SUMMARY
SAG/RBX/ROC protein is an essential RING component of SCF E3 ubiquitin ligase. The role of SAG during embryogenesis remains unknown. We report here a critical role for SAG in controlling vascular and neural development by modulating RAS activity via promoting degradation of neurofibromatosis type 1 (NF1). Mice mutant for Sag died at embryonic day 11.5-12.5 with severe abnormalities in vascular and nervous system. Sag inactivation caused Nf1 accumulation and Ras inhibition, which blocks embryonic stem (ES) cells from undergoing endothelial differentiation and inhibits angiogenesis and proliferation in teratomas. Simultaneous Nf1 deletion fully rescues the differentiation defects in Sag−/− ES cells, and partially rescues vascular and neural defects in Sag−/− embryos, suggesting that the effects of Sag deletion may not be solely explained by Nf1 misregulation. Collectively, our study identifies NF1 as a physiological substrate of SAG-CUL1-FBXW7 E3 ligase and establishes a ubiquitin-dependent regulatory mechanism for the NF1-RAS pathway during embryogenesis.
Keywords: Apoptosis, Endothelial differentiation, mouse knockout, NF1-RAS, SAG-CUL1-FBXW7 E3 ubiquitin ligase, vascular and neural development
INTRODUCTION
The SCF E3 ubiquitin ligase is the largest family of E3 ligases, consisting of SKP1, Cullins, F-box proteins, and a RING protein, RBX (RING Box protein-1), also known as ROC (Regulators of Cullins). The substrate specificity of SCF complex is determined by the F box proteins that bind to SKP1 and Cullins through its F-box domain and to substrates through its WD40 or LRR domains (Jin et al., 2004; Zheng et al., 2002), whereas the core SCF E3 ubiquitin ligase is a complex of Cullins-RBX, in which RBX binds to E2 and facilitates ubiquitin transfer from E2 to the substrates (Wu et al., 2000). By promoting the ubiquitination of various regulatory proteins for degradation by 26S proteasome, SCF ligases regulate many biological processes, including apoptosis, cell cycle progression, signal transduction and DNA replication (Deshaies and Joazeiro, 2009; Nakayama and Nakayama, 2006). Although a large number of F-box proteins were found in the human genome (Jin et al., 2004) that selectively target various protein substrates, there are only two family members of RING proteins in human or mouse, RBX1 and RBX2, also known as SAG (Sensitive to Apoptosis Gene) (hereafter referred to as SAG) (Duan et al., 1999; Kamura et al., 1999; Ohta et al., 1999; Tan et al., 1999; Wei and Sun, 2010). Both family members, having a functional RING domain at the carboxyl terminus, are evolutionally conserved with a similar tissue expression pattern (Sun et al., 2001). While either RBX1 or SAG is capable of binding to six members of cullin family (CUL 1-3, CUL4A, B and CUL-5) and has in vitro E3 ubiquitin ligase activity when complexed with cullin-1 (Furukawa et al., 2002; Swaroop et al., 2000), RBX1 is constitutively expressed and prefers to bind with CUL2/VHL, whereas SAG is stress-inducible and preferably binds to CUL5/SOCS (Gu et al., 2007; Kamura et al., 2004).
The role of SAG during development has been previously studied in several model organisms. The Hrt1, the only yeast homologue of RBX1/SAG, is a growth-essential gene whose targeted disruption causes yeast death, which can be fully rescued by either human RBX1 (Ohta et al., 1999; Seol et al., 1999) or SAG (Swaroop et al., 2000). In C. elegans, siRNA knockdown of Rbx1 (ZK287.5) induced the death during embryogenesis as well as in adult animals (Jia et al., 2011; Sasagawa et al., 2003), whereas knockdown of Rbx2/Sag (R10A10.2) did not cause any significant phenotypic changes (Moore and Boyd, 2004). In Drosophila, disruption of Roc1a or Roc1b caused lethality or male sterility, respectively, whereas disruption of Roc2/Sag caused no overt developmental phenotype (Donaldson et al., 2004; Reynolds et al., 2008). These studies suggest that RBX2 homologues in C. elegans and Drosophila are functionally redundant and RBX2 loss can be compensated by its family member, RBX1 or ROC1a/1b during embryogenesis. Our recent knockout study revealed that the in vivo functions of Rbx1 and Sag are non-redundant in mice. In Sag wild-type background, the Rbx1 inactivation caused early embryonic lethality at embryonic day 7.5 (E7.5) due to proliferation defects, which is partially caused by p27 accumulation, as simultaneous deletion of p27 extended the life span of Rbx1 deficient embryos from E6.5 to E9.5. Thus, one in vivo physiological function of Rbx1 is to ensure cell proliferation by preventing p27 accumulation during the early stage of embryogenesis (Tan et al., 2009). However, the role of Sag during mouse embryogenesis is unknown.
The NF1 tumor suppressor gene is frequently mutated in many types of sporadic human cancers including glioblastoma multiforme, malignant peripheral nerve sheath tumor and epithelial cancers in the ovary and lung (Ding et al., 2008; Parsons et al., 2008; Sangha et al., 2008; TCGA Research Network, 2008; Woodruff, 1999). Individuals with NF1 mutations are predisposed to developing a variety of benign and malignant tumors, many of which affect the peripheral and central nervous system (Cichowski and Jacks, 2001). NF1 contains a functional domain that shows homology to the members of the RAS GTPase Activating Protein (GAP) family. As a GAP, NF1 negatively regulates RAS proto-oncogene by accelerating the conversion of active RAS-GTP to inactive RAS-GDP (Le and Parada, 2007). Upon growth factor stimulation, NF1 is rapidly degraded by the ubiquitin-proteasome pathway (Cichowski et al., 2003; McGillicuddy et al., 2009) to ensure a proper activation of RAS signals for proliferation. Previous studies on NF1 ubiquitination and degradation only focused on its GAP-related domain (GRD). One study found that a fragment containing NF1 GRD domain and its adjacent 80 amino acids (codons 1096-1534) was degradation-sensitive, but did not identify corresponding E3 ubiquitin ligase (Cichowski et al., 2003). A recent study reported that ETEA, a UBA and UBX domains-containing protein, bound to NF1 GRD domain, and negatively regulated NF1 levels (Phan et al., 2010). However, ETEA is not a bona fide E3 ubiquitin ligase. Thus, the E3 ubiquitin ligase responsible for targeted degradation of NF1 remains unknown.
In this study, we used a knockout approach and revealed the critical roles of Sag in embryonic vascular and neural development, and endothelial differentiation of ES cells. We found that Sag disruption caused Nf1 accumulation to inhibit Ras-Mapk signals. Simultaneous Nf1 deletion rescues the defects fully in endothelial differentiation of Sag−/− ES cells and partially in developing vascular and nervous system of Sag−/− embryos, suggesting Nf1 misregulation contributes partly to the defects derived from Sag deletion . We further showed that NF1 is a physiological substrate of SAG-CUL1-FBXW7 E3 ubiquitin ligase. Upon recognized and bound by FBXW7, NF1 is targeted for ubiquitination and degradation by SAG E3. Thus, our study reveals the mechanisms that regulate the stability of NF1 proteins, which might provide insights on designing novel therapy for NF1-related diseases.
RESULTS
The Sag disruption causes embryonic death at E11.5-12.5
To understand the in vivo physiological function(s) of SAG, we inactivated Sag in mice via a gene-trap approach. Mouse Sag (NM_011279) was mapped onto chromosome 9 with three exons and two introns (Fig. S1A). One ES clone (XE423) with a gene-trap vector inserted in the second intron of the Sag gene was identified (Nord et al., 2006) and characterized by PCR genome walking (Fig. S1A). The insertion disrupts the Sag transcript resulting in a truncated fusion mRNA that encodes partial Sag N-terminal sequence without the RING domain and Sag function (Sun, 1999). The confirmed ES clone was injected into C57BL/6 blastocysts to obtain chimeras, which were backcrossed with C57BL/6 mice to obtain germline transmission. Offspring carrying the Sag genetrap allele were identified by PCR genotyping (Fig. S1B), and further confirmed by genomic Southern blot analysis (Fig. S1C&D). Expression of Sag protein in Sag+/+, but not in Sag−/− embryos was detected at E10.5 (Fig S1E).
Intercrossing of heterozygous mice, which survived to term without any observable abnormalities, failed to generate Sag-disrupted homozygous mice among a total of 624 genotyped offsprings. The ratio of wild-type mice to heterozygous mice was about 1:2, exactly as expected for a condition with homozygous lethal phenotype (Fig. S1F). To define at which stage of development the Sag−/− embryos die, we dissected uteri of pregnant mice between E9.5 and E15.5. No viable Sag−/− embryos were detected after E13.5 days, whereas a few viable Sag−/− embryos were detected at E11.5-12.5 with reduced frequency. At E9.5-10.5, a Mendelian ratio of 1:2:1 (+/+:+/−:−/−) was observed (Fig. S1F). Thus, homozygous deletion of Sag causes embryonic lethality at E11.5-12.5.
