Abstract
The ability to control the placement of individual protein molecules on surfaces could enable advances in a wide range of areas, from the development of nanoscale biomolecular devices to fundamental studies in cell biology. Such control, however, remains a challenge in nanobiotechnology due to the limitations of current lithographic techniques. Herein we report an approach that combines scanning probe block copolymer lithography with site-selective immobilization strategies to create arrays of proteins down to the single-molecule level with arbitrary pattern control. Scanning probe block copolymer lithography was used to synthesize individual sub-10-nm single crystal gold nanoparticles that can act as scaffolds for the adsorption of functionalized alkylthiol monolayers, which facilitate the immobilization of specific proteins. The number of protein molecules that adsorb onto the nanoparticles is dependent upon particle size; when the particle size approaches the dimensions of a protein molecule, each particle can support a single protein. This was demonstrated with both gold nanoparticle and quantum dot labeling coupled with transmission electron microscopy imaging experiments. The immobilized proteins remain bioactive, as evidenced by enzymatic assays and antigen-antibody binding experiments. Importantly, this approach to generate single-biomolecule arrays is, in principle, applicable to many parallelized cantilever and cantilever-free scanning probe molecular printing methods.
Keywords: dip-pen nanolithography, scanning probe lithography, single molecule array, gold nanoparticles, protein nanoarray
Protein immobilization on solid substrates with nanoscale control has been utilized in a variety of applications, including chip-based bioassays (1), proteomics (2, 3), drug discovery (3), and cellular biology studies (4). In cellular biology research, the ability to fabricate protein nanostructures on surfaces has enabled the study of many basic cellular functions including growth, signaling, and differentiation (4–7). For combinatorial molecular biology, the miniaturization of protein nanoarrays allows for smaller and higher density chips and the need for smaller sample volumes; in certain cases, this can translate into diagnostic systems with higher sensitivity and the ability to track disease and biological processes more efficiently (2, 3). The ability to site-specifically isolate single biomolecules can also facilitate molecular level studies of such structures (8, 9). Therefore, being able to nanofabricate biomolecular features at a resolution of 10 nm or less is of significant interest because this length scale approaches the dimensions of single protein molecules and offers an opportunity to address many previously unexplored biological phenomena.
The use of dip-pen nanolithography (DPN) (4, 10) for the generation of arrays of biomolecules either by a direct deposition of proteins (11–13) or indirect methods (14, 15), through DPN writing of patterns followed by capture of proteins onto the patterns, has been widely demonstrated. However, while these methods are powerful and applicable to biomolecular patterning, their resolution is essentially limited to that of conventional DPN, which is significantly larger than most soluble protein molecules that are generally smaller than 10 nm in diameter. There are also some examples of using traditional methods such as electron beam lithography to prepare small collections of particles that can support individual protein attachment; the ability to control the placement of biomolecules with this degree of resolution and precision over large areas remains a challenge for current nanolithographic processes (16–18). Indeed, the conventional methods are expensive, inherently low throughput, and difficult to implement on the sub-10-nm scale.
Herein, we demonstrate the use of scanning probe block copolymer lithography (SPBCL) (19) as a technique to control the number of protein molecules immobilized at specific locations on a surface down to the single-molecule level. The relevant surface chemistries employed in conjunction with SPBCL are validated for the purposes of bioconjugation, and the effect of the size of the SPBCL-generated gold nanoparticles (AuNPs) on the number of attached proteins is described. The biological activity of proteins immobilized on deposited AuNPs is also shown. In combination, these “soft” methods enable the high throughput generation of functional protein arrays over large areas, down to a single biomolecule level, with arbitrary pattern control under essentially ambient conditions.
SPBCL relies on scanning probe-based methods such as meniscus-based dip-pen nanolithography (DPN) (20, 21) or polymer pen lithography (PPL) (22) to pattern attoliter volumes of block copolymers complexed with metal ions in a parallelized manner over large areas. The block copolymer acts as a synthetic “nanoreactor” that confines the atoms involved in the synthesis of nanoparticles on a substrate. Once the block copolymer–metal ion ink has been patterned, plasma treatment and subsequent thermal annealing are used to reduce the metal ions and remove the polymer, ultimately yielding a single nanoparticle per patterned feature. The feature size, and consequently nanoparticle dimensions, can be controlled by adjusting the dwell time of the scanning probe. In the case of PPL, the contact force applied to the polymer pens also offers control over feature and nanoparticle size. Because SPBCL is able to generate nanoparticles less than 10 nm in diameter, it allows one to prepare particles, which in principle can isolate individual proteins.
