Abstract
Higher-order structure influences critical functions in nearly all non-coding and coding RNAs. Most single-nucleotide resolution RNA structure determination technologies cannot be used to analyze RNA from scarce biological samples, like viral genomes. To make quantitative RNA structure analysis applicable to a much wider array of RNA structure-function problems, we developed and applied high-sensitivity selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE) to structural analysis of authentic genomic RNA of the xenotropic murine leukemia virus-related virus (XMRV). For analysis of fluorescently labeled cDNAs generated in high-sensitivity SHAPE experiments, we developed a two-color capillary electrophoresis approach with zeptomole molecular detection limits and sub-femtomole sensitivity for complete SHAPE experiments involving hundreds of individual RNA structure measurements. High-sensitivity SHAPE data correlated closely (R = 0.89) with data obtained by conventional capillary electrophoresis. Using high-sensitivity SHAPE, we determined the dimeric structure of the XMRV packaging domain, examined dynamic interactions between a packaging domain RNA and viral nucleocapsid protein inside virion particles, and identified the packaging signal for this virus. Despite extensive sequence differences between XMRV and the intensively studied Moloney murine leukemia virus, architectures of the regulatory domains are similar and reveal common principles of gammaretrovirus RNA genome packaging.
Introduction
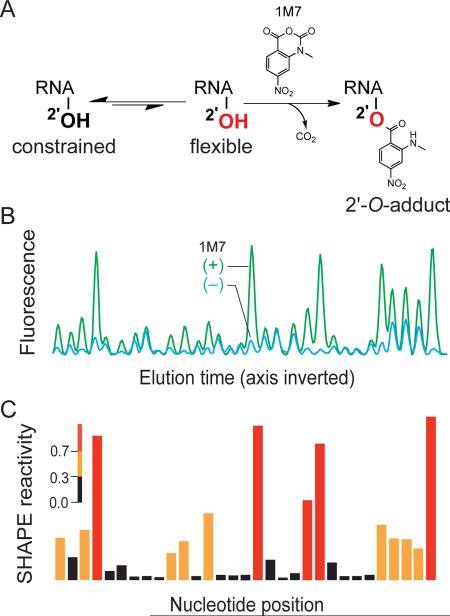
As is likely to be the case for essentially all coding and non-coding RNAs, higher-order secondary and tertiary structures in viral genomes play numerous roles in replication.1–3 High-throughput selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE) makes possible characterization of RNA structure at single-nucleotide resolution on the kilobase scale and has proven to be a powerful approach for understanding structure-function relationships in RNA.4–7 In a SHAPE experiment, conformationally flexible (generally single-stranded) nucleotides react with 1-methyl-7-nitroisatoic anhydride (1M7),8 or another electrophilic SHAPE reagent,9,10 to create a covalent 2'-O-ribose adduct (Figure 1A). Sites of adduct formation are detected by primer extension using fluorescently labeled primers. SHAPE primer extension products can be separated on commercially available capillary electrophoresis sequencing instruments11,12 resulting in an electropherogram in which peak intensities correlate with local flexibility at each individual nucleotide in the RNA (Figure 1B).13 These data are processed to yield a reactivity profile for every RNA nucleotide (Figure 1C). SHAPE has been validated by analysis of RNAs with known structures and was used to discover structure-function relationships in an entire HIV-1 genome,5,11 and in regulatory domains in the Moloney murine leukemia virus (MuLV).14,15
Figure 1. SHAPE Overview.
(A) Mechanism of RNA SHAPE chemistry. Hydroxyl-selective electrophiles modify the 2'-OH group of flexible RNA nucleotides (red) to create a 2'-O-adduct. (B) Subsequent detection of 2'-O-adducts by primer extension and separation by capillary electrophoresis yields an electropherogram of fluorescent peaks. Peak volumes correspond to SHAPE reactivity. (C) Integrated peak intensities after background subtraction.
The sensitivity of SHAPE is limited by the generally poor detection limits of conventional capillary electrophoresis instrumentation and requires ~3 pmol of RNA per experiment.4 This limitation is significant: Tens of liters of a highly optimized cell culture system were required to obtain sufficient genome copies (~100 pmol) for analysis of an entire HIV-1 genome.5 High levels of background noise from light scattering, leakage of excitation light, and electronic noise reduce the ability of conventional instruments to detect fluorescently-labeled primer extension products.16 When optimized for low-abundance species, capillary electrophoresis is, in principal, capable of detecting single molecules.17
To address the challenge of analyzing rare, biologically important RNAs, we have developed an ultra-sensitive RNA structure analysis platform that employs a two-color capillary electrophoresis instrument with zeptomole (10−21) instrument detection limits and sub-femtomole sensitivity for complete SHAPE experiments comprised of hundreds of individual structure measurements. Here we describe application of this technology to analysis of a low-abundance gammaretrovirus, xenotropic murine leukemia virus related virus (XMRV).18 Although XMRV is not likely to be a human pathogen,19,20 as originally reported,18,21 this virus is an excellent example of a recently identified infectious agent currently available in only low amounts. XMRV is evolutionarily related to the well-characterized MuLV,22–25 suggesting that critical features of its structure and biology are conserved. Here, we used high-sensitivity SHAPE to analyze scarce XMRV RNA gently extracted from virions and inside viral particles. Despite extensive sequence differences between XMRV and the MuLV, the regulatory domain architectures are similar, revealing universal principles of retroviral RNA genome packaging.