The Sag disruption causes vascular defects in yolk-sacs and embryos
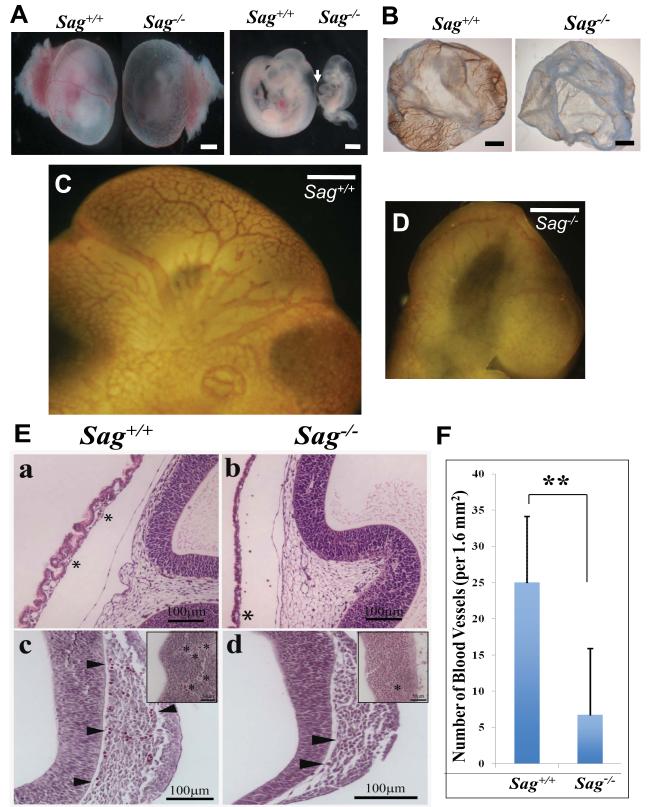
Dissection of Sag−/− embryos at E10.5 revealed that Sag−/− embryos were growth-retarded with frequent pericardial edema (Fig. 1A, arrow). Furthermore, the yolk sacs of Sag−/− embryos have dramatic reduction of blood vessels, which was confirmed by CD31/PECAM whole-mount staining for endothelial cells (Fig. 1B). Compared to Sag+/+ embryos with extensive vascular network of both large and small vessels in the head region (Fig. 1C), Sag−/− embryos exhibited disrupted vasculature with missing major blood vessels and disorganized secondary branches (Fig. 1D). Similar vascular defects were also found in Sag−/− E9.5 embryos (data not shown). H&E stained sagittal sections revealed a substantial reduction in blood vessels and the number of nucleated red blood cells within blood islands in the yolk-sac of Sag−/− embryos (Fig. 1E, top). Vascular defects in the head region were also evident with a reduction in the number of blood vessels surrounding and within the neuroepithelium (Fig. 1E&1F).
Figure 1. Sag inactivation disrupts vascular development in yolk-sac and embryo brains.
(A) Appearance of mouse embryos at E10.5 with (left) and without (right) yolk-sacs. Arrow points to pericardial edema. Bar size=0.75 mm. (B) Appearance of mouse yolk sac at E10.5 after whole-mount staining with CD31. Bar size=1 mm. (C&D) Appearance of mouse embryo brain regions at E10.5 after whole-mount CD31 staining of embryos. Bar size = 3 mm. (E) Reduced number of blood vessles in Sag−/− embryos: Haematoxylin and eosin stained embryo sections of Sag+/+ (a, c) and Sag−/− (b,d) at E10.5. * indicates network of small blood vessels with cellular contents. Arrow heads indicate blood vessels (c, d) around brain region. Inserts indicate number and size of blood vessels (*) within neuroepithelium. Number of blood vessels around the entire brain region was counted (bars, s.e.m., n=5 counted areas). (F) Data was plotted. **, p=0.00269. See also Figure S1.
The Sag disruption abrogates endothelial differentiation of ES cells and inhibits teratoma growth and angiogenesis
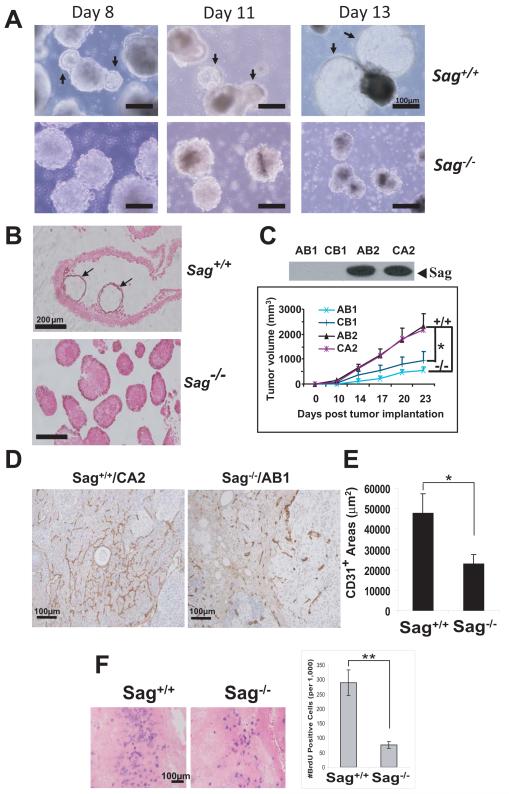
To dissect the underlying mechanisms responsible for the observed vascular defects in Sag−/− embryos, we turned into an in vitro cell culture system to circumvent early lethality caused by Sag deficiency. We generated Sag−/− embryonic stem (ES) cells from blastocysts, obtained from Sag+/− inter-crosses. Sag+/+ ES cells underwent endothelial differentiation to form cystic embryoid bodies (cEBs) (Fig. 2A) with blood island structures, lined with endothelial cells (Fig 2B, arrows). In contrast, Sag−/− ES cells completely failed to differentiate into cEBs, though they did form EBs (Fig. 2A&B). These results suggest that Sag is required for EBs to undergo endothelial differentiation to form cEBs under in vitro cultured conditions. We next determined the growth rate and incidence of teratoma derived from ES cells upon injected into in vivo nude mice. We used two independent pairs of Sag−/− vs. Sag+/+ ES cells and found that teratomas derived from Sag−/− ES cells grew much slower and formed significantly smaller tumors with a lower tumor incidence (AB1 teratomas developed in 2 out of 7 mice) (Fig. 2C, Fig. S2A). Teratomas derived from Sag−/− ES cells also had a significantly reduced blood vessel density (Fig. 2D&E). Immuno-costaining experiment revealed that these blood vessels were derived from transplanted ES cells, not from host progenitor cells, since the endothelial cells co-expressed both CD31 and β-gal (targeting vector contains β-gal cDNA) in teratomas derived from Sag−/−, but not from Sag+/+ ES cells (Fig. S2B). Furthermore, we observed a reduced proliferation rate in teratomas from Sag−/− ES cells, compared to those from Sag+/+ ES cells (Fig. 2F). Thus, Sag disruption remarkably inhibits angiogenesis and growth/proliferation of teratomas in vivo.
Figure 2. SAG disruption inhibits ES cell endothelial differentiation in vitro and teratoma formation in vivo.
(A&B) Endothelial differentiation of embryoid bodies: ES cells underwent endothelial differentiation for indicated periods and photographed. Arrows indicate cystic EBs that occurred in Sag+/+ ES cells only (A). The EBs were whole-mounted with CD31, and counterstained with eosin (200X) (B). (C-F) Growth and angiogenesis of teratomas: Two pairs of ES cell lines, AB2/CA2 (Sag+/+) vs. AB1/CB1 (Sag−/−) (C, top) were injected into nude mice (7 per group). Tumor growth was monitored for 23 days. The mean tumor volume (mm3) of seven tumors (except AB1 line with two tumors generated) was plotted (mean ± s.e.m, *, p=0.0044, between AB1/CB1 and AB2/CA2, using an F-test from the mixed model) (C, bottom). Tumor sections were immunostained with CD31 for microvessels (D), and were quantified in multiple representative areas of at least two tumors (mean ± s.e.m; *, p=0.0117) (E). Mice were injected with BrdU and tumor tissues were processed for BrdU staining. BrdU positive cells (blue) were counted in multiple areas (mean ± s.e.m; **, p=0.0002) (F). See also, Figure S2.
Sag is required for active Ras/Mapk signals during endothelial differentiation and teratoma proliferation
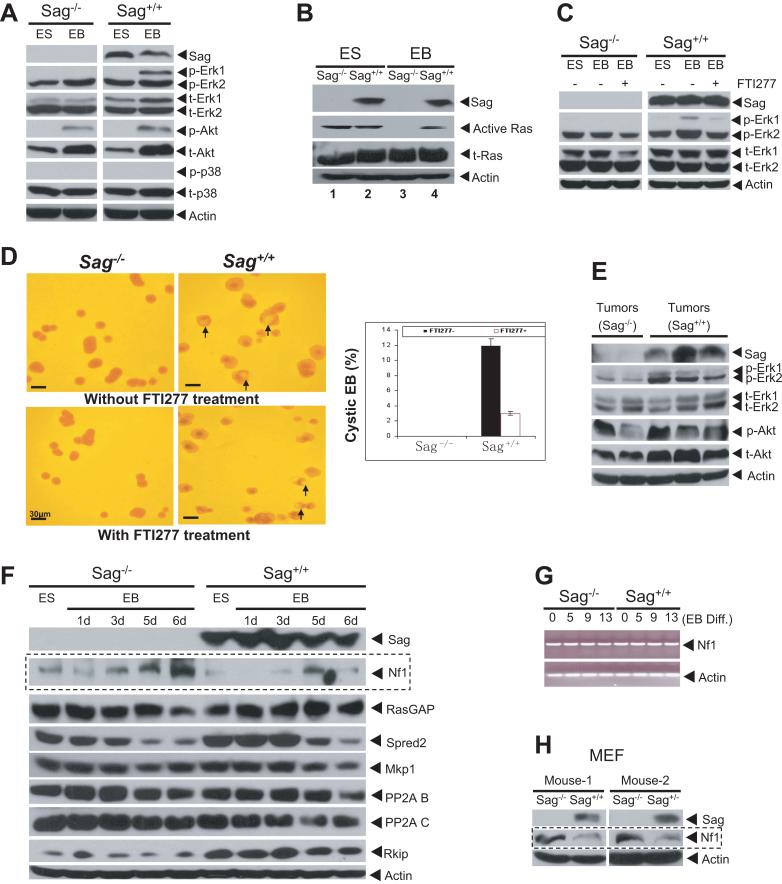
Because RAS/MAP kinase pathways are known to regulate proliferation, angiogenesis and endothelial cell differentiation (Kawasaki et al., 2008; Kranenburg et al., 2004), we sought to determine whether the Ras signaling pathway is inactivated during Sag−/− ES cell differentiation. As shown in Figure 3A, phosphorylation of p44/Erk1 was significantly induced during differentiation of Sag+/+ EBs, but not in Sag−/− EBs. No difference was seen in the levels of total or phosphorylated forms of Akt and p38. A Ras activity assay further showed that Ras was active in ES cells, regardless of Sag status, but was completely inactive in EBs derived from Sag−/− ES cells (Fig. 3B). Similar results were obtained from another independent set of ES clones (not shown). Thus, Ras activation and Erk1 phosphorylation were maintained during cystic EB formation, whereas they were inactivated in the absence of Sag. We further found that FTI277, an inhibitor of Ras farnesyltransferase (Kawasaki et al., 2008) that inhibited p44/Erk1 phosphorylation (Fig. 3C), could significantly reduce cystic EB formation from Sag+/+ ES cells (Fig. 3D). A similar result was obtained with PD098059, a Mek inhibitor (Fig S3). Consistently, a reduced Erk phosphorylation, an indicator of Mek inactivation, was found in Sag−/− teratoma tissues, as compared to Sag+/+ teratomas (Fig 3E). Taken together, the findings demonstrated that the inability to maintain active Ras/Erk signals, upon Sag disruption, contributes significantly to the failure of endothelial differentiation in vitro, angiogenesis, and possibly reduced proliferation in teratomas in vivo.