Results and Discussion
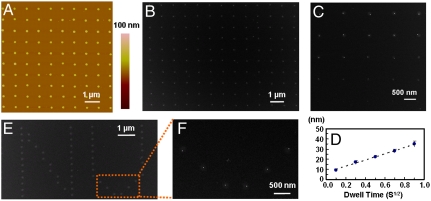
In a typical SPBCL experiment, chloroauric acid (HAuCl4) is added to an aqueous solution of poly(ethylene oxide)-b-poly(2-vinylpyridine) (PEO-b-P2VP). After stirring for 24 h, the metal ion-coordinated block copolymer mixture was coated onto scanning probes, which were then used for lithography. The block copolymer features are directly deposited on a Si substrate or a Si3N4 TEM membrane (Fig. 1A), and subsequent removal of the organic polymer and reduction of the metal ions result in the formation of single nanoparticles within the original patterned locations (Fig. 1B). Importantly in SPBCL, the AuNP that forms is tailorable and can be much smaller than the original polymer feature size and even the tip radius (4, 10). In this report, arrays of AuNPs with diameters between 4 and 40 nm were studied (examples between 10 and 40 nm are shown in Fig. 1 C and D). Custom patterns of single nanoparticles can also be made in arbitrary formats. For example, arrays of approximately 10-nm particles in the form of the letters “NU” were made from approximately 100-nm-diameter polymer features (Fig. 1 E and F).
Fig. 1.
SPBCL patterning of AuNP arrays. (A) An AFM topographical image of a dot array of PEO-b-P2VP/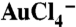 ink on a Si3N4 TEM membrane patterned by SPBCL. (B) A TEM image of single AuNP arrays produced after polymer removal by plasma treatment. (C) Controlled formation of AuNPs (white dots) across a range of sizes within the block copolymer matrix (gray background) after brief plasma exposure. (D) A graph of the size distribution of AuNPs synthesized as a function of tip-substrate dwell time. (E) An arbitrary pattern made of individual PEO-b-P2VP/
ink on a Si3N4 TEM membrane patterned by SPBCL. (B) A TEM image of single AuNP arrays produced after polymer removal by plasma treatment. (C) Controlled formation of AuNPs (white dots) across a range of sizes within the block copolymer matrix (gray background) after brief plasma exposure. (D) A graph of the size distribution of AuNPs synthesized as a function of tip-substrate dwell time. (E) An arbitrary pattern made of individual PEO-b-P2VP/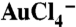 dot features printed in the form of “NU.” (F) A magnified SEM image of the AuNPs after polymer removal.
dot features printed in the form of “NU.” (F) A magnified SEM image of the AuNPs after polymer removal.
