droplet size as an indication of FIT2 activity. To test this hypothesis, we made baculoviruses for murine FIT1-His6-StrepII and FIT2-FLL(157–9)AAA-His6-StrepII and purified these recombinant proteins in Hi5 insect cells.
Fig. 4.
Lipid droplet size correlates with FIT TAG- and DAG-binding affinity. (A) HEK293 cells were transfected with empty pcDNA3.1 vector (Mock), or expression vectors for FIT1-V5, FIT2-V5, or FIT2-FLL(157-9)AAA-V5. Cells were stained for lipid droplets (LDs, green) using BODIPY 493/503 and nuclei using Hoechst 33342 (blue). Images are representative of five independent experiments. Scale bar, 10 µm. (B) Coomassie blue-stained gel of FITM1-His6-StrepII, FIT2-His6-StrepII, and FIT2-FLL(157-9)AAA-His6-StrepII. Recombinant proteins were purified as described in Materials and Methods, subjected to 15% SDS-PAGE, and visualized using Coomassie blue. (C) CD spectroscopy of purified FIT proteins. The average of six spectra for each protein is shown. (D) Dose-response curves of FIT1-His6-StrepII, FIT2-His6-StrepII, and FIT2-FLL(157-9)AAA-His6-StrepII display differential [9,10-3H(N)]-triolein ([3H]-TAG) binding affinities and capacities. Data are represented as mean ± SD. This experiment was replicated five times with similar results. (E) Dose-response curves of FIT1-His6-StrepII, FIT2-His6-StrepII, and FIT2-FLL(157-9)AAA-His6-StrepII display differential [1,2,3-3H]-dioleoyl-rac-glycerol ([3H]-DAG) binding affinities and capacities. Data are represented as mean ± SD. This experiment was replicated three times with similar results.
Murine FIT1-His6-StrepII purified from insect cells migrated as an approximately 30 kDa doublet on reducing SDS gels, and FIT2-FLL(157–9)AAA-His6-StrepII migrated as an approximately 26 kDa doublet similar to FIT2-His6-StrepII (Fig. 1B, Fig. 4B). Gel filtration showed that FIT1-His6-StrepII and FIT2-FLL(157–9)AAA-His6-StrepII were monodisperse when solubilized in Fos-choline 13 (Fig. S6). Furthermore, far UV CD demonstrated that FIT1-His6-StrepII and FIT2-FLL(157–9)AAA-His6-StrepII both have similar secondary structures to FIT2-His6-StrepII (Fig. 4C) and are also primarily α-helical in nature.
To test if LD sizes produced by FIT1, FIT2, and FIT2-FLL(157–9)AAA correlated with triglyceride binding, FIT1-His6-StrepII, FIT2-His6-StrepII, and FIT2-FLL(157–9)AAA-His6-StrepII were purified side-by-side and subjected to dose-response binding assays identical to those in Fig. 2. FIT1-His6-StrepII, FIT2-His6-StrepII, and FIT2-FLL(157–9)AAA-His6-StrepII all showed saturation binding kinetics; however, FIT2-FLL(157–9)AAA-His6-StrepII showed a significant increase in capacity for triglyceride binding compared to FIT2-His6-StrepII (Fig. 4D). In addition, half-maximal binding by FIT2-FLL(157–9)AAA-His6-StrepII was reached at lower triolein concentration than FIT2-His6-StrepII, implying a higher affinity for triglyceride than wild-type FIT2. Half-maximal triolein binding by FIT1-His6-StrepII was reached at higher triolein concentration than FIT2-His6-StrepII (see Fig. 4D), suggestive of a lower binding affinity of FIT1-His6-StrepII for triglyceride than FIT2-His6-StrepII. Similar differences in binding characteristics were observed for direct binding to [3H]-DAG by FIT1-His6-StrepII, FIT2-His6-StrepII, and FIT2-FLL(157–9)AAA-His6-StrepII as shown for binding to [3H]-TAG (Fig. 4E). These data once again highlight the specificity of FIT proteins for binding TAG and DAG and indicate that TAG- and DAG-binding capacity and/or affinity are lower for FIT1 and higher for FIT2-FLL(157–9)AAA relative to wild-type FIT2, correlating LD size with TAG- and DAG-binding capacity and/or affinity. These data indicate that TAG-binding, and possibly DAG-binding, by FIT proteins is important and functionally relevant for the biochemical mechanism by which they generate triglyceride-rich LDs.
