Abstract
Solution NMR provides a powerful approach for detecting complex formation involving weak to moderate intermolecular affinity. However, solution NMR has only rarely been used to detect complex formation between two membrane proteins in model membranes. The impact of specific binding on the NMR spectrum of a membrane protein can be difficult to distinguish from spectral changes that are induced by non-specific binding and/or by changes that arise from forced co-habitation of the two proteins in a single model membrane assembly. This is particularly the case when solubility limits make it impossible to complete a titration to the point of near-saturation of complex formation. In this work experiments are presented that provide the basis for establishing whether specific complex formation occurs between two membrane proteins under conditions where binding is not of high avidity. Application of these methods led to the conclusion that the membrane protein CD147 (also known as EMMPRIN or basigin) forms a specific heterodimeric complex in the membrane with the 99 residue transmembrane C-terminal fragment of the amyloid precursor protein (C99 or APP-βCTF), the latter being the immediate precursor of the amyloid-β polypeptides that are closely linked to the etiology of Alzheimer’s disease.
Introduction
Because NMR chemical shifts for any given compound are often very sensitive to local molecular environment, NMR methods represent a powerful approach for detecting complex formation between two molecules in solution. This has proven especially important for the case of pairs of biomolecules that form specific complexes with only low to moderate affinity. For such complexes the on/off rates for complex formation and dissociation are usually rapid on the NMR time scale, such that observed chemical shifts represent the population weighted averages between the intrinsic spectra of free and complex species. Assuming rapid exchange on the NMR time scale and that “molecule A” can be titrated with “molecule B” from 0% saturation of molecule A up through near-saturation, the changes induced in the spectrum of molecule A can be quantitated as a function of the concentration of molecule B to confirm specificity of complex formation and to determine the dissociation constant, which reflects the free energy difference between free and complexed states. Use of NMR often confers an added advantage in that the nature of the changes observed between the free and complexed state spectra sometimes provides direct insight into the structure and dynamics of the complex. For example NMR has been used to map the binding interface between two membrane-associated molecules.1–5
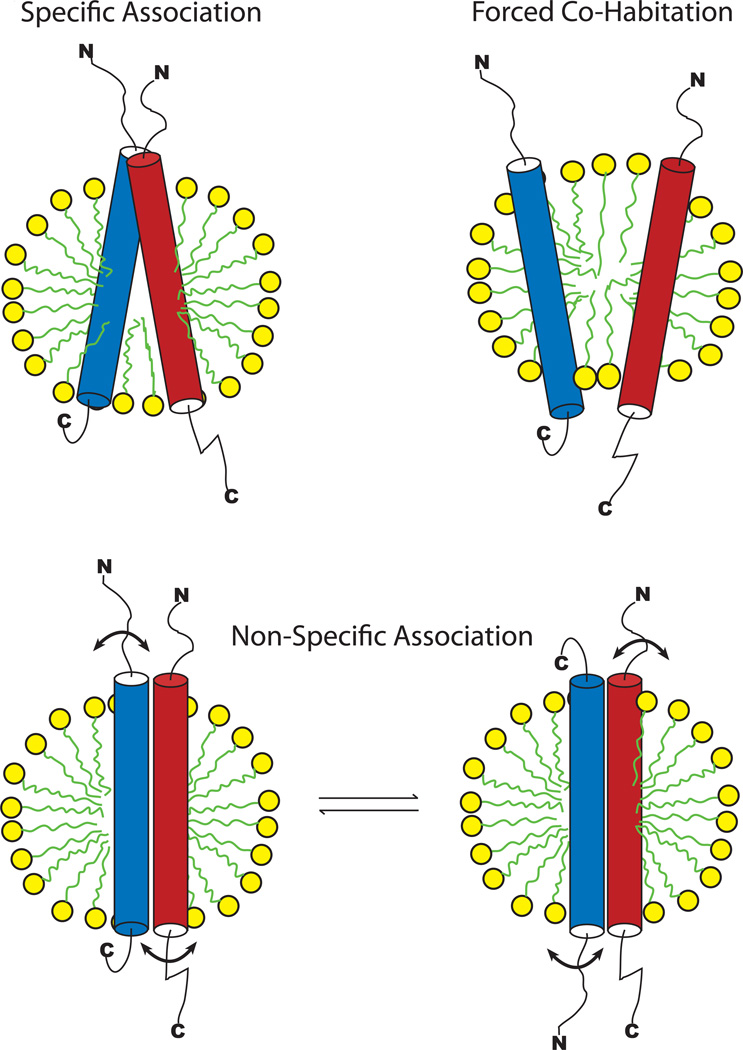
Complex formation between integral membrane proteins in bilayers or in model membranes such as detergent micelles can be difficult to detect, particularly when affinity is modest such that complexes do not have lifetimes of sufficient duration to allow them to be “trapped” using chemical or biochemical methods (such as pull-down assays). NMR can sometimes be used to monitor such titrations. However, when affinity is only modest it can sometimes be difficult carry out the titration to the point of near saturation before the solubility limit of the titrant is reached. In this case, changes are observed in the spectrum of “protein A” during titration by “protein B”, but the shift vs. [B] concentration curves may be nearly linear. In such cases, it is unclear whether the observed shifts represent specific formation of a stoichiometric complex or whether the shifts result from non-specific binding or forced co-habitation of two proteins in a single model membrane unit (e.g., a detergent micelle) (Figure 1). “Specific association” is here defined as stoichiometric complex formation that involves a specific intermolecular interface. Non-specific association means that there is affinity between molecules, but that this affinity does not lead to formation of complexes of specific stoichiometry involving structurally well-defined intermolecular interfaces. An example is provided by “promiscuous inhibitors”, colloidal aggregates of drug-like molecules that will form complexes with many different proteins, mostly via hydrophobic interactions.6 Forced co-habitation means the entrapment of two hydrophobic molecules in the same micelle simply because there are not enough micelles present to allow all of the hydrophobic molecules to populate their own micelles.7 While the two molecules may have no affinity for each other they may nevertheless be forced to interact by virtue of co-entrapment in a very small space (i.e., within the same detergent micelle). In all three cases, intermolecular interactions may be NMR-detectable. Here, we developed methods to distinguish between these possibilities.
Figure 1.
Possible modes of interactions of two membrane proteins in model membranes. Detergent micelles are shown, but the same principles apply to other classes of model membranes. (Top left) Specific Association. (Top right) Forced Co-Habitation: two membrane proteins with no affinity for each other are forced to share a single model membrane unit by statistical mechanics and/or because the number of membrane proteins molecules exceeds the number of available model membrane units present in the solution. (Bottom) Non-Specific Association. By way of example we have depicted two exchanging modes of transient 1:1 association in which the interaction interfaces are not fixed but are heterogeneous (indicated by the motion arrows). Many other possible modes of interaction and non-1:1 stoichiometries would also fit into this class.