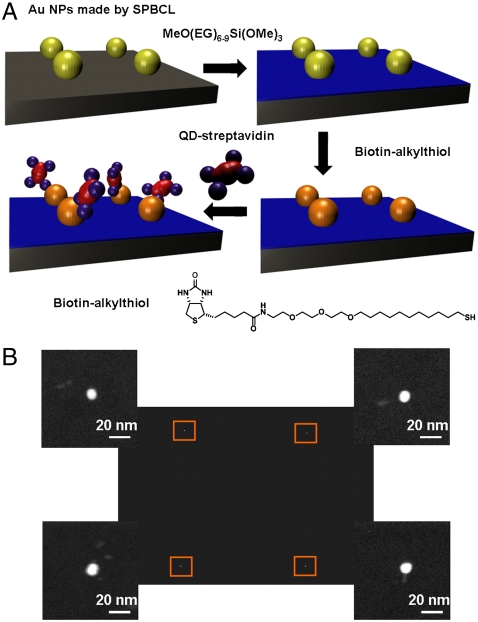
To determine if proteins can be immobilized on the SPBCL-generated AuNPs, we studied it in the context of streptavidin binding to biotin. The biotin-streptavidin interaction is known to be very strong (Kd ≈ 10-15 M) and represents an example of site-specific binding (23). To this end, SPBCL was used to write a square array of approximately 10-nm AuNPs with a 2-μm pitch. The bulk silicon wafer surface bearing the patterned AuNPs was then modified with a polyethylene glycol (PEG)-functionalized silane in order to prevent nonspecific adsorption of the protein onto unpatterned areas. SAMs of biotin-alkylthiols were subsequently formed on the AuNPs by immersing the nanoparticle arrays in a biotin-alkylthiol solution (Fig. 2A). These biotin-bearing AuNP arrays were then used to immobilize CdSe/ZnS core-shell quantum dots (QDs) that were modified with streptavidin. Here, the QDs acted as an easily observable label that can confirm the binding of streptavidin to the biotin-AuNPs. Transmission electron microscopy (TEM) images of the substrate acquired postimmobilization demonstrated the binding of QD-labeled streptavidin on every patterned gold nanoparticle functionalized with biotin (Fig. 2B). In these TEM images, the white dots represent the AuNPs from the SPBCL pattern, while the associated gray rods are the CdSe/ZnS QDs. Energy-dispersive X-ray spectroscopy (EDX) elemental mapping of Au and Cd also confirmed the identity of the QDs immobilized onto patterned AuNPs (Fig. S1). No QDs were found on the PEG-passivated surface, confirming that streptavidin was exclusively attached to the Au pattern and there was no significant nonspecific adsorption.
Fig. 2.
Streptavidin immobilization on Au patterns. (A) Schematic diagram of streptavidin-CdSe/ZnS QD conjugate immobilization onto SPBCL-defined AuNPs functionalized with biotin-thiol conjugates. (B) A TEM image of a square array pattern of Au nanoparticles with associated CdSe/ZnS QDs. The magnified image of each particle (defined by the orange box at each corner) shows that all of the AuNPs patterned by SPBCL are associated with the gray rods of the QD labels.
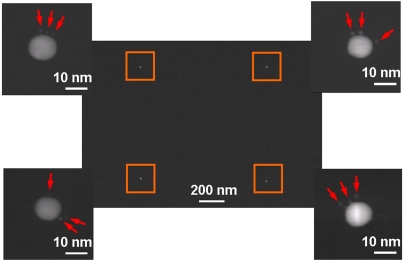
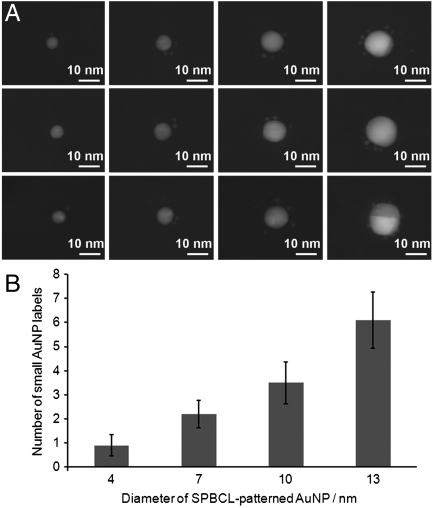
In order to determine the number of streptavidin molecules per single AuNP in the pattern, an AuNP array that was produced in a similar manner was then treated with a solution of streptavidin that had been separately labeled with 1.4-nm AuNPs. Importantly, the commercially supplied AuNP-streptavidin conjugates used in this case were purified such that the material was predominantly a 1∶1 ratio of nanoparticles to streptavidin (NANOGOLD® streptavidin, Nanoprobes) (24, 25). These conjugates thus enabled the quantification of the number of streptavidin molecules immobilized onto the arrays by observation and counting of the smaller streptavidin-bound AuNPs that were bound to the SPBCL-generated AuNPs. Furthermore, the smaller streptavidin-conjugated 1.4-nm AuNPs could be distinguished from the larger approximately 10-nm AuNPs defined by SPBCL (Fig. 3). On average, each of the approximately 10-nm AuNPs was associated with three of the smaller AuNP labels, which in turn indicated the immobilization of three streptavidin molecules per particle. Because the size of a streptavidin molecule is approximately 4 nm, the accommodation of three protein molecules onto a single 10-nm AuNP was reasonable. The relationship between the size of the AuNPs patterned by SPBCL and the number of protein molecules attached to them was further investigated by deliberately adjusting the dwell time of the inked tip to produce arrays of AuNPs with different diameters (Fig. 4A). As expected, the larger SPBCL-patterned AuNPs had more 1.4-nm AuNPs and corresponding streptavidin molecules attached (Fig. 4B). When the printed AuNP particle size was reduced to approximately 4 nm, which correlated with the dimensions of the streptavidin molecule, our analysis demonstrated that approximately 80% of the printed AuNPs had only one bound streptavidin, leading to an array of single biomolecules.