Results
Design and construction of a highly sensitive capillary electrophoresis instrument
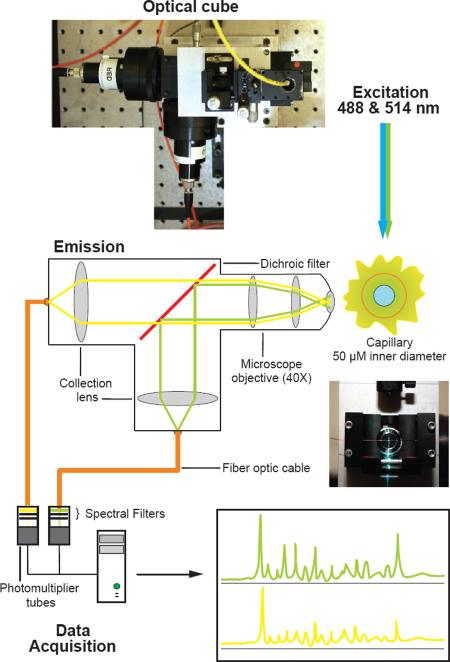
We sought to improve the detection limits of SHAPE experiments by designing and building a robust, yet highly sensitive, capillary electrophoresis instrument. A single-mode fiber optic cable with an integrated lens was used to focus the light from a dual color argon laser (488 and 514 nm) onto a capillary (Figure 2). The optical fiber enabled the laser to be placed at a non-interfering distance and ensured robust alignment of the laser beam. A high-numerical aperture microscope objective oriented at a right angle to the laser beam collected light from the capillary and minimized collection of scattered excitation light. The microscope objective was mated to an optical cube (Figure 2) with a dichroic filter to reflect green wavelengths while transmitting yellow light. Collection lenses mounted into the cube focused the light onto a multimode optical fiber that acted as a spatial filter to remove scattered light and transfer fluoresced light to the detector. Once the optical path was aligned, the cube rigidly maintained the positions of the various components so that realignment was required infrequently. Filters specific for each fluorophore were placed between the optical cube and the detector photomultiplier tubes. The multimode fibers permitted the detectors to be placed at a distance from the optical cube facilitating alignment of the optical system and minimizing interference from high voltage capillary electrophoresis. The melding of these individual innovations yielded a robust and reliable optical system for the study of RNA structure.
Figure 2. Robust, high sensitivity, capillary electrophoresis instrument design.
Photograph and schematic of the high-sensitivity instrument. The optical cube design maximizes light detection and minimizes background noise within each of the three main capillary electrophoresis instrument components: excitation, emission and data acquisition.
Detection limits for free fluorophores and for a complex SHAPE library
The limits of detection for the capillary electrophoresis instrument for the free 5-FAM and 6-JOE fluorophores used in our SHAPE experiments were determined by analysis of serially diluted free dyes. Dilutions were analyzed until a signal-to-noise ratio of 3:1 could no longer be maintained (Supplementary Figure 1). For 5-FAM, the detection limit was 20 zeptomoles (2 × 10−20 mol), or 10,000 molecules. For 6-JOE, the limit was ten-fold higher at 200 zmol (2 × 10−19 mol).
We then validated the ability of the system to separate, visualize and quantify hundreds of fluorescent peaks simultaneously as required in a SHAPE experiment. We performed dideoxy sequencing reactions26 using an in vitro transcript corresponding to an RNA regulatory signal in the MuLV RNA.25 One fmol of input RNA, or 1,000 times less than that required for a conventional instrument, was used. After primer extension, 59 ± 5% of the input RNA was converted to fluorescently labeled cDNA, (see Methods). Throughout this work, amounts are reported in terms of the input RNA. Triplicate measurements were highly reproducible with standard deviations less than 5% per nucleotide position (Supplementary Figure 2A). We also compared data from a SHAPE reaction on the MuLV RNA obtained with a conventional instrument using 1 pmol RNA versus with the high-sensitivity instrument using 1 fmol of the same sample (Supplementary Figure 2B). The two electropherograms are highly similar (R = 0.90) (Supplementary Figure 2C).
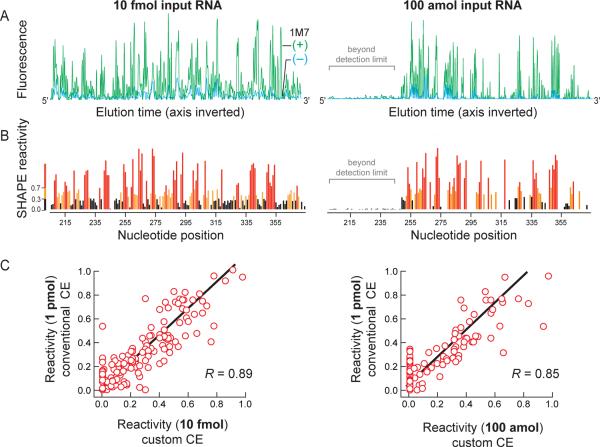
We next used the in vitro MuLV transcript to determine the SHAPE limit of detection. We probed the structure of this RNA by SHAPE using 1M7,8 and the resulting fluorescently labeled cDNA fragments were resolved by both conventional and high-sensitivity capillary electrophoresis. For a cDNA pool derived from 10 fmol of input RNA, we resolved peaks corresponding to all 170 nucleotides in the MuLV RNA using the high-sensitivity instrument (Figure 3A, left). Experiments corresponding to the (+) reagent and no-reagent control reactions were resolved in separate capillary electrophoresis runs, each also containing the same sequencing marker. These data were combined and processed as a single experiment (see Methods) to obtain a SHAPE reactivity profile (Figure 3B, left) and compared to data from a conventional instrument using 100 times more input RNA. The integrated intensities from the high sensitivity instrument correlated closely with data from a conventional instrument; the R-value is ≥0.89 (Figure 3C, left). Next, we sequentially diluted the SHAPE-produced cDNA library until a 3:1 signal-to-noise ratio was reached. This occurred at a dilution corresponding to 100 amol input RNA (Figure 3A and B, right panels). At this level of input RNA, the usable data eventually decays to a level below the instrument detection level due to drop-off12,23 associated with the reverse transcription step (Figure 3A, right panel).