As a test case for the methods being developed in this study we examined interactions between two single-span membrane proteins: the C-terminal 99 residue transmembrane domain of the amyloid precursor protein (C99, also known as APP-βCTF) and the C-terminal transmembrane and cytosolic domains of CD147 (see Figure 2A). C99 is a very important protein, serving as the immediate precursor for the amyloid-β (Aβ) polypeptides that are widely believed to be central to the etiology of Alzheimer’s disease. CD147 (also known as EMMPRIN or basigin) is a protein that has been shown to reduce levels of amyloid-β under cellular conditions,8–10 a potentially beneficial activity in the sense that such activity could disfavor Alzheimer’s disease. However, the mechanism by which CD147 lowers Aβ production remains unclear. Full length CD147 is a 27 kDa cell surface protein with two extracellular N-terminal immunoglobulin domains, a single transmembrane segment, and a small intracellular domain (Figure 2A).11 The removal of CD147 by RNA interference causes dose-dependent increases in the levels of secreted Aβ.8,10 Although CD147 was co-immunoprecipitated with the γ-secretase complex that cleaves C99 to release Aβ10,12 and observed to co-purify with γ-secretase,13 additional studies suggested that any direct CD147 association with γ-secretase is transient.13 Moreover, CD147 expression had no effects on the expression and stability of γ-secretase core subunits8,10 or on the association of the γ-secretase complex with lipid rafts.8 Based on these observations, it was suggested that CD147 does not directly modulate γ-secretase cleavage of APP and that the decrease in Aβ levels induced by CD147 might be due to stimulation of expression of an as yet to be identified matrix metalloprotease.8 This hypothesis does not preclude the possibility that amyloid-β lowering activity of CD147 might involve direct association with APP or C99. The work of this paper provides a direct test for complex formation between C99 and the transmembrane/cytosolic C-terminal domain of CD147 (CD147-CTD).
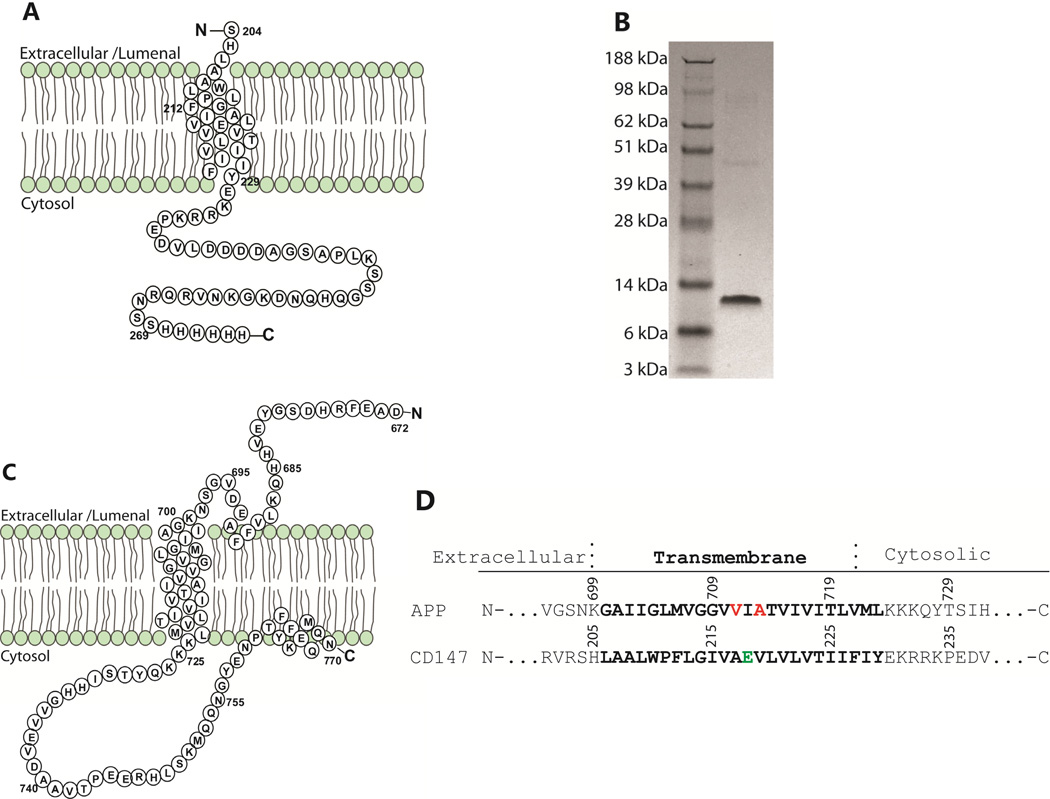
Figure 2.
(A) Sequence and topology plot for the CD147 transmembrane/cytosolic domain. (B) SDS-PAGE gel with Coomassie staining of the purified CD147-CTD. (C) Sequence and topology plot for C99. (D) Sequence alignments for the transmembrane domains of human C99 and human CD147-CTD. The γ-secretase cleavage sites at C99 for production of Aβ40 and Aβ42 are highlighted in red. The unusual glutamic acid in the CD147 transmembrane domain is highlighted in green. The transmembrane sequence of CD147 is highly conserved among all species.
Finally, we note that a complication to this study is the fact that the transmembrane domain of C99 contains GXXXG motifs that confer a propensity for homodimerization.1,14–15 Accordingly, in developing methods for testing for specific heterodimerization of C99 with CD147-CTD we have to consider the possibility that homodimerization of C99 may compete with this process.
Results
Expression and Purification of the Transmembrane/Cystosolic Domain of CD147
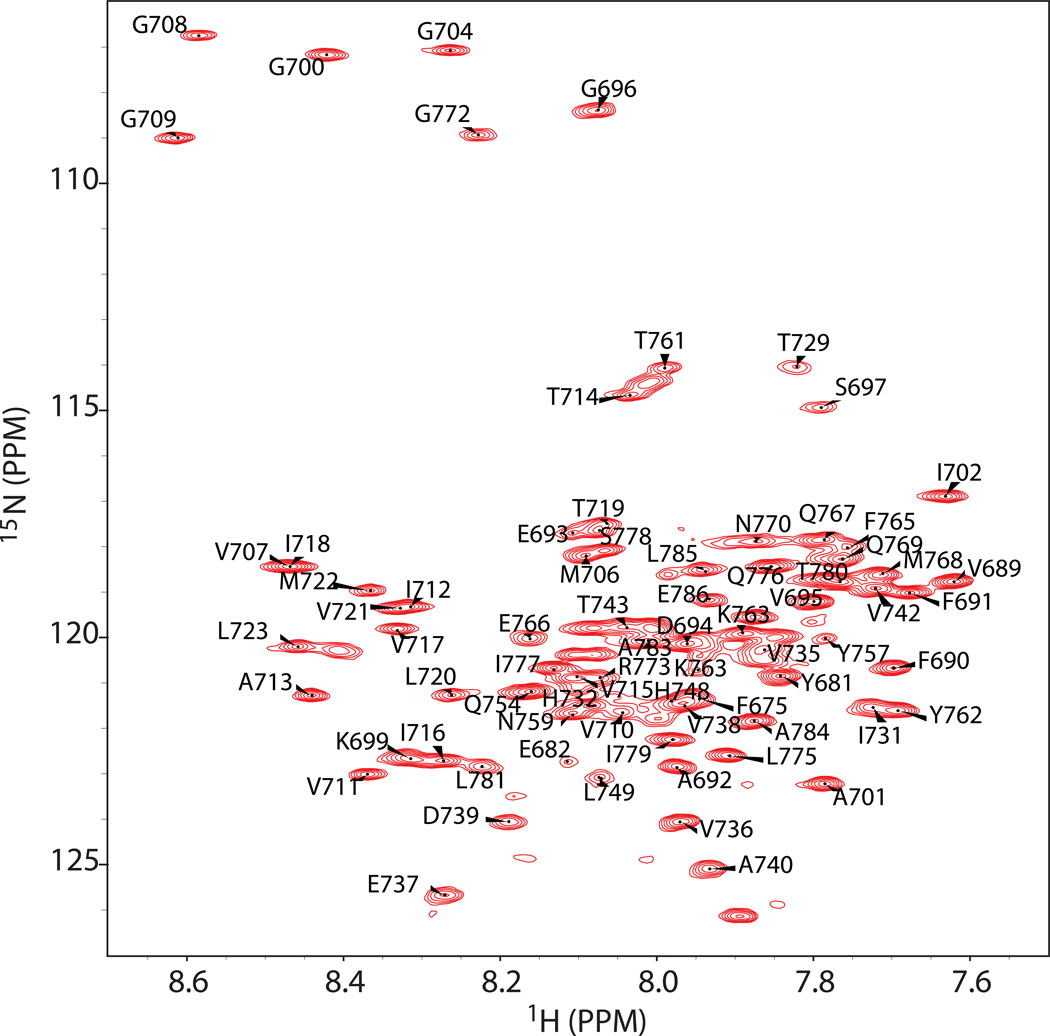
Human CD147-CTD (Figure 2A) was expressed in E. coli and then purified into lysomyristoylphosphatidylglycerol (LMPG) micelles, a detergent that we have previously shown to be well-suited for NMR studies of C99.1 The purity of the 10 kDa protein was confirmed with SDS-PAGE (Figure 2B). Far-UV CD spectroscopy (not shown) indicated that CD147-CTD is a mostly-helical protein in LMPG micelles, consistent with the fact that CD147 is a type-I integral membrane protein. It was found that lowering the solution pH below 7.4 resulted in significantly reduced solubility, so the experiments of this paper were carried out at pH 7.4. At this pH and in LMPG micelles, C99 yields a much better NMR spectrum than CD147-CTD (not shown). Accordingly, studies of interactions between C99 and CD147-CTD were conducted by monitoring the 1H,15N-TROSY NMR spectrum of uniformly 15N-labeled C99, with the unlabeled CD147-CTD being spectroscopically silent. In previous studies near-complete amide 1H-15N resonance assignments of U-15N-C99 in LMPG were determined1 at pH 6.5. For this work it was possible to directly correlate a number of the peaks from the pH 6.5 TROSY spectrum to the corresponding peaks in the pH 7.4 spectrum, permitting assignment of the latter, as shown in Figure 3.
