Abstract
Dihydrouridine (D) is a highly conserved modified base found in tRNAs from all domains of life. Dihydrouridine synthase (Dus) catalyzes the D formation of tRNA through reduction of uracil base with flavin mononucleotide (FMN) as a cofactor. Here, we report the crystal structures of Thermus thermophilus Dus (TthDus), which is responsible for D formation at positions 20 and 20a, in complex with tRNA and with a short fragment of tRNA (D-loop). Dus interacts extensively with the D-arm and recognizes the elbow region composed of the kissing loop interaction between T- and D-loops in tRNA, pulling U20 into the catalytic center for reduction. Although distortion of the D-loop structure was observed upon binding of Dus to tRNA, the canonical D-loop/T-loop interaction was maintained. These results were consistent with the observation that Dus preferentially recognizes modified rather than unmodified tRNAs, indicating that Dus introduces D20 by monitoring the complete L-shaped structure of tRNAs. In the active site, U20 is stacked on the isoalloxazine ring of FMN, and C5 of the U20 uracil ring is covalently cross linked to the thiol group of Cys93, implying a catalytic mechanism of D20 formation. In addition, the involvement of a cofactor molecule in uracil ring recognition was proposed. Based on a series of mutation analyses, we propose a molecular basis of tRNA recognition and D formation catalyzed by Dus.
Keywords: protein-tRNA complex, RNA modification, substrate recognition, X-ray crystallography
Posttranscriptional modification produces a diverse array of RNA molecules, and has a number of important cellular roles, such as stabilization of the RNA structure, control of translation fidelity, and metabolic response to the environment. More than 100 chemically modified nucleosides have been identified to date, the largest numbers of which are found in tRNAs. Dihydrouridine (D) modification is one of the most ubiquitous modifications of tRNA, and is found in bacteria, eukaryotes, and some archaea (1). Dihydrouridine is formed by reduction of the carbon-carbon double bond at positions 5 and 6 of the uridine base by dihydrouridine synthases (Dus), and is mostly found in the D-loop of tRNAs for which it is named. Individual tRNAs contain varying numbers of dihydrouridines. It was suggested that dihydrouridine promotes structural flexibility in RNA by destabilizing the C3′-endoribose conformation associated with base-stacked RNA (2). The content of dihydrouridine in psychrophilic organisms is significantly higher than in mesophiles and thermophiles (3). In addition, it has long been known that dihydrouridine levels are increased in cancerous tissues (4). Recently, human Dus has been reported to be involved in pulmonary carcinogenesis (5), and suggested to regulate dsRNA-activated protein kinase in cells (6).
The family of Dus has been identified in Saccharomyces cerevisiae and Escherichia coli (7, 8). Biochemical analyses showed that Dus is a flavin-dependent enzyme, which requires NADPH or NADH for its enzymatic activity. The results of genomic analyses showed that a wide number of species possess the gene encoding Dus. Moreover, several homologs of Dus genes have been found in many organisms. The four Dus from S. cerevisiae are responsible for reduction of uridines at specific positions; i.e., Dus1 for uridine at positions 16 and 17, Dus2 for position 20, Dus3 for position 47, and Dus4 for positions 20a and 20b (7). Similarly, the site specificity and nonredundant catalytic functions were also confirmed in three Dus from E. coli (YjbN, YhdG, and YohI) (8). The crystal structure of Dus from Thermotoga maritima has been reported (9), and mutation analysis of Dus from E. coli revealed important residues for dihydrouridine formation (10). One of the most remarkable biochemical features of Dus is that other modifications of tRNA are required for the enzymatic activity (11). However, the details of the reaction, including the tRNA recognition mechanism and its catalysis of dihydrouridine formation, are still unknown.
In the present study, we investigated the tRNA recognition and catalytic mechanisms of Dus from a structural viewpoint. Based on the crystal structures of TthDus in complex with tRNA and tRNA fragment, and mutation analyses, we propose a tRNA recognition mechanism of Dus to discriminate modified tRNA, and a unique substrate recognition mechanism in which a cofactor molecule is used. Furthermore, the roles of each important residue in the reaction are presented.
Results
Crystal Structure of TthDus.
First, we determined the crystal structure of TthDus by the Se-SAD (single-wavelength anomalous dispersion) method (see Materials and Methods) at a resolution of 1.7 Å. Analogous to the Dus from T. maritima (9), the structure of TthDus consists of two domains; i.e., an N-terminal α/β barrel domain and a helical domain (Fig. S1A). An extension of 24 residues at the C terminus in TthDus was disordered. In addition, the region of Ala171-Ile180 located above the active site (see below) was disordered. The flavin mononucleotide (FMN) cofactor was captured at the bottom of the pocket located at the center of the N-terminal domain, indicating that this pocket is an active site. The surface including the active site formed a large positively charged groove (Fig. S1B).
Crystal Structure of TthDus-tRNA Complex.
