Abstract
Arenaviruses form a noncytolytic infection in their rodent hosts, yet can elicit severe hemorrhagic disease in humans. How arenaviruses regulate gene expression remains unclear, and further understanding may provide insight into the dichotomy of these disparate infection processes. Here we reconstitute arenavirus RNA synthesis initiation and gene expression regulation in vitro using purified components and demonstrate a direct role of the viral Z protein in controlling RNA synthesis. Our data reveal that Z forms a species-specific complex with the viral polymerase (L) and inhibits RNA synthesis initiation by impairing L catalytic activity. This Z–L complex locks the viral polymerase in a promoter-bound, catalytically inactive state and may additionally ensure polymerase packaging during virion maturation. Z modulates host factors involved in cellular translation, proliferation, and antiviral signaling. Our data defines an additional role in governing viral RNA synthesis, revealing Z as the center of a network of host and viral connections that regulates viral gene expression.
Keywords: transcription factor, Machupo virus, matrix protein, negative-strand RNA virus, gene regulation
Transcription initiation is a fundamental focal point of gene expression regulation in all orders of life. Modulation of RNA synthesis affords an important response to stress, alterations in growth conditions, and environmental cues. Although initiation of RNA synthesis is often indirectly controlled through gene promoters and accessory signaling elements, regulation also occurs through factors directly interacting with and altering the catalytic potential of the RNA synthesis machinery itself (1). Studies with phage T7 and the T7 lysozyme repressor system have provided an important paradigm for control of viral RNA transcription (2, 3), but these processes remain a mystery for many eukaryotic viruses and medically relevant pathogens. Gene expression regulation is particularly critical for negative-sense RNA viruses of the family Arenaviridae, which must respond to input from the host environment to maintain a noncytolytic infection in their rodent reservoir (4). In stark contrast to this persistent infection, arenavirus infection in humans can lead to severe hemorrhagic fever disease and many arenaviruses represent important emerging pathogens (5). Intriguingly, recent evidence indicates that the balance between persistent and acute viral infection can be altered by a single mutation within the viral polymerase (6). Mechanistic insight into how arenaviruses tune gene expression and control viral RNA synthesis is required to further our understanding of the response of arenavirus infection to various host and cellular environments.
Arenaviruses are the simplest of all hemorrhagic fever viruses, with only four viral proteins. The viral genome consists of two segments of single-stranded negative-sense RNA, where each segment encodes two proteins in an ambisense orientation. The small segment encodes for the viral glycoprotein, essential for entry into the host cell, and the nucleocapsid protein (NP), which coats the genomic RNA and protects it from degradation and detection by host antiviral sensors (7, 8). The large segment encodes for the large polymerase protein (L) as well as the smallest viral protein, Z. All of the enzymatic machinery required for viral RNA synthesis and mRNA transcription is contained within L, a 250-kDa multifunctional protein that is organized as a central ring-like RNA-dependent RNA-polymerase (RdRP) domain with three accessory appendages (9). As ectopic overexpression of Z induces potent inhibition of replicon-based reporter protein expression (10, 11), Z has been hypothesized as an important regulator of L and arenaviral gene expression. However, Z is known to interact with several host factor binding partners, and it remains unclear whether Z directly regulates the viral RNA synthesis machinery or acts through alteration of an essential host process.
The Z gene is highly conserved across all arenavirus species and encodes a protein organized as an unstructured amino and carboxyl terminus bracketing a really interesting new gene (RING) domain that coordinates a pair of structural zinc ions (12). Z has been implicated in many facets of the arenavirus replication cycle and is the primary driving force of virion maturation (13). In addition to recruiting the cellular endosomal sorting complex required for transport (ESCRT) machinery to mediate virion budding (13, 14), Z has been demonstrated to interact with the translation initiation factor eIF4E (15) as well as retinoic acid inducible gene I (RIG-I), promyelocytic leukemia protein (PML), and proline-rich homeodomain protein (PRH) (16–18). Thus, Z is positioned at the center of a network of host and viral connections and may serve as a crucial viral sensor of the host cellular environment.
To unravel the complex network of Z functions and host interactions, and to determine whether Z plays a direct role in regulating arenavirus gene expression, we reconstituted arenavirus RNA synthesis and regulation by Z in vitro using purified components. Here we use purified L from Machupo virus (MACV), the causative agent of Bolivian hemorrhagic fever, to establish an in vitro model of arenavirus RNA synthesis that maintains authentic promoter recruitment and RNA synthesis initiation while relying solely on recombinant L and short naked RNA templates. With this minimal system, we demonstrate that Z functions as a direct regulator of arenaviral RNA synthesis and forms a direct complex with L governed by species-specific protein interactions. Z controls gene expression by locking a promoter-bound polymerase complex in a catalytically inactive state. These findings offer mechanistic insight into Z mediation of L catalytic activity and uncover how a single viral protein coordinates both modulation of host processes and regulation of viral gene expression.
Results
Z Is a Direct Inhibitor of L Catalytic Activity.