Figure 3.
600 MHz 1H,15N-TROSY spectrum of C99 in 2.5% (w/v) LMPG micelles acquired at 45 C. The pH of the sample was 7.4. The resonances were assigned by tracing the resonance chemical shift changes through a series of pH titration experiments between pH 6.5 and 7.5. The 1H,15N-TROSY spectrum of C99 was previously assigned at pH 6.5.1
Complex formation between C99 and CD147-CTD is not detected at low protein mole fractions in the micelles
NMR has been widely used to map ligand binding interfaces of proteins by monitoring titration-induced NMR peak chemical shift changes or intensity variations.16 For the titrations of this work it is important to consider the thermodynamic nature of protein concentration in a membrane-mimetic environment (i.e., detergent micelles). For molecular association occurring in model membranes, bulk concentration units are not thermodynamically appropriate.17–18 Instead, mole fraction percentage units should be used. For example, while the bulk concentration of a 1 mM membrane protein is the same whether the sample contains 100 or 200 mM detergent, from a thermodynamic standpoint the “interfacial concentration” of the protein is twice as high in 100 mM detergent (ca. 1 mol%) as in 200 mM detergent (ca. 0.5 mol%).
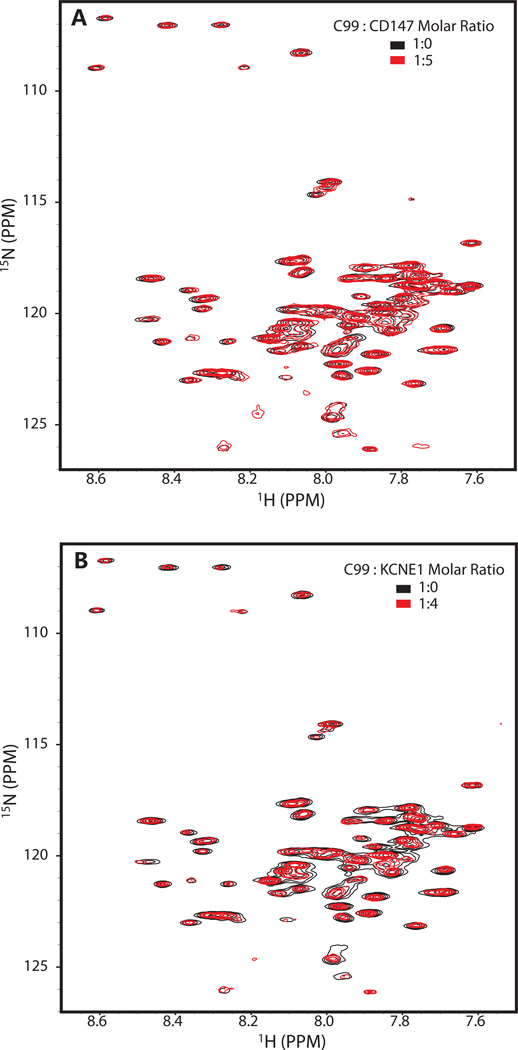
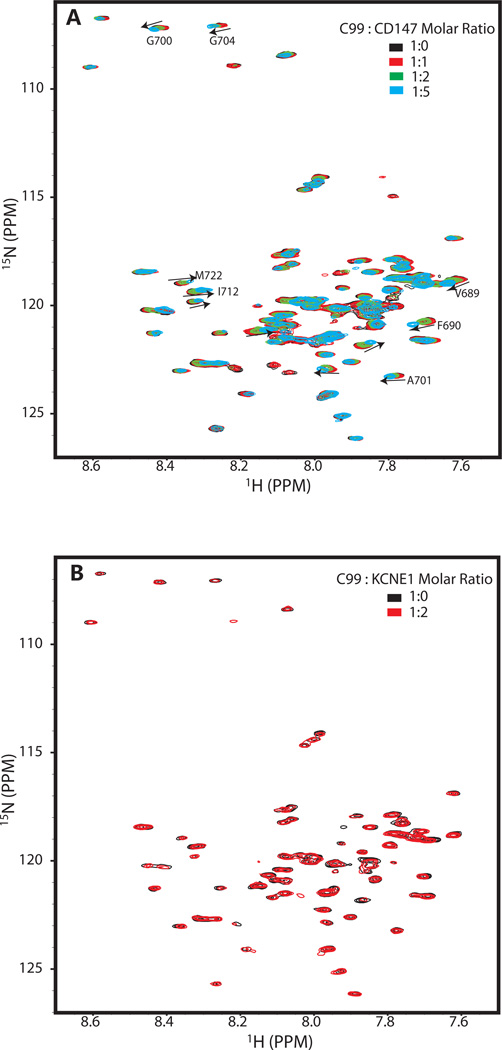
We monitored the 1H,15N-TROSY spectrum from 0.2 mM U-15N-C99 as it was titrated with unlabeled CD147-CTD to test whether direct association of these proteins can be detected under conditions of a high (190 mM, 9%) and constant LMPG concentration, conditions in which the mol fraction of C99 in the micelles is low such that little homodimer is present (see below). As a negative control, we also titrated U-15N-C99 with unlabeled KCNE1, a single span membrane protein that modulates potassium channel function19–20 and that appears to be completely unrelated to C99, CD147, and their associated cell biology. Figure 4 shows the results for these experiments. No chemical shift changes were observed in the spectra of C99 even after addition of a 4-fold molar excess of either CD147-CTD or KCNE1. We conclude that under very dilute conditions (only 0.1 mol% C99 in the membrane-mimetic phase) no direct interaction between CD147-CTD and C99 can be detected.
Figure 4.
Titrations of 0.2 mM (0.11 mol%) U-15N-C99 with either unlabeled CD147-CTD (A) or KCNE1 (negative control, B) at high detergent concentration as monitored by 600 MHz 1H-15N TROSY NMR spectra at 45 C. The sample buffer in both cases contained 250 mM imidazole and 9% LMPG (188 mM), pH 7.4 .