We recently found that TthDus expressed in E. coli forms a stable complex with tRNA in cells that cannot be dissociated even on SDS-PAGE (12). Rider, et al. also reported similar results (11). In the present study, TthDus and Tth-tRNAPhe were coexpressed in E. coli and their complex was purified for structure analysis. Analysis of the extracted tRNA by liquid chromatography/mass spectrometry (LC/MS) showed that Tth-tRNAPhe was contained in the Dus-tRNA complex, although tRNAIle1, tRNAArg2, and tRNAHis from E. coli were also detected (Fig. S2). These observations suggested that TthDus forms a complex with a variety of tRNAs regardless of the source.
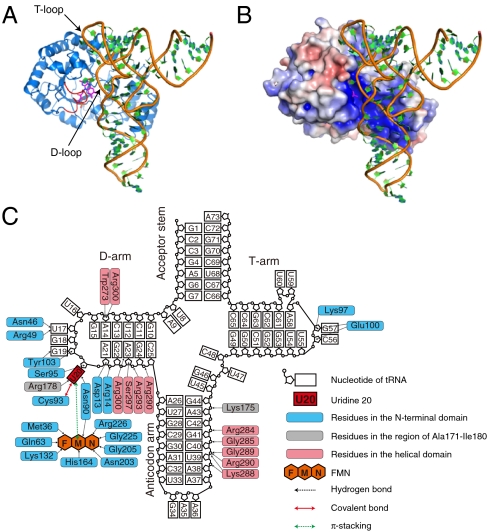
The crystal structure of TthDus and Tth-tRNAPhe complex was determined by the molecular replacement method in combination with single-wavelength anomalous dispersion method (MRSAD) at a resolution of 3.51 Å. The tRNA was captured in the positively charged groove, in which the D-, T-, and anticodon-arms are accommodated (Fig. 1 A and B). The D-arm containing substrate nucleotide was recognized by both the N-terminal domain and the helical domain (Fig. 1C). The D-loop lay on the active site pocket, and bound with Asn46, Arg49, Asn90, Cys93, Ser95, Tyr103, and Arg178 in the N-terminal domain of TthDus and FMN. U20, the substrate nucleotide, flipped out from the D-loop and intruded deeply into the active site, where the base of U20 was directly recognized by Cys93, Arg178, Asn90, and FMN (Fig. 1 A and C). Interestingly, the region of Ala171-Ile180, which was disordered in apo TthDus, formed a short helix and contributed to U20 recognition by hydrogen bonding using Arg178 (Fig. 1 A and C). In addition, the ribose moiety of U20 was recognized by Ser95. The base and ribose moieties of G19 were recognized by Tyr103. In addition, the base of U17 was recognized by Arg49 and Asn46 through hydrogen bonding. On the other hand, the D-stem was mainly recognized by residues in the helical domain, although Asp13 and Arg14 in the N-terminal domain also contributed. The backbone structure of the T-loop was recognized by two residues in the N-terminal domain. The anticodon-arm of the tRNA was bound with residues from the helical domain and the region of Ala171-Ile180.
Fig. 1.
Overall structure of TthDus-tRNA complex (A) Ribbon diagram. The region of Ala171-Ile180, which was disordered in the RNA-free TthDus, is shown in red. FMN is shown as sticks. (B) Electrostatic surface potential of TthDus. Positively and negatively charged surfaces are colored blue and red, respectively (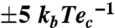 ). (C) Overall interaction scheme among TthDus, FMN, and tRNA. Hydrogen bonds, π-stacking interaction, and covalent bond between TthDus and tRNA are shown as black, green, and red arrows, respectively.
). (C) Overall interaction scheme among TthDus, FMN, and tRNA. Hydrogen bonds, π-stacking interaction, and covalent bond between TthDus and tRNA are shown as black, green, and red arrows, respectively.
Conformation Changes upon Complex Formation.
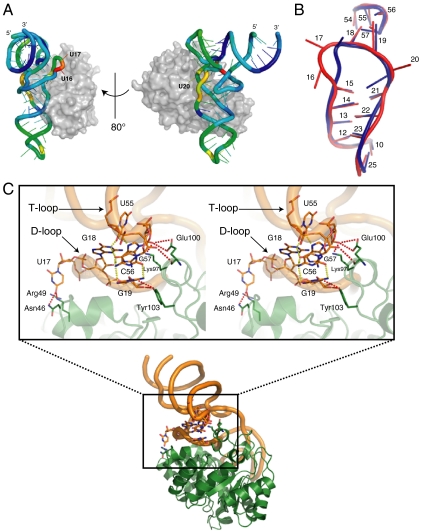
Structure comparison of the bound tRNA with yeast tRNAPhe (PDB: 1ehz) (13) showed remarkable conformational changes in U16 and U17 in addition to the target U20 (Fig. 2 A and B). In both nucleotides, the backbone structure was distorted, and the bases turned toward different directions (Fig. 2B). U17 was recognized in a nucleotide-specific manner by Arg49 and Asn46 (Fig. 2C), whereas U16 was not (Fig. 1C). Despite the marked conformational changes on both sides, G18 and G19 still formed base pairs with U55 and C56 of the T-loop, respectively (Fig. 2 B and C). In addition, G18 formed polar interactions with G57, A58, and U60, and contributed to stabilization of the elbow region. The base and ribose moieties of G19 were recognized by Tyr103. Although G18 was not directly recognized by TthDus, the backbone of C56 and G57, which interacted with G18, was recognized by Lys97 and Glu100. Therefore, G18 was recognized by TthDus through the interaction between T- and D-loops. In contrast, no significant conformational change occurred in other regions bound by TthDus; i.e., the D-stem, T-loop, and anticodon-stem (Fig. 2A). These observations indicated that the distortion of U16 and U17 is independent of recognition in other regions, and is therefore caused by the flipping out of U20 to intrude into the active site.