Figure 3. SHAPE limit of detection using MuLV.
(A) Processed electropherograms and (B) quantitative SHAPE reactivities for MuLV RNA treated with 5 mM 1M7 are shown for cDNA pools corresponding to 10 fmol and 100 amol of input RNA. Regions labeled in gray in the 100 amol experiment represent data that fall below the statistical detection limit at the end of the electropherogram due to signal decay during primer extension.11,12 (C) Correlation between SHAPE reactivities, measured at 10 fmol and 100 amol of input RNA, with an identical experiment resolved by conventional capillary electrophoresis instrumentation at 1 pmol input RNA. The full datasets span a range from 0 to ~2 but only data to 1.0 are shown to emphasize the strong best-fit correlation for the most information-rich measurements. R-values for the full data range are higher, at 0.95 and 0.89, respectively.
We estimated the amount of cDNA injected into the instrument at the 100 amol RNA dilution using electrokinetic injection calculations.27 The quantity injected, Q (in moles), onto the capillary electrophoresis instrument is proportional to the sum of electrophoretic (μep) and electroosmotic (μeo) velocities of the DNA in the polymer-filled capillary
| (1) |
scaled by the capillary area (πr2), the electric field strength (E), the input RNA concentration (C) and the injection time (ti). The electro-osmotic mobility is approximately zero in this system (data not shown) and cDNA electrophoretic mobility is approximately equal to the length of the capillary divided by the separation time, ts (the time between cDNA injection and fluorescent primer peak detection),28 thus:
| (2) |
For a solution containing 100 amol input RNA, approximately 1 amol of cDNA was physically injected into the instrument [L = 36 cm, E = 217 V/cm, ts = 875 s (measured for the primer DNA, and thus a lower limit), C = 10 pM (100 amol / 10 μL) and ti = 30 s]. Given that 20 zmol of cDNA are required to visualize a single peak (nucleotide) in the electropherogram (Supplementary Figure 1), 3.4 amol (170 nucleotides × 20 zmol) represents the minimal sample quantity sufficient to report a SHAPE experiment for the MuLV packaging domain.
Thus, at 1 amol of cDNA injected onto the instrument, roughly one-third too few molecules are present to fully represent the entire SHAPE profile. Consistent with this calculation for the 100 amol SHAPE limit of detection, ~60 nucleotides of SHAPE data in the electropherogram integrate to zero or near-zero values, reflecting signal decay that results from imperfect processivity of the reverse transcriptase enzyme used to generate the cDNA pool and a two-fold reduction in injection efficiency for the longest cDNAs (Figure 3A and B, right panels, gray bar). These positions also exhibit an accumulation of zero magnitude points along the x-axis in a correlation plot (Figure 3C, right). Strikingly, at this SHAPE limit of detection, the data are nevertheless still highly correlated with data obtained using conventional instrumentation at 1 pmol RNA (R ≥ 0.85). Ultra-sensitive SHAPE thus yields accurate RNA structural information up to, and even beyond, the limit of fluorophore detection.
Accurate, nucleotide-resolution secondary structure prediction by ultra-sensitive SHAPE
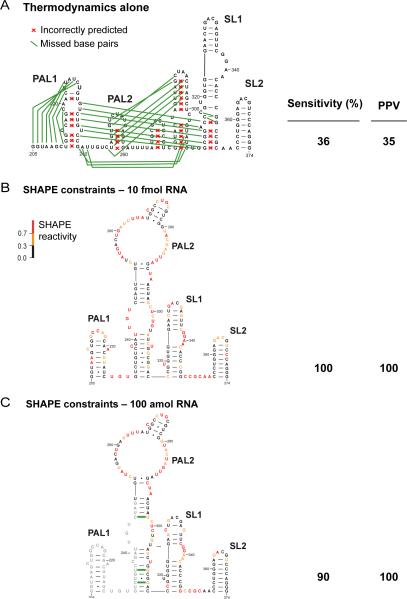
SHAPE reactivities are inversely correlated with the probability that a nucleotide is base paired, and it is therefore possible to use SHAPE information to predict RNA secondary structures accurately.29,30 We used the nucleotide-resolution SHAPE data obtained at 10 fmol and 100 amol, equal to 100 and 10,000 fold lower concentrations than measurable using conventional instrumentation, to direct RNA secondary structure predictions for the MuLV packaging domain. This 170-nucleotide RNA corresponds to a regulatory domain at the 5' end of the MuLV RNA genome31 that links two viral genome strands into a dimeric structure and comprises the “packaging signal” responsible for directing the full genome into nascent viral particles.32 Point mutations in this domain can reduce packaging to non-specific levels.25 The MuLV regulatory domain has been extensively characterized both structurally and biologically and forms a well-defined monomeric structure in the absence of magnesium ion.