Interaction of C99 with CD147-CTD is evident at higher protein mole fractions in the micelles
To examine the detergent effect on possible heterodimerization of CD147-CTD with 0.2 mM C99, we repeated titration experiments at 2.5% (50 mM) LMPG, conditions in which the C99 concentration is 0.4 mol%. Significant CD147-CTD-induced chemical shift changes were observed in the 1H,15N-TROSY NMR spectrum of U-15N-C99 as shown in Figure 5A (see also site-specific shifts in Supporting Figure 2). The assignable peaks that exhibited the largest chemical shift changes are located primarily within the transmembrane or juxtamembrane domains. Significant chemical shift perturbations were also observed for residues V689 and F690, which are located in a membrane surface-penetrating amphipathic helix on the N-terminus.1 However no chemical shift changes in C99’s spectrum were observed upon negative control titration of KCNE1 under identical conditions in 2.5% LMPG, as shown in Figure 5B. These results suggest C99 undergoes heterodimerization with CD147 (but not with KCNE1) at higher mol% concentrations of the proteins in the model membranes. The plot of chemical shift changes versus the mole% of CD147-CTD appears to exhibit the beginning of hyperbolic curvature (Supporting Figure 2) which suggests their binding is specific. However, the avidity of association between C99 and CD147-CTD appears to be modest as evidenced by the fact that saturation was not approached even upon reaching a 4-fold molar excess of CD147-CTD over C99 in the titration.
Figure 5.
Titrations of 0.2 mM (0.4 mol%) U-15N-C99 with either unlabeled CD147-CTD (A) or KCNE1 (negative control, B) at moderate detergent concentration as monitored by 600 MHz 1H-15N TROSY NMR spectra at 318ºK. The sample buffer in both cases contained 250 mM imidazole and 2.5% LMPG (52 mM) pH 7.4. Site-specific changes in chemical shifts observed over the course of the CD147-CTD titration are presented as a bar graph in Supporting Figure 2.
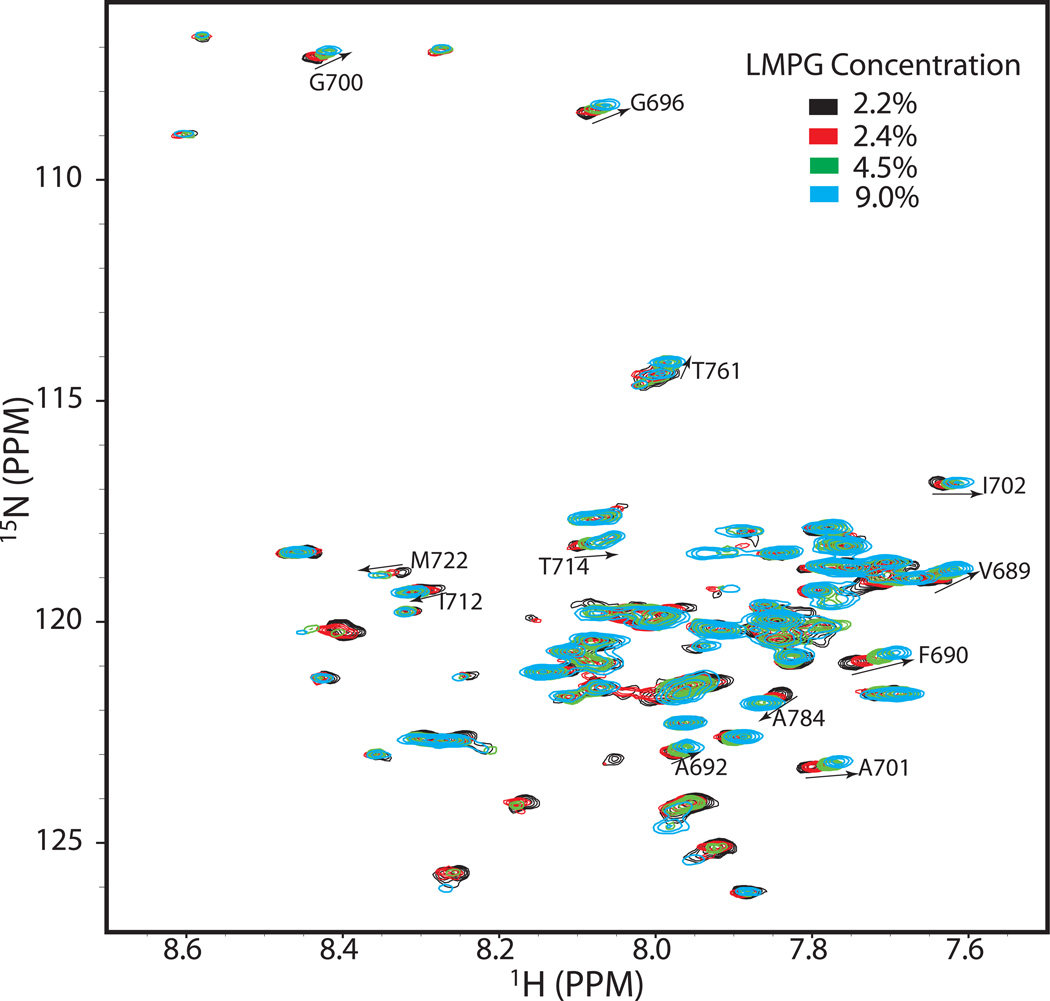
To further test how detergent concentration affects the heterodimerization, C99 and CD147-CTD were mixed at constant concentrations (0.2 mM and 1 mM, respectively) and NMR spectra for U-15N-C99 were collected as the LMPG concentration was varied from 2.2% to 9%. (The high LMPG concentration in the CD147-CTD protein stock solution made it impossible to generate samples with LMPG concentrations lower than 2.2%.) Even though this range of accessible LMPG concentration is modest, significant LMPG concentration-dependent chemical shift changes were observed (Figure 6, see also site-specific shifts in Supporting Figure 3). Resonances exhibiting the largest chemical shift changes generally match those shown in Figure 5A (compare also Supporting Figs 2–3) and reflect a shift in the monomer-heterodimer equilibrium towards monomer at the higher LMPG concentrations. This supports the notion that heterodimerization is taking place at higher protein concentrations in the mixed micelles. That dimerization reflects specific avidity of C99 and CD147-CTD for each other and not just the consequence of forcing these two molecules to co-habitate the same micelles is supported by the fact that a 2X molar excess of the negative control KCNE1 protein does not perturb C99’s spectrum, even at the lower LMPG concentrations (Figure 5B).
Figure 6.
600 MHz 1H-15N TROSY spectra of 0.2 mM U-15N-C99 plus 1 mM unlabeled CD147-CTD at varying concentrations of LMPG (2.2%--black, 2.4%--red, 4.5%--green, and 9.0%--cyan) at 45 C. The buffer contained 250 mM imidazole, pH 7.4. Site-specific changes in chemical shifts observed over the course of this titration are presented as a bar graph in Supporting Figure 3.