Fig. 2.
T- and D-loop recognition mechanisms (A) Conformational differences between the bound Tth-tRNAPhe and yeast tRNAPhe. The cartoon model is colored according to the rmsd between two tRNA molecules from blue (0.59 Å) to red (6.49 Å). All atoms in the ribose and phosphate moieties were used for calculation. TthDus is shown as a white surface. (B) Superposition of the T-arms and D-loops of Tth-tRNAPhe (red) and yeast tRNAPhe (blue). The numbers represent the tRNA base positions. (C) Recognition of the elbow region. Polar interactions between TthDus residues and tRNA nucleotides (red dashed line) and those within tRNA nucleotides (yellow dashed line) are shown as stereo views. The cartoon models of TthDus and tRNA are also shown in orange and green, respectively.
On the other hand, the structures of TthDus and its tRNA complex were well superposed with a root mean square deviation (rmsd) of 0.57 Å for 304 Cα atoms, showing that the conformation did not change upon complexation except in the region of Ala171-Ile180.
Structure Around the Active Site.
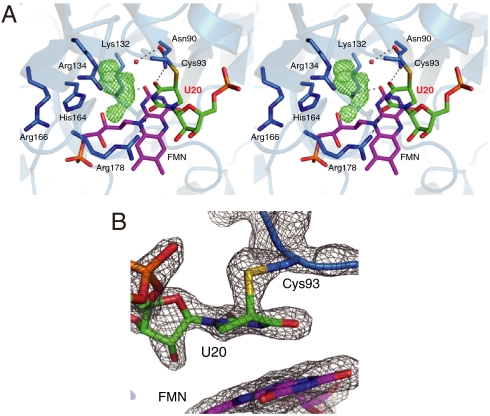
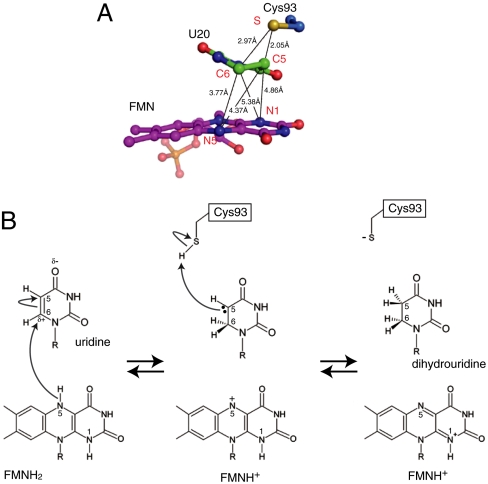
To identify the active site structure of the tRNA complex in detail, we determined the crystal structure of TthDus in complex with a short tRNA fragment at a resolution of 1.95 Å. The structure of the RNA fragment from G18 to A21 was constructed (Fig. S3A). The target uridine was located between Cys93 and FMN cofactor with the uracil ring parallel to the isoalloxazine ring of FMN (Fig. 3). O4 and O2 of U20 formed hydrogen bonds with Nδ2 of Asn90 and Nη2 of Arg178, respectively. The relative locations of the uracil ring, FMN, and coordinating residues were similar to those of dihydroorotate dehydrogenase (DHOD) (14) and dihydropyrimidine dehydrogenase (DHPDH) (15), which also catalyze pyrimidine ring redox reaction (see Discussion) (Fig. S3 C and D).
Fig. 3.
Structure of TthDus-tRNA fragment complex. (A) Close-up view of the U20 binding site (stereo views). Electron density (Fo-Fc map) of the unknown cofactor molecule is also shown as green mesh (contoured at 3σ). Dotted lines represent hydrogen bonds. Water molecule is shown as a red ball. (B) Covalent bond between Cys93 and U20. The 2Fo-Fc map is also shown (contoured at 2σ).
There is a space between U20 and the inner wall of the active site. A flat obvious electron density filled this space. Interestingly, this unknown molecule interacted with U20 in a manner similar to base pairing, and was surrounded by Lys132, Arg134, and His164 (Fig. 3A). Arg166 was located behind Arg134 and His164. All four of these residues are completely conserved in Dus family proteins (Fig. S4A), and were suggested to be important for the enzymatic activity (10). These residues would contribute to the reaction by recognizing U20 indirectly via this molecule. No molecules in the crystallizing buffer could be fitted to this density, suggesting that this molecule was captured in E. coli cells during cultivation. These observations suggest an important role of this unknown molecule as a cofactor for target base recognition.