Identification of a FIT2 Partial Loss-of-Function Mutant.
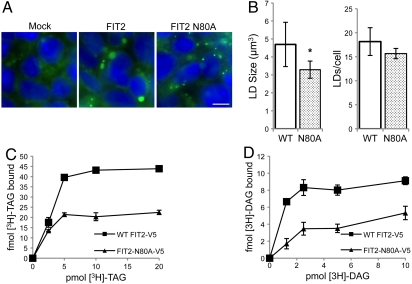
To further support a role of triglyceride binding in FIT2 function, we sought to identify a loss-of-function mutant of FIT2 and determine its ability to bind to triglyceride. FIT2 tagged at the C terminus with a V5-His6 tag (FIT2-V5-His6) was subjected to alanine scanning mutagenesis. Mutagenesis efforts were focused on transmembrane domains, as these domains contain the most highly conserved residues. HEK293 cells were used as a model system to identify FIT2 mutants that were defective in LD formation upon overexpression. Using this approach, we identified FIT2-N80A-V5 as a mutant that was defective in LD formation (Fig. 5A). In these experiments, FIT2-V5, and FIT2-N80A-V5 expression levels were similar, and both were localized to the endoplasmic reticulum (Fig. S4, Fig. S5). The N80A mutant of FIT2 displayed a ≈30% reduction in LD size and only a small, but insignificant reduction in LD number per cell compared to wild-type FIT2-V5, indicating partial loss-of-function (Fig. 5B). We next tested the hypothesis that the N80A mutant of FIT2 had reduced binding to triglyceride. We developed a simple and rapid cell culture-based approach to screen for [3H]-TAG binding (see Materials and Methods for details). This method screens for triglyceride binding defects by expressing and purifying a mutant FIT2 from HEK293 cells via immunoprecipitation in Fos-choline 13 buffer. Using this approach, binding assays were performed on immunoprecipitated FIT2 in buffer containing Fos-choline 13 detergent. Dose-response binding assays with the N80A mutant of FIT2 displayed reduced triglyceride-binding capacity relative to wild-type FIT2-V5 (Fig. 5C). The maximum specific binding of [3H]-TAG to FIT2-N80A-V5 was reduced by ≈55% compared to wild-type FIT2-V5 (Fig. 5C). Similarly, in dose-response binding assays using [3H]-DAG, maximum specific binding of [3H]-DAG to FIT2-N80A-V5 was reduced by ≈58% compared to wild-type FIT2-V5. (Fig. 5D). These data further support the conclusion that triglyceride and diacylglycerol binding to FITs is specific and important for FIT-mediated triglyceride LD formation.
Fig. 5.
FIT2 partial loss-of-function mutant has decreased TAG- and DAG-binding affinity. (A) HEK293 cells were transfected with empty pcDNA3.1 vector (Mock), or expression vectors for wild-type FIT2-V5 or FIT2-N80A-V5. Cells were stained for LDs using BODIPY 493/503 (green) and nuclei using Hoechst 33342 (blue) and imaged by fluorescence microscopy. Images are representative of five independent experiments. Scale bar, 10 µm. (B) HEK293 cells transfected with FIT2-V5 or FIT2-N80A-V5 were stained as in A and subjected to confocal fluorescence microscopy. Z-stacks were captured and three-dimensional renderings were constructed and analyzed using Volocity software (Perkin Elmer). Analysis was performed on 10 independent three-dimensional renderings from two independent experiments. Each rendering contained approximately 200–300 cells. (Left) Quantification of LD size is shown (wild-type versus N80A*, p < 5.5 × 10-3). (Right) Quantification of LD number per cell is shown (wild-type versus N80A). (C) Dose-response curves generated using cell culture-based assay display reduced [9,10-3H(N)]-triolein ([3H]-TAG) binding capacity for FIT2-N80A-V5 compared to wild-type FIT2-V5. Data are represented as mean ± SD. This experiment was replicated three times with similar results. (D) Dose-response curves display reduced [1,2,3-3H]-dioleoyl-rac-glycerol ([3H]-DAG) binding capacity for FIT2-N80A-V5 compared to wild-type FIT2-V5. Data are represented as mean ± SD. This experiment was replicated four times with similar results.