Fig. 3.
A TEM image of a square array pattern of approximately 10-nm AuNPs with associated smaller 1.4-nm AuNPs. The magnified images defined by the orange box at each corner show that each of the large AuNPs has three smaller AuNP labels (indicated by the red arrows) on average, indicating the immobilization of three streptavidin molecules at each site.
Fig. 4.
Quantification of protein immobilization. (A) TEM images showing the association of 1.4-nm AuNPs labels with the SPBCL-defined large AuNPs. The number of the AuNP labels and, in turn the number of the immobilized streptavidin molecules is dependent on the size of the SPBCL-defined AuNPs. (B) Histogram of the number of the associated small AuNP labels as a function of the size of the AuNPs patterned by SPBCL.
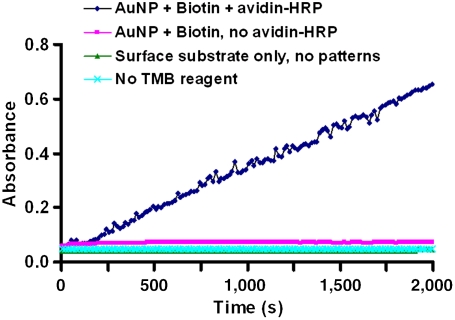
To demonstrate that proteins immobilized in this way retained their biological activity, SPBCL was used to generate a uniform array of 10-nm AuNPs presenting biotin. These arrays were then used to immobilize avidin-horseradish peroxidase (HRP) conjugates. The enzymatic activity of the HRP could then be detected by the 3,3′,5,5′-tetramethylbenzidine (TMB) assay with hydrogen peroxide, where the enzymatic reaction resulted in a blue color that could be quantified by UV-Vis absorbance measurements of the test solution at 652 nm (26). Thus, by immersing the printed substrates bearing avidin-HRP in the assay solution, an increase in absorbance was observed over time, which confirmed the activity of the immobilized protein (Fig. 5). In contrast, in the control experiments using surface substrates that were not SPBCL-patterned, or where avidin-HRP was not immobilized on the AuNPs, or if the TMB reagent was omitted, no increase in absorbance above the baseline was observed.
Fig. 5.
Graph of UV-Vis absorbance at 652 nm over time for the TMB assay, measured for the various surface substrates including the AuNP-immobilized arrays of avidin-HRP.