Surprisingly, the electron density of Cys93 was connected to the C5 atom of the U20 (Fig. 3B). In addition, the coplanarity of the uridine base collapsed at the C5 and C6 atoms (Fig. 3B). These observations indicated that the C5 atom formed a covalent bond with the sulfur atom of Cys93, and consequently the double bond between C5 and C6 atoms of the uridine base was transformed into a single bond. This conclusion was supported by the results of time-of-flight mass spectrometry (TOF-MS) analysis indicating that the molecular mass of the purified TthDus-tRNA fragment complex was 42130 Da, whereas that of apo TthDus was 39559 Da (Fig. S3B).
Identifying Critical Residues for tRNA Complex Formation.
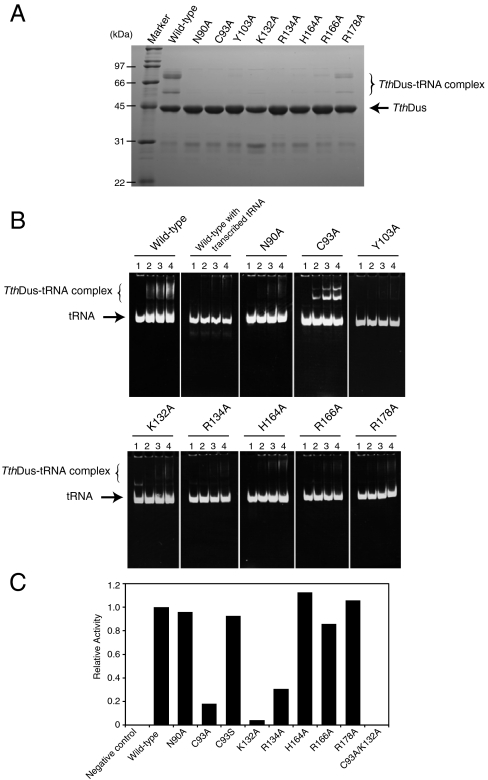
To identify the residues essential for tRNA recognition, 24 mutants were prepared and their tRNA binding activities were investigated. As Dus does not bind to transcribed tRNA (Fig. 4B), we first evaluated the interaction of each mutant with tRNA based on the presence of the covalent complex on SDS-PAGE (Fig. 4A, Fig. S5). Surprisingly, substitution of residues surrounding the unknown cofactor; i.e., K132A, R134A, H164A, and R166A, resulted in complete inactivation, although they had no direct interaction with U20. In addition, N90A, C93A, and Y103A were incapable of complex formation. In contrast, R178A could form a covalent complex, although it was located in the active site and interacted with U20 directly. These results indicated the significant contribution of the cofactor to complex formation. Tyr103 interacted with the D-loop of tRNA, suggesting the importance of recognizing the elbow region for stable complex formation.
Fig. 4.
Mutation analyses (A) Evaluation of the covalent complex formation of TthDus mutants by SDS-PAGE. Results for the mutants in the active site are shown. The results for the Y103A mutant, which cannot form a covalent complex, are also shown. Other mutants are shown in Fig. S5. (B) Native PAGE of the TthDus mutants incubated with tRNAArg extracted from E. coli dus-deficient strain. The results for wild type with transcribed tRNAArg are also shown. Lane 1: 20 pmol tRNA, lane 2; 20 pmol tRNA + 20 pmol TthDus mutant, lane 3; 20 pmol tRNA + 40 pmol TthDus mutant, lane 4; 20 pmol tRNA + 60 pmol TthDus mutant. (C) Enzymatic activity of each EcYjbN mutant. The amounts of dihydrouridine introduced into E. coli tRNAArg2 and tRNAArg3 are shown relative to the wild type.
Next, to explore the contribution of the unknown cofactor, mutants surrounding the active site were prepared and their affinities were evaluated by native PAGE, in which E. coli tRNAArg extracted from a dus-deficient strain of E. coli was used (Fig. 4B). In addition, Y103A mutant was evaluated. Complex formation was observed in the wild type. In contrast, all mutants other than C93A showed markedly decreased binding activity (Fig. 4B). These results indicated the significant contribution of the cofactor to complex formation. In addition, N90A and Y103A were incapable of forming a complex as demonstrated on both SDS-PAGE and native PAGE, suggesting that the direct interactions of Asn90 and Tyr103 are also important. The absorption spectrum of the purified mutant protein showed that K132A did not possess FMN (Fig. S6A). As the uracil ring of U20 showed π-stacking interaction with FMN in the complex structure (Figs. 1C and 3), it is likely that FMN contributes to complex formation as well as residues surrounding the active site.
Effects of Mutations on Dus Activity.