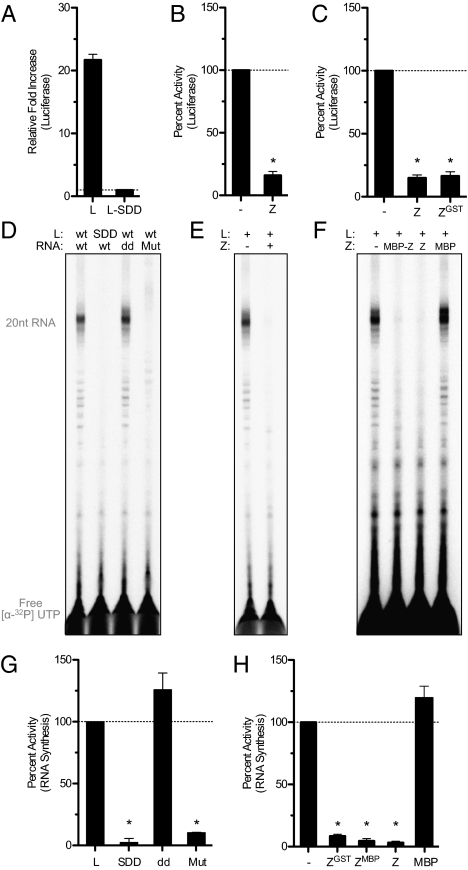
To probe how arenaviruses control viral gene expression, we reconstituted Z regulation of arenavirus RNA synthesis in cells and in vitro with purified components. We designed a MACV S-segment replicon where the viral glycoprotein gene has been replaced with gaussia luciferase to quantitatively assess activity of the viral polymerase complex within transfected cells. Luciferase signal was only observed in the presence of MACV L, and a mutation to the predicted RdRP catalytic active site (SDD1328AAA) abolished reporter protein expression (Fig. 1A). Ectopic overexpression of MACV Z resulted in the inhibition of the MACV replicon-based reporter protein expression (Fig. 1B), and we observed equivalent levels of inhibition with a GST-tagged version of Z (Fig. 1C). Previous results have revealed a similar phenotype using overexpressed Z in other arenaviral replicon systems (10, 11, 19), yet it has been impossible to determine whether Z directly impacts the viral RNA synthesis machinery or functions through alteration of an essential host cellular process. We therefore established an in vitro system to examine a potential direct effect of Z on viral RNA synthesis.
Fig. 1.
Z is a direct inhibitor of L catalytic activity. (A) Cellular-based replicon assay for MACV RNA synthesis where functional viral polymerase results in the production of gaussia luciferase. Luciferase activity was measured from the supernatant of cells expressing pMCm-GLuc replicon in the presence of L (L) or a catalytically inactivated mutant (L-SDD). Error bars represent the SD from the mean of at least three independent experiments. (B and C) Replicon assay as in A with cells transfected with the viral replicon and L alone (−) or with MACV Z (Z) or GST-tagged MACV Z (ZGST) plasmids. Luciferase values were normalized to cells expressing only the replicon plasmid and wild-type L. Error bars represent the SD as in A and conditions that are statistically significant from the wild-type L control are designated with the symbol * (P < 0.001 or for ZGST P = 0.0016). (D) MACV in vitro RNA synthesis. Purified L (L: WT) or a catalytically inactivated mutant (L: SDD) was incubated with a wild-type template (template RNA: WT), a 3′ terminal dideoxy template (template RNA: dd) or an RNA template with a mutated promoter binding site (template RNA: Mut) in the presence of GpC primer as described in the text. RNA products were labeled with [α-32P]-UTP and separated by denaturing gel electrophoresis. (E and F) RNA products generated from RNA synthesis reactions supplemented with purified GST-tagged MACV Z (+), MBP-tagged MACV Z (MBP-Z), untagged MACV Z (Z), or free MBP (MBP) and analyzed as in D. (G and H) Total RNA synthesis was quantified with a PhosphorImager and graphed as values normalized to reactions containing wild-type L and RNA template. Error bars represent the SD from the mean of at least three independent experiments (*P < 0.001). See also Figs. S1 and S2.
The large size (∼250 kDa) and flexible multidomain organization of arenaviral L proteins have limited biochemical characterization of viral RNA synthesis. Previously, we demonstrated purification of MACV L protein and formation of an L–RNA complex with the conserved 19-nt RNA promoter element (9). Using this recombinant L–RNA complex, we established an in vitro transcription system to study the initial events of viral RNA synthesis. The current model for initiation of arenavirus RNA synthesis is a “prime-and-realign” model, where the short dinucleotide primer (GpC) is first synthesized and realigned with the template RNA resulting in the insertion of an extra 5′ guanosine (20). Consistent with this model, when recombinant L–RNA complexes were incubated with NTPs and [α-32P]-UTP, we observed the formation of a predominant ∼20-nt RNA product from the 19-nt RNA template as well as minor populations of ∼21-nt RNA and shorter products likely resulting from premature termination (Fig. 1D). GpC dinucleotide primer was included in RNA synthesis reactions as it has previously been demonstrated to stimulate viral RNA synthesis (9, 20); but similar products are observed with and without exogenous GpC primer addition (discussed below).
Analogous to the cellular-based replicon assay, a mutation (SDD1328AAA) within the RdRP active site of purified L abolished all RNA synthesis in vitro (Fig. 1D). Purified viral polymerases have been shown to possess terminal transferase activity (21, 22). To demonstrate that the RNA synthesis observed in vitro was not dependent upon a terminal transferase, we used a template in which the 3′ terminal guanosine is replaced by a dideoxy cytosine nucleotide (template RNA: dd). We previously demonstrated that the G1C RNA mutation does not impact L–RNA complex formation (9). In the presence of GpC dinucleotide primer, we observed similar RNA synthesis with this template, verifying that in vitro RNA synthesis is the result of de novo initiation and not an L-associated terminal transferase activity (Fig. 1D). In the absence of exogenous GpC primer, RNA synthesis with the G1C template resulted in slightly altered ratios of the predominant RNA products, which may indicate an effect of this mutation in normal prime-and-realign initiation events (discussed below). To further demonstrate that the products reflect de novo initiation of RNA synthesis, we examined whether they contained a reactive 5′ triphosphate. We took advantage of the vaccinia virus guanyltransferase, which can add a capping guanylate to free 5′ triphosphate but not 5′ monophosphate. The products of in vitro transcription were capped by the vaccinia virus guanyltransferase (discussed below), demonstrating that they retained a 5′ tri- or diphosphate RNA terminus, as observed previously with purified arenavirus genomic RNA (23). We next examined the products of RNA synthesis using an RNA template containing mutations to the high-affinity 3′ RNA promoter binding site. We previously demonstrated that the absolutely conserved 3′ C2G3U4G5 motif is required for L–RNA complex formation (9) and, as further verification of the authenticity of this reconstituted RNA synthesis system, we observed that mutating this 3′ binding site to C2C3G4G5 (template RNA: Mut) eliminated in vitro RNA synthesis activity (Fig. 1D).