The lowest free energy structures for the MuLV packaging domain were calculated using either thermodynamic nearest-neighbor constraints alone or thermodynamic nearest-neighbor constraints and SHAPE reactivity data included as pseudo-free energy constraints.29 Structure predictions were evaluated by their sensitivity (the percent correctly predicted base pairs) and positive predictive value (PPV, the percentage of predicted pairs that are present in the accepted structure). In the absence of SHAPE data, the secondary structure of the MuLV RNA was predicted with sensitivity and PPV values of 36% and 35%, respectively. There were thus significant errors relative to the accepted structure,14 as shown in Figure 4A. We then predicted the structure using SHAPE data as pseudo-free energy change constraints.29,33 The resulting base pair sensitivities were 100 and 90%, respectively, and no incorrect base pairs were proposed (PPV = 100%) (Figure 4B and 4C). Consistent with the strong correlation observed between the ultra-sensitive SHAPE data and data obtained by using pmol RNA amounts, ultra-sensitive SHAPE yielded accurate secondary structure predictions with as few as 100 amol, or 10,000 molecules, of RNA.
Figure 4. SHAPE-directed folding of the MuLV genomic RNA monomer.
(A) Secondary structure of the MuLV monomer in the packaging domain as predicted using thermodynamic constraints alone. Missed base pairs and incorrectly predicted base pairs relative to the accepted structure14 are denoted by green lines and red x's, respectively. (B and C) MuLV monomer structures predicted using SHAPE constraints to direct RNA secondary structure predictions. Data were obtained at 10 fmol and 100 amol input RNA.
Ultra-sensitive SHAPE analysis of authentic XMRV
We then analyzed a 184-nucleotide region near the 5' end of the XMRV RNA genome that is functionally homologous to the MuLV dimerization and packaging regulatory domain. SHAPE reactivity measurements were obtained using 50 fmol aliquots of XMRV genomic RNA that had been gently extracted from virions purified from cell culture,34 using the 22RV1 virus-producing cell line.35 Integrated SHAPE reactivities (Figure 5A), taken together with comparison to prior structural analysis of MuLV, were used to develop a model for the structure of this regulatory domain. SHAPE probing data of RNA isolated from virions indicated clearly that the XMRV genome was present as a dimer in mature viral particles. The overall architecture is similar to that of MuLV and includes four key dimer interaction elements. Two palindromic sequences, termed PAL1 and PAL2, form intermolecular duplexes in the dimeric structure, as evidenced by their uniformly low SHAPE reactivities (Figure 5B). In contrast, when these structures are forced to fold in a monomeric conformation, the lowest free energy models contain several structures with reactivities that are incompatible with the SHAPE data (Supp. Figure 5). The apical loops in SL1 and SL2 form intermolecular loop-loop kissing interactions, as these nucleotides are unreactive towards SHAPE and by analogy with previous work on MuLV (Figure 5B).23
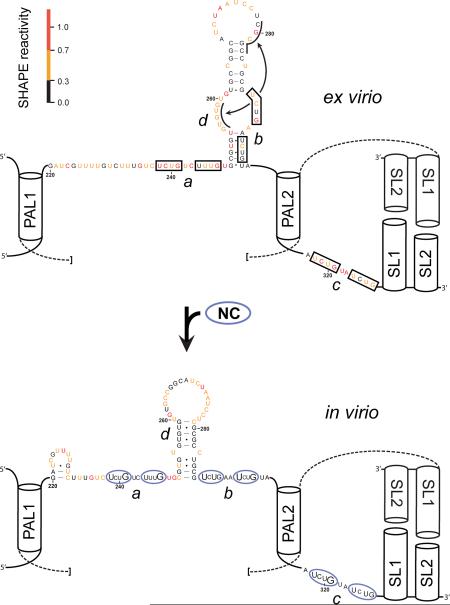
Figure 5. Structural analysis of authentic XMRV genomic RNA.
(A) Integrated SHAPE data obtained by treatment of XMRV genomic RNA with 5 mM 1M7. (B) Secondary structure model for the XMRV RNA genome in the dimer state. Experimental SHAPE data from panel A are superimposed as nucleotide colors. Key interaction elements that stabilize the dimer structure are labeled. NC binding motifs identified in this work are boxed. (C) SHAPE reactivity differences between the RNA gently extracted from virions versus the genomic RNA modified inside virion particles. Positive amplitudes indicate nucleotides that are protected from SHAPE reaction in virio relative to the protein-free ex virio RNA.
We next analyzed the structure of the XMRV genome inside viral particles, again using fmol quantities of RNA. Intact viral particles were treated with the 1M7 SHAPE reagent, the modified RNA was extracted from the virions and primer extension products were quantified by ultra-sensitive SHAPE. Because SHAPE data are highly reproducible, the net effect of different environments – including a complex in virio environment – can be readily characterized by subtracting one SHAPE profile from another to yield a difference plot (Figure 5C). The difference analysis revealed three major regions that were less reactive, or protected, in virions relative to the gently extracted genomic RNA and one segment that was more reactive in virions (in blue, Figure 5C). These experiments, performed on fmol amounts of a structurally uncharacterized authentic retroviral RNA genome, provided strong clues as to the mechanism of genome packaging in XMRV.
Discussion
Most biologically important RNAs are present at very low levels. As a result, studies on these RNAs have been limited to indirect functional studies or direct experimental studies using in vitro transcribed RNA as models. Recent work blending SHAPE with next-generation sequencing has lowered detection limits by a factor of ten, to the tenth-picomole (10−13) level.7 Analysis of scarce RNAs might be further optimized by RT-PCR approaches but care must be taken so that amplification steps do not reduce the quantitative accuracy of the resulting structural information.36 In this study, we increased the molecular sensitivity of the SHAPE nucleotide-resolution RNA structure probing technology to sub-femtomole (10−16) levels, or four orders of magnitude, by implementing multiple innovations to achieve highly sensitive capillary electrophoresis detection of the fluorescently-labeled cDNA primer extension products.