Figure 3. SAG disruption causes inactivation of Ras/Mapks and accumulation of NF1.
(A&B) ES cells were cultured without LIF in static suspension to induce the formation of EBs and cystic EBs up to 13 days. The EBs were lysed for immunoblotting (IB) (A) or assayed for Ras activity (B) (C&D) ES cells were differentiated for 13 days in the absence or presence (days 2 to 4) of FTI277 (1 μM), and lysed for IB (C) or photographed to show cystic EBs (arrows). The % of cEBs in a total of ~300 EBs was plotted, mean ± s.d. (n=3) (D, bottom). (E) Inhibition of Erk phosphoryation: Independent termatoma tissues derived from Sag−/− or Sag+/+ ES cells were homogenized for IB. (F) NF1 accumulation in Sag−/− EBs. ES cells and EBs (differentiated for up to 6 days) were lysed for IB. (G) No change in Nf1 mRNA during ES cell differentiation. ES or EBs were collected at indicated time points for RT-PCR analysis. (H) NF1 accumulation in Sag−/− MEFs. Primary MEFs were prepared from E10.5 embryos for IB. See also Figure S3.
Accumulation of neurofibromin (Nf1) during ES cell differentiation and in MEFs upon Sag disruption
RAS signaling pathways are inhibited by naturally occurring inhibitory proteins or inactivating phosphatases (Karnoub and Weinberg, 2008; Kolch, 2005). These proteins, upon Sag disruption, would accumulate to inactivate the RAS pathway during endothelial differentiation if they were the direct substrates of SAG-SCF E3 ubiquitin ligases. We therefore examined the levels of several Ras signaling inhibitory proteins in ES cells and in EBs formed during differentiation. As shown in Figure 3F, the basal level of neurofibromin (encoded by Nf1) was slightly higher in Sag−/− ES cells than that in Sag+/+ ES cells. The Nf1 level gradually increased, reaching the peak at 6 day of Sag−/− EB differentiation, but remained low in Sag+/+ EBs (panel 2). No significant difference was observed between the two groups in the levels of p120RasGap, a family member of Nf1 with a similar Ras inhibiting activity (Henkemeyer et al., 1995), nor of other Ras/Raf inhibitors or phosphatases of the Ras pathway tested (Dhillon et al., 2007b; Kolch, 2005), including Spred2, Mkp-1, and PP-2A (subunits B and C), except Rkip, a Raf inhibitor (Hagan et al., 2006), which showed higher levels in Sag+/+ ES cells and EBs, but was not regulated by ES cell differentiation (Fig. 3F). RT-PCR analysis revealed that Nf1 mRNA remained consistent during ES cell differentiation, regardless of Sag status (Fig. 3G), indicating that Nf1 accumulation upon Sag disruption likely results from a reduced degradation. We further measured the Nf1 levels in mouse embryonic fibroblasts (MEF) derived from E10.5 embryos with three Sag genotypes and found an elevated Nf1 level in Sag−/− MEFs, as compared to that in Sag+/+ or Sag+/− MEFs (Fig. 3H). Taken together, our results suggested that Nf1 could be a substrate of Sag-SCF E3 ubiquitin ligase, which accumulates upon Sag disruption, to inhibit Ras/Erk signals.
SAG-CUL1-FBXW7 binds to NF1 and promotes its ubiquitination
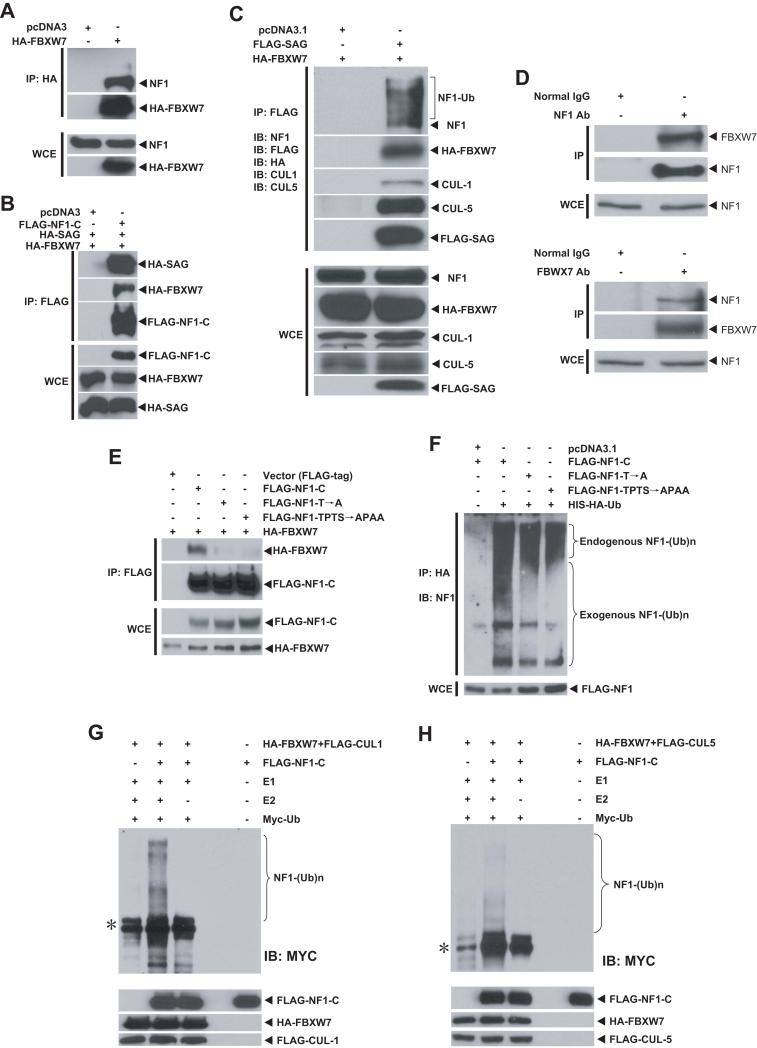
The crystal structure of SCF complex (Zheng et al., 2002) reveals that the substrate is recognized and recruited by an F-box protein to the ligase complex for ubiquitination. Thus, Sag-mediated Nf1 degradation must involve an F-box protein that recognizes Nf1 for binding. Examination of the NF1 sequence for the consensus binding motifs of several F-box proteins revealed an evolutionarily conserved perfect binding site for FBXW7 (also known as CDC4) (L-I/L/P-pT-P-XXXX) at the C-terminus (Fig. S4A), which is consistent with the binding motif found in other known substrates of FBXW7 (Fig. S4B). No FBXW7 binding site was identified in p120RasGAP, a family protein not subject to proteasome-dependent proteolysis (Cichowski et al., 2003). We therefore determined the potential binding of FBXW7 and NF1. Using an immunoprecipitation assay, we found that transiently transfected FBXW7 pulled down endogenous NF1 (Fig. 4A). In a reciprocal experiment, NF1C, a C-terminal portion of NF1 (codons 2180-2840) containing the FBXW7 binding site, pulled down both FBXW7 and SAG when co-transfected, indicating in vivo binding of SAG-FBXW7-NF1 (Fig. 4B). Furthermore, SAG, when co-transfected with FBXW7, pulled down FBXW7, as well as endogenous NF1 and CUL1 (Fig. 4C), indicating an in vivo formation of SAG-CUL1-FBXW7 E3 ligase. More significantly, SAG cotransfection promoted FBXW7-mediated ubiquitination of endogenous NF1, as demonstrated by slower migrating bands (Fig. 4C, top panel), which was not observed when FBXW7 was transfected alone (Fig. 4A, top). In order to detect endogenous FBXW7-NF1 binding, we examined several commerically available antibody against FBXW7 and identified one from Sigma that was capable of detecting overexpressed FBXW7 (data not shown). Using this antibody, we were able to detect endogenous FBXW7-NF1 binding, as evidenced by detection of FBXW7 (likely being enriched when complexed with NF1) in NF1 immunoprecipitates (Fig 4D, top panels). Reciprocally, NF1 can also be detected in FBXW7 immunoprecipitates (Fig 4D, bottom panels).
Figure 4. FBXW7 and SAG bind to NF1 and promote its ubiquitination.