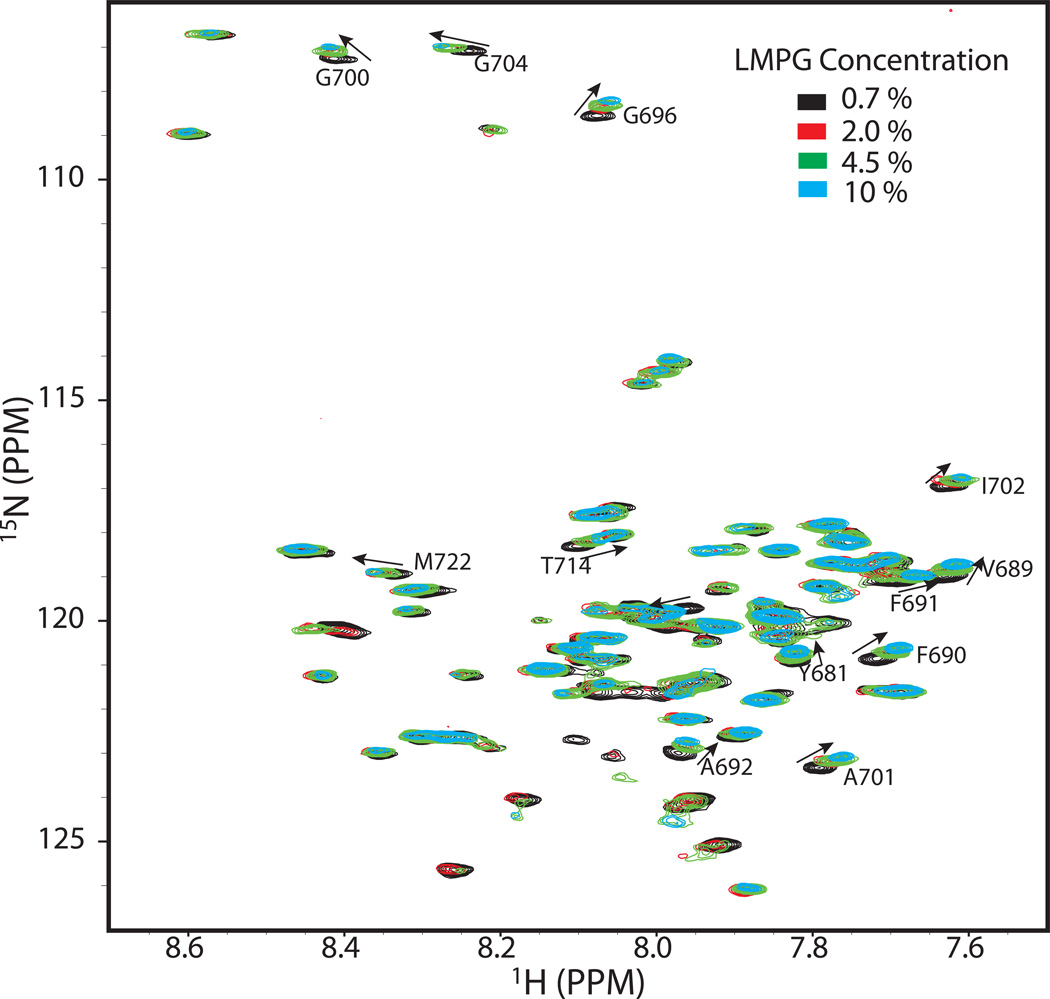
Homodimerization of C99 can also be detected at higher mole fractions in micelles
There is a variety of evidence that C99 or its derived transmembrane fragment form homodimers under membrane or model membrane conditions.1,14,21–25 Here we examined the avidity and reversibility of C99 homodimerization by acquiring 1H, 15N-TROSY spectra of 0.2 mM U-15N-C99 as the LMPG concentration was varied over a range of concentrations: 0.7%, 2.0%, 4.5% and 10% (15 to 210 mM) LMPG. As expected, significant chemical shift changes were observed, which is likely due to a shift in the monomer-dimer equilibrium towards the monomer at higher LMPG concentrations. Interestingly, the peaks exhibiting the largest chemical shift changes (Figure 7 and Supporting Figure 4) exhibited some modest degree of overlap with those observed to shift the most in response to titration by CD147-CTD (Figure 5 and Supporting Figure 2). This suggests that the location of the homodimerization interface on C99 overlaps with that involved in heterodimerization with CD147. However, the directions of the shift changes for these most-affected peaks were sometimes different for the C99-only titration relative to their corresponding responses to titration by CD147-CTD. This reflects the chemically-distinct nature of the interface for the homodimer versus the heterodimer. As in the case of heterodimerization, shift versus C99 protein concentration curves did not exhibit saturation at high protein-to-detergent ratios (Supporting Figure 4), indicating that conversion to homodimer remains incomplete even at the highest accessible C99 mole fraction concentration. Evidently homodimerization, like heterodimerization, is not very avid in LMPG micelles. These data also indicate that heterodimerization of C99 with CD147-CTD is more favorable than C99 homodimerization since the CD147-induced chemical shift changes “override” the homodimerization-induced chemical shift changes seen when detergent concentration is varied in the C99-only case.
Figure 7.
600 MHz 1H-15N TROSY spectra of 0.2 mM U-15N-C99 at varying concentrations of LMPG (0.7%--black, 2.0%--red, 4.5%--green, and 10%--cyan) at 45 C. The buffer contained 250 mM imidazole, pH 7.4. Site-specific changes in chemical shifts observed over the course of this titration are presented as a bar graph in Supporting Figure 4.
Paramagnetic relaxation enhancement (PRE) measurements verify specific complex formation between CD147-CTD and C99
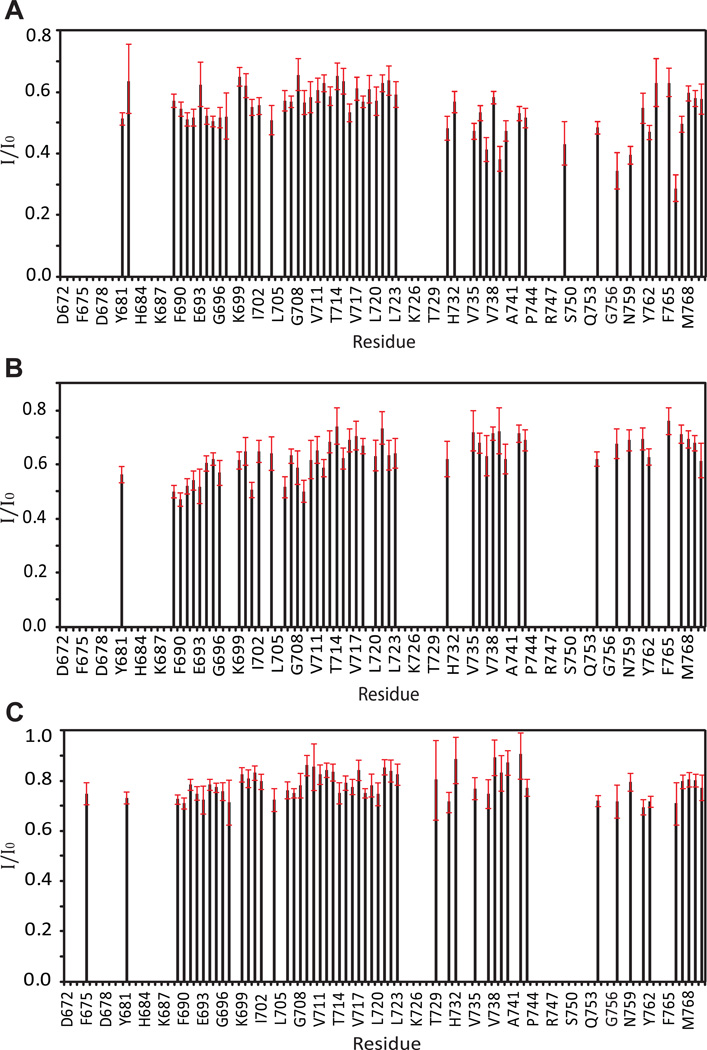
Paramagnetic relaxation enhancement (PRE) NMR experiments have received increasing application as a route to long-range distance information (up to 25 Å) for use in structure determination or refinement.26–27 Distances between a spin label fixed at a site in a protein and NMR active nuclei in the protein (usually amide 1H) can be extracted from the paramagnet-induced increased NMR transverse relaxation rates, manifested as line broadening. Spin labels placed on proteins have also been used to screen for the binding of small molecules on the basis that PRE from a spin label on the protein will result in dramatic line broadening in the spectra of small molecules that bind to proximal sites on the protein, provided that on/off exchange is rapid on the NMR time scale.27–29 This approach was adapted for our study of heterodimerization between C99 and CD147-CTD. Two different single-cysteine forms of CD147-CTD were prepared (F212C and Y229C). Each was then spin labeled at their cysteine sites using the thiol-reactive nitroxide reagent, MTSL. To address the relative protein orientations during heterodimerization, one cysteine mutant (F212C) was introduced into the transmembrane domain of CD147-CTD close to the extracellular interface (see Figure 2A). The other spin label was attached to a site (Y229C) also located in the transmembrane segment, but close to the cytosolic domain. For use as a negative control, spin-labeled KCNE1 was prepared using a single-cysteine mutant form of this protein (S64C on the juxtamembrane close to the cytoplasmic domain).