To determine whether Z directly inhibits the viral RNA synthesis machinery, we purified recombinant GST-tagged MACV Z (Fig. S1). Supplementing RNA synthesis reactions with purified GST-Z resulted in potent inhibition of L catalytic activity (Fig. 1E). To verify this inhibition was not related to GST or the presence of a large solubility tag, we purified Z tagged with maltose binding protein (MBP) and removed the tag using tobacco etch virus protease (Fig. S1). Purified untagged Z, as well as MBP-Z, potently inhibited in vitro RNA synthesis (Fig. 1F). A time-course analysis of Z addition revealed that ongoing in vitro RNA synthesis reactions do not become resistant to Z inhibition (Fig. S2). Together, these results demonstrate that Z functions as a direct inhibitor of the viral RNA synthesis machinery.
Viral RNA Synthesis Regulation by Z Is Species Specific.
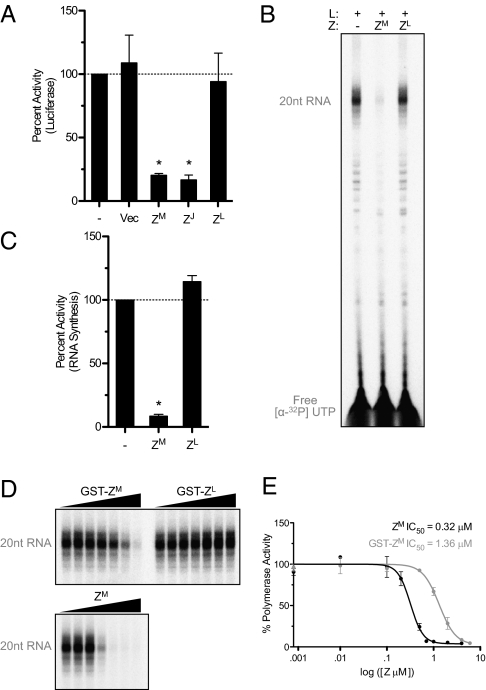
Z from several individual arenavirus species limits viral gene expression, and Lassa virus (LASV) Z is capable of cross-inhibiting a more closely related arenavirus lymphocytic choriomeningitis virus (LCMV) (19). We examined whether Z inhibited viral RNA synthesis through a species-specific interaction or a more broadly generalized mechanism. Two distinct phylogenetic and geographic virus groups exist within the family Arenaviridae: a New World complex of viruses found throughout the Americas, which includes MACV and the closely related Junín virus (JUNV), and an Old World complex of viruses found primarily in Africa, including LASV and LCMV. Similar to MACV Z, expression of the closely related Z protein from JUNV resulted in inhibition of MACV replicon-based RNA synthesis (Fig. 2A). In contrast, expression of the more distantly related LCMV Z protein had no effect (Fig. 2A). Additionally, whereas purified GST-tagged MACV Z (ZM) potently inhibits RNA synthesis by MACV L in vitro, purified GST-tagged LCMV Z (ZL) has no effect (Fig. 2 B and C), confirming the specificity of MACV Z inhibition in vitro. Therefore, arenavirus Z proteins do not function as broadly active inhibitors and must instead use a species-specific interaction.
Fig. 2.
Viral RNA synthesis regulation by Z is species specific. (A) In-cell replicon assay as in Fig. 1 with cells expressing an empty cassette (Vec), MACV Z (ZM), JUNV Z (ZJ), or LCMV Z (ZL). (B) RNA products resulting from in vitro RNA synthesis reactions supplemented with GpC primer and purified GST-MACV Z (ZM) or GST-LCMV (ZL) were analyzed and (C) quantified as in Fig. 1 (*P < 0.001). (D) In vitro RNA synthesis reactions supplemented with GpC primer and an increasing amount of untagged MACV Z (ZM), GST-MACV Z (GST-ZM), or GST-LCMV (GST-ZL). RNA products were analyzed and (E) quantified as in Fig. 1. Untagged MACV Z and GST-MACV Z data were analyzed with the GraphPad software package. Error bars for each point correspond to the SD from the mean of independent experiments.
As the cellular levels of Z vary throughout infection (10), and Z is also present in mature virions, we next sought to examine the levels of Z required to inhibit polymerase function. Untagged MACV Z inhibited viral RNA synthesis in the midnanomolar range (IC50 of 0.32 μM) (Fig. 2 D and E). GST-tagged MACV Z had a slightly elevated IC50 (1.36 μM), likely due to the presence of the large solubility tag, and GST-tagged LCMV Z again had no impact (Fig. 2D). On the basis of previous approximations of the ratio of arenavirus virion proteins (1 L:160 NP:60 GP:20 Z) (24), and the number of packaged NP molecules (∼1,530 copies of NP) (25), the concentration of Z for an approximately spherical arenavirus virion (50–100 nm radius) (26) is estimated to be ∼80–600 μM (SI Materials and Methods). The intravirion concentration of Z is significantly higher than the calculated IC50 (Fig. 2 D and E), indicating that the integral virion-packaged Z may also function along with the physical constraints of a mature virion to maintain polymerase quiescence.
Z and L Form a Direct Heterodimeric Complex.