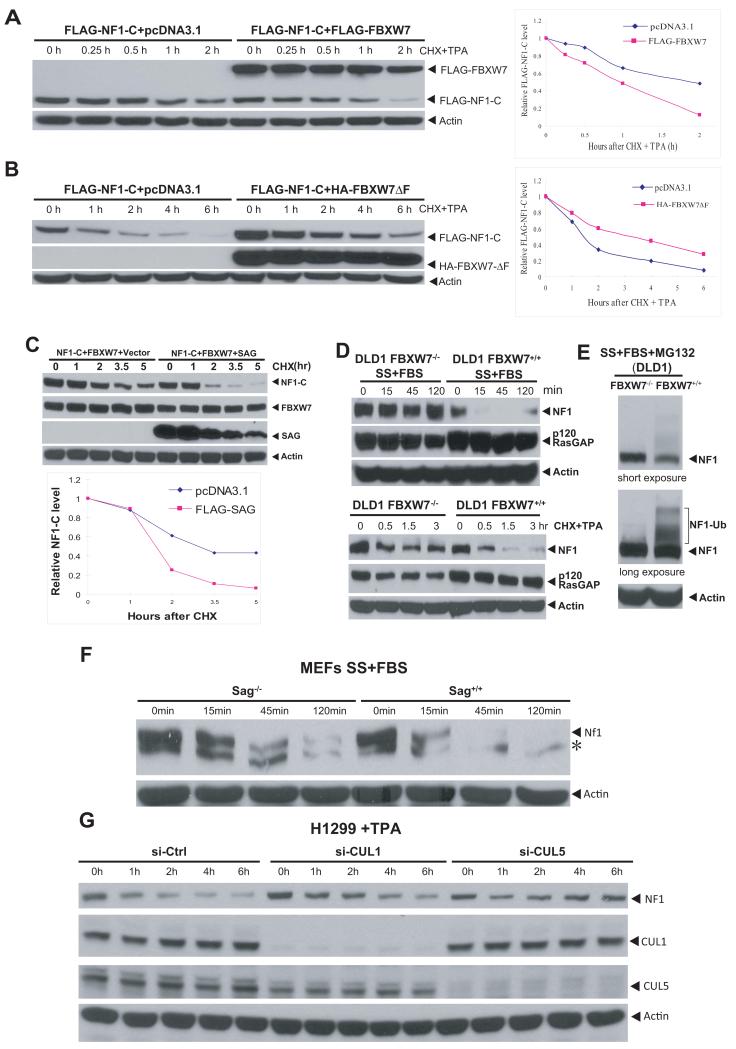
(A-C) SAG-FBXW7-NF1 forms a complex in vivo: The 293 cells were transfected and treated with MG-132 (10 μM) 4 hrs prior to harvesting. Cell lysates were immunoprecipitated (IP) with bead-conjugated FLAG or HA Abs, followed by IB. The bottom panels are the direct IB using WCE (whole cell extract) as input. (D) Endogenous FBXW7-NF1 binding: Cell lysates from 293 cells were immunoprecipated using Ab against NF1 (top) or FBXW7 (bottom), along with normal IgG control, followed by IB. Direct IB was performed using WCE. (E&F) NF1-FBXW7 binding and NF1 ubiquitination: FLAG-tagged NF1-C and its two mutants were co-transfected into H1299 cells. Cells were treated with MG-132 (10 μM) 4 hrs before harvesting for IP and IB with NF1 Ab to show ubiquitination of both endogenous (top portion) and exogenously expressed NF1-C (bottom portion), F). (G&H) CUL1-FBXW7 or CUL5-FBXW7 promotes NF1 ubiqutination in vitro: SCF E3 was prepared by FLAG bead IP using 293 cells transfected with CUL1 and FBXW7 (G) or CUL5 and FBXW7 (H). NF1 substrate was prepared by transfecting FLAG-NF1-C into 293 cells, followed by FLAG-bead IP and 3xFLAG peptide elution. SCF E3, NF1-C substrate were added into a reaction mixture containing ATP, Myc-Ub, E1 and E2. E2 was omitted in some reactions. The reaction mixture after 60 min incubation was subjected to IB using c-Myc-tag Ab. *: Non-specific bands. See also Figure S4.
We next determined if FBXW7-NF1 binding and NF1 ubiquitination were dependent on the FBXW7 consensus-binding motif on NF1. We generated two NF1 mutants on the FBXW7 binding motif (Fig. S4C) and found that these mutations had a significant reduction in binding to FBXW7 (Fig. 4E). Consequently, FBXW7-induced ubiquitination of exogenously expressed NF1-C was remarkably inhibited in these mutants, without affecting ubiquitination of endogenous NF1 (Fig. 4F). We further determined NF1 ubiquitination using an in vitro purified system and found that FBXW7-CUL1 promoted NF1 ubiqutination in a manner dependent on E2 ubiquitin conjugating enzyme (Fig. 4G). The involvement of CUL1 in promoting NF1 ubiquitination was further demonstrated as follows: (1) endogenous NF1 could be pulled down by exogenously expressed CUL1 (Fig S4D), (2) siRNA knockdown of CUL1 significantly inhibited NF1 ubiquitination (Fig. S4E), and (3) endogenous SKP1 were present in SAG immunoprecipitates (Fig. S4F). Thus, SAG-SKP1-CUL1-FBXW7 could form an active E3 ubiquitin ligase in vivo that regulates cellular levels of NF1 by promoting its ubiquitination and degradation. To a lesser extent, CUL-5 appears to be also involved in NF1 degradation (Fig 4C&H, Fig S4D&E), although it generally belongs to an E3 complex with SOCS-Sag (Kamura et al., 2004).
SAG-FBXW7 shortens NF1 protein half-life and is required for serum- or mitogen-induced NF1 ubiquitination and degradation
We next determined whether FBXW7 shortens protein half-life of NF1, and whether SAG could further accelerate it. The protein half-life of transfected NF1C was about 2 hrs upon TPA stimulation, which was reduced to ~1 hr when co-transfected with wt FBXW7 (Fig. 5A), but extended to ~4 hrs when co-transfected with an F-box deleted dominant negative FBXW7 mutant (Fig. 5B). The protein half-life of two NF1 mutants was much extended under the same conditions (Fig. S5A&B). Moreover, cotransfection of SAG with FBXW7 further shortened the protein half-life of NF1C from 3 hr (FBXW7 alone) to ~1.5 hrs (when combined) in the absence of TPA stimulation (Fig. 5C). We further determined the effect of FBXW7 on protein half-life of endogenous NF1. Under normal growth conditions, endogenous NF1 is relatively stable with a protein half-life much longer than 3 hrs, regardless of FBXW7 status (Fig. S5C). NF1 was, however, subject to mitogen-induced reduction in an FBXW7 dependent manner. In serum-starved FBXW7−/− DLD1 cells (Rajagopalan et al., 2004), serum addition had no effect on NF1 levels up to 120 min. In contrast, serum addition to starved FBXW7+/+ DLD-1 cells induced a rapid elimination of NF1 within 15 min, which lasted up to 120 min before it started to recover (Fig. 5D, top). Likewise, the protein half-life of endogenous NF1 after exposure to TPA was about 30 min, which was extensively extended in FBXW7−/− cells (Fig. 5D, bottom). No change at the level of p120RasGAP was observed, regardless of FBXW7 status (Fig 5D). We further confirmed that FBXW7-dependent NF1 reduction was via the ubiquitin-proteasome pathway, since concurrent MG-132 treatment blocked NF1 degradation, along with accumulation of ubiquitinated NF1 in FBXW7+/+ cells, but not in FBXW7−/− cells (Fig. 5E). Finally, we found that both SAG and CUL1/5 were involved in controling the NF1 turn-over. The Nf1 half-life was much extended in Sag−/− MEF cells, as compared to Sag+/+ cells under the conditions of serum starvation, followed by serum addtion to trigger Nf1 degradation (Fig. 5F). Similarly, TPA-induced NF1 degradation was much delayed upon siRNA knockdown of either CUL1, or to lesser extent, CUL5 (Fig. 5G). Thus, NF1 is a physiological substrate of SAG-CUL1-FBXW7 (and possibly a SAG-CUL5 based E3 ligase) for targeted degradation.
Figure 5. FBXW7 and SAG shorten NF1 protein half-life.
(A&B) FBXW7 shortens, but FBXW7ΔF extends the NF1 protein half-life: The 293 cells were transfected and treated with CHX (cycloheximide) to block new protein synthesis and with TPA to trigger degradation. Samples were harvested for IB. Densitometry quantification was performed with ImageJ. (C) SAG further shortens NF1 protein half-life: FLAG-NF1-C was co-transfected with FBXW7, with or without FLAG-SAG into 293 cells. Cells were harvested after CHX treatment for IB and quantified by ImageJ . (D&E) FBXW7-dependent degradation of endogenous NF1: DLD-1 cells with or without FBXW7 deleted were serum starved for 24 hrs, followed by serum addition. Cells were then harvested for IB (D, top). Cells were treated with TPA and CHX, and harvested for IB (D, bottom left). DLD-1 cells were serum starved for 24 hrs, followed by serum addition in combination with MG-132. Cells were harvested 15 min later for IB (E). (F&G) Inactivation of Sag or CUL1/5 extended Nf1 protein half-life: MEFs (Sag+/+ or Sag−/−) were serum starved up to 48 hrs, followed by serum addition. Cells were harvested at indicated periods for IB. *: non-specific band (F). H1299 cells were transfected with siRNA targeting CUL1 or CUL5, along with control siRNA. Cells were treated with TPA (10 nM) 48 hrs later for indicated periods and harvested for IB (G). See also Figure S5.
