Abstract
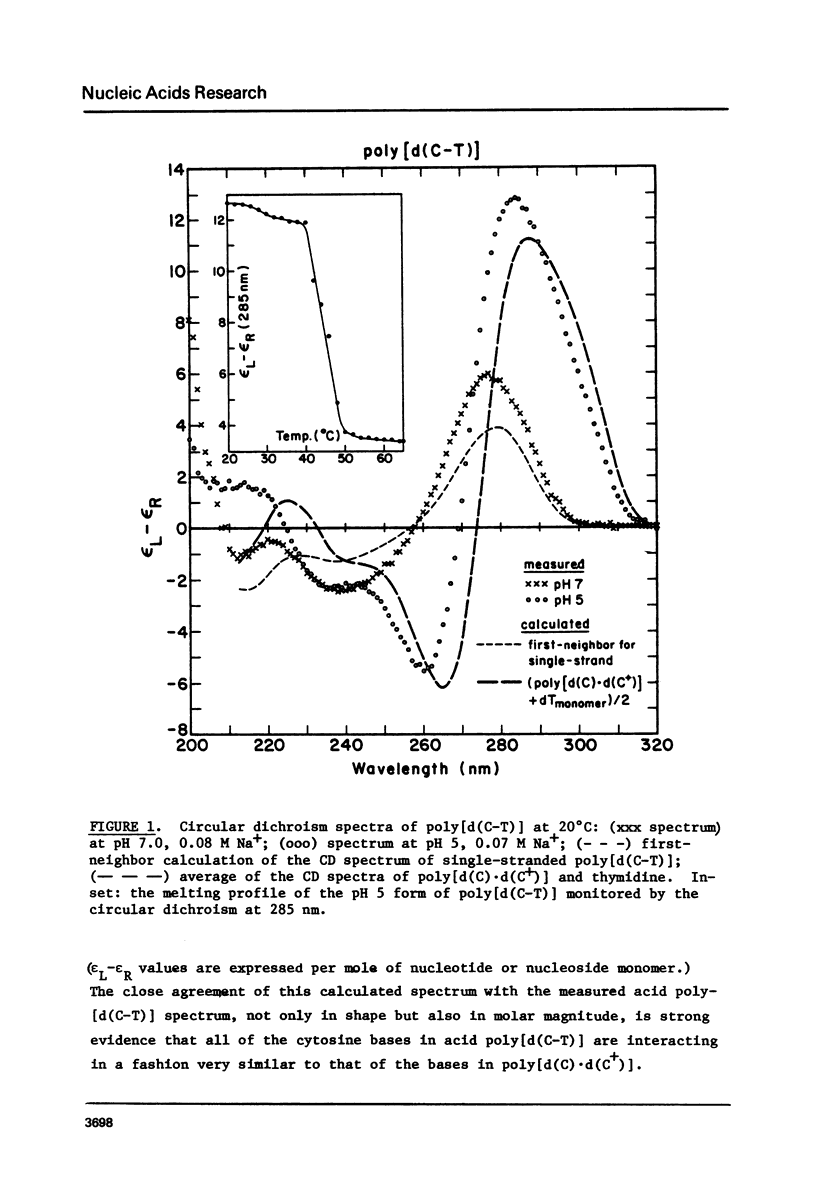
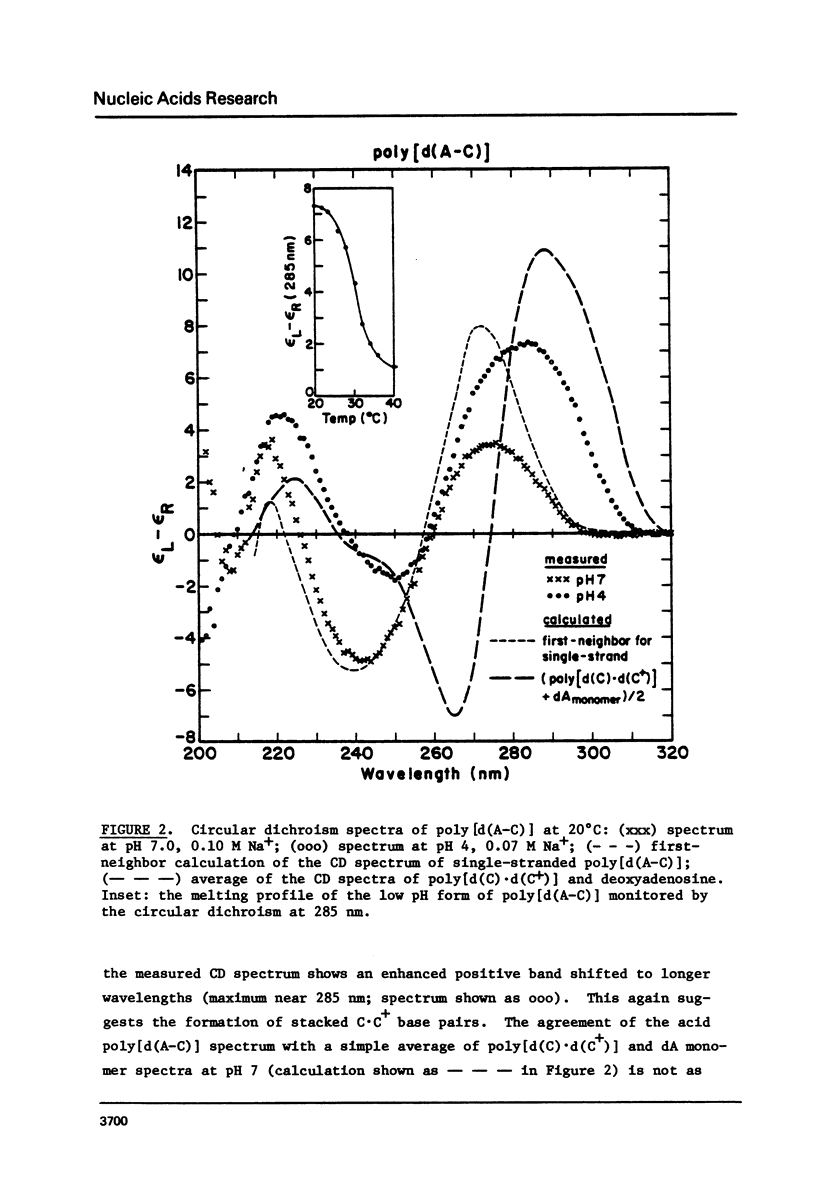
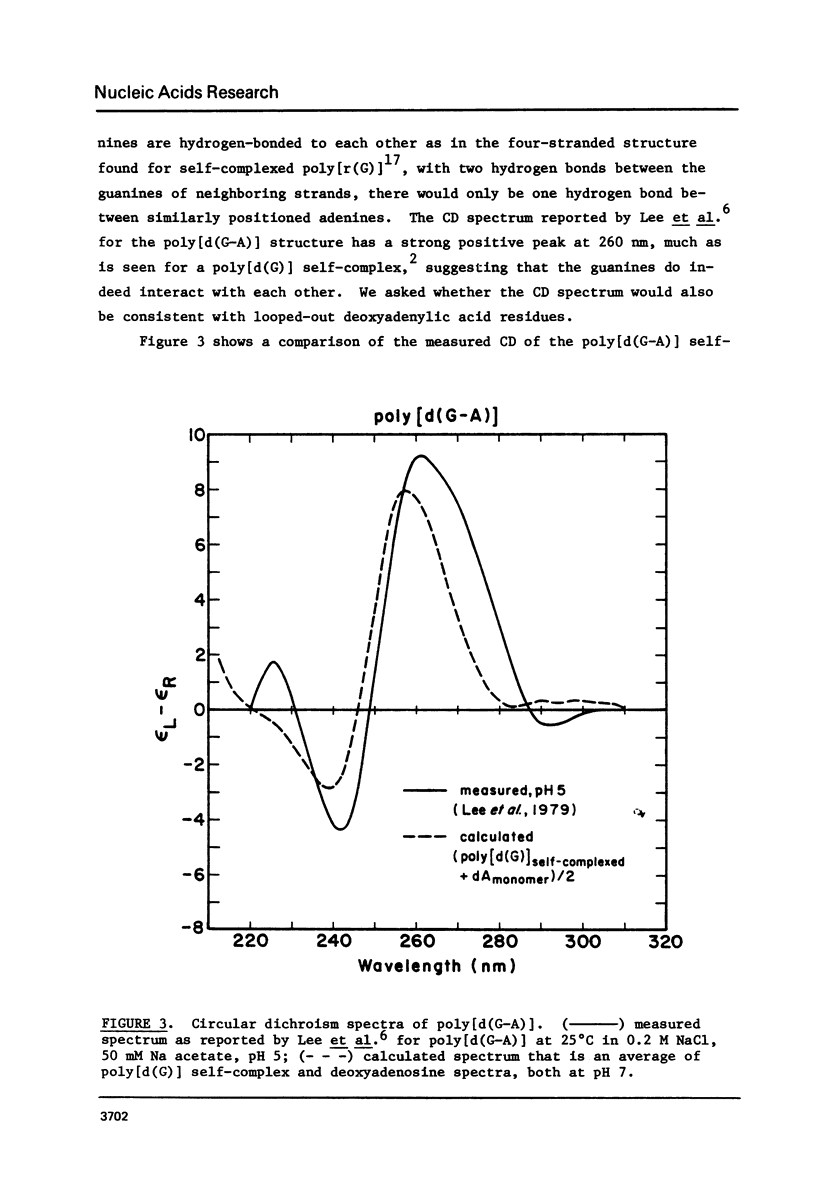
We have found that the CD spectrum of poly [d(C-T)] undergoes a marked change upon protonation, taking on the unmistakable features of the spectrum of poly[d(C) x d(C+)]. The acid poly[d9C-T)] structure appears to be one in which each of the T residues is individually looped out of a helix formed by stacked C x C+ base pairs. We also present data for poly[d(C-A)] and review data for the other four possible repeating DNA dinucleotide sequences. These data show that self-complexes in which up to 50% of the residues are extrahelical may be unexceptional.
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler A. J., Grossman L., Fasman G. D. Polyriboadenylic and polydeoxyriboadenylic acids. Optical rotatory studies of pH-dependent conformations and their relative stability. Biochemistry. 1969 Sep;8(9):3846–3859. doi: 10.1021/bi00837a051. [DOI] [PubMed] [Google Scholar]
- Arnott S., Chandrasekaran R., Birdsall D. L., Leslie A. G., Ratliff R. L. Left-handed DNA helices. Nature. 1980 Feb 21;283(5749):743–745. doi: 10.1038/283743a0. [DOI] [PubMed] [Google Scholar]