To discuss the roles of residues located around the active site in the enzymatic activity, the effects of mutations in these residues were evaluated. We used Dus from E. coli (EcYjbN) for mutation analysis, as all tRNA modifications in E. coli have been determined. The amount of the introduced D modification in tRNA extracted from the dus-deficient strain of E. coli (E. coli ΔyjbN strain) complemented with the mutant gene was analyzed by LC/MS. E. coli ΔyjbN strain complemented with the empty vector (negative control) could introduce D modification at neither position 20 nor 20A, whereas adequate D modification was confirmed in the positive control strain complemented with the wild-type yjbN (Fig. 4C).
Conserved residues located around the active site were substituted with Ala, and the dihydrouridine activities were compared between mutants and wild type (residue numbers of TthDus were used for E. coli mutants for clarity; the correspondences between TthDus and EcYjbN are shown in Fig. S4B). Fig. 4C shows the relative dihydrouridine formation activities of the mutants. K132A, C93A, and R134A showed significantly decreased activity. As purified K132A mutant showed no absorption in the visible range, its marked inactivation could be explained by the absence of the FMN (Fig. S6B). Although weak residual activity was still observed, this was likely due to the residual FMN binding activity of K132A, which was confirmed by thermodynamic analysis of K132A (Fig. S6C). The activity of C93A was also significantly decreased, although it was not completely inhibited. However, substitution with Ser recovered the activity to a level comparable to that of the wild type. Thus, serine and cysteine may be compatible in Dus. These results demonstrated the importance of the hydrogen atom at Cys93 as well as FMN. The activity of the R134A mutant was decreased to 31% of the wild type, although Arg134 did not interact directly with the target nucleotide. Arg134 is located around the unknown cofactor and is one of the completely conserved residues. The decrease in activity may reflect the contribution of the unknown cofactor to the enzymatic activity as well as tRNA recognition. Although the single mutants described above demonstrated the importance of Lys132 and Cys93, each mutant still possessed weak enzymatic activity. A C93A/K132A double mutant, however, showed complete loss of activity.
Discussion
tRNA Recognition Mechanism of TthDus.
In the structure of TthDus-tRNA complex, the D-arm, T-loop, and anticodon-stem were bound by TthDus. The conformation of the D-loop was significantly distorted upon TthDus binding (Fig. 2B), which is probably induced by the intrusion of U20 into the active site. Despite the significant conformational change at U16-U17 and U20, interactions between D- and T-loops by G18-G19 were maintained, suggesting that the structure of the elbow region was highly stabilized. The backbone structure of the elbow region was recognized in both T- and D- loops (Fig. 2C). Moreover, substitution of Tyr103, which recognized G19 stabilizing D-loop/T-loop interaction, resulted in complete incapability of complex formation (Fig. 4 A and B). It should be noted that substitution of nucleotides involved in D-loop/T-loop interaction completely blocks D modification (16). These observations strongly suggest that Dus recognizes the elbow region stabilized by D-loop/T-loop interactions.
The structure of tRNA, especially in D- and T-loops, is stabilized by modifications (17). Moreover, G19-C56 base pairing is prone to disruption in the transcripts as compared to modified tRNAs (18). Several previous studies indicated that Dus targets tRNA containing modifications (11, 16). In this study, we showed that Dus can bind with tRNA that already has modifications other than D, but cannot recognize transcribed tRNA despite having the identical sequence (Fig. 4B). These observations indicated that the poor enzymatic activity for transcripts is due to the inability of complex formation. Dus should discriminate the modified tRNA in the binding process, in which the canonical structure of the D-loop/T-loop highly stabilized by the modification would be recognized. Archaeal Trm5, which targets modified tRNA, also binds tRNA in a similar manner using the elbow region as a “hallmark” (19). As both Dus and Trm5 are promiscuous enzymes that modify multiple tRNAs, this would be a general tRNA binding mechanism to distinguish correctly modified tRNA from immature tRNAs. It is interesting to note that the proposed manner of tRNA binding is the opposite of the mechanism adopted by ArcTGT, which prefers the immature λ-form tRNA through recognition of the flexible D-loop (20).
The levels of sequence conservation of residues that interact with tRNA except U20 were remarkably low as compared to those interacting with U20. Furthermore, no conformation changes were observed in TthDus upon complex formation except for the Ala171-Ile180 region. The tRNA was captured in the positively charged groove (Fig. 1B). These observations suggest that the interaction occurs in a space-filling manner in combination with charge complementarity. The modifications in tRNA were reported to induce a conformation change of the overall L-shape structure of tRNA (21). TthDus formed a significant number of interactions with backbone structures of anticodon- and D-arms (Fig. 1C). Dus may recognize the distorted backbone structure of modified tRNA as well as the rigid D-loop/T-loop.