Simultaneous deletion of Nf1 rescues the vascular defects in Sag−/− ES cells and embryos
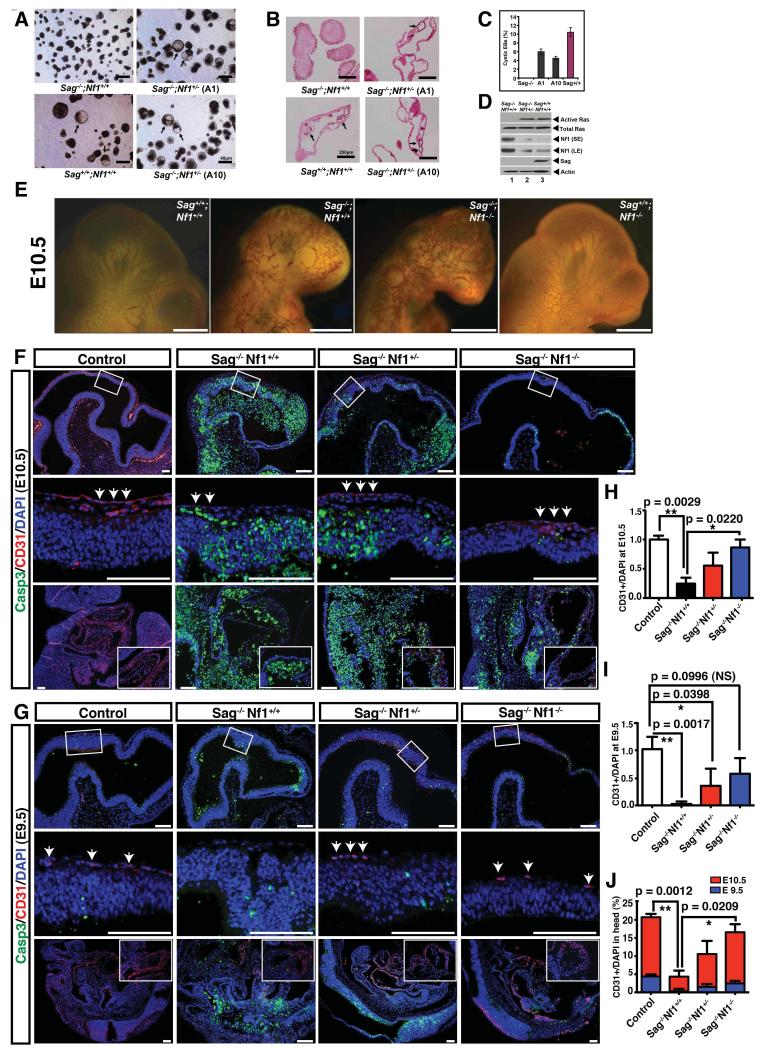
We next attempted to define the physiological importance of Nf1 accumulation upon Sag disruption in endothelial differentiation of ES cells and vascular development of embryos. We reasoned that if Nf1 accumulation inhibits endothelial differentiation of EBs by inhibiting Ras/Erk activation, then simultaneous deletion of Nf1 would restore the formation of cystic EBs in the Sag−/− background. We generated Sag/Nf1 double null ES clones from blastocysts obtained by the mating of Sag+/− and Nf1+/− mice. Among 64 ES lines established, we identified a single clone (ISG8) with the Sag−/−;Nf1−/− genotype. However, this clone did not grow well and was readily attached onto the Petri dish during suspension culture for differentiation, leading to much fewer numbers of embryoid bodies, although we did observe the formation of cEBs at a rate comparable to that of wild type (Fig. S6A&C) with full restoration of Ras activity (Fig. S6B). We then focused our attention to three independent Sag−/−;Nf1+/− ES cell lines and found that all three were able to undergo endothelial differentiation to form cystic EBs, although the frequency was relatively lower than that of wild type (Fig. 6A, two clones, A1 and A10 were shown). The blood island structures with endothelial lining were apparent in these rescued cEBs (Fig. 6B), indicating that heterozygous loss of Nf1 is sufficient to rescue the defect in endothelial differentiation caused by Sag inactivation. We further confirmed that the Nf1 status on the Sag+/+ background did not affect endothelial differentiation, since both Nf1+/− and Nf1−/− ES cells are capable of forming cEBs (Fig S6C). We next determined if Ras activity was also rescued upon Nf1 inactivation and found that while Sag−/−;Nf1+/+ EBs showed a high level of Nf1 with a undetectable Ras activity (Fig 6D, lane 1), Sag−/−;Nf1+/− EBs had a significant lower level of Nf1, but a high level of activated Ras, similar to that seen in wild type EBs (Fig. 6D, lanes 2 vs. 3). These results suggested that loss of one Nf1 allele is sufficient to reduce abnormally high levels of Nf1 (caused by Sag disruption) to a level that permits cystic EB formation during ES cell differentiation. Taken together, our results demonstrated that Ras inactivation, as a result of Nf1 accumulation upon Sag disruption, is the major cause for the failure of Sag−/− ES cells to undergo endothelial differentiation.
Figure 6. Rescuing Sag−/− defective phenotypes by simultaneous deletion of Nf1.
(A-C) Heterozygous deletion of Nf1 rescued the differentiation defects. ES cells underwent endothelial differentiation for 12 days and were photographed to show cystic EBs (A, arrows). The proportion of cEBs in a total of ~300 EBs was plotted, mean ± s.d. (n=3) (C). The EBs were whole-mounted with CD31, and counterstained with eosin (200x) (B). (D) Heterozygous deletion of Nf1 rescued Ras activity: EBs formed after differentiation for 5 days were measured for Ras activity by a pull-down assay, and IB for total Ras, Nf1 and Sag. (SE: short exposure, LE: longer exposure). (E) Nf1 deletion partially rescued vascular defects in embryonic brains: Sag+/−;Nf1+/− mice were intercrossed, embryos at E10.5 were dissected and subjected to CD31 whole-mount staining. Representative embryos of four genotypes were photographed. Bar size = 3 mm (E). (F&G) Deletion of Nf1 partially rescued a reduction of endothelial cells in Sag deficient embryos at E10.5 (F) and E9.5 (G). Sagittal sections of control and mutant embryos were stained with CD31, cleaved caspase 3 and DAPI. Images were captured at low and high-magnifications for cranial neuroepithelial cells (top and middle panels) as well as for trunk areas (bottom panels). Scale bar, 100 μm. (H&I) Quantification of CD31 positive cells in the head areas at E10.5 (H) and E9.5 (I). The data were normalized to the total number of DAPI and represented as fold change relative to the number of CD31 positive cells in Sag+/+ Nf1+/+, which were set to 1 (mean±s.e.m, n=3). (J) The percentage of CD31 positive cells in head areas of control and mutant embryos at E9.5 and E10.5. The number of cells expressing CD31 was normalized to the total number of DAPI positive cells. (mean±s.e.m, n=3). See also Figure S6.
We then determined whether simultaneous deletion of Nf1 would rescue vascular defects in Sag−/− embryos (Fig. 1D) by crossing of Sag+/− mice with Nf1+/− mice (Zhu et al., 2005). Although homozygous deletion of Nf1 caused embryonic lethality by E13.5 due to cardiac defects, mutant embryos appeared normal through E10.5 (Brannan et al., 1994; Jacks et al., 1994). We examined the blood vessel networks in head areas of E10.5 embryos with 4 genotypes, using CD31 whole-mount staining (Fig 6E). While Sag−/−;Nf1+/+ brains showed reduced primary vascular networks with missing major trunk vessels, Sag+/+;Nf1−/− brains showed relatively normal vasculature, similar to that of Sag+/+;Nf1+/+. Significantly, the vasculature in the head areas of Sag−/−;Nf1−/− embryos showed a partial restoration with reappearance of some trunk blood vessels (Fig. 6E), even though these Sag−/−;Nf1−/− embryos remained growth-retarded (Fig. 6E&F). Furthermore, CD31-positive endothelial cells in control embryos formed a chain-like structure that covered the entire neuroepithelium, whereas only residual CD31-positive cells were found in the Sag−/− embryos (arrows, Fig. 6F). Importantly, the chain-like structure formed by CD31-positive endothelial cells was partially restored by both Nf1 heterozygous and homozygous mutations (Fig. 6F&H).
We further observed that Sag disruption caused a widespread apoptosis in embryos evidenced by cleaved caspase 3 staining (Fig. 6F). To rule out the possibility that excessive apoptosis observed in Sag-deficient embryos could potentially exert non-specific effects on normal development of endothelial cells, we further analyzed control and Sag-deficient embryos at earlier stages. At E9.5, compared to controls, Sag-deficient embryos exhibited relatively normal morphology with only a minor increase in apoptosis within developing nervous system (Fig. 6G). However, the number of CD31-positive endothelial cells was greatly reduced in Sag-deficient embryos (arrows, Fig. 6G). Particularly, compared to control embryos, Sag-deficient embryos only contained less than 10% of CD31-positive endothelial cells in the head areas (Fig. 6I). Strikingly, this defect was significantly rescued by loss of one or two alleles of Nf1 (arrows, Fig. 6G, 6I). When comparing the relative number of CD31-positive endothelial cells at E9.5 and E10.5 among control, Sag single, and Sag/Nf1 double mutant embryos, we found that CD31-positive endothelial cells in all the mutant embryos, albeit at a much lower level, expanded proportionally from E9.5 to E10.5, as compared to those in controls (Fig. 6J). Taken together, our studies, using both in vitro ES cells and in vivo embryos, suggest that Sag-mediated Nf1 degradation is critical for the genesis of endothelial cells in the vascular system. This notion was further supported by the fact that CD31-positive endothelial cells in Sag-deficient embryos at both E9.5 and E10.5 stages did not colocalize with caspase-3 positive cells (Fig. 6F&G), excluding the involvement of apoptosis.
Nf1 inactivation partially rescues apoptotic, but not proliferation defects in developing Sag-deficient neural precursor cells
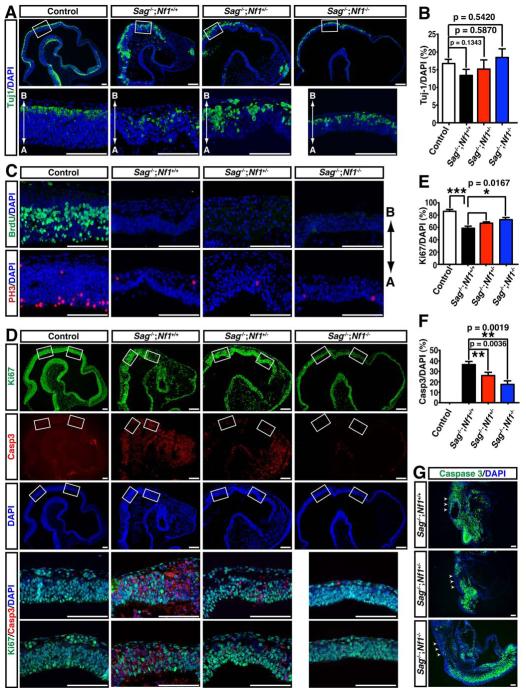
To further determine genetic interaction between Sag and Nf1 in vivo, we studied Nf1-dependent Sag function in the developing nervous system, as 40% to 80% of NF1 children exhibit learning deficits (Hyman et al., 2005). During development, the onset of neurogenesis in the neocortex occurs at the basal surface of the neuroepithelium at E10.5 (Farkas and Huttner, 2008; Gotz and Huttner, 2005). We found that neuronal differentiation, revealed by Tuj1 expression, was not significantly different between control and Sag-deficient embryos with or without additional Nf1 mutations (Fig. 7A, B). These results indicate that Sag−/− neural precursor cells in the neocortex undergo neuronal differentiation according to a normal developmental schedule, albeit in overall growth-retarded embryos. We next examined whether there are proliferation defects in Sag−/− neural precursor cells. During cell cycle, neural precursor cells undergo DNA replication (S phase) in the basal part of the neuroepithelium (Farkas and Huttner, 2008), which were labeled by a single BrdU pulse (Fig. 7C, upper left panel). The nuclei of neural precursor cells migrate apically through G2 phase to undergo M phase in apical position along the ventricle, which were revealed by an M phase marker, phospho-histone H3 (PH3) (Fig. 7C, bottom left panel). Strikingly, little or no BrdU- or PH3-positive neural precursor cells were identified in the Sag−/− neocortex (Fig. 7C). More importantly, none of the defects in cell cycle progression was rescued by Nf1 inactivation (Fig. 7C). Thus, Sag-deficient neural precursor cells in the developing neocortex exhibited severe defects in entering S and M phases of the cell cycle in an Nf1-independent manner. Consequently, compared to control neocortex, the number of neural precursor cells labeled by Ki67 expression was significantly reduced in the Sag−/− neuroepithelium, consistent with the fact that these cells had proliferation defects (Fig. 7D&E). Given that Nf1 deficiency does not rescue cell-cycle progression defects in Sag−/− neural precursor cells, we unexpectedly observed a significant level of rescue of Ki67-positive neural precursor cells upon Nf1 deletion (Fig. 7D&E). We, therefore, determined whether apoptosis additionally contributed to Nf1-dependent reduction of neural precursor cells in Sag−/− neocortex. While control neocortex had little or no caspase3-positive apoptotic cells, nearly 40% of Sag−/− cells in E10.5 neocortex were undergoing apoptosis, which was significantly rescued by Nf1 inactivation in a dose-dependent manner (Fig. 7D&F). In addition to the developing brain, Nf1 inactivation also partially rescued apoptosis in the Sag-deficient spinal cord (arrows, Fig.7G), though apoptosis in surrounding mesoderm-derived tissues was not affected (Fig. 7G). Moreover, neuronal apoptosis of Sag−/− E9.5 embryos could also be rescued by Nf1 deletion (Fig. 6G&S6D). Taken together, our observations demonstrate that during neocortical development, Sag plays critical roles in regulating apoptosis and cell cycle progression in an Nf1-dependent and Nf1-independent manner, respectively. Furthermore, Nf1-dependent Sag function in suppressing apoptosis is specifically required for developing nervous system, but not for surrounding mesenchymal tissues.