Discussion
The experiments in the present study indicate that FIT2 is a triglyceride-binding protein with specificity for neutral glycerolipids. This conclusion is based on a series of in vitro biochemical studies showing direct, and specific binding of triglyceride and diacylglycerol to purified FIT2. FIT2 is a unique triglyceride-binding membrane protein. This finding is consistent with the function of FIT2 in LD biogenesis.
To study FIT2 binding to triglyceride, we modeled our experimental design on the work of Radhakrishnan et al. (17, 18), in which assays were established to measure direct binding of the sterol-sensing domain of SCAP to cholesterol and Insig-2 to oxysterols. The problems we encountered in measuring direct binding of FIT2 to triglyceride were similar to the obstacles to measuring direct binding of SCAP(TM1-8) and Insig-2 to their respective, highly water-insoluble ligands—cholesterol and oxysterols. TAG has very low solubility in aqueous phase, and has a maximal estimated bilayer concentration of 2.8 mol % (20). None of the commonly used detergents yielded monodisperse FIT2. Furthermore, only the Fos-choline series of detergents was capable of solubilizing FITM2 and retaining its TAG-binding characteristics, indicating that FIT2 is only stable in detergents that are similar in structure to biological lipids. Because of these limitations, we needed to solubilize the TAG used in our direct binding experiments in buffer containing micelles of Fos-choline 13. An obvious limitation of studying ligand interactions using the in vitro system presented here is the observed substoichiometric binding of triolein to FIT2, making quantification of exact kinetic constants of the binding reaction complex.
The conclusion that FIT2 binds specifically to TAG is consistent with previous studies that demonstrated that expression of FIT1, FIT2, and FIT2-FLL(157–9)AAA results in increased cellular TAG content without significantly altering other lipid species such as DAG, cholesteryl ester, or phospholipids (14, 15). Given these findings, the biological significance of FIT2 binding to DAG is unclear. However, several recent studies in which DAG levels were increased in mammalian cells (21) and S. cerevisiae (22) suggests that DAG may have a role in LD formation.
A notable finding from the present study is that the FIT2 homologue, FIT1, binds TAG and DAG with reduced affinity and/or capacity. Oppositely, a gain-of-function mutant of FIT2, FIT2-FLL(157–9)AAA, binds TAG and DAG with greater capacity and/or affinity. Importantly, the average sizes of LDs formed by these three proteins are distinctly different. FIT2 expression resulted in large LDs that are ≈80% larger than FIT1 LDs, which are similar in size to LDs produced by DGAT2 overexpression (15). Overexpression of FIT1 or FIT2 did not result in a significant difference in the number of LDs per cell. On average, FIT2-FLL(157–9)AAA expression resulted in fivefold larger LDs than FIT2 (15). In addition, we identified a FIT2 partial loss-of-function mutant, N80A, which produced LDs that were on average 30% smaller than LDs produced by wild-type FIT2. Importantly, maximum specific binding of TAG and DAG to N80A was reduced by ≈55% and ≈58%, respectively, compared to wild-type FIT2. In the absence of structural information of FIT2, it is not known if N80 coordinates TAG or TAG binding. Taken together, these data indicate that LD size is correlated to TAG and DAG-binding capacity and/or affinity, and supports that idea that TAG and/or DAG binding is important to the biochemical mechanism by which FIT proteins form LDs in cells.