The high-resolution structure of TthDus-tRNA fragment complex showed the involvement of a cofactor in U20 recognition as well as FMN and directly coordinating residues, such as Asn90 (Fig. 3A). Substitution of the residues surrounding the unknown cofactor; i.e., K132A, R134A, H164A, and R166A, blocked tRNA complex formation (Fig. 4 A and B). However, these residues do not interact with U20 directly. In addition, a significant decrease in the enzymatic activity was observed in the R134A mutant (Fig. 4C). These residues are all completely conserved among Dus family proteins (Fig. S4A), and they have all been suggested to be important for the activity under conditions in which the expression of Dus is highly suppressed (10). These results strongly imply an important role of the unknown cofactor in U20 recognition. The role of the unknown cofactor can also be explained from the viewpoint of the structure. The relative orientations of the pyrimidine ring, FMN, and Cys93 are similar to those of related enzymes, DHOD and DHPDH, suggesting that the location of U20 in the structure is appropriate (Figs. S3 C and D). However, the inner wall of the active site of TthDus is obviously far from the substrate. Although the substrates are directly coordinated by residues in the other two enzymes, this cannot be achieved in TthDus without the cofactor molecule. It should be noted that a sulfate ion was captured at the same position in the crystal structure of Dus from T. maritima (9). These structural features also emphasize the necessity of the cofactor, which is buried in the space between U20 and the completely conserved residues.
Taken together, these observations suggest that Dus recognizes the properly stabilized elbow region composed of D-loop/T-loop interaction and the distorted backbone structure of the modified tRNA, which enables Dus to discriminate between mature and immature tRNAs. At the active site, the target uridine is recognized by completely conserved residues indirectly through the unknown cofactor molecule in addition to the direct interactions by Asn90 and FMN.
Proposed Reaction Mechanism.
Through the reaction, two hydrogen atoms in the Dus-FMN complex are transferred to C5 and C6 of the uracil ring of the target nucleotide. The arrangement of FMN and the pyrimidine ring in the TthDus-substrate complex was similar to those of DHOD and DHPDH, which catalyze similar redox reaction of pyrimidine rings. In DHOD and DHPDH, cysteine acts as a key general-acid/base catalyst. Cys93, which is completely conserved in Dus family proteins (Fig. S4A), was located at the corresponding position in TthDus. In mutation analysis of EcYjbN, alanine substitution of Cys93 caused a significant decrease in the activity, which was restored by substitution with serine. Serine was reported to be used as the catalytic residue in DHOD family 2 (22). These results showed that Cys93 is an active residue of Dus. In addition, the importance of FMN was confirmed by the marked inactivation and lack of FMN binding of K132A (Fig. 4, Fig. S6). Taken together, these results indicated that FMN and Cys93 provide two hydrogens.
The reduced form of FMN (FMNH2) can provide hydrides from two nitrogen atoms; i.e., N5 and N1. The distance between C6 of U20 and N5 in FMN was 3.77 Å, which was shorter than the distance of 4.37 Å between C5 and N5 (Fig. 5A). In addition, N1 of FMN was located 4.86 Å and 5.38 Å from C5 and C6 of the pyrimidine ring, respectively, and these distances are too great to provide hydrogen. Therefore, it was concluded that the hydride is supplied from N5 of FMNH2 to C6 of the pyrimidine ring. This conclusion also means that the distal hydrogen atom of Cys93 is transferred to C5. The distance between C5 of the pyrimidine ring and the sulfur atom of Cys93 is 2.05 Å, sufficiently close for hydrogen transfer. By supplying the hydride to C6 from FMNH2, C5 will show nucleophilicity with the resultant electron pair. The distal hydrogen of Cys93 would be attacked by the nucleophilic electron pair of C5. As C5 does not have nucleophilicity before the hydride is provided, the reaction must start from the transfer of the hydride ion from FMNH2 to C6. After transferring two hydrogen atoms, the structures of both FMN and pyrimidine ring should differ from those before the reaction; i.e., the FMN and uridine adopt planar and distorted structures, respectively. These conformational changes would cause the release of product from the active site. This mechanism may be used in the step of complex formation to exclude tRNA, which already has D modification. Taken together, we propose the following reaction mechanism based on the crystal structure and the results of mutation analysis (Fig. 5B): (i) a hydride is transferred from N5 of FMNH2 to C6 of the pyrimidine ring, (ii) an electron pair is consequently transferred to C5 of the pyrimidine ring, (iii) followed by nucleophilic attack by C5 to the distal hydrogen atom of Cys93, and (iv) release of the product.
Fig. 5.
Proposed reaction mechanism of Dus (A) Distances between Cys93, target U20, and FMN in the structure of TthDus-tRNA fragment complex. (B) Schematic representation of proposed mechanism for uridine reduction of Dus. Hydride attacks C6 of uridine with a slight positive charge (left). The generated electron pair at C5 attacks the distal hydrogen of Cys93 (center). Dihydrouridine is generated (right).
The final issue to be discussed is the covalent bond formed between C5 of the uridine base and the sulfur atom in Cys93 (Fig. 3B). The reasonable configuration of functional molecules; i.e., the target uridine intrudes deeply into the active site with C5 and C6 atoms adjacent to FMN and Cys93, and its similarity with the relevant enzymes, shows that the revealed structure can be considered as the structural analog of the reaction intermediate. Formation of the covalent bond with the sulfur atom suggests that a leaving group has been attached to the sulfur atom and there has been detached coupling with nucleophilic attack of the sulfur atom by the electron pair formed at C5. The substitution of the distal hydrogen atom of Cys93 into the leaving group may be caused by a nonoptimal environment for the enzyme, such as temperature and redox conditions due to the heterologous overexpression. Indeed, when EcYjbN was overexpressed in E. coli, this covalent complex was not observed (Fig. S7). Due to the covalent complex formation, we could obtain crystals and as a consequence could discuss the tRNA recognition and reaction mechanism of Dus.