Figure 7. Inactivation of Nf1 partially rescued apoptotic phenotype but not proliferation defects in developing Sag−/− neural precursor cells.
(A) Neuronal differentiation detected by Tuj-1 (an early neuronal marker). Sagittal sections of embryonic brains (E10.5) with different genotypes were stained with Tuj-1 and DAPI (Scale bar = 100 μm). A: apical and B: basal. (B) Quantification of Tuj-1 positive cells in the neuroepithelium were normalized to total DAPI positive cells (mean±s.e.m, n=3). (C) Nf1 deletion failed to rescue proliferation defects in Sag−/− neural precursor cells. Brain sections were stained with BrdU (an S phase marker) or Phospho-histone H3 (PH3, an M phase marker) as well as DAPI (Scale bar = 100 μm). (D-F) Nf1 deletion partially rescued neural apoptosis: Sagittal sections of embryonic brains were stained with Ki67, caspase 3 and DAPI (Scale bar = 100 μm). The number of cells expressing Ki67 and caspase3 was normalized to the total number of DAPI positive cells (*** p < 0.0001, mean±s.e.m, n=3) (E&F). (G) Nf1 deletion partially rescued apoptosis in the spinal cord, but not surrounding mesenchymal cells. Embryonic sections were stained with caspase 3 and DAPI. Arrows point to spinal cord (Scale bar =100 μm).
DISCUSSION
Despite Sag being dispensable during development in Drosophila and C. elegan (Moore and Boyd, 2004; Reynolds et al., 2008), we demonstrate that Sag is absolutely required for mouse development. Sag disruption causes embryonic lethality at E11.5-12.5, which is associated with severe developmental defects including those in the vascular and nervous system. Since SCF E3 ligases are essential for many developmental processes (Petroski and Deshaies, 2005), it is not surprising that Sag, one of the two Rbx/Roc family members essential for E3 ligase activity, plays critical roles in many aspects of embryonic development. Although accumulation of many critical Sag/SCF-targeted substrates could contribute to severe developmental defects observed in Sag−/− embryos, our biochemical and genetic experiments indicate a causal involvement of Nf1, which, upon accumulation as a result of Sag disruption, inactivates Ras-Mapk signals to block endothelial differentiation and induce apoptosis in the nervous system.
The RAS-MAPK signaling pathway is actively involved in cell proliferation, angiogenesis, and tumorigenesis (Dhillon et al., 2007a; Kranenburg et al., 2004) as well as endothelial specification of VEGFR2-expressing vascular progenitor cells (Kawasaki et al., 2008). Here we showed that active Ras/Erk signals are also required for endothelial differentiation of mouse ES cells. Blockade of this pathway by inhibitors of RAS (FTI-277) or ERK (PD098059) repressed ES cell differentiation into endothelial cells to form blood islands. We further showed in vivo that reduced Ras/Erk activation was associated with reduced angiogenesis and decreased proliferation in teratomas derived from Sag−/− ES cells. Our mechanistic study linked Ras inactivation, upon Sag disruption, to accumulation of Nf1, a naturally occurring Ras inhibitor (Cichowski and Jacks, 2001).
Our study also defined SAG-SKP1-CUL1-FBXW7 as a physiological E3 ligase for targeted ubiquitination and degradation of NF1. First, NF1 contains an evolutionarily conserved FBXW7 binding motif at the C-terminus (codons 2755-2758), which is required for FBXW7-NF1 binding and NF1 ubiquitination. Second, SAG-CUL1-FBXW7-NF1 forms a complex in vivo that promotes NF1 ubiquitination. Third, FBXW7 shortens NF1 protein half-life in a binding motif dependent manner, which is further promoted by SAG. Fourth, ubiquitination of exogenously expressed NF1-C is promoted by FBXW7-CUL1, but inhibited by siRNA knockdown of CUL1 or genetic deletion of FBXW7. Finally and most importantly, endogenous FBXW7 binds to endogenous NF1 under physiological conditions and SAG-CUL1-FBXW7 is required for ubiquitination and degradation of endogenous NF1 upon mitogen stimulation. Our study also demonstrates that SAG could form an active E3 complex with SKP1, FBXW7 and CUL1, in addition to well-known CUL5 (Kamura et al., 2004), for targeted degradation of NF1.
FBXW7 is a p53-dependent haploinsufficient tumor suppressor (Kimura et al., 2003; Mao et al., 2004). Similar to Sag disruption, Fbxw7 gene ablation also causes embryonic lethality at E10.5-E11.5 with remarkable abnormalities in vascular development in the brain and yolk sac (Tetzlaff et al., 2004; Tsunematsu et al., 2004). These phenotypic similarities suggested that Sag and Fbxw7 might have a similar function, such as preventing Nf1 accumulation, during vascular development. However, unlike Fbxw7 whose deletion caused accumulation of Notch-1 and Notch-4 (Masuda et al., 2010; Tetzlaff et al., 2004; Tsunematsu et al., 2004), we found that neither Notch-1 nor Notch-4 was accumulated in Sag−/− ES, EB, or embryos (data not shown), suggesting that accumulation of Notch family proteins do not contribute to observed defects in Sag−/− ES cells and embryos. Instead, we showed a causal involvement of Nf1 in some aspects of vascular and neural development, regulated by Sag. First, Sag is required for endothelial cell differentiation, which is Nf1 dependent. Second, Sag plays an essential role in suppressing apoptosis during embryogenesis, with a partial involvement of Nf1 in neural precursor cells in developing brains and spinal cords, but not in mesoderm-derived tissues. Third, despite the presence of a significant number of neural precursor cells in Sag−/− neocortex, these Ki67-positive precursor cells completely lack the ability to undergo DNA replication and mitosis, which is completely independent of Nf1. It will be interesting to determine the specific stage(s) in which Sag−/− neural precursor cells are arrested during cell cycle. These observations suggest that Nf1- independent apoptosis and cell cycle failure, potentially as a result of accumulation of other critical Sag substrates, could be the major cause for growth retarded and lethal phenotypes observed in Sag−/−;Nf1−/− embryos with an embryonic life not extended beyond E11.5-12.5. Nevertheless, our genetic rescue data, by demonstrating that Nf1 inactivation rescued vascular defects, particularly at earlier developmental stages when Sag deficiency causes no overt morphological defects, strongly argue that genetic interaction between Nf1 and Sag is specific and primary to vascular development and not secondary to overall developmental retardation. Furthermore, it is worth noting that despite the presence of excessive and widespread apoptosis at E10.5, CD31-positive endothelial cells expanded proportionally from E9.5 to E10.5 in Sag-deficient embryos compared to controls. This observation further supports the notion that Sag/Nf1 regulatory network controls the generation of CD31-positive endothelial cells, but not subsequent proliferation or apoptosis.
In summary, our study demonstrates an in vivo physiological function of Sag in regulation of vascular and neural development by modulating Ras-Mapk signaling pathway through targeted degradation of Nf1. Upon induction by different stimuli (Duan et al., 1999; Gu et al., 2007; Tan et al., 2008), SAG recruits FBXW7 and CUL1 to promote ubiquitination and degradation of NF1, thus activating RAS-ERK signals to induce differentiation and proliferation, and to suppress apoptosis during vascular and neural development, respectively (see graphical abstract). Thus, under physiological conditions, SAG regulates NF1 levels to maintain active RAS/ERK signals for effective vascular and neural development. However, under pathological condition, such as in cancer, SAG, which is overexpressed with a poor patient prognosis (Jia et al., 2010; Sasaki et al., 2001), would inhibit apoptosis, promote proliferation and angiogenesis (Duan et al., 1999; Gu, 2007; Sun et al., 2001). SAG, therefore, could be a valid anti-cancer and anti-angiogenesis target (Jia et al., 2010; Nalepa et al., 2006; Sun, 2006; Wei and Sun, 2010). Moreover, 40 to 80% of individuals with NF1 have cognitive deficits, which are caused by loss of one allele of the NF1 gene - haploinsufficiency in the brain (Costa et al., 2002; Cui et al., 2008; Hyman et al., 2006). Thus, targeting the SAG/FBXW7 E3 may provide an attractive therapeutic strategy to increase NF1 expression for treating NF1-associated diseases caused by haploinsufficiency.