In addition to triglyceride binding by FIT proteins, we speculate that FIT proteins undergo a conformational change to form LDs in cells. This notion is supported by our previous findings that the gain-of-function mutant FIT2-FLL(157–9)AAA has an altered conformation relative to wild-type FIT2 (15). The present study suggests that FIT2-FLL(157–9)AAA may have a lower Kd and higher capacity for binding TAG and DAG compared to FIT2. Collectively, these data suggest that FIT2-FLL(157–9)AAA may have a more favorable conformation for making LDs, and may imply that the reaction mechanism of FIT-mediated droplet formation might first require TAG/DAG binding that induces an activating conformational change. We attempted to detect an activating conformational change in purified FIT2-His6-StrepII in the presence of TAG and DAG; however, limited proteolysis, CD spectroscopy, and thermal denaturation-CD spectroscopy failed to detect any differences in conformation or secondary structure of FIT2 in the presence of these lipids. The inability to detect conformational changes using these methods is most likely due to the substoichiometric binding of TAG/DAG to FIT2, or to the possibility that TAG/DAG binding to FIT2 has a rapid on-off rate. Given the TAG- and DAG-binding function of FIT proteins, it is possible that the multispanning FIT proteins act via a TAG-transport mechanism by translocating TAG from the ER membrane into a nascent LD. Testing of these ideas awaits the development of in vitro reconstitution assays.
In addition to their differing TAG- and DAG-binding features, an important distinction between FIT1 and FIT2 is their respective tissue expression pattern. FIT2 is ubiquitously expressed at low levels in mouse and human tissues, but transcripts and protein levels are extremely high in adipose depots (14). The high level of FIT2 in adipose is likely explained by its direct regulation by the nuclear hormone receptor peroxisome proliferator-activated receptor γ (23, 24). FIT1 is almost exclusively expressed in skeletal muscle, with lower levels found in heart (14). This distinct tissue distribution of FIT1 and FIT2 and the finding that FIT2 strongly binds TAG, whereas FIT1 binds weakly, suggest distinct physiological roles for each FIT protein in lipid metabolism. What might their distinct functions be? FIT2-driven LD formation may have a role in long-term TAG storage in adipose, whereas FIT1 forms small LDs that are characteristic of the rapidly turning over LDs found in skeletal muscle (25–28), suggesting that FIT1-directed LD formation may play a role in linking myocellular TAG energy reservoirs to mitochondrial respiration. The dynamics of LD turnover for the release of fatty acids would be expected to play an important role in the maintenance of energy homeostasis in tissues such as skeletal muscle that rely heavily on fatty acid oxidation for ATP production.
In conclusion, we have demonstrated that FIT proteins directly bind triglyceride and diacylglyceride in detergent micelles, and that lipid binding capacity correlates with LD size. The precise molecular mechanism by which this evolutionarily conserved family of fat-storing proteins functions to form triglyceride LDs is the goal of future studies.
Materials and Methods
Reagents and Buffers.
Reagents are shown in SI Materials and Methods. Buffer A, 50 mM Tris·HCl pH 7.4 and 150 mM NaCl. Buffer C, buffer A with 0.1% (wt/vol) Fos-choline 13 (unless otherwise indicated). Buffer F, buffer C with 10% (wt/vol) glycerol. All other buffer compositions are detailed in SI Material and Methods.
Solubilization of Lipids.
Lipids were dried down and resuspended in ethanol to a final concentration of 50 nM to 100 μM prior to initiating binding assays. See SI Materials and Methods for details.
Purification of Recombinant FIT1, FIT2, and FIT2 FLL(157–9)AAA.
See SI Materials and Methods for details of baculovirus construction. Detergent extracts of Hi5 insect cells expressing recombinant proteins were passed over a column containing Strep-tactin resin, followed by 200–300 column bed volume of buffer to wash. Protein was eluted from the column and concentrated to 1 mg/mL before use. See SI Materials and Methods for details of this procedure.