To 0.2 mM U-15N-C99 in 2.5% LMPG was added spin-labeled CD147-CTD or KCNE1. 1H,15N-TROSY spectra were recorded under both paramagnetic and diamagnetic conditions. The reductions in peak intensities resulting from paramagnetic relaxation enhancement-based line broadening are shown in Figure 8. For titrations of C99 by both spin-labeled CD147-CTD and spin-labeled KCNE1 (negative control), widespread peak broadening in the spectrum of U-15N-C99 was evident. However the line broadening induced by spin-labeled KCNE1 was observed to be rather uniform throughout the spectrum (Figure 8C and Supporting Figure 5), indicating that while KCNE1 and C99 may co-habitate the same micelle, which forces them to be in close proximity, there is no preferred mode of interaction that reflects complex formation. However in the cases of spin-labeled Y229C and F212C mutant forms of CD147-CTD, we observed non-uniform line broadening in C99’s NMR spectrum. Spin-labeled F212C-CD147-CTD induced the greatest degree of line broadening in the 1H,15N-TROSY resonances from C99 residues in the 689–709 range, sites located in the extracellularly-disposed juxtamembrane or transmembrane domains (Figure 8B). In the case of spin-labeled Y229C-CD147-CTD, only residues located in the cytosolic C-terminus exhibited signficant line broadening beyond the average (Figure 8A and Supporting Figure 5A). Combined with the earlier observation that the peaks exhibiting the largest chemical shift changes in the spectrum of C99 in response to homodimerization are largely the same set of C99 peaks that shift in response to heterodimerization with CD147-CTD, these results confirm that C99 and CD147-CTD form a specific heterodimeric complex that involves largely the same set of C99 residues that are also involved in homodimerization of this protein. This dimer interface on C99 includes both sites located in the short amphipathic helix (residue 689–692) found just before the start of the transmembrane domain and also glycine residues that are part of tandem GXXXG motifs found in the extracellular end of the transmembrane domain (see Figure 1C).
Figure 8.
Site-specific PRE reductions in intensities of 600 MHz 1H-15N TROSY peaks from 0.2 mM U-15N-C99 resulting from the presence of either (A) a 4X molar excess of nitroxide spin-labeled Y229C CD147-CTD, (B) a 4X molar excess of nitroxide spin-labeled F212C CD147-CTD, or (C) a 2X molar excess of nitroxide spin-labeled S64C KCNE1. I/I0 values represent the difference between C99 TROSY peak heights in the presence of either spin-labeled CD147-CTD or KCNE1 relative to the corresponding C99 peak intensities (which are higher) under diamagnetic conditions (see Methods for additional information). All samples contained 2.5% LMPG at pH 7.4, with the NMR data being acquired at 45 C. The residues for which measurements are not reported represent those with unassigned or unresolved peaks.
Discussion
Distinguishing between specific association, non-specific association, and forced co-habitation under micellar conditions
The experiments of this work establish a general methodology that can be used to classify interactions occurring between a pair of membrane proteins as being the consequence of either specific association, non-specific association, or forced co-habitation of two proteins of the same model membrane units (Figure 1). The first of these experiments was to carry out a pair of 1H,15N-TROSY-monitored titrations of protein A (i.e., C99) with both its potential partner, protein B (i.e. CD147-CTD), and with a negative control membrane protein (i.e., KCNE1) that has the same membrane topology as protein B, but that should have no affinity for protein A. In this work it was seen that at higher protein-to-detergent ratios the negative control protein KCNE1 induced essentially no shifts in the spectrum of C99, a very different result from what was seen when C99 was titrated by CD147-CTD. This eliminated the possibility that the CD147-CTD-induced shifts were due to forced co-habitation of protein CD147-CTD and C99 in the same micelle. The approach used to reach this conclusion should be generally applicable, with judicious choice and preparation of a suitable membrane protein for use as the negative control being essential. We note that thanks to recently-developed structural genomics repositories and other resources (c.f. http://psimr.asu.edu), E. coli expression systems are now available for numerous membrane proteins, facilitating choice and access to a suitable negative control membrane protein.
The second set of experiments introduced in this work allowed specific binding to be distinguished from non-specific binding. This experiment requires that protein B (i.e., CD147-CTD) be prepared in two different spin labeled forms—one with the spin label at or near the extracellular domain and one with the spin label at or near the cytosolic domain. If specific binding is taking place then when these spin-labeled forms of protein B are titrated into a protein A solution, the amide sites on protein A for which TROSY/HSQC peaks are most profoundly broadened should be those that are located on the same side of the native membrane as the spin labeled site in protein B. On the other hand, titration of membrane protein A with a spin-labeled negative control membrane protein should reveal patterns of line broadening in the spectrum protein A that reflect only non-specific interactions. For example, if the spin label is located on the extracellular domain of the negative control protein, roughly equal broadening should be observed in peaks from both the extra- and intra-cellular domains of protein A. In this work we used these experiments to confirm that the interactions of C99 and CD147-CTD involve specific association, whereas the interaction between C99 and KCNE1 was seen to be of a non-specific nature.
The two sets of experiments introduced in this work provide only qualitative insight into affinity. Quantitation of Kd would require that other (possibly non-NMR) methods be used. However, qualitative insight is not inconsequential. Here we found that binding could not readily be detected via chemical shift perturbation when the C99 and CD147 concentrations were on the order of 0.1 mol%, but could be detected at a ≥4-fold higher concentration, where changes in the NMR spectrum of C99 are roughly linear with CD147-CTD concentration up through 2 mol% CD147-CTD. This indicates that Kd for these two proteins in LMPG micelles must be ≥ 2 mol%.
A final set of experiments that proved very useful in this work was to monitor the NMR spectrum of C99-only as a function of the protein-to-detergent ratio. Data from this experiment provided the data consistent a propensity of C99 to form specific homodimers, as previously observed.1,14,21–25 Moreover, when homodimerization-induced resonance changes were compared with data in which the dissociation of the C99/CD147-CTD complex was gradually induced by raising the detergent-to-protein ratio, it was possible to establish that heterodimerization is more avid than homodimerization.
Heterodimerization of C99 with CD147-CTD
As summarized in the Introduction, CD147 is found to promote reduced levels of amyloid-β under physiological conditions. However, there has been controversy regarding the mechanism for this phenomenon. While early studies pointed to a direct regulatory role for CD147 in cleavage of C99 by the gamma-secretase complex10, later studies have disputed this and proposed that CD147 instead lowers amyloid-β levels by promoting extracellular cleavage and clearance of this polypeptide.8 The results of this work do not directly address this controversy or provide direct support for either model. However, this work does establish that under micellar model membrane conditions C99 forms a complex with the combined transmembrane and cytosolic domains of CD147. While the affinity associated with formation of this complex appears to be only modest, it could be much higher under native membrane conditions. Affinity under physiological conditions might also be modulated (up or down) by the presence of CD147’s ectodomain in the full-length protein and/or by the presence of other interacting proteins. In any case, the conclusion of this work that these two proteins have a propensity to form a complex may prove useful in future studies to fully unravel the relationship of CD147 to the amyloid precursor protein and its downstream proteolytic processing/degradation.