Materials and Methods
Preparation of TthDus, TthDus-tRNA Complex, and TthDus-tRNA Fragment Complex.
The methods used to prepare TthDus and TthDus-tRNA complex were reported previously (12). TthDus in complex with tRNA fragment was prepared by treating TthDus-tRNA complex with RNaseA (Sigma-Aldrich) followed by purification by Ni affinity chromatography and size exclusion chromatography. Full details of the purification procedures are presented in the SI Text.
Crystallization and Data Collection.
Crystallization and data collection of SeMet-labeled TthDus, native TthDus, and TthDus-tRNA complex were reported previously (12). The crystals of TthDus-tRNA fragment complex were obtained from buffer containing 0.1 M Hepes (pH 7.0) and 18% (wt/vol) PEG 12000. The X-ray diffraction dataset was collected at beamline BL-5A of Photon Factory.
Structure Determination and Refinement.
The structure of TthDus was determined at a resolution of 1.70 Å by the Se-SAD method. The crystal structure of TthDus in complex with Tth-tRNAPhe was determined at a resolution of 3.51 Å by the MRSAD method using the structure of TthDus as a search model and Se atoms as anomalous scatterers. The crystal structure of the TthDus-tRNA fragment complex was determined at 1.95 Å resolution by the molecular replacement method using the structure of TthDus as a search model. The statistics for data collection and refinement are shown in Table S1. The details of the structure determination procedures are presented in the SI Text.
Confirmation of Covalent Complex Formation for TthDus Mutants by SDS-PAGE.
TthDus mutants and Tth-tRNAPhe were coexpressed in E. coli strain B834(DE3). The expressed Dus mutants were purified from the soluble fraction after heat treatment at 70 °C for 20 min by Ni Sepharose 6 Fast Flow (GE Healthcare). The concentration of TthDus mutant was determined using a Quant-iT Protein Assay Kit (Invitrogen), and adjusted to 0.13 mg mL-1. Aliquots of 6 μL of the samples were subjected to SDS-PAGE. All vectors for mutation analyses were prepared with a KOD-Plus Mutagenesis Kit (Toyobo).
Gel Shift Assay.
Gel shift assay was performed as described previously (23).
Evaluation of Dihydrouridine Formation in tRNA by E. coli yjbN.
The wild-type and mutant yjbN-pMW118 (Nippon Gene) vector was used to complement the E.coli yjbN knockout strain (E. coli ΔyjbN). The extracted total tRNA from cells was digested with RNase T1, and then analyzed by capillary liquid chromatography nano-electrospray ionization/mass spectrometry (24). Among the fragments obtained from total tRNA by RNase T1 treatment, the ADAGp fragment derived from D-loops of tRNAArg2 and tRNAArg3 were used to estimate the amount of introduced dihydrouridine. The fraction of introduced dihydrouridine was estimated from the relative amounts of ADAGp and AUAGp fragments. The rates of the individual mutants were compared with that of the wild-type enzyme. The details are described in the SI Text.
Supplementary Material
Acknowledgments.
We thank Dr. Kenjyo Miyauchi for his support in preparation of tRNAArg2. This work was supported by Grants-in-Aid from the Ministry of Education, Science, Sports, and Culture of Japan (Y.T., I.T., and M.Y.). F.Y. was supported by the International Graduate Program for Research Pioneers in Life Sciences (IGP-RPLS).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates of TthDus, TthDus-tRNA fragment complex, and TthDus-tRNA complex have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3B0P, 3B0U, and 3B0V).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112352108/-/DCSupplemental.