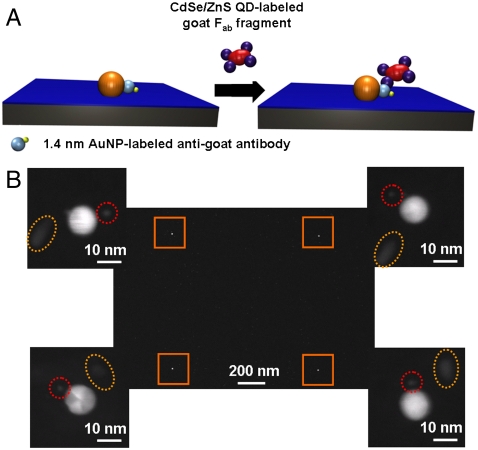
In order to generalize this procedure to other types of proteins, a more widely applicable method of protein immobilization was required. In this respect, the use of metal affinity binding is extensively used both for the purification and immobilization of proteins that possess an innate metal binding sequence or a peptide sequence that is capable of binding divalent metal ions such as the polyhistidine “His-tag” (23, 27). In order to evaluate this protein attachment method in the context of the SPBCL platform, an alkylthiol with the nitrilotriacetic acid (NTA) group was used to form the SAMs on the AuNPs (Fig. 6A). The AuNP patterns with these NTA groups were then used for the site-selective binding of native (unmodified) IgG by metal affinity because these antibodies possess a histidine-rich sequence near the C terminal of the FC region (28, 29). To determine if single-molecule immobilization was possible, these approximately 10-nm SPBCL-generated AuNP arrays were treated with IgG molecules labeled with 1.4-nm AuNPs (at a 1∶1 ratio of IgG:AuNP). They were subsequently rinsed with PBS buffer and imaged by TEM, which showed a single 1.4-nm particle label per approximately 10 nm particle substrate (Fig. 6B). This observation is consistent with the dimensions of the IgG molecules (15 × 9.5 × 6.5 nm) (30). Finally, in order to demonstrate that the anti-goat IgG also retained its biological activity after immobilization, these arrays were treated with a solution of goat-derived Fab antibody fragments that were conjugated with QD rods. In a representative TEM image (Fig. 6B), each of 10-nm substrate particles has a single 1.4-nm particle label and at least one QD rod label. EDX analysis was used to further confirm the identity of the AuNPs and QDs (Fig. S2).
Fig. 6.
Antibody immobilization on Au pattern. (A) Schematic diagram of antibody–antigen binding on the patterned AuNPs bearing NTA-functionalized SAMs. (B) TEM image of a square array pattern of large AuNPs formed by SPBCL. The magnified images on each corner shows that each of the large AuNPs capped with NTA groups is in close proximity to a single small AuNP (as indicated by the red circles) from the IgG and a QD (as indicated by the yellow ovals) from the Fab.
In summary, we have shown that SPBCL can be used to prepare gold nanostructures small enough to control the individual placement of protein molecules on surfaces. Importantly, this technique takes advantage of the ability of SPBCL to create nanostructures smaller than the tip diameter, thereby bypassing some of the limitations of the highest resolution molecular printing techniques. In addition, the ability to adjust particle size based upon polymer feature allows rapid prototyping structures tailored for different sized and numbers of proteins. The method is compatible with highly parallelized multiprobe nanolithography methods such as polymer pen lithography (PPL) and hard-tip soft-spring lithography (HSL) (31), making it very attractive for high throughput use. Unlike conventional block copolymer micelle nanolithography (32, 33), which is only capable of making repetitive geometric arrays, SPBCL allows for the deposition of individual block copolymer features at user-defined positions with nanoscale resolution, enabling the generation of complex patterns. Such feature size and pattern geometry for larger structures have proven important in understanding cell surface interactions (34, 35) and the ability to now achieve such control down to the dimensions of single biomolecules, including proteins (36), viruses (37), and oligonucleotides (38) will enable new insights in this area.
Materials and Methods
SPBCL Fabrication of AuNP Arrays.
AuNP arrays were fabricated on Si or Si3N4 substrates by SPBCL according to previously published procedures (19). In brief, the diblock copolymer PEO-b-P2VP (Mn 2,800-b-1,500, Mw/Mn 1.11; Polymer Source Inc,) was dissolved in an aqueous solution (concentration of 0.5% w/w) followed by the addition of HAuCl4 (Sigma-Aldrich) where the molar ratio of [AuCl4]- to pyridine is 1∶3. After vigorous stirring for at least 24 h, Si AFM tips or pen arrays (Nanoink) were dipped into the solution and blown dry with nitrogen. In a typical DPN experiment, tips are brought in contact with the substrate, which is a Si3N4 TEM grid (Ted Pella) for the experiments described herein. Thus, the block copolymer features are directly deposited onto Si3N4 TEM membranes (15 or 50 nm thick) and can be put in a TEM for imaging. Importantly, there is no need to transfer nanoparticles. The patterning process was performed in an NScriptor system (Nanoink) at room temperature and at least 70% relative humidity. After patterning, the samples were exposed to oxygen plasma for about 15 min at 60 W under a pressure of 100 mTorr, followed by thermal annealing at 650 °C for 2 h to remove the polymer and reduce the Au ions into AuNPs.