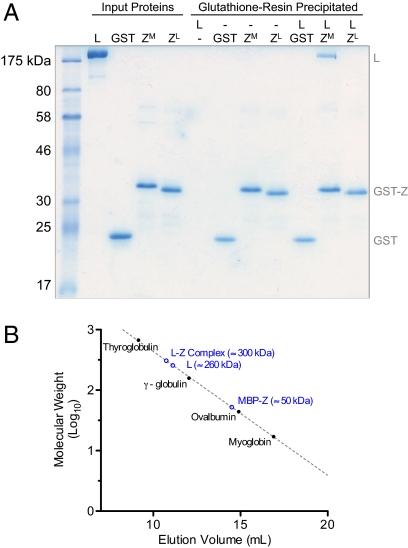
As Z potently inhibits L catalytic activity in vitro using purified components, we asked whether Z is capable of directly interacting with L. Using a GST pull-down approach, we observed direct complex formation between MACV L and Z (Fig. 3A). Although all of the GST-tagged arenavirus and control proteins interacted with the glutathione resin, purified MACV L was only retained in the presence of GST-tagged MACV Z, and not when incubated with free GST, GST-tagged LCMV Z, or resin alone (Fig. 3A). To approximate the stoichiometry of this interaction, we estimated the molecular weight of the L–Z complex by preincubating L and MBP-tagged Z and compared the gel-filtration elution profile of the complexed proteins with purified L, MBP-tagged Z, and known molecular weight standards. Following preincubation, the elution profiles of L and Z were shifted, and the L–Z complex eluted with an apparent molecular weight of ∼300 kDa, consistent with a 1:1 complex of MACV L and MBP-tagged MACV Z (Fig. 3B and Fig. S3).
Fig. 3.
Z and L form a direct heterodimeric complex. (A) GST pull-down assay using purified components. Purified MACV L (L), free GST (GST), GST-tagged MACV Z (ZM), or GST-tagged LCMV Z (ZL) were incubated in the designated combinations before purification with glutathione resin. Pelleted resin was washed three times and bound proteins were eluted and analyzed by 12% SDS/PAGE and colloidal Coomassie stain. Input proteins were loaded as markers and indicate one-third of the total input protein from each assay. (B) The molecular weight of the L–Z complex was estimated by preincubating L and MBP-tagged Z and comparing the gel-filtration elution profile of the complexed proteins with purified MACV L (L), MBP-tagged MACV Z (MBP-Z), and known molecular weight standards. See also Figs. S3 and S4.
In addition to a direct interaction with L, Z has been implicated in binding a variety of host factors including eIF4E, RIG-I, PML, and PRH (15–18). To verify that purified GST-tagged LCMV Z retains functional activity, we assessed the ability of purified MACV Z and LCMV Z to interact with eIF4E. Both viral Z proteins were able to interact with eIF4E, demonstrating that purified LCMV Z retains functional activity and that the ability to interact with eIF4E also extends to MACV Z and New World arenaviruses (Fig. S4 A and B). However, eIF4E interaction was only detectable by immunoblot, consistent with previously published results (12), and eIF4E alone was not sufficient to affect Z–L complex formation or Z-induced regulation of viral RNA synthesis (Fig. S4C). These results reveal that Z forms a direct complex with the viral polymerase that is resistant to eIF4E competition, and that species specificity of Z regulation is limited by the ability to interact with L.
Z Blocks an Early Step of RNA Synthesis Initiation.
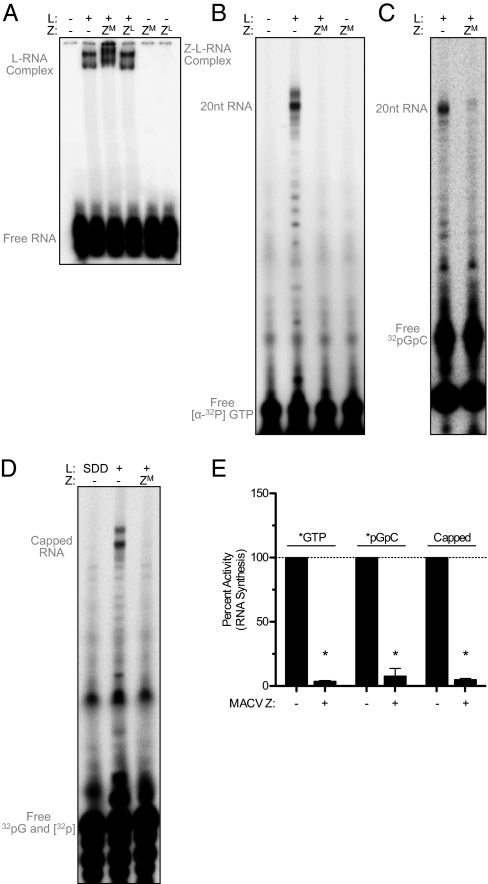
To assess the functional consequence of Z–L complex formation, we next asked whether Z disrupted the ability of L to engage the conserved C2G3U4G5 promoter element. Using a MACV L–RNA electrophoretic gel shift assay (9), we observed that MACV L–RNA complex formation is unaffected by the addition of either MACV or LCMV GST-tagged Z (Fig. 4A). This result demonstrates that Z does not inhibit L from engaging the viral RNA promoter, but instead inhibits a subsequent step of RNA synthesis. Notably, in the presence of MACV Z we observed a supershift of the L–RNA complex, further supporting a direct interaction between Z and L (Fig. 4A). This supershift was not observed in the presence of LCMV Z, and the purified Z proteins did not interact directly with RNA when incubated in the absence of L (Fig. 4A).
Fig. 4.