Gel Filtration Chromatography of FIT1, FIT2, and FIT2 FLL(157–9)AAA.
Recombinant FIT proteins were chromatographed on a Superdex 200 HR 10/30 column (Amersham) at a flow rate of 0.5 mL/ min in a buffer F. Absorbance was monitored at 280 nm to detect elution of proteins.
CD Spectroscopy.
Spectra of FIT proteins in buffer C were collected on a Jasco J-815 CD Spectrophotometer using a 2-mm path length cuvette at room temperature. See SI Materials and Methods for details.
Immunoblot Analysis.
See SI Materials and Methods for details.
Coelution Assay.
Recombinant FIT2 protein and radioactive triolein were coincubated in binding reactions, followed by elution with desthiobiotin. Fractions were collected and analyzed for radioactivity and FIT2 protein. See SI Materials and Methods for details.
Radiolabeled-Lipid Binding Assays with Purified StrepII-Tagged Proteins.
Purified protein was mixed with Ni-nitrilotriacetate beads or Strep-tactin slurry (GE Healthcare or IBA, respectively) in buffer C, and 3H-lipids. After approximately 4 h incubation, beads were extensively washed in buffer C and assayed for retained radioactivity. See SI Materials and Methods for details.
Competition Binding Assays.
Competition experiments in Fig. 3 were conducted as standard binding assays, but contained excess unlabeled (cold) lipid competitor. See SI Materials and Methods for details.
Cell Culture-Based Binding Assays.
HEK293 cells were transfected with V5-tagged FIT2 constructs. Cells were extracted in Fos-choline 13 at 4 °C, immunoprecipitated, and standard binding assays were performed. See SI Materials and Methods for details.
Cell Culture, Microscopy, and Quantification of LD Size and Number.
HEK293 cells transfected with FIT constructs were imaged using confocal microscopy and LD size and number was quantified using Volocity software (Perkin Elmer). Calnexin was used as an ER marker to determine ER localization of FIT proteins in cells. See SI Materials and Methods for details.
Supplementary Material
Acknowledgments.
We thank Albe Man Kid Chan for invaluable assistance in baculovirus generation and insect cell culture. This work was supported in part by National Institutes of Health (NIH) Grant R01HL082697 (to D.L.S.) and NIH Training Program in Cellular and Molecular Biology and Genetics Grant T32 GM007491 (to D.A.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.Z. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1110817108/-/DCSupplemental.
References
- 1.Farese RV, Jr., Walther TC. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell. 2009;139:855–860. doi: 10.1016/j.cell.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolins NE, Brasaemle DL, Bickel PE. A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett. 2006;580:5484–5491. doi: 10.1016/j.febslet.2006.08.040. [DOI] [PubMed] [Google Scholar]
- 3.Guo Y, et al. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 2008;453:657–661. doi: 10.1038/nature06928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashrafi K, et al. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- 5.Szymanski KM, et al. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc Natl Acad Sci USA. 2007;104:20890–20895. doi: 10.1073/pnas.0704154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolins NE, et al. S3-12, Adipophilin, and TIP47 package lipid in adipocytes. J Biol Chem. 2005;280:19146–19155. doi: 10.1074/jbc.M500978200. [DOI] [PubMed] [Google Scholar]
- 7.Wolins NE, et al. Adipocyte protein S3-12 coats nascent lipid droplets. J Biol Chem. 2003;278:37713–37721. doi: 10.1074/jbc.M304025200. [DOI] [PubMed] [Google Scholar]
- 8.Bulankina AV, et al. TIP47 functions in the biogenesis of lipid droplets. J Cell Biol. 2009;185:641–655. doi: 10.1083/jcb.200812042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brasaemle DL. Thematic review series: Adipocyte biology. The perilipin family of structural lipid droplet proteins: Stabilization of lipid droplets and control of lipolysis. J Lipid Res. 2007;48:2547–2559. doi: 10.1194/jlr.R700014-JLR200. [DOI] [PubMed] [Google Scholar]