Using ultra-sensitive SHAPE, we examined the structure of the dimerization and packaging regulatory domain of XMRV genomic RNA gently extracted from virions and also within authentic virions. The ultra-sensitive SHAPE data obtained on authentic viral RNA strongly support a model in which XMRV is present as a dimer in mature virions (compare Figures 5B and Supp. Figure 5). Genomic dimerization is mediated by four key intermolecular interactions at PAL1, PAL2, SL1 and SL2 sequences (Figure 5B). Remarkably, even though there are extensive sequence differences and nucleotide insertions in XMRV relative to MuLV (Supplementary Figure 3), the structures of their respective packaging domains are conserved. It is likely that a similar dimeric architecture is evolutionarily conserved among many gammaretroviruses.
The dimerization and packaging domain in the MuLV virus has been intensively studied and packaging of the RNA genome involves, in part, a binding interaction between the nucleocapsid (NC) domain of Gag and a specific sequence of nucleotides in the RNA.37 The MuLV NC protein binds tandem repeats of the sequence UCUG. The strongest interactions occur at the first U and final G of the motif and this single-stranded element must be flanked by helical regions for highest-affinity binding.25 There are two such elements in the MuLV RNA genome and these elements constitute the RNA packaging signal for this virus.25
In contrast, inspection of the SHAPE reactivity differences in the XMRV packaging domain for the ex virio and in virio RNA revealed three strong sets of protections. By analogy with previous work on MuLV25, virion-specific protection from SHAPE reactivity is likely due to protein binding. Virion-specific increases in reactivity are most directly attributed to RNA conformational changes. Two of these three motifs are unique to XMRV as compared to MuLV. The first unique motif (Figure 5C, motif a) has the sequence UCUGUCUUUG and is positioned between the structured elements PAL1 and SL0 (see boxed nucleotides 238–247, Figure 5B). The second motif, UCUGAAUCUG, has no analog in MuLV (Figure 5C, motif b) and is partially occluded by the SL0 helix in the model for the deproteinized RNA (Figure 5B, nucleotides 289–298). Difference plot analysis suggested that the base of helix SL0 is disrupted upon NC binding and has a different conformation inside the virion. (Figure 5C, region d). This model is supported by differences within SL0 in the in virio state compared to the ex virio state (Figure 6, doubleheaded arrow). It is also consistent with the known nucleic acid chaperone properties of NC.38 The third motif in (Figure 5C, motif c), UCUGUAUCUG, is identical in both position and sequence to that in MuLV (Figure 5B, nucleotides 318–327). These three flexible regions in XMRV, consisting of some form of the sequence UCUGRRURUG (Figure 6, see regions a, b and c), are flanked by well-defined RNA helices and interact most strongly with NC at the first U and last G nucleotides within these motifs (capitalized in Figure 6, lower panel).
Figure 6. Model for XMRV retroviral genome packaging.
Secondary structure model for the NC domain of Gag bound to the XMRV packaging signal RNA. Regions with the largest changes in SHAPE reactivity for the RNA analyzed inside virions, relative to the ex virio RNA, are identified (a–d, bottom panel) with the same labels used in Figure 5C. Within each NC binding site (ovals), nucleotides showing the strongest protections inside virions are emphasized with large lettering. Conserved helical elements that stabilize the dimer state and do not differ in conformation when in virio and ex virio data are compared are shown schematically as cylinders; these helices flank single-stranded regions bound by NC and are components of the protein binding site.25
In conclusion, high sensitivity SHAPE has made possible structural analysis of a rare, recently discovered, authentic genomic viral RNA. SHAPE characterization of the XMRV RNA within virion particles revealed functional determinants of a structure critical for packaging. XMRV appears to use a genome packaging strategy that is similar to that employed by MuLV, but has distinct and more complex features (Figure 6). We anticipate that by making analysis of femtomole quantities of RNA possible, high sensitivity SHAPE technologies will accelerate functional and structural characterization of many low abundance RNAs, including viral genomes, rare biological mutants, and, with further refinement, complex mixtures like messenger RNAs and clinical samples.
Experimental Procedures
Instrument design and construction
The high-sensitivity capillary electrophoresis laser-induced fluorescence instrument is of custom design and construction (see Supplementary Figure 4).39 The detector utilizes a right-angle excitation design (Figure 2). A single-mode fiber-optic cable with an integrated lens (3.5-cm focal length, Oz optics) was used to focus the beam from a multiline argon laser (488 and 514 nm, JDS Uniphase) into the lumen of a capillary. Light from the capillary was collected using a microscope objective (40×, 0.75 na, Plan Fluor, Nikon). The laser and capillary were positioned with respect to each other using an XYZ stage (Newport). A dichroic filter (FF550-Di01-25×36, Semrock) was used to divide the collected light into the 5-FAM and 6-Joe wavelengths. Collection lenses (two collimators for single-mode fiber/ HPUCO-23-400/700-M-50AC-30OD, Oz Optics) were used to direct the light into multimode optical cables (200/230 μm multimode IRVIS fiber/ QMMJ-33-IRVIS-200/230-3-1, Oz Optics). The entry-way into the fiber optic cable also acted as a pin hole to spatially filter the light and remove residual scattered excitation light. Bandpass filters (525 and 565 nm; FF01-525/20-25, FF01-565/24-25, Semrock) appropriate for each fluorophore were used to further remove excitation light prior to entry into the detector. The light was detected, amplified and converted to current using a photomultiplier tube (PMT, H7732, Hamamatsu) mounted in a custom housing with shutters and filter slots. Current from the PMT was amplified and converted to voltage using a preamplifier (Advanced Research Instruments). The signal was then digitized using an analog to digital board (National Instruments) and the instrument was controlled via a custom Labview program designed in conjunction with the UNC Chemistry department electronics shop (available upon request). Prefabricated 310 Genetic Analyzer capillaries 47 cm in length with a 50-μm inner diameter (Applied Biosystems) were constructed to have an optical window 36 cm from the inlet. Two nonconductive columns were mounted on either side of the optical unit to accommodate the capillary inlet, outlet, and custom-built capillary washing system. The capillary electrophoresis system was powered by a high voltage power supply (Spellman). A full set of mechanical drawings is available upon request.