Z blocks an early step of RNA synthesis initiation. (A) Gel shift assay of complex formation between L, RNA, and Z. Radiolabeled 3′ RNA was incubated with buffer alone (−) or the indicated combinations of purified L (L), GST-tagged MACV Z (ZM), and GST-tagged LCMV Z (ZL). The resulting complexes were separated by nondenaturing gel electrophoresis. (B) In vitro RNA synthesis reactions in the absence of GpC primer were labeled with [α-32P]-GTP or (C) 5′ end-labeled [α-32P]-GpC and supplemented with untagged MACV Z (ZM). (D) Analysis of MACV in vitro transcripts subsequently labeled after RNA synthesis. Transcripts from unlabeled RNA synthesis reactions in the absence of GpC primer were purified by phenol-chloroform extraction and ethanol precipitation and then subsequently labeled by capping the 5′ end with vaccinia virus guanyltransferase and [α-32P]-GTP and separated as in B. (E) RNA products were analyzed and quantified as in Fig. 1 (*P < 0.001 or for [α-32P]-GpC P = 0.0042). See also Fig. S5.
As Z inhibits viral RNA synthesis (Fig. 1) but does not inhibit recruitment of L to the promoter RNA (Fig. 4A), we examined whether truncated RNA products were generated in the presence of Z. RNA synthesis reactions labeled with [α-32P]-GTP and carried out in the absence of any exogenously added GpC dinucleotide primer, produced the same predominant 20-nt RNA product as well as some minor truncated species (Fig. 4B). Inclusion of MACV Z inhibited the production of all detectable RNA synthesis, including the minor shorter RNA products (Fig. 4 B and E). In the presence of Z, no premature termination products were observed as would be predicted if Z functioned similarly to factors that block transcriptional elongation (Fig. 4B and Fig. S2 D–F) (3). We cannot eliminate the possibility, however, that short GpC primer formation could still occur in the presence of Z and that this product may be obscured by the presence of the [α-32P-GTP] visible on the autoradiogram (Fig. 4 B and E). However, we have not detected pppGpC or any other small RNAs produced in the presence of Z, indicating that if such short products are synthesized, they are below the limit of detection. RNA synthesis reactions carried out in the presence of [α-32P]-GTP and in which CTP and the GpC primer were omitted, led to the appearance of a minor RNA product of 20 nt (Fig. S5B). This product reflects terminal transferase activity of L, an activity maintained by many other viral RNA polymerases (21, 22), as the minor labeled RNA product was ablated by the 3′ dideoxy template mutation and also in the presence of a catalytically inactive MACV L (Fig. S5 B and C). Consistent with the conclusion that L is catalytically inactive when complexed with Z, Z also blocked the terminal transferase activity of L (Fig. S5 B and C). Collectively, these data support the model that Z renders L catalytically inactive.
To determine whether exogenously supplied GpC permits any ability for L to escape Z inhibition, we prepared in vitro RNA synthesis reactions using GpC primer directly labeled at the 5′ end. We observed that the GpC primer is incorporated into elongated RNA synthesis products (Fig. 4C), confirming that GpC can be used to prime arenavirus RNA synthesis. As with unprimed reactions, Z directly inhibited all detectable [α-32P]-GpC primed polymerase activity (Fig. 4 C and E), demonstrating that L is incapable of initiating de novo or primed RNA synthesis in the presence of Z. As an additional method to confirm that smaller products are not generated in the presence of Z, we labeled all de novo initiated transcripts after RNA synthesis through subsequent capping of the 5′ end with vaccinia virus guanyltransferase and [α-32P]-GTP (Fig. 4 D and E). Together these results demonstrate the formation of a Z–L–RNA complex and define the mechanism of Z regulation as inhibition of the initial catalytic steps of RNA synthesis following promoter engagement.
Discussion
Here we use purified components to reconstitute individual steps of arenavirus RNA synthesis initiation and demonstrate that the viral Z protein inhibits RNA synthesis by locking the polymerase in a catalytically inactive promoter-bound state. Z–L complex formation provides a direct link between viral gene expression, host cellular modulation, and virion packaging, revealing the multifarious activities of Z as an important network that regulates the dynamics of arenaviral infection.
Mechanism of Z Inhibition.
Arenavirus genomic replication requires L and an appropriate NP-encapsidated RNA template (27, 28). Although NP is not essential for the catalytic activity of L itself (Fig. 1), it is likely that NP is critical for activities not recapitulated by the initial steps of RNA synthesis initiation in vitro, including integrity of the full-length genome and possibly reduction of RNA secondary structure (29, 30). Here we demonstrate that Z is a direct modulator of the viral RNA synthesis machinery. In the cell, overexpression of Z results in inhibition of replicon-based reporter protein expression (Fig. 1A), and Z similarly inhibits RNA synthesis reactions using purified components in vitro (Fig. 1 E and F). Z has been identified as a binding partner to both host and viral proteins (15–18, 31–34). However, by establishing an in vitro model system of arenavirus RNA synthesis (Fig. 1D), we demonstrate that host interactions are not required for Z to inhibit L catalytic activity (Fig. 1).
Z inhibits L catalytic function, but interaction with Z does not restrict recruitment of L to the viral RNA promoter (Fig. 4A). Our data reveal that Z interacts directly with L (Fig. 3 and Fig. S3) and the promoter-bound L–RNA complex (Fig. 4A) and that Z alone does not interact with viral RNA (Fig. 4A). In the presence of Z, L maintains interaction with the high-affinity 3′ RNA promoter binding site (Fig. 4A) (9), but Z–L–RNA complex formation results in an inhibited polymerase incapable of RNA synthesis (Figs. 1 and 4 and Fig. S5). This block in an early step of RNA synthesis initiation prevents both de novo and GpC-primed transcription (Figs. 1 and 4), indicating that the polymerase is frozen in a catalytically inactive conformation or occluded complex. In our in vitro RNA synthesis system, L never becomes resistant to Z inhibition (Fig. S2), but determining the effect of Z on an actively elongating complex will require further analysis of single polymerase molecules.