PEG Passivation.
The surfaces patterned with AuNP as described above were immersed in a 6-mM solution of 2-[methoxy(polyethyleneoxy)propyl]trimethoxysilane (MeO-PEG-Si(OMe)3, Mw 460–590; Gelest) and 1% v/v triethylamine in anhydrous toluene. The solution is kept at 80 °C on a hot plate for at least 24 h. To remove any noncovalently attached siloxane, substrates were rinsed extensively with ethyl acetate, methanol, and ethanol. After drying under a stream of nitrogen, the PEG-silane passivated sample was immediately used for subsequent steps.
Streptavidin Immobilization.
After PEG passivation, the substrates patterned with AuNPs were immersed in a 1-mM ethanol solution of biotin-alkylthiol (HS-C11-EG3-Biotin, Prochimia) for 4 h at room temperature and then rinsed with ethanol and dried with nitrogen. A 50-nM streptavidin solution was prepared by diluting commercially available streptavidin conjugates, which were labeled with CdSe/ZnS core/shell (approximately 7 nm) quantum dots (Qdot® 605 Streptavidin Conjugate, Invitrogen), or 1.4-nm AuNPs (NANOGOLD® streptavidin, Nanoprobes), with phosphate buffered saline (PBS). Then, samples were immersed in the streptavidin solution for 2 h at room temperature just after the biotin-alkylthiol SAM formation step, followed by rinsing with PBS. Under all these conditions, gentle rinsing does not dislodge the nanoparticles from the substrate.
Peroxidase Substrate (TMB) Assay.
The biotin-thiol functionalized AuNP arrays were incubated in a 30-nM avidin-horseradish peroxidase (Invitrogen) solution in PBS at room temperature for 2 h. The samples were then placed into the wells of a 96-well microtiter plate and 100 μL of premixed 1% 3,3′,5,5′ Tetrmethylbenzidine (TMB, 0.4 g/L) aqueous solution is added. Immediately afterward 100 μL hydrogen peroxide (0.02%) in buffer (26) (Pierce Protein Research Products) is added to each sample well. The UV-visible light absorbance of the solution in each well is monitored and quantified over 2,000 s at 652 nm by using Synergy H4 Hybrid Multiwell Plate Reader (BioTek, Winooski, VT).
Antibody Immobilization by Metal Coordination.
Patterned AuNP substrates after PEG passivation (not streptavidin functionalized) were immersed in a 1-mM solution of an alkylthiol terminated with nitrilotriacetic acid (NTA) groups (HS-C11-EG3-NTA) in ethanol for 4 h, followed by rinsing with ethanol, and then immersed in a 40-mM aqueous solution of NiCl2 for 1 h. The Ni2+-charged NTA-thiol-AuNP substrates were then incubated in a PBS solution of antigoat IgG (80 μg/mL) labeled with 1.4-nm AuNPs (Nanoprobes) for 4 h. The AuNP arrays were subsequently incubated with a solution of goat-derived Fab labeled with CdSe/ZnS quantum dots (Qdot® 655 goat Fab antirabbit IgG conjugate, Invitrogen) and imaged by TEM.
Supplementary Material
Acknowledgments.
C.A.M. acknowledges the US Air Force Office of Scientific Research, the Defense Advanced Research Projects Agency, and the National Institutes of Health (Center of Cancer Nanotechnology Excellence) for support of this research. C.A.M is grateful for a National Security Science and Engineering Faculty Fellowship from the Department of Defense. J.C. acknowledges the Natural Sciences and Engineering Research Council of Canada for a Postdoctoral Fellowship. L.S.W. is grateful to the Engineering and Physical Sciences Research Council for a Life Science Interface Fellowship (EP/F042590/1).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1116099108/-/DCSupplemental.