Conclusion
The NMR-based approach developed in this work for elucidating the nature of intermolecular interactions between C99 and CD147-CTD should be generally adaptable to numerous other potentially binding pairs of membrane proteins, some of which are of high biomedical significance. For any give pair of proteins, confirmation of specific binding represents a critical first step for further characterization of the complex. For pairs that are found to interact only through non-specific interactions or forced co-habitation, the negative results provided by this approach may provide the basis either for pursuit of more optimal model membrane conditions (in which specific binding might take place) or for a timely cessation of unpromising studies.
Methods
Expression and Purification of C99
Wild-type human C99 with a C-terminal purification tag was expressed in E. coli and purified into LMPG micelles as previously described1 essentially without any modification. To facilitate the acquisition of 1H-15N TROSY correlation spectra, 15N enrichment was provided by supplementing M9 minimal medium with 1 g/L 15NH4Cl (Sigma,MO). Pure C99 protein was eluted from a Ni-NTA column using 2–4 column volumes of elution buffer that contains 250 mM Imidazole, 0.05% LMPG, pH=7.8. Samples were concentrated to using centrifugal filter units (Millipore, MA) of 15 kDa molecular weight cut-off (MWCO). This ultrafiltration step also concentrates the LMPG in the sample since it has a very low critical micellar concentration and the micelles are much larger than 15 kDa. Final NMR samples were prepared to be 0.2 mM U-15N-C99 in a buffer containing 250 mM imidazole, 1 mM EDTA, 5% D2O plus the desired concentration of LMPG at pH 7.4. The LMPG concentrations in stock C99 samples was determined using 1D proton NMR in which the integral of the signal from 5 mM DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid) was used as a concentration standard (see section below and Supporting Figure 1).
Expression and Purification of CD147-CTD
The gene encoding CD147-CTD (residues 204 to 269) was derived from the full length human gene and amplified by polymerase chain reaction (PCR), including introduction of a BamH1 cleavage site at 3-end and a Xho1 site at 5-end. The double-digested PCR product was then ligated into a pET25b E. coli expression vector to include a C-terminal hexaHis-tag for purification using a Ni-NTA column. The construct was sequenced by the Vanderbilt University DNA sequencing core facility.
For PRE measurement, single cysteine residues were introduced into CD147-CTD, which contains no wild type cysteines. Mutations were at either 212 (F212C) or 229 (Y229C) using a Qiagen QuickChange mutagenesis kit (Valencia,CA). The cDNAs of CD147-CTD and its single-cysteine mutants were then ligated into expression vectors that were transformed into BL21(DE3) E. coli (Novagen, NJ). A single colony was picked from a Luria broth-agar/ampicillin plate and used to inoculate a 10ml overnight culture. The overnight culture was then used to inoculate 1 L of Luria broth medium supplemented with 0.1 g/L ampicillin. The cells were grown at 37 C with vigorous shaking at 225 rpm. CD147-CTD expression was induced by 1 mM IPTG when OD600 reached 0.8. Cells were harvested by centrifugation after induction for 3–4 hrs and could then be stored in −80 C freezer.
CD147-CTD and its single-cysteine mutants were purified as described for E. coli diacylglycerol kinase30 with some modifications. The cell pellet was resuspended in lysis buffer (75 mM Tris, 300 mM NaCl, 0.2 mM EDTA, pH 7.8) at a 20-fold dilution (20 ml lysis buffer for each gram of cell pellet). To the lysate 0.2 mg/ml lysozyme, 0.02 mg/ml of DNase, 0.02 mg/ml RNase and 5 mM magnesium acetate were added, followed by tumbling the lysate at room temperature for 0.5 hr. The lysate was then sonicated for 5 min (alternating 5 s on and 5 s off). After sonication, the solution was cooled on ice and Empigen detergent (Fluka) was added to 3% to extract the protein from the membrane. The lysate was then tumbled at 4 C for 0.5 hr, followed by centrifugation at 20,000 rpm for 20 minutes to remove debris. The supernatant was then mixed with Qiagen (Valencia,CA) Ni-NTA-Superflow resin (1.2 ml of resin for every gram of cell pellet) which had been pre-equilibrated with buffer A (40 mM HEPES, 300 mM NaCl, pH 7.5). The mixture was tumbled for 0.5 hr at 4 C to maximize the chelating of detergent-solubilized 6xHis-tagged CD147-CTD to the Ni(II) resin. The resin was then transferred to a column and washed with 5 bed volumes of cold Emp/A buffer (40 mM HEPES, 300 mM NaCl, 3% Empigen, pH 7.5). The resin was then washed with cold buffer B (40 mM HEPES, 300 mM NaCl, 1.5% Empigen, 40 mM imidazole, pH 7.8) until all impurities were eluted, as judged by monitoring A280. The detergent was then exchanged from 1.5% Empigen to 0.05% LMPG by rinsing the column with 12 X 1 bed volumes of cold rinse buffer (25 mM sodium phosphate, 0.05% LMPG, pH 7.5). Finally, CD147-CTD was eluted from the column using elution buffer (250 mM Imidazole, 0.05% LMPG, pH 7.8). The purity of the protein was assessed by SDS-PAGE. Usually 15–20 mg of pure protein was obtained from 1 L of LB culture. CD147-CTD stock solutions used for titrations were then prepared by concentrating the protein to 1.5 mM.
Expression and Purification of KCNE1
In negative control experiments we titrated C99 with human KCNE1, a 129 residue single span membrane protein that modulates the function of certain potassium channels. KCNE1 and its single-cysteine mutant S64C were expressed and purified as described19,31 without any modification. S64C was constructed based on use of a previously constructed cysteine-free KCNE1 mutant (C105A). Final NMR samples were prepared from a stock solution of 1.5 mM KCNE1 in buffer containing 250 mM imidazole, pH 7.4 with the desired level of LMPG.
Determination of Detergent Concentrations
Detergent concentrations required for accurate determination of the mol% concentration of membrane proteins were determined using 1D proton NMR. The integral of the NMR signal from a known concentration of DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid) was used as a concentration reference to which the integral of a LMPG peak can be compared in order to determine the overall LMPG concentration (Supporting Figure 1A). In order to validate this method we verified the linearity of a plot of the peak integral ratio vs. LMPG concentration in a series of samples with fixed (5 mM) DSS concentration and varying LMPG (0.05%, 0.5%, 1% and 2%; Supporting Figure 1B). For these experiments 32 transients were acquired for each sample using a Bruker 1D proton pulse sequence with water presaturation. The recovery delay was set to 10 s. The same pulse sequence and parameter settings were used for protein samples with unknown LMPG concentrations.