- Cantor C. R., Warshaw M. M., Shapiro H. Oligonucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9(9):1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- Early T. A., Olmsted J., 3rd, Kearns D. R., Lezius A. G. Base pairing structure in the poly d(G-T) double helix: wobble base pairs. Nucleic Acids Res. 1978 Jun;5(6):1955–1970. doi: 10.1093/nar/5.6.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray D. M. A circular dichroism study of poly dG, poly dC, and poly dG:dC. Biopolymers. 1974;13(10):2087–2102. doi: 10.1002/bip.1974.360131011. [DOI] [PubMed] [Google Scholar]
- Gray D. M., Morgan A. R., Ratliff R. L. A comparison of the circular dichroism spectra of synthetic DNA sequences of the homopurine . homopyrimidine and mixed purine- pyrimidine types. Nucleic Acids Res. 1978 Oct;5(10):3679–3695. doi: 10.1093/nar/5.10.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray D. M., Ratliff R. L. Circular dichroism evidence for G-U and G-T base pairing in poly[r(G-U)] and poly[d(G-T)]. Biopolymers. 1977 Jun;16(6):1331–1342. doi: 10.1002/bip.1977.360160613. [DOI] [PubMed] [Google Scholar]
- Gray D. M., Ratliff R. L. Circular dichroism spectra of poly[d(AC):d(GT)], poly[r(AC):r(GU)], and hybrids poly[d(AC):r(GU)] and poly[r(AC):d(GT)] in the presence of ethanol. Biopolymers. 1975 Mar;14(3):487–498. doi: 10.1002/bip.1975.360140305. [DOI] [PubMed] [Google Scholar]