References
- 1.Lee KB, Kim EY, Mirkin CA, Wolinsky SM. The use of nanoarrays for highly sensitive and selective detection of human immunodeficiency virus type 1 in plasma. Nano Lett. 2004;4:1869–1872. [Google Scholar]
- 2.Wingren C, Borrebaeck CAK. Progress in miniaturization of protein arrays—a step closer to high-density nanoarrays. Drug Discov Today. 2007;12:813–819. doi: 10.1016/j.drudis.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Chen H, Li J. Nanotechnology: Moving from microarrays toward nanoarrays. Methods Mol Biol. 2007;381:411–436. doi: 10.1007/978-1-59745-303-5_22. [DOI] [PubMed] [Google Scholar]
- 4.Théry M. Micropatterning as a tool to decipher cell morphogenesis and functions. J Cell Sci. 2010;123:4201–4213. doi: 10.1242/jcs.075150. [DOI] [PubMed] [Google Scholar]
- 5.Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol. 2006;7:265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 6.Lutolf MP, Gilbert PM, Blau HM. Designing materials to direct stem-cell fate. Nature. 2009;462:433–441. doi: 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geblinger D, Addadi L, Geiger B. Nano-topography sensing by osteoclasts. J Cell Sci. 2010;123:1503–1510. doi: 10.1242/jcs.060954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishijima A, Yanagida T. Single molecule nanobioscience. Trends Biochem Sci. 2001;26:438–444. doi: 10.1016/s0968-0004(01)01860-6. [DOI] [PubMed] [Google Scholar]
- 9.Gräslund A, Rigler R, Widengren J. Single Molecule Spectroscopy in Chemistry, Physics and Biology. Berlin: Springer; 2010. [Google Scholar]
- 10.Piner RD, Zhu J, Xu F, Hong SH, Mirkin CA. “Dip-pen” nanolithography. Science. 1999;283:661–663. doi: 10.1126/science.283.5402.661. [DOI] [PubMed] [Google Scholar]
- 11.Lee KB, Lim JH, Mirkin CA. Protein nanostructures formed via direct-write dip-pen nanolithography. J Am Chem Soc. 2003;125:5588–5589. doi: 10.1021/ja034236p. [DOI] [PubMed] [Google Scholar]
- 12.Wu C-C, et al. Porous multilayer-coated AFM tips for dip-pen nanolithography of proteins. J Am Chem Soc. 2009;131:7526–7527. doi: 10.1021/ja901756a. [DOI] [PubMed] [Google Scholar]
- 13.Irvine EJ, Hernandez-Santana A, Faulds K, Graham D. Fabricating protein immunoassay arrays on nitrocellulose using dip-pen lithography techniques. Analyst. 2011;136:2925–2930. doi: 10.1039/c1an15178a. [DOI] [PubMed] [Google Scholar]
- 14.Lee SW, et al. Biologically active protein nanoarrays generated using parallel Dip-pen nanolithography. Adv Mater. 2006;18:1133–1136. [Google Scholar]
- 15.Lee S, et al. Quantitative analysis of human serum leptin using a nanoarray protein chip based on single-molecule sandwich immunoassay. Talanta. 2009;78:608–612. doi: 10.1016/j.talanta.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 16.Palma M, et al. Selective biomolecular nanoarrays for parallel single-molecule investigations. J Am Chem Soc. 2011;133:7656–7659. doi: 10.1021/ja201031g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schvartzman M, et al. Nanolithographic control of the spatial organization of cellular adhesion receptors at the single-molecule level. Nano Lett. 2011;11:1306–1312. doi: 10.1021/nl104378f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nair PM, Salaita K, Petit RS, Groves JT. Using patterned supported lipid membranes to investigate the role of receptor organization in intercellular signaling. Nat Protoc. 2011;6:523–539. doi: 10.1038/nprot.2011.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chai J, et al. Scanning probe block copolymer lithography. Proc Natl Acad Sci USA. 2010;107:20202–20206. doi: 10.1073/pnas.1014892107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piner RD, Mirkin CA. Effect of water on lateral force microscopy in air. Langmuir. 1997;13:6864–6868. [Google Scholar]