Instrument limit of detection
Experiments to determine instrumental detection limits were performed using free 5-FAM and 6-JOE fluorophores. Samples, diluted from a 10 μM stock, were hydrodynamically injected into the system by elevating the capillary at the sample inlet by 10 cm for 12 s to permit sample introduction via siphoning. The volume of sample injected at 0.17 nL/sec27 into the 50-μm capillary using gravity injection was thus approximately 2 nL. Experiments were carried out in triplicate (Supplementary Figure 1). The capillary was filled with 100 mM borate (pH 8) and samples were separated at 6 kV for 10 min. Samples were diluted until an integrated fluorescence signal-to-noise ratio of 3:1 was no longer maintained. The signal-to-noise ratio was calculated by dividing the integrated fluorescence by standard deviation of background signal from the first 100 s after injection.
Retroviral RNA transcripts
The in vitro MuLV RNA construct is 331 nucleotides and spans the dimerization and packaging regulatory domain (~170 nts) including 5' and 3' flanking sequences of 46 and 115 nucleotides, respectively.14,17,24 The transcript was generated and purified as described.25
Isolation of XMRV
XMRV was obtained from the constitutively producing XMRV cell line, 22Rv1 (ATCC),35 cultured in RPMI media with 10% (vol/vol) fetal bovine serum. Culture fluids were collected and filtered, first through 1-μm then 0.45-μm filters, before virus was isolated by continuous-flow ultracentrifugation.34 Virus was subtilisin-treated as described40 and collected by centrifugation through a 3.6-mL 20% (wt/vol) sucrose (in PBS without Ca2+ or Mg2+) cushion at 390,000 × g for 2 h at 4 °C. The supernatant was removed, the viral pellet was overlaid with ice-cold PBS (without Ca2+ or Mg2+) and the sample was centrifuged for an additional 1 hr at 4 °C. The supernatant was aspirated and the pellets were resuspended in 600 μL HNS buffer [50 mM HEPES (pH 8.0), 200 mM NaCl, 0.1 mM EDTA, 10% (v/v)] for further manipulations, described below.
SHAPE analysis
XMRV RNA, gently extracted from XMRV virions as described previously,15 was incubated in 1× folding buffer [50 mM HEPES (pH 7.5), 200 mM potassium acetate (pH 7.5), and 5 mM MgCl2] at 37 °C for 20 min prior to modification. The resuspended virus in HNS buffer (300 μL) was pipetted into 33.3 μL 50 mM 1M7 (1-methyl-7-nitroisatoic anhydride) in DMSO (plus reagent) or neat DMSO (minus reagent), mixed by pipetting and incubated at 37 °C for 15 min to modify RNA. Virions were collected by ultracentrifugation as described.40 Virion resuspensions were preincubated at 37 °C for 5 min and 300 μL of virus solution was pipetted into 33 μL solutions of either 1M7 (50 mM) or DMSO at 37 °C. Solutions were mixed by pipetting and incubated for 15 min at 37 °C. Samples turn from faint yellow to dark orange upon reaction with 1M7. Samples were resuspended in 100 μL of 1× lysis buffer [50 mM Hepes (pH 7.5), 200 mM NaCl, 3 mM MgCl2] and lysed with 1% (wt/vol) SDS and 20 mg/mL proteinase K at 22 °C for 30 min. The lysate was extracted four times with phenol/chloroform/isoamyl alcohol (25:24:1), followed by four extractions with pure chloroform. Samples were then precipitated with ethanol and stored in 70% ethanol at −20 °C until primer extension.
In vitro generated MuLV RNA monomer transcripts (2 pmol) were renatured by heating at 95 °C for 3 min in 12 μL water, cooled on ice for 3 min and equilibrated at 37 °C for 3 min in a folding buffer without magnesium [50 mM HEPES (pH 7.5), 200 mM potassium acetate (pH 7.5)]. SHAPE modification was performed by treating 9 μL MuLV RNA with 1 μL 1M7 (50 mM in DMSO), followed by incubation at 37 °C for 2 min. RNA was recovered using a precipitation approach optimized for the quantitative recovery of low abundance RNAs [2.5 vol ethanol, 1 vol isopropanol, 1 μL glycogen (20 mg/mL); incubation at –20 °C for 60 min; and centrifugation at 20,000× g]. Pellets were resuspended in 6 μL 1/2× TE [5 mM Tris, 0.5 mM EDTA (pH 8.0)] buffer.