- 10.Fei W, et al. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J Cell Biol. 2008;180:473–482. doi: 10.1083/jcb.200711136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W, et al. The human lipodystrophy gene product Berardinelli-Seip congenital lipodystrophy 2/seipin plays a key role in adipocyte differentiation. Endocrinology. 2009;150:4552–4561. doi: 10.1210/en.2009-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Payne VA, et al. The human lipodystrophy gene BSCL2/seipin may be essential for normal adipocyte differentiation. Diabetes. 2008;57:2055–2060. doi: 10.2337/db08-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian Y, et al. Tissue-autonomous function of Drosophila seipin in preventing ectopic lipid droplet formation. PLoS Genet. 2011;7:e1001364. doi: 10.1371/journal.pgen.1001364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kadereit B, et al. Evolutionarily conserved gene family important for fat storage. Proc Natl Acad Sci USA. 2008;105:94–99. doi: 10.1073/pnas.0708579105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gross DA, Snapp EL, Silver DL. Structural insights into triglyceride storage mediated by fat storage-inducing transmembrane (FIT) protein 2. PLoS One. 2010;5:e10796. doi: 10.1371/journal.pone.0010796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris CA, et al. DGAT enzymes are required for triacylglycerol synthesis and lipid droplets in adipocytes. J Lipid Res. 2011;52:657–667. doi: 10.1194/jlr.M013003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Radhakrishnan A, Sun LP, Kwon HJ, Brown MS, Goldstein JL. Direct binding of cholesterol to the purified membrane region of SCAP: Mechanism for a sterol-sensing domain. Mol Cell. 2004;15:259–268. doi: 10.1016/j.molcel.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 18.Radhakrishnan A, Goldstein JL, McDonald JG, Brown MS. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: A delicate balance. Cell Metab. 2008;8:512–521. doi: 10.1016/j.cmet.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Motamed M, et al. Identification of luminal loop 1 of scap protein as the sterol sensor that maintains cholesterol homeostasis. J Biol Chem. 2011;286:18002–18012. doi: 10.1074/jbc.M111.238311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spooner PJ, Small DM. Effect of free cholesterol on incorporation of triolein in phospholipid bilayers. Biochemistry. 1987;26:5820–5825. doi: 10.1021/bi00392a036. [DOI] [PubMed] [Google Scholar]
- 21.Skinner JR, et al. Diacylglycerol enrichment of endoplasmic reticulum or lipid droplets recruits perilipin 3/TIP47 during lipid storage and mobilization. J Biol Chem. 2009;284:30941–30948. doi: 10.1074/jbc.M109.013995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adeyo O, et al. The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J Cell Biol. 2011;192:1043–1055. doi: 10.1083/jcb.201010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lefterova MI, et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22:2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Villanueva CJ, et al. TLE3 is a dual-function transcriptional coregulator of adipogenesis. Cell Metab. 2011;13:413–427. doi: 10.1016/j.cmet.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiens B. Skeletal muscle lipid metabolism in exercise and insulin resistance. Physiol Rev. 2006;86:205–243. doi: 10.1152/physrev.00023.2004. [DOI] [PubMed] [Google Scholar]
- 26.van Loon LJ. Use of intramuscular triacylglycerol as a substrate source during exercise in humans. J Appl Physiol. 2004;97:1170–1187. doi: 10.1152/japplphysiol.00368.2004. [DOI] [PubMed] [Google Scholar]
- 27.Watt MJ, Heigenhauser GJ, Dyck DJ, Spriet LL. Intramuscular triacylglycerol, glycogen and acetyl group metabolism during 4 h of moderate exercise in man. J Physiol. 2002;541:969–978. doi: 10.1113/jphysiol.2002.018820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thyfault JP, et al. Metabolic profiling of muscle contraction in lean compared with obese rodents. Am J Physiol Regul Integr Comp Physiol. 2010;299:R926–934. doi: 10.1152/ajpregu.00093.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.