EXPERIMENTAL PROCEDURES
Generation and maintenance of ES cell lines with Sag and/or Nf1 disruption
Blastocysts were isolated from inter-crossing of Sag+/− mice or Sag+/−;Nf1+/− mice, and placed in culture on irradiated mouse embryonic feeder cells in high glucose DMEM (Invitrogen, Carlsbad, CA) supplemented with 15% FBS (Harlan, Indianopolis, IN), 0.1 mM β-mercaptoethanol (Sigma, St. Louis, MO), 103U/ml LIF (ESGRO, Millipore),25 μM PD098059 (Sigma) and penicillin/streptomycin. Inner cell mass outgrowths were trypsinized and passaged until ES cell lines were established in 35 mm cell culture dishes (Hughes et al., 2007).
Whole-mount immunostaining on yolk sac and embryos
Whole-mount CD31 immunostaining was performed on E9.5 and E10.5 yolk sacs and embryos (Shen et al., 2005). Briefly, embryos or yolk sacs were fixed in 4% PFA/PBS, dehydrated by methanol, quenched by H2O2, and blocked in 4% BSA. The samples were stained by incubating with anti-CD31 (rat monoclonal MEC13.3, BD Biosciences) at 4°C overnight, followed by peroxidase-conjugated secondary antibodies. The embryos were developed in 0.25% DAB with H2O2 in PBS, and photographed on a dissecting microscope (model S6D; Leica) with a progressive 3CCD camera (Sony).
Endothelial differentiation of ES cells and whole-mount staining of embryoid bodies
Single suspended mouseES cells were plated at density of 1 × 106 cells/10 cm Petri dish containing 15% DMEM. Cells were cultured for up to 13 days with media change every other day. Embryoid bodies were first fixed in the 4% PFA, blocked with 3% milk and 0.1% Triton X-100 in PBS, and incubated with CD31 antibody overnight. After washing, the samples were incubated with secondary antibody overnight. Color was developed using DAB. The samples were then dehydrated, embedded and sectioned. The sections were counter-stained with Eosin-Y.
Immunofluorescence
Paraffin sections were deparaffinized, rehydrated and analyzed by immunofluorescence (Wang et al., 2009; Zhu et al., 2001). Briefly, sections were incubated with primary antibodies in blocking solution overnight. The antibodies used are as follows: Ki 67 (1:500, mouse, BD Biosciences), Tuj-1 (1:500, Rabbit, Covance), Cleaved Caspase3 (1:500, Rabbit, Cell signaling), Phospho-histone H3 (1:500, Rabbit, Abcam), BrdU (1:500, Rat, Abcam) and CD31 (1:200, Mouse, Dako). The secondary antibodies were conjugates of Alexa Fluor 488 or Alexa Fluor 555 (1:500, Invitorgen). DAPI (1:1000, Invitrogen) was used as nuclear counter staining. Sections were examined under a fluorescence microscope (Olympus).
Statistical analysis
The paired Student t test was used for statistical analysis, using SAS software for two paired samples. A mixed model was fit to the tumor volumes (excluding mice with no tumors). Presence or absence of Sag knockout was included as a fixed effect. ES cell line was included as a random effect in the model to account for correlation between observations within a cell line. An F-test from the mixed model fit was used to test whether the mean value for tumors from cell lines with Sag knockout was the same as that for the tumors with wild type Sag.
Supplementary Material
HIGHLIGHTS.
Sag E3 KO causes embryonic lethality at E11.5-12.5 with vascular and neural defects
Sag E3 KO causes endothelial differentiation defects by Nf1-mediated Ras inhibition
NF1 is a physiological substrate of SAG-CUL1-FBXW7 E3 ligase for targeted degradation
Simultaneous deletion of Nf1 partially rescues Sag−/− defects in embryos and ES cells
Acknowledgements
We thank Dr. B. Vogelstein for providing us DLD-1 and HCT-116 cells with FBXW7 gene deleted, and Drs. K. Nakayama and B. E. Clurman for plasmids expressing FBXW7. We also thank Dr. Matthew Schipper for his help in statistical analysis of the teratoma data. This work is supported by NCI grants (CA111554, CA118762 and CA156744) to YS and (NS053900 and W81XWH-11-1-0251) to YZ.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brannan CI, Perkins AS, Vogel KS, Ratner N, Nordlund ML, Reid SW, Buchberg AM, Jenkins NA, Parada LF, Copeland NG. Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crest-derived tissues. Genes Dev. 1994;8:1019–1029. doi: 10.1101/gad.8.9.1019. [DOI] [PubMed] [Google Scholar]
- Cichowski K, Jacks T. NF1 tumor suppressor gene function: narrowing the GAP. Cell. 2001;104:593–604. doi: 10.1016/s0092-8674(01)00245-8. [DOI] [PubMed] [Google Scholar]
- Cichowski K, Santiago S, Jardim M, Johnson BW, Jacks T. Dynamic regulation of the Ras pathway via proteolysis of the NF1 tumor suppressor. Genes Dev. 2003;17:449–454. doi: 10.1101/gad.1054703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa RM, Federov NB, Kogan JH, Murphy GG, Stern J, Ohno M, Kucherlapati R, Jacks T, Silva AJ. Mechanism for the learning deficits in a mouse model of neurofibromatosis type 1. Nature. 2002;415:526–530. doi: 10.1038/nature711. [DOI] [PubMed] [Google Scholar]
- Cui Y, Costa RM, Murphy GG, Elgersma Y, Zhu Y, Gutmann DH, Parada LF, Mody I, Silva AJ. Neurofibromin regulation of ERK signaling modulates GABA release and learning. Cell. 2008;135:549–560. doi: 10.1016/j.cell.2008.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007a;26:3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- Dhillon AS, von Kriegsheim A, Grindlay J, Kolch W. Phosphatase and feedback regulation of Raf-1 signaling. Cell Cycle. 2007b;6:3–7. doi: 10.4161/cc.6.1.3593. [DOI] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson TD, Noureddine MA, Reynolds PJ, Bradford W, Duronio RJ. Targeted disruption of Drosophila Roc1b reveals functional differences in the Roc subunit of Cullin-dependent E3 ubiquitin ligases. Mol Biol Cell. 2004;15:4892–4903. doi: 10.1091/mbc.E04-03-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H, Wang Y, Aviram M, Swaroop M, Loo JA, Bian J, Tian Y, Mueller T, Bisgaier CL, Sun Y. SAG, a novel zinc RING finger protein that protects cells from apoptosis induced by redox agents. Mol Cell Biol. 1999;19:3145–3155. doi: 10.1128/mcb.19.4.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas LM, Huttner WB. The cell biology of neural stem and progenitor cells and its significance for their proliferation versus differentiation during mammalian brain development. Curr Opin Cell Biol. 2008;20:707–715. doi: 10.1016/j.ceb.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Furukawa M, Ohta T, Xiong Y. Activation of UBC5 ubiquitin-conjugating enzyme by the RING finger of ROC1 and assembly of active ubiquitin ligases by all cullins. J Biol Chem. 2002;277:15758–15765. doi: 10.1074/jbc.M108565200. [DOI] [PubMed] [Google Scholar]
- Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Gu Q, Bowden TG, Normolle D, Sun Y. SAG/ROC2 E3 ligase regulates skin carcinogenesis by stage dependent targeting of c-Jun/AP1 and IkB/NF-kB. J Cell Biol. 2007;178:1009–1023. doi: 10.1083/jcb.200612067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Q, Tan M, Sun Y. SAG/ROC2/Rbx2 is a novel activator protein-1 target that promotes c-Jun degradation and inhibits 12-O-tetradecanoylphorbol-13-acetate-induced neoplastic transformation. Cancer Res. 2007;67:3616–3625. doi: 10.1158/0008-5472.CAN-06-4020. [DOI] [PubMed] [Google Scholar]
- Hagan S, Garcia R, Dhillon A, Kolch W. Raf kinase inhibitor protein regulation of raf and MAPK signaling. Methods Enzymol. 2006;407:248–259. doi: 10.1016/S0076-6879(05)07021-7. [DOI] [PubMed] [Google Scholar]
- Henkemeyer M, Rossi DJ, Holmyard DP, Puri MC, Mbamalu G, Harpal K, Shih TS, Jacks T, Pawson T. Vascular system defects and neuronal apoptosis in mice lacking ras GTPase-activating protein. Nature. 1995;377:695–701. doi: 10.1038/377695a0. [DOI] [PubMed] [Google Scholar]
- Hughes ED, Qu YY, Genik SJ, Lyons RH, Pacheco CD, Lieberman AP, Samuelson LC, Nasonkin IO, Camper SA, Van Keuren ML, et al. Genetic variation in C57BL/6 ES cell lines and genetic instability in the Bruce4 C57BL/6 ES cell line. Mamm Genome. 2007;18:549–558. doi: 10.1007/s00335-007-9054-0. [DOI] [PubMed] [Google Scholar]
- Hyman SL, Arthur Shores E, North KN. Learning disabilities in children with neurofibromatosis type 1: subtypes, cognitive profile, and attention-deficit-hyperactivity disorder. Dev Med Child Neurol. 2006;48:973–977. doi: 10.1017/S0012162206002131. [DOI] [PubMed] [Google Scholar]
- Hyman SL, Shores A, North KN. The nature and frequency of cognitive deficits in children with neurofibromatosis type 1. Neurology. 2005;65:1037–1044. doi: 10.1212/01.wnl.0000179303.72345.ce. [DOI] [PubMed] [Google Scholar]
- Jacks T, Shih TS, Schmitt EM, Bronson RT, Bernards A, Weinberg RA. Tumour predisposition in mice heterozygous for a targeted mutation in Nf1. Nat Genet. 1994;7:353–361. doi: 10.1038/ng0794-353. [DOI] [PubMed] [Google Scholar]