Primer extension
The general protocol was described in detail previously.4,14 DNA primers incorporated locked nucleic acid nucleotides (LNA, underlined below) for use on low-copy RNA to increase binding affinity and stringency.41–43 The efficiency of primer extension using LNA-containing primers was calculated to be 59 ± 5% based on the fraction full-length extension product relative to the sum of this value and the unextended primer peak after correcting for the two-fold excess of primer over RNA target, using electropherograms from a no-reagent control reactions. MuLV primers (5'-GGUGC ACCAA AGAGU CCAAA AGC-3', 5'-end labeled with 5-FAM or 6-JOE) annealed 3' of the MuLV dimerization domain (nucleotides 422 to 445) XMRV primers (5'-GAAGT CTCTG TCTCT CGTCC CCC-3') annealed to nucleotides 443 to 468 in the genome sequence. Primers (0.5 μL; 2 fmol) were annealed to XMRV RNA (6 μL; 1 fmol in 1/2× TE buffer) by heating at 65 °C and 45 °C for 5 min and then snap-cooled on ice. Reverse transcription buffer [3 μl; 200 mM Tris (pH 8.0), 250 mM KCl, 10 mM MgCl2, 2 mM each dNTP, 20 mM DTT] was added at 0 °C, and primer extension was performed with Superscript III (Invitrogen) reverse transcriptase (0.5 μl; 100 U) at 45 °C for 1 min, followed by incubation at 52 °C and 65 °C, for 10 min each. MuLV transcript RNA was subjected to primer extension exactly as described above except that primer and RNA concentrations were 2 and 1 pmol, respectively. Reactions were quenched by cooling to 4 °C and addition of 3 M sodium acetate (pH 5.2). A sequencing marker was generated by adding 0.5 μL dideoxy-GTP (10 mM) to the primer extension reaction mixture using unmodified RNA. The 1M7 and DMSO reaction mixes were each combined with equal amounts of dideoxy-GTP-terminated sequencing ladders, precipitated with ethanol, and resuspended in deionized formamide (20 μl).
Capillary electrophoresis
For conventional capillary electrophoresis, cDNA fragments were resolved on an Applied Biosystems 3130 instrument. This instrument is fully automated for use with high abundance cDNA samples of ~1 pmol or greater. The samples were loaded in microtiter plates and injected onto the instrument using standard instrument protocols. MuLV and XMRV cDNA fragments, ranging in concentration from 100 amol to 50 fmol of input RNA, were resolved by custom high-sensitivity capillary electrophoresis. For ultra-sensitive SHAPE, samples of 1 to 10 μL were electrokinetically injected (10.2 kV for 30 s) onto a 47 cm (50-μm inner diameter) uncoated capillary containing Pop7 (Applied Biosystems) as the internal polymer. The detection window was placed at 36 cm. Voltage was applied using positive polarity (10.2 kV for 60 min) and immersing the capillary ends in a 1× running buffer (Applied Biosystems). The PMT voltages were 0.9 kV and the laser power was 2 mW. Excitation was achieved using an argon laser at 488 and 514 nm wavelengths focused within the capillary at the detection window. 5-FAM was used to detect cDNAs corresponding to SHAPE structure probing and the lower sensitivity dye, 6-JOE, was used for sequencing. A single high-sensitivity SHAPE experiment was thus resolved in two separate capillary electrophoresis runs, one each for the plus- and no-reagent reactions. Sensitivity for sequencing is not a critical issue because sequencing can be performed with an in vitro transcript or with DNA rather than with a scarce biological sample.
Data processing and structure prediction
Raw sequence traces were corrected for dye variation and signal decay; peak intensities were integrated using ShapeFinder.12 Traces were analyzed by combining data from two separate capillary runs. Each capillary electrophoresis separation contained (i) a reaction performed in the presence or absence of 1M7 (detected using a FAM-labeled primer) and (ii) a sequencing reaction (labeled with JOE) performed using dideoxy GTP. Data from the two runs were aligned using the identical sequencing lanes and then analyzed as described.12 SHAPE reactivities were normalized by dividing by the average intensity of the 10% most reactive nucleotides, after excluding outliers found by box plot analysis.29 SHAPE reactivity information was used to impose a pseudo-free energy change constraint in conjunction with nearest neighbor thermodynamic parameters in the prediction program RNAstructure.29,30,33
Supplementary Material
Acknowledgments
We are indebted to Freddy Pinero in the UNC Chemistry Instrument shop for mechanical drawings and construction of custom instrument pieces, Alex Villa for creation of the Labview software, Jenny Mauger for helpful discussions on Labview programming, and Julian W. Bess, Jr., (AIDS and Cancer Virus Program, Biological Products Core) for providing and purifying XMRV stocks for this work. This work was supported by grants from the US National Institutes of Health (GM064803 to KMW and CA139599 to NLA) and has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E with SAIC-Frederick, Inc.