- 21.Rozhok S, Piner R, Mirkin CA. Dip-pen nanolithography: What controls ink transport? J Phys Chem B. 2003;107:751–757. [Google Scholar]
- 22.Huo F, et al. Polymer pen lithography. Science. 2008;321:1658–1660. doi: 10.1126/science.1162193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong LS, Khan F, Micklefield J. Selective covalent protein immobilization: Strategies and applications. Chem Rev. 2009;109:4025–4053. doi: 10.1021/cr8004668. [DOI] [PubMed] [Google Scholar]
- 24.Zehbe I, et al. Sensitive in situ hybridization with catalyzed reporter deposition, streptavidin-nanogold, and silver acetate autometallography—detection of single-copy human papillomavirus. Am J Pathol. 1997;150:1553–1561. [PMC free article] [PubMed] [Google Scholar]
- 25.Weipoltshammer K, Schöfer C, Almeder M, Wachtler F. Signal enhancement at the electron microscopic level using nanogold and gold-based autometallography. Histochem Cell Biol. 2000;114:489–495. doi: 10.1007/s004180000217. [DOI] [PubMed] [Google Scholar]
- 26.Josephy PD, Eling T, Mason RP. The horseradish peroxidase-catalyzed oxidation of 3,5,3′,5′-tetramethylbenzidine free radical and charge-transfer complex intermediates. J Biol Chem. 1982;257:3669–3675. [PubMed] [Google Scholar]
- 27.Hochuli E, Bannwarth W, Dobeli H, Gentz R, Stuber D. Genetic approach to facilitate purification of recombinant proteins with a novel metal chelate adsorbent. Nat Biotechnol. 1988;6:1321–1325. [Google Scholar]
- 28.Hale JE, Beidler DE. Purification of humanized murine and murine monoclonal antibodies using immobilized metal-affinity chromatography. Anal Biochem. 1994;222:29–33. doi: 10.1006/abio.1994.1449. [DOI] [PubMed] [Google Scholar]
- 29.Hale JE. Irreversible, oriented immobilization of antibodies to cobalt-iminodiacetate resin for use as immunoaffinity media. Anal Biochem. 1995;231:46–49. doi: 10.1006/abio.1995.1501. [DOI] [PubMed] [Google Scholar]
- 30.Guddat LW, Herron JN, Edmundson AB. Three-dimensional structure of a human immunoglobulin with a hinge deletion. Proc Natl Acad Sci USA. 1993;90:4271–4275. doi: 10.1073/pnas.90.9.4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shim W, et al. Hard-tip, soft-spring lithography. Nature. 2011;469:516–520. doi: 10.1038/nature09697. [DOI] [PubMed] [Google Scholar]
- 32.Arnold M, et al. Activation of integrin function by nanopatterned adhesive interfaces. Chem Phys Chem. 2004;5:383–388. doi: 10.1002/cphc.200301014. [DOI] [PubMed] [Google Scholar]
- 33.Wolfram T, Belz F, Schoen T, Spatz JP. Site-specific presentation of single recombinant proteins in defined nanoarrays. Biointerphases. 2007;2:44–48. doi: 10.1116/1.2713991. [DOI] [PubMed] [Google Scholar]
- 34.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 35.Kilian KA, Bugarija B, Lahn BT, Mrksich M. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc Natl Acad Sci USA. 2010;107:4872–4877. doi: 10.1073/pnas.0903269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim J-H, et al. Direct-write dip-pen nanolithography of proteins on modified silicon oxide surfaces. Angew Chem Int Ed Engl. 2003;42:2309–2312. doi: 10.1002/anie.200351256. [DOI] [PubMed] [Google Scholar]
- 37.Vega RA, Maspoch D, Salaita K, Mirkin CA. Nanoarrays of single virus particles. Angew Chem Int Ed Engl. 2005;44:6013–6015. doi: 10.1002/anie.200501978. [DOI] [PubMed] [Google Scholar]
- 38.Mirkin CA, et al. Direct patterning of modified oligonucleotides on metals and insulators by dip-pen nanolithography. Science. 2002;296:1836–1838. doi: 10.1126/science.1071480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.