As we observe here with Z regulation of L RNA synthesis initiation, modulating gene expression through control of RNA transcription initiation is a common theme found in all orders of life. The eukaryotic transcription factor Maf1 directly interacts with RNA polymerase III to repress RNA synthesis, but unlike the Z–L–RNA complex observed in vitro (Fig. 4A), Maf1 prevents promoter engagement and closed complex formation (35). Similarly, RNA synthesis initiation in phage T7 is controlled by interaction with T7 lysozyme that causes noncatalytic inhibition of T7 RNA polymerase resulting in short abortive transcript production (36). In contrast, Z prevents all detectable RNA synthesis, suggesting a direct impairment to the catalytic activity of L (Figs. 1 and 4 and Fig. S5). This situation mirrors repression of RNA synthesis initiation maintained by the bacterial transcription factor Gre-family homolog 1, which disrupts catalytic activity by physically occluding the NTP channel and locking the RNA polymerase in an inactive ratcheted state (37). Although the exact molecular mechanism of Z inhibition will require further biochemical and structural analysis, our data are consistent with a model where the catalytic potential of L is physically compromised within the Z–L complex.
Regulation of Arenavirus Gene Expression.
Z interacts with several host factors and has been implicated in modulating a variety of host processes including mRNA translation, cell proliferation, and antiviral signaling (15–18). Our data now reveal a direct role of Z in regulating viral gene expression, placing Z at a crossroads that links coordination of viral RNA synthesis with pathways essential for cellular viability. These results confirm a direct feedback loop where increased viral gene expression and higher levels of Z will act to suppress RNA synthesis and balance the infection process (10) (Figs. 2 and 5).
Fig. 5.
Model of arenavirus RNA synthesis regulation by Z. Low concentrations of Z permit ongoing RNA synthesis, whereas high concentrations of Z result in an inhibited Z–L–RNA complex bound to the viral promoter. As described in the text, the Z–L–RNA complex may serve as an important intermediate ensuring L is packaged into mature virions.
Z is highly conserved across all arenavirus species, although only Z from MACV and other New World arenaviruses has been demonstrated to interact with RIG-I (16). In contrast, we now demonstrate that MACV Z interacts with eIF4E (Fig. S4B), indicating that at least some of the binding partners of Z are shared between both New and Old World arenaviruses. However, interaction with eIF4E was not sufficient to regulate Z-induced inhibition of L in vitro (Fig. S4). JUNV Z inhibition of MACV RNA synthesis (Fig. 2A), and previous reports of cross-regulation by Z of LASV and LCMV (19), support that the Z–L interface is sufficiently conserved such that closely related arenaviruses share similar binding and regulatory interactions within their polymerase machinery. However, we demonstrate both in cells and in vitro that LCMV Z is unable to interact with or inhibit MACV L (Figs. 2–4A). These results show that Z inhibition of arenaviral RNA synthesis is species specific and unique to the distinct geographic and phylogenetic arenavirus clades.
Polymerase Packaging.
As the viral genome itself is not an infectious unit, all reverse transcribing, double-stranded, and negative-sense RNA viruses must package their polymerase machinery to generate an infectious particle. Whereas many viruses solve this problem by including their polymerase protein as an integral structural component essential for virion formation, negative-sense RNA viruses do not appear to invoke this strategy. In addition to complex formation with L, Z interacts with the cellular ESCRT machinery and is the major driving force of viral budding and virion formation (13). Importantly, inhibition by Z does not disrupt the recruitment of L to the genomic promoter (Fig. 4A), and the resulting Z–L–RNA complex may thus serve as a critical intermediate to ensure genomic RNA is packaged along with a polymerase poised to reinitiate viral RNA synthesis in the newly infected cell (Fig. 5). Although the individual mechanisms remain undetermined, inhibition of viral RNA gene expression by overexpression of the matrix, or matrix-like, protein has been observed for influenza A virus and in a multitude of nonsegmented negative-sense RNA viral systems (38–41). It is therefore possible that the Z–L–RNA complex and Z–L inhibition mechanism identified here may serve as an important paradigm for the link between transcription regulation and polymerase packaging of negative-sense RNA viruses.
Methods
In Vitro RNA Synthesis.
Arenavirus RNA synthesis was reconstituted using recombinant MACV L and a 19-nt template corresponding to the conserved 3′ terminus of the viral genome. L–RNA complexes were established as described previously (9). Briefly, 0.3 μg of purified L (final reaction concentration of 0.12 μM) was incubated with 2 μM of RNA in transcription buffer (50 mM Tris pH 7, 40 mM NaCl, 5 mM MnCl2, 10 mM KCl, 1 mM DTT, and 0.1 mg/mL BSA (New England Biolabs) at 25 °C for 30 min. After initial incubation, 0.5 μL of [α-32P]-UTP (∼10 μCi), and cold NTPs (1 mM ATP/CTP/GTP and 25 μM UTP) were added and reactions were incubated at 30 °C for 2 h in a final volume of 10 μL. Reactions included 40 μM GpC primer (Dharmacon) as indicated in the text. Alternatively, reactions were labeled with [α-32P]-GTP and cold NTPs (1 mM ATP/CTP/UTP and 100 μM GTP), or with 1 mM cold NTPs and 5′ [32P]-GpC primer labeled with [γ-32P]-ATP and T4 polynucleotidyl kinase (New England Biolabs). Where indicated, RNA synthesis reactions were supplemented with recombinant Z or MBP (2 μM final concentration) during L–RNA complex assembly. After 2 h incubation, reactions were terminated with 10 μL of stop solution (95% deionized formamide, 20 mM EDTA) and incubation at 95–100 °C for 2.5 min. Reactions were analyzed by denaturing gel electrophoresis on a 36 cm long, 20% polyacrylamide-urea (7 M) sequencing gel (with 0.5× TBE). RNA products were visualized using a PhosphorImager, sized according to chemically synthesized RNA markers, and quantified with ImageQuant (Amersham). Statistical significance was calculated using an unpaired, two-tailed t test.
GST Pull-Down and L–RNA Gel Shift Assays.