References
- (1).Coffin JM, Hughes SH, Varmus HE. Retroviruses. Cold Spring Harbor Press; Plainview, NY: 1997. [PubMed] [Google Scholar]
- (2).D'Souza V, Summers MF. Nat Rev Microbiol. 2005;3:643. doi: 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- (3).Roberts L, Holcik M. EMBO Rep. 2009;10:449. doi: 10.1038/embor.2009.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Wilkinson KA, Merino EJ, Weeks KM. Nat Protoc. 2006;1:1610. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- (5).Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr., Swanstrom R, Burch CL, Weeks KM. Nature. 2009;460:711. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Mauger K. M. W. a. D. M. Acc. Chem. Res. 2011;44 doi: 10.1021/ar200051h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Lucks JB, Mortimer SA, Trapnell C, Luo S, Aviran S, Schroth GP, Pachter L, Doudna JA, Arkin AP. Proc Natl Acad Sci U S A. 2011;108:11063. doi: 10.1073/pnas.1106501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Mortimer SA, Weeks KM. J Am Chem Soc. 2007;129:4144. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- (9).Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. J Am Chem Soc. 2005;127:4223. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- (10).Mortimer SA, Weeks KM. J Am Chem Soc. 2008;130:16178. doi: 10.1021/ja8061216. [DOI] [PubMed] [Google Scholar]
- (11).Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. RNA. 2008;14:1979. doi: 10.1261/rna.1166808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Gherghe CM, Shajani Z, Wilkinson KA, Varani G, Weeks KM. J Am Chem Soc. 2008;130:12244. doi: 10.1021/ja804541s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Gherghe C, Weeks KM. J Biol Chem. 2006;281:37952. doi: 10.1074/jbc.M607380200. [DOI] [PubMed] [Google Scholar]
- (15).Gherghe C, Leonard CW, Gorelick RJ, Weeks KM. J Virol. 2010;84:898. doi: 10.1128/JVI.01602-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).MacTaylor CE, Ewing AG. Electrophoresis. 1997;18:2279. doi: 10.1002/elps.1150181218. [DOI] [PubMed] [Google Scholar]
- (17).Borland LM, Kottegoda S, Phillips KS, Allbritton NL. Annu Rev Anal Chem (Palo Alto Calif) 2008;1:191. doi: 10.1146/annurev.anchem.1.031207.113100. [DOI] [PubMed] [Google Scholar]
- (18).Urisman A, Molinaro RJ, Fischer N, Plummer SJ, Casey G, Klein EA, Malathi K, Magi-Galluzzi C, Tubbs RR, Ganem D, Silverman RH, DeRisi JL. PLoS Pathog. 2006;2:e25. doi: 10.1371/journal.ppat.0020025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- (19).Weiss RA. BMC Biol. 2010;8:124. doi: 10.1186/1741-7007-8-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Paprotka T, Delviks-Frankenberry KA, Cingoz O, Martinez A, Kung HJ, Tepper CG, Hu WS, Fivash MJ, Jr., Coffin JM, Pathak VK. Science. 2011;333:97. doi: 10.1126/science.1205292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Lombardi VC, Ruscetti FW, Das Gupta J, Pfost MA, Hagen KS, Peterson DL, Ruscetti SK, Bagni RK, Petrow-Sadowski C, Gold B, Dean M, Silverman RH, Mikovits JA. Science. 2009;326:585. doi: 10.1126/science.1179052. [DOI] [PubMed] [Google Scholar]
- (22).Ly H, Parslow TG. J Virol. 2002;76:3135. doi: 10.1128/JVI.76.7.3135-3144.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Badorrek CS, Weeks KM. Biochemistry. 2006;45:12664. doi: 10.1021/bi060521k. [DOI] [PubMed] [Google Scholar]
- (24).Badorrek CS, Gherghe CM, Weeks KM. Proc Natl Acad Sci U S A. 2006;103:13640. doi: 10.1073/pnas.0606156103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Gherghe C, Lombo T, Leonard CW, Datta SA, Bess JW, Jr., Gorelick RJ, Rein A, Weeks KM. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1006897107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Sanger F, Nicklen S, Coulson AR. Biotechnology. 1992;24:104. [PubMed] [Google Scholar]
- (27).Weinberger R. Practical Capillary Electrophoresis. Vol. 2. Academic Press; San Diego: 2000. [Google Scholar]
- (28).Ghosal S. Annu Rev Fluid Mech. 2006;38:309. [Google Scholar]
- (29).Deigan KE, Li TW, Mathews DH, Weeks KM. Proc Natl Acad Sci U S A. 2009;106:97. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Low JT, Weeks KM. Methods. 2010;52:150. doi: 10.1016/j.ymeth.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Badorrek CS, Weeks KM. Nat Chem Biol. 2005;1:104. doi: 10.1038/nchembio712. [DOI] [PubMed] [Google Scholar]
- (32).Adam MA, Miller AD. J Virol. 1988;62:3802. doi: 10.1128/jvi.62.10.3802-3806.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Proc Natl Acad Sci U S A. 2004;101:7287. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Chertova E, Bess JW, Jr., Crise BJ, Sowder IR, Schaden TM, Hilburn JM, Hoxie JA, Benveniste RE, Lifson JD, Henderson LE, Arthur LO. J Virol. 2002;76:5315. doi: 10.1128/JVI.76.11.5315-5325.2002. Jr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Sramkoski RM, Pretlow T. G., 2nd; Giaconia, J. M., Pretlow TP, Schwartz S, Sy MS, Marengo SR, Rhim JS, Zhang D, Jacobberger JW. In Vitro Cell Dev Biol Anim. 1999;35:403. doi: 10.1007/s11626-999-0115-4. [DOI] [PubMed] [Google Scholar]
- (36).Weeks KM. Proc Natl Acad Sci U S A. 2011;108:10933. doi: 10.1073/pnas.1107835108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).D'Souza V, Summers MF. Nature. 2004;431:586. doi: 10.1038/nature02944. [DOI] [PubMed] [Google Scholar]
- (38).Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Prog Nucleic Acid Res Mol Biol. 2005;80:217. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- (39).Sims CE, Meredith GD, Krasieva TB, Berns MW, Tromberg BJ, Allbritton NL. Anal Chem. 1998;70:4570. doi: 10.1021/ac9802269. [DOI] [PubMed] [Google Scholar]
- (40).Ott DE, Coren LV, Johnson DG, Sowder R. C., 2nd; Arthur, L. O., Henderson LE. AIDS Res Hum Retroviruses. 1995;11:1003. doi: 10.1089/aid.1995.11.1003. [DOI] [PubMed] [Google Scholar]
- (41).Latorra D, Arar K, Hurley JM. Mol Cell Probes. 2003;17:253. doi: 10.1016/s0890-8508(03)00062-8. [DOI] [PubMed] [Google Scholar]
- (42).Levin JD, Fiala D, Samala MF, Kahn JD, Peterson RJ. Nucleic Acids Res. 2006;34:e142. doi: 10.1093/nar/gkl756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Fratczak A, Kierzek R, Kierzek E. Biochemistry. 2009;48:514. doi: 10.1021/bi8021069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.