- Jia L, Bickel JS, Wu J, Morgan MA, Li H, Yang J, Yu X, Chan RC, Sun Y. RBX1 (RING-box protein 1) E3 ubiquitin ligase is required for genomic integrity by modulating DNA replication licensing proteins. J Biol Chem. 2011;286:3379–3386. doi: 10.1074/jbc.M110.188425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L, Yang J, Hao X, Zheng M, He H, Xiong X, Xu L, Sun Y. Validation of SAG/RBX2/ROC2 E3 Ubiquitin Ligase as an Anticancer and Radiosensitizing Target. Clin Cancer Res. 2010;16:814–824. doi: 10.1158/1078-0432.CCR-09-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kaelin WG, Elledge SJ, Conaway RC, et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- Kamura T, Maenaka K, Kotoshiba S, Matsumoto M, Kohda D, Conaway RC, Conaway JW, Nakayama KI. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 2004;18:3055–3065. doi: 10.1101/gad.1252404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki K, Watabe T, Sase H, Hirashima M, Koide H, Morishita Y, Yuki K, Sasaoka T, Suda T, Katsuki M, et al. Ras signaling directs endothelial specification of VEGFR2+ vascular progenitor cells. J Cell Biol. 2008;181:131–141. doi: 10.1083/jcb.200709127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Gotoh M, Nakamura Y, Arakawa H. hCDC4b, a regulator of cyclin E, as a direct transcriptional target of p53. Cancer Sci. 2003;94:431–436. doi: 10.1111/j.1349-7006.2003.tb01460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolch W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat Rev Mol Cell Biol. 2005;6:827–837. doi: 10.1038/nrm1743. [DOI] [PubMed] [Google Scholar]
- Kranenburg O, Gebbink MF, Voest EE. Stimulation of angiogenesis by Ras proteins. Biochim Biophys Acta. 2004;1654:23–37. doi: 10.1016/j.bbcan.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Le LQ, Parada LF. Tumor microenvironment and neurofibromatosis type I: connecting the GAPs. Oncogene. 2007;26:4609–4616. doi: 10.1038/sj.onc.1210261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao JH, Perez-Losada J, Wu D, Delrosario R, Tsunematsu R, Nakayama KI, Brown K, Bryson S, Balmain A. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature. 2004;432:775–779. doi: 10.1038/nature03155. [DOI] [PubMed] [Google Scholar]
- Masuda K, Ishikawa Y, Onoyama I, Unno M, de Alboran IM, Nakayama KI, Nakayama K. Complex regulation of cell-cycle inhibitors by Fbxw7 in mouse embryonic fibroblasts. Oncogene. 2010;29:1798–1809. doi: 10.1038/onc.2009.469. [DOI] [PubMed] [Google Scholar]
- McGillicuddy LT, Fromm JA, Hollstein PE, Kubek S, Beroukhim R, De Raedt T, Johnson BW, Williams SM, Nghiemphu P, Liau LM, et al. Proteasomal and genetic inactivation of the NF1 tumor suppressor in gliomagenesis. Cancer Cell. 2009;16:44–54. doi: 10.1016/j.ccr.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R, Boyd L. Analysis of RING finger genes required for embryogenesis in C. elegans. Genesis. 2004;38:1–12. doi: 10.1002/gene.10243. [DOI] [PubMed] [Google Scholar]
- Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- Nalepa G, Rolfe M, Harper JW. Drug discovery in the ubiquitin-proteasome system. Nat Rev Drug Discov. 2006;5:596–613. doi: 10.1038/nrd2056. [DOI] [PubMed] [Google Scholar]
- Nord AS, Chang PJ, Conklin BR, Cox AV, Harper CA, Hicks GG, Huang CC, Johns SJ, Kawamoto M, Liu S, et al. The International Gene Trap Consortium Website: a portal to all publicly available gene trap cell lines in mouse. Nucleic Acids Res. 2006;34:D642–648. doi: 10.1093/nar/gkj097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T, Michel JJ, Schottelius AJ, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Phan VT, Ding VW, Li F, Chalkley RJ, Burlingame A, McCormick F. The RasGAP Proteins Ira2/Neurofibromin Are Negatively Regulated by Gpb1 in Yeast and ETEA in Humans. Mol Cell Biol. 2010;30:2264–2279. doi: 10.1128/MCB.01450-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan H, Jallepalli PV, Rago C, Velculescu VE, Kinzler KW, Vogelstein B, Lengauer C. Inactivation of hCDC4 can cause chromosomal instability. Nature. 2004;428:77–81. doi: 10.1038/nature02313. [DOI] [PubMed] [Google Scholar]
- Reynolds PJ, Simms JR, Duronio RJ. Identifying determinants of cullin binding specificity among the three functionally different Drosophila melanogaster Roc proteins via domain swapping. PLoS One. 2008;3:e2918. doi: 10.1371/journal.pone.0002918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangha N, Wu R, Kuick R, Powers S, Mu D, Fiander D, Yuen K, Katabuchi H, Tashiro H, Fearon ER, et al. Neurofibromin 1 (NF1) defects are common in human ovarian serous carcinomas and co-occur with TP53 mutations. Neoplasia. 2008;10:1362–1372. doi: 10.1593/neo.08784. following 1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasagawa Y, Urano T, Kohara Y, Takahashi H, Higashitani A. Caenorhabditis elegans RBX1 is essential for meiosis, mitotic chromosomal condensation and segregation, and cytokinesis. Genes Cells. 2003;8:857–872. doi: 10.1046/j.1365-2443.2003.00682.x. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Yukiue H, Kobayashi Y, Moriyama S, Nakashima Y, Kaji M, Fukai I, Kiriyama M, Yamakawa Y, Fujii Y. Expression of the sensitive to apoptosis gene, SAG, as a prognostic marker in nonsmall cell lung cancer. Int J Cancer. 2001;95:375–377. doi: 10.1002/1097-0215(20011120)95:6<375::aid-ijc1066>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Seol JH, Feldman RMR, Zachariae WZ, Shevchenko A, Correll CC, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, et al. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes & Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen TL, Park AY, Alcaraz A, Peng X, Jang I, Koni P, Flavell RA, Gu H, Guan JL. Conditional knockout of focal adhesion kinase in endothelial cells reveals its role in angiogenesis and vascular development in late embryogenesis. J Cell Biol. 2005;169:941–952. doi: 10.1083/jcb.200411155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. Alteration of SAG mRNA in human cancer cell lines: Requirement for the RING finger domain for apoptosis protection. Carcinogenesis. 1999;20:1899–1903. doi: 10.1093/carcin/20.10.1899. [DOI] [PubMed] [Google Scholar]
- Sun Y. E3 ubiquitin ligases as cancer targets and biomarkers. Neoplasia. 2006;8:645–654. doi: 10.1593/neo.06376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Tan M, Duan H, Swaroop M. SAG/ROC/Rbx/Hrt, a zinc RING finger gene family: molecular cloning, biochemical properties, and biological functions. Antioxid Redox Signal. 2001;3:635–650. doi: 10.1089/15230860152542989. [DOI] [PubMed] [Google Scholar]
- Swaroop M, Wang Y, Miller P, Duan H, Jatkoe T, Madore S, Sun Y. Yeast homolog of human SAG/ROC2/Rbx2/Hrt2 is essential for cell growth, but not for germination: Chip profiling implicates its role in cell cycle regulation. Oncogene. 2000;19:2855–2866. doi: 10.1038/sj.onc.1203635. [DOI] [PubMed] [Google Scholar]
- Tan M, Davis SW, Saunders TL, Zhu Y, Sun Y. RBX1/ROC1 disruption results in early embryonic lethality due to proliferation failure, partially rescued by simultaneous loss of p27. Proc Natl Acad Sci U S A. 2009;106:6203–6208. doi: 10.1073/pnas.0812425106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Gu Q, He H, Pamarthy D, Semenza GL, Sun Y. SAG/ROC2/RBX2 is a HIF-1 target gene that promotes HIF-1alpha ubiquitination and degradation. Oncogene. 2008;27:1404–1411. doi: 10.1038/sj.onc.1210780. [DOI] [PubMed] [Google Scholar]
- Tan P, Fuchs SY, Chen A, Wu K, Gomez C, Ronai Z, Pan Z-Q. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of IkBa. Mol Cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- TCGA Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetzlaff MT, Yu W, Li M, Zhang P, Finegold M, Mahon K, Harper JW, Schwartz RJ, Elledge SJ. Defective cardiovascular development and elevated cyclin E and Notch proteins in mice lacking the Fbw7 F-box protein. Proc Natl Acad Sci U S A. 2004;101:3338–3345. doi: 10.1073/pnas.0307875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunematsu R, Nakayama K, Oike Y, Nishiyama M, Ishida N, Hatakeyama S, Bessho Y, Kageyama R, Suda T, Nakayama KI. Mouse Fbw7/Sel-10/Cdc4 is required for notch degradation during vascular development. J Biol Chem. 2004;279:9417–9423. doi: 10.1074/jbc.M312337200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yang J, Zheng H, Tomasek GJ, Zhang P, McKeever PE, Lee EY, Zhu Y. Expression of mutant p53 proteins implicates a lineage relationship between neural stem cells and malignant astrocytic glioma in a murine model. Cancer Cell. 2009;15:514–526. doi: 10.1016/j.ccr.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D, Sun Y. Small RING finger proteins RBX1 and RBX2 of SCF E3 ubiquitin ligases: the role in cancer and as cancer targets. Genes & Cancer. 2010;1:700–707. doi: 10.1177/1947601910382776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff JM. Pathology of tumors of the peripheral nerve sheath in type 1 neurofibromatosis. Am J Med Genet. 1999;89:23–30. doi: 10.1002/(sici)1096-8628(19990326)89:1<23::aid-ajmg6>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Wu K, Fuchs SY, Chen A, Tan P, Gomez C, Ronai Z, Pan ZQ. The SCF(HOS/beta-TRCP)-ROC1 E3 ubiquitin ligase utilizes two distinct domains within CUL1 for substrate targeting and ubiquitin ligation. Mol Cell Biol. 2000;20:1382–1393. doi: 10.1128/mcb.20.4.1382-1393.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Guignard F, Zhao D, Liu L, Burns DK, Mason RP, Messing A, Parada LF. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer Cell. 2005;8:119–130. doi: 10.1016/j.ccr.2005.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Romero MI, Ghosh P, Ye Z, Charnay P, Rushing EJ, Marth JD, Parada LF. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 2001;15:859–876. doi: 10.1101/gad.862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.