Recombinant L (5 μg) and/or eIF4E (8 μg) was incubated with GST-tagged Z or GST alone (3 μg) in binding buffer (PBS with 250 mM KCl and 0.2% Nonidet P-40) (12) in a total volume of 50 μL at room temperature for 30 min. Reaction volumes were raised to 500 μL with binding buffer and 15 μL of packed glutathione sepharose resin (GE Healthcare) and incubated at room temperature for 1 h while rotating. Glutathione resin and bound proteins were gently pelleted by centrifugation at 400 × g for 30 s and washed three times with wash buffer (PBS with 500 mM KCl and 1.0% Nonidet P-40). Proteins were eluted by boiling in 25 μL of SDS loading buffer, and 10 μL was analyzed by 12% denaturing SDS/PAGE and staining with colloidal Coomassie (Sigma). L–RNA complex formation and the electrophoretic shift assay were performed as described previously (9), and reactions were supplemented with 0.35 μg GST-tagged MACV Z or LCMV Z as indicated.
Supplementary Material
Acknowledgments
We acknowledge Robin Ross and Benjamin Seiler [New England Center of Excellence in Biodefense and Emerging Diseases (NERCE-BEID)] for support in recombinant protein production and Christine Anderson (NERCE-BEID) for initial sequence verification of JUNV-Candid#1. This study was supported by National Institutes of Health Grants AI057159 and AI059371. S.P.J.W. is a recipient of a Burroughs Wellcome Investigators in the Pathogenesis of Infectious Disease Award.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112742108/-/DCSupplemental.
References
- 1.Tjian R. The biochemistry of transcription in eukaryotes: A paradigm for multisubunit regulatory complexes. Philos Trans R Soc Lond B Biol Sci. 1996;351:491–499. doi: 10.1098/rstb.1996.0047. [DOI] [PubMed] [Google Scholar]
- 2.Kumar A, Patel SS. Inhibition of T7 RNA polymerase: Transcription initiation and transition from initiation to elongation are inhibited by T7 lysozyme via a ternary complex with RNA polymerase and promoter DNA. Biochemistry. 1997;36:13954–13962. doi: 10.1021/bi971432y. [DOI] [PubMed] [Google Scholar]
- 3.Zhang X, Studier FW. Mechanism of inhibition of bacteriophage T7 RNA polymerase by T7 lysozyme. J Mol Biol. 1997;269:10–27. doi: 10.1006/jmbi.1997.1016. [DOI] [PubMed] [Google Scholar]
- 4.Childs JE, Peters CJ. Ecology and epidemiology of arenaviruses and their hosts. In: Salvato MS, editor. The Arenaviridae. New York: Plenum; 1993. pp. 331–373. [Google Scholar]
- 5.Geisbert TW, Jahrling PB. Exotic emerging viral diseases: Progress and challenges. Nat Med. 2004;10(12, Suppl):S110–S121. doi: 10.1038/nm1142. [DOI] [PubMed] [Google Scholar]
- 6.Bergthaler A, et al. Viral replicative capacity is the primary determinant of lymphocytic choriomeningitis virus persistence and immunosuppression. Proc Natl Acad Sci USA. 2010;107:21641–21646. doi: 10.1073/pnas.1011998107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qi X, et al. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature. 2010;468:779–783. doi: 10.1038/nature09605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hastie KM, Kimberlin CR, Zandonatti MA, MacRae IJ, Saphire EO. Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3′ to 5′ exonuclease activity essential for immune suppression. Proc Natl Acad Sci USA. 2011;108:2396–2401. doi: 10.1073/pnas.1016404108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kranzusch PJ, et al. Assembly of a functional Machupo virus polymerase complex. Proc Natl Acad Sci USA. 2010;107:20069–20074. doi: 10.1073/pnas.1007152107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornu TI, de la Torre JC. RING finger Z protein of lymphocytic choriomeningitis virus (LCMV) inhibits transcription and RNA replication of an LCMV S-segment minigenome. J Virol. 2001;75:9415–9426. doi: 10.1128/JVI.75.19.9415-9426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jácamo R, López N, Wilda M, Franze-Fernández MT. Tacaribe virus Z protein interacts with the L polymerase protein to inhibit viral RNA synthesis. J Virol. 2003;77:10383–10393. doi: 10.1128/JVI.77.19.10383-10393.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Volpon L, Osborne MJ, Capul AA, de la Torre JC, Borden KL. Structural characterization of the Z RING-eIF4E complex reveals a distinct mode of control for eIF4E. Proc Natl Acad Sci USA. 2010;107:5441–5446. doi: 10.1073/pnas.0909877107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez M, Craven RC, de la Torre JC. The small RING finger protein Z drives arenavirus budding: Implications for antiviral strategies. Proc Natl Acad Sci USA. 2003;100:12978–12983. doi: 10.1073/pnas.2133782100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shtanko O, Watanabe S, Jasenosky LD, Watanabe T, Kawaoka Y. ALIX/AIP1 is required for NP incorporation into Mopeia virus Z-induced virus-like particles. J Virol. 2011;85:3631–3641. doi: 10.1128/JVI.01984-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell Dwyer EJ, Lai H, MacDonald RC, Salvato MS, Borden KL. The lymphocytic choriomeningitis virus RING protein Z associates with eukaryotic initiation factor 4E and selectively represses translation in a RING-dependent manner. J Virol. 2000;74:3293–3300. doi: 10.1128/jvi.74.7.3293-3300.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan L, Briese T, Lipkin WI. Z proteins of New World arenaviruses bind RIG-I and interfere with type I interferon induction. J Virol. 2010;84:1785–1791. doi: 10.1128/JVI.01362-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borden KL, Campbell Dwyer EJ, Salvato MS. An arenavirus RING (zinc-binding) protein binds the oncoprotein promyelocyte leukemia protein (PML) and relocates PML nuclear bodies to the cytoplasm. J Virol. 1998;72:758–766. doi: 10.1128/jvi.72.1.758-766.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Topcu Z, Mack DL, Hromas RA, Borden KL. The promyelocytic leukemia protein PML interacts with the proline-rich homeodomain protein PRH: A RING may link hematopoiesis and growth control. Oncogene. 1999;18:7091–7100. doi: 10.1038/sj.onc.1203201. [DOI] [PubMed] [Google Scholar]