- Hayes F. N., Lilly E. H., Ratliff R. L., Smith D. A., Williams D. L. Thermal transitions in mixtures of polydeoxyribodinucleotides. Biopolymers. 1970;9(9):1105–1117. doi: 10.1002/bip.1970.360090911. [DOI] [PubMed] [Google Scholar]
- INMAN R. B. TRANSITIONS OF DNA HOMOPOLYMERS. J Mol Biol. 1964 Sep;9:624–637. doi: 10.1016/s0022-2836(64)80171-6. [DOI] [PubMed] [Google Scholar]
- LANGRIDGE R., RICH A. Molecular structure of helical polycytidylic acid. Nature. 1963 May 25;198:725–728. doi: 10.1038/198725a0. [DOI] [PubMed] [Google Scholar]
- Lee J. S., Johnson D. A., Morgan A. R. Complexes formed by (pyrimidine)n . (purine)n DNAs on lowering the pH are three-stranded. Nucleic Acids Res. 1979 Jul 11;6(9):3073–3091. doi: 10.1093/nar/6.9.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezius A. G., Domin E. A wobbly double helix. Nat New Biol. 1973 Aug 8;244(136):169–170. doi: 10.1038/newbio244169a0. [DOI] [PubMed] [Google Scholar]
- Marck C., Thiele D., Schneider C., Guschlbauer W. Protonated polynucleotides structures - 22.CD study of the acid-base titration of poly(dG).poly(dC). Nucleic Acids Res. 1978 Jun;5(6):1979–1996. doi: 10.1093/nar/5.6.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D. J., Canuel L. L., Pohl F. M. "Alternating B-DNA" conformation for the oligo(dG-dC) duplex in high-salt solution. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2508–2511. doi: 10.1073/pnas.76.6.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl F. M., Jovin T. M. Salt-induced co-operative conformational change of a synthetic DNA: equilibrium and kinetic studies with poly (dG-dC). J Mol Biol. 1972 Jun 28;67(3):375–396. doi: 10.1016/0022-2836(72)90457-3. [DOI] [PubMed] [Google Scholar]
- Pohl F. M. Polymorphism of a synthetic DNA in solution. Nature. 1976 Mar 25;260(5549):365–366. doi: 10.1038/260365a0. [DOI] [PubMed] [Google Scholar]
- Thiele D., Marck C., Schneider C., Guschlbauer W. Protonated polynucleotides structures - 23. The acid-base hysteresis of poly(dG).poly(dC). Nucleic Acids Res. 1978 Jun;5(6):1997–2012. doi: 10.1093/nar/5.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topal M. D., Fresco J. R. Base pairing and fidelity in codon-anticodon interaction. Nature. 1976 Sep 23;263(5575):289–293. doi: 10.1038/263289a0. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Zimmerman S. B., Cohen G. H., Davies D. R. X-ray fiber diffraction and model-building study of polyguanylic acid and polyinosinic acid. J Mol Biol. 1975 Feb 25;92(2):181–192. doi: 10.1016/0022-2836(75)90222-3. [DOI] [PubMed] [Google Scholar]