References
- 1.Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dalluge JJ, Hashizume T, Sopchik AE, McCloskey JA, Davis DR. Conformational flexibility in RNA: the role of dihydrouridine. Nucleic Acids Res. 1996;24:1073–1079. doi: 10.1093/nar/24.6.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dalluge JJ, et al. Posttranscriptional modification of tRNA in psychrophilic bacteria. J Bacteriol. 1997;179:1918–1923. doi: 10.1128/jb.179.6.1918-1923.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuchino Y, Borek E. Tumor-specific phenylalanine tRNA contains two supernumerary methylated bases. Nature. 1978;271:126–129. doi: 10.1038/271126a0. [DOI] [PubMed] [Google Scholar]
- 5.Kato T, et al. A novel human tRNA-dihydrouridine synthase involved in pulmonary carcinogenesis. Cancer Res. 2005;65:5638–5646. doi: 10.1158/0008-5472.CAN-05-0600. [DOI] [PubMed] [Google Scholar]
- 6.Mittelstadt M, et al. Interaction of human tRNA-dihydrouridine synthase-2 with interferon-induced protein kinase PKR. Nucleic Acids Res. 2008;36:998–1008. doi: 10.1093/nar/gkm1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xing F, Hiley SL, Hughes TR, Phizicky EM. The specificities of four yeast dihydrouridine synthases for cytoplasmic tRNAs. J Biol Chem. 2004;279:17850–17860. doi: 10.1074/jbc.M401221200. [DOI] [PubMed] [Google Scholar]
- 8.Bishop AC, Xu J, Johnson RC, Schimmel P, de Crécy-Lagard V. Identification of the tRNA-dihydrouridine synthase family. J Biol Chem. 2002;277:25090–25095. doi: 10.1074/jbc.M203208200. [DOI] [PubMed] [Google Scholar]
- 9.Park F, et al. The 1.59 Å resolution crystal structure of TM0096, a flavin mononucleotide binding protein from Thermotoga maritima. Proteins. 2004;55:772–774. doi: 10.1002/prot.20086. [DOI] [PubMed] [Google Scholar]
- 10.Savage DF, de Crécy-Lagard V, Bishop AC. Molecular determinants of dihydrouridine synthase activity. FEBS Lett. 2006;580:5198–5202. doi: 10.1016/j.febslet.2006.08.062. [DOI] [PubMed] [Google Scholar]
- 11.Rider LW, Ottosen MB, Gattis SG, Palfey BA. Mechanism of dihydrouridine synthase 2 from yeast and the importance of modifications for efficient tRNA reduction. J Biol Chem. 2009;284:10324–10333. doi: 10.1074/jbc.M806137200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu F, et al. Crystallization and preliminary X-ray crystallographic analysis of dihydrouridine synthase from Thermus thermophilus and its complex with tRNA. Acta Crystallogr F. 2011;67:685–688. doi: 10.1107/S1744309111012486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi H, Moore PB. The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: a classic structure revisited. RNA. 2000;6:1091–1105. doi: 10.1017/s1355838200000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rowland P, Björnberg O, Nielsen FS, Jensen KF, Larsen S. The crystal structure of Lactococcus lactis dihydroorotate dehydrogenase A complexed with the enzyme reaction product throws light on its enzymatic function. Protein Sci. 1998;7:1269–1279. doi: 10.1002/pro.5560070601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobritzsch D, Schneider G, Schnackerz KD, Lindqvist Y. Crystal structure of dihydropyrimidine dehydrogenase, a major determinant of the pharmacokinetics of the anti-cancer drug 5-fluorouracil. EMBO J. 2001;20:650–660. doi: 10.1093/emboj/20.4.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cavaille J, Chetouani F, Bachellerie JP. The yeast Saccharomyces cerevisiae YDL112w ORF encodes the putative 2′-O-ribose methyltransferase catalyzing the formation of Gm18 in tRNAs. RNA. 1999;5:66–81. doi: 10.1017/s1355838299981475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derrick WB, Horowitz J. Probing structural differences between native and in vitro transcribed Escherichia coli valine transfer RNA: evidence for stable base modification-dependent conformers. Nucleic Acids Res. 1993;21:4948–4953. doi: 10.1093/nar/21.21.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perret V, et al. Conformation in solution of yeast tRNA(Asp) transcripts deprived of modified nucleotides. Biochimie. 1990;72:735–743. doi: 10.1016/0300-9084(90)90158-d. [DOI] [PubMed] [Google Scholar]
- 19.Goto-Ito S, Ito T, Kuratani M, Bessho Y, Yokoyama S. Tertiary structure checkpoint at anticodon loop modification in tRNA functional maturation. Nat Struct Mol Biol. 2009;16:1109–1115. doi: 10.1038/nsmb.1653. [DOI] [PubMed] [Google Scholar]
- 20.Ishitani R, et al. Alternative tertiary structure of tRNA for recognition by a posttranscriptional modification enzyme. Cell. 2003;113:383–394. doi: 10.1016/s0092-8674(03)00280-0. [DOI] [PubMed] [Google Scholar]
- 21.Byrne RT, Konevega AL, Rodnina MV, Antson AA. The crystal structure of unmodified tRNAPhe from Escherichia coli. Nucleic Acids Res. 2010;38:4154–4162. doi: 10.1093/nar/gkq133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Björnberg O, Jordan DB, Palfey BA, Jensen KF. Dihydrooxonate is a substrate of dihydroorotate dehydrogenase (DHOD) providing evidence for involvement of cysteine and serine residues in base catalysis. Arch Biochem Biophys. 2001;391:286–294. doi: 10.1006/abbi.2001.2409. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura A, Yao M, Chimnaronk S, Sakai N, Tanaka I. Ammonia channel couples glutaminase with transamidase reactions in GatCAB. Science. 2006;312:1954–1958. doi: 10.1126/science.1127156. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki T, Ikeuchi Y, Noma A, Suzuki T, Sakaguchi Y. Mass spectrometric identification and characterization of RNA-modifying enzymes. Methods Enzymol. 2007;425:212–229. doi: 10.1016/S0076-6879(07)25009-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.