- 19.Capul AA, de la Torre JC, Buchmeier MJ. Conserved residues in Lassa fever virus Z protein modulate viral infectivity at the level of the ribonucleoprotein. J Virol. 2011;85:3172–3178. doi: 10.1128/JVI.02081-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcin D, Kolakofsky D. Tacaribe arenavirus RNA synthesis in vitro is primer dependent and suggests an unusual model for the initiation of genome replication. J Virol. 1992;66:1370–1376. doi: 10.1128/jvi.66.3.1370-1376.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ranjith-Kumar CT, et al. Terminal nucleotidyl transferase activity of recombinant Flaviviridae RNA-dependent RNA polymerases: Implication for viral RNA synthesis. J Virol. 2001;75:8615–8623. doi: 10.1128/JVI.75.18.8615-8623.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graham SC, et al. The N-terminus of the RNA polymerase from infectious pancreatic necrosis virus is the determinant of genome attachment. PLoS Pathog. 2011;7:e1002085. doi: 10.1371/journal.ppat.1002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcin D, Kolakofsky D. A novel mechanism for the initiation of Tacaribe arenavirus genome replication. J Virol. 1990;64:6196–6203. doi: 10.1128/jvi.64.12.6196-6203.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strecker T, et al. Lassa virus Z protein is a matrix protein and sufficient for the release of virus-like particles [corrected] J Virol. 2003;77:10700–10705. doi: 10.1128/JVI.77.19.10700-10705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vezza AC, Gard GP, Compans RW, Bishop DH. Structural components of the arenavirus Pichinde. J Virol. 1977;23:776–786. doi: 10.1128/jvi.23.3.776-786.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neuman BW, et al. Complementarity in the supramolecular design of arenaviruses and retroviruses revealed by electron cryomicroscopy and image analysis. J Virol. 2005;79:3822–3830. doi: 10.1128/JVI.79.6.3822-3830.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee KJ, Novella IS, Teng MN, Oldstone MB, de La Torre JC. NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J Virol. 2000;74:3470–3477. doi: 10.1128/jvi.74.8.3470-3477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.López N, Jácamo R, Franze-Fernández MT. Transcription and RNA replication of tacaribe virus genome and antigenome analogs require N and L proteins: Z protein is an inhibitor of these processes. J Virol. 2001;75:12241–12251. doi: 10.1128/JVI.75.24.12241-12251.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruigrok RW, Crépin T, Kolakofsky D. Nucleoproteins and nucleocapsids of negative-strand RNA viruses. Curr Opin Microbiol. 2011;14:504–510. doi: 10.1016/j.mib.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 30.Kerber R, et al. Cross-species analysis of the replication complex of Old World arenaviruses reveals two sites in nucleoprotein involved in L protein function. J Virol. 2011;85:12518–12528. doi: 10.1128/JVI.05091-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Capul AA, et al. Arenavirus Z-glycoprotein association requires Z myristoylation but not functional RING or late domains. J Virol. 2007;81:9451–9460. doi: 10.1128/JVI.00499-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilda M, Lopez N, Casabona JC, Franze-Fernandez MT. Mapping of the tacaribe arenavirus Z-protein binding sites on the L protein identified both amino acids within the putative polymerase domain and a region at the N terminus of L that are critically involved in binding. J Virol. 2008;82:11454–11460. doi: 10.1128/JVI.01533-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levingston Macleod JM, et al. Identification of two functional domains within the arenavirus nucleoprotein. J Virol. 2011;85:2012–2023. doi: 10.1128/JVI.01875-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shtanko O, et al. A role for the C terminus of Mopeia virus nucleoprotein in its incorporation into Z protein-induced virus-like particles. J Virol. 2010;84:5415–5422. doi: 10.1128/JVI.02417-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vannini A, et al. Molecular basis of RNA polymerase III transcription repression by Maf1. Cell. 2010;143:59–70. doi: 10.1016/j.cell.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 36.Jeruzalmi D, Steitz TA. Structure of T7 RNA polymerase complexed to the transcriptional inhibitor T7 lysozyme. EMBO J. 1998;17:4101–4113. doi: 10.1093/emboj/17.14.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tagami S, et al. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature. 2010;468:978–982. doi: 10.1038/nature09573. [DOI] [PubMed] [Google Scholar]
- 38.Clinton GM, Little SP, Hagen FS, Huang AS. The matrix (M) protein of vesicular stomatitis virus regulates transcription. Cell. 1978;15:1455–1462. doi: 10.1016/0092-8674(78)90069-7. [DOI] [PubMed] [Google Scholar]
- 39.Finke S, Mueller-Waldeck R, Conzelmann KK. Rabies virus matrix protein regulates the balance of virus transcription and replication. J Gen Virol. 2003;84:1613–1621. doi: 10.1099/vir.0.19128-0. [DOI] [PubMed] [Google Scholar]
- 40.Iwasaki M, et al. The matrix protein of measles virus regulates viral RNA synthesis and assembly by interacting with the nucleocapsid protein. J Virol. 2009;83:10374–10383. doi: 10.1128/JVI.01056-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watanabe K, Handa H, Mizumoto K, Nagata K. Mechanism for inhibition of influenza virus RNA polymerase activity by matrix protein. J Virol. 1996;70:241–247. doi: 10.1128/jvi.70.1.241-247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.