Spin-Labeling of Single-Cysteine Mutant Forms of CD147-CTD and KCNE1
Nitroxide spin labeling was introduced using a cysteine-specific reaction of the protein with MTSL (2,2,5,5-tetramethyl-3-pyrroline-3-methyl methanethiosulfonate; Toronto Research Chemicals, Canada). This reaction requires cysteine to be in its reduced state. Therefore, prior to reaction with MTSL dithiothreitol (DTT) was added to 2 mM into 0.5 mM protein samples at pH 7.8, followed by incubation at 37 C for 20–30 min in order to completely reduce the disulfide bonds. The reduced protein sample was immediately passed through a Bio-Rad Econo-Pac 10 desalting column to remove the DTT and imidazole. The elution buffer contained 25 mM sodium phosphate and 0.05% LMPG at pH 7.8. A 10-fold molar excess (over protein) of MTSL was then added from a 250 mM stock solution in methanol (the latter of which could be stored in the dark and in the freezer under argon). The solution was gently agitated for 1 hr at room temperature, followed by 2 hrs at 37 C and then overnight incubation at room temperature. In our experience MTSL or residual MTSL-modified DTT has a significant affinity for micelle surfaces, such that buffer exchange using ultrafiltration or a desalting column does not always completely remove the free spin label. Therefore the spin-labeled protein sample was re-loaded onto a Ni-NTA column, followed by extensive washing using 20 bed-volume of 25 mM sodium phosphate, 0.05% LMPG, pH 7.8. The protein was then eluted using 250 mM imidazole, 0.05% LMPG, pH 7.8 and concentrated using Millipore Ultra-15 centrifugal filter units (MWCO 15 kDa), followed by adjustment of the pH to 7.4. The spin-labeling efficiency was evaluated by continuous-wave EPR. EPR Signals from spin-labeled protein samples were integrated and quantitated through the comparison with the signal integral of 100 uM TEMPO (2,2,6,6-tetramethyl-piperidine-1-oxyl). For Y226C-CD147, the spin-labeling efficiency was >95%, while for F212C-CD147 and S64C-KCNE1, efficiency was 90%.
Use of NMR to Monitor Protein-Protein Titrations
For all titration experiments, the concentration of U-15N-C99 was fixed at 0.2 mM. Final NMR samples were prepared in 250 mM imidazole with the desired concentration of LMPG. The LMPG concentration was adjusted by directly adding LMPG powder or a 10% LMPG stock solution, followed by adjusting the pH to 7.4. All NMR spectra were acquired at 45 C using a Bruker Avance 600 MHz spectrometer (Bruker Biospin, Germany) equipped with Z-gradient triple-channel cryogenic probe. A sensitivity-enhanced TROSY pulse sequence32 was employed. Spectra were processed using nmrPipe33 with linear prediction and zero-filling. Peak intensities were obtained using nmrDraw peak-picking followed by non-linear spectral line shape modeling (nLinLS).33 To the greatest extent possible, resonance assignments for C99 at pH 7.4 were carried out by correlating peaks with previously-assigned peaks for samples at pH 6.5.1 However, this is possible only for peaks that are in relatively well-resolved parts of the spectra and that do not undergo large pH-dependent shifts.
NMR Measurement of Paramagnetic Relaxation Enhancement
All paramagnetic samples were prepared by mixing 0.2 mM U-15N-C99 with spin-labeled CD147-CTD or spin-labeled KCNE1 at the desired ratio. Diamagnetic forms of the same samples were generated by reducing the nitroxide by adding 10 mM ascorbic acid. The LMPG concentration of all samples was 2.5% (ca. 50 mM). 1H,15N-TROSY NMR spectra were acquired using identical parameters for both paramagnetic and the corresponding diamagnetic samples. Each spectrum was processed twice using nmrPipe using either Gaussian or cosine-squared apodization with zero fillings. The reported paramagnetic conditions to diamagnetic conditions peak intensity ratios represent the average of the intensity ratios measured following processing using these two apodization functions.
Supplementary Material
Acknowledgement
This work was supported by Alzheimer Association Grant IIRG-07-59379 and by NIH grants PO1 GM080513 and U54 GM094608.
Footnotes
Supporting Information Available. Complete Ref. 8 and supporting figures illustrating the method used to determine detergent concentration in NMR samples, the chemical shift changes for C99 induced by titration with CD147 at low LMPG, the chemical shift changes for C99 at a fixed ratio with excess CD147 during titration by LMPG, the chemical shift changes for C99 as it is titrated with LMPG, and the concentration dependence of residue-specific NMR peak line broadening for C99 as it was titrated by spin-labeled CD147 or KCNE1. The material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Beel AJ, Mobley CK, Kim HJ, Tian F, Hadziselimovic A, Jap B, Prestegard JH, Sanders CR. Biochemistry-Us. 2008;47:9428–9446. doi: 10.1021/bi800993c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buffy JJ, Buck-Koehntop BA, Porcelli F, Traaseth NJ, Thomas DD, Veglia G. J Mol Biol. 2006;358:420–429. doi: 10.1016/j.jmb.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Kim MJ, Park SH, Opella SJ, Marsilje TH, Michellys PY, Seidel HM, Tian SS. J Biol Chem. 2007;282:14253–14261. doi: 10.1074/jbc.M611616200. [DOI] [PubMed] [Google Scholar]
- 4.Malia TJ, Wagner G. Biochemistry-Us. 2007;46:514–525. doi: 10.1021/bi061577h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Horn WD, Kim HJ, Ellis CD, Hadziselimovic A, Sulistijo ES, Karra MD, Tian C, Sonnichsen FD, Sanders CR. Science. 2009;324:1726–1729. doi: 10.1126/science.1171716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shoichet BK, McGovern SL, Caselli E, Grigorieff N. J Med Chem. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- 7.Mcdonnell PA, Opella SJ. J Magn Reson Ser B. 1993;102:120–125. [Google Scholar]
- 8.Vetrivel KS, et al. J Biol Chem. 2008;283:19489–19498. doi: 10.1074/jbc.M801037200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou SX, Zhou H, Walian PJ, Jap BK. Drug News Perspect. 2006;19:133–138. doi: 10.1358/dnp.2006.19.3.985932. [DOI] [PubMed] [Google Scholar]
- 10.Zhou SX, Zhou H, Walian PJ, Jap BK. P Natl Acad Sci USA. 2005;102:7499–7504. doi: 10.1073/pnas.0502768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nabeshima K, Iwasaki H, Koga K, Hojo H, Suzumiya J, Kikuchi M. Pathol Int. 2006;56:359–367. doi: 10.1111/j.1440-1827.2006.01972.x. [DOI] [PubMed] [Google Scholar]
- 12.Winblad B, Nahalkova J, Volkmann I, Aoki M, Bogdanovic N, Tjernberg LO, Behbahani H. Neurochem Int. 2010;56:67–76. doi: 10.1016/j.neuint.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 13.Steiner H, Winkler E, Hobson S, Fukumori A, Dumpelfeld B, Luebbers T, Baumann K, Haass C, Hopf C. Biochemistry-Us. 2009;48:1183–1197. doi: 10.1021/bi801204g. [DOI] [PubMed] [Google Scholar]
- 14.Wang H, Barreyro L, Provasi D, Djemil I, Torres-Arancivia C, Filizola M, Ubarretxena-Belandia I. J Mol Biol. 2011;408:879–895. doi: 10.1016/j.jmb.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khalifa NB, Van Hees J, Tasiaux B, Huysseune S, Smith SO, Constantinescu SN, Octave JN, Kienlen-Campard P. Cell Adh Migr. 2010;4:268–272. doi: 10.4161/cam.4.2.11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuiderweg ERP. Biochemistry-Us. 2002;41:1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]
- 17.Otzen DE, Sehgal P, Mogensen JE. Bba-Biomembranes. 2005;1716:59–68. doi: 10.1016/j.bbamem.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 18.Carman GM, Deems RA, Dennis EA. J Biol Chem. 1995;270:18711–18714. doi: 10.1074/jbc.270.32.18711. [DOI] [PubMed] [Google Scholar]
- 19.Tian CL, Vanoye CG, Kang CB, Welch RC, Kim HJ, George AL, Sanders CR. Biochemistry-Us. 2007;46:11459–11472. doi: 10.1021/bi700705j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Horn WD, Vanoye CG, Sanders CR. Curr Opin Struct Biol. 2011;21:283–291. doi: 10.1016/j.sbi.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munter LM, Voigt P, Harmeier A, Kaden D, Gottschalk KE, Weise C, Pipkorn R, Schaefer M, Langosch D, Multhaup G. Embo J. 2007;26:1702–1712. doi: 10.1038/sj.emboj.7601616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorman PM, Kim S, Guo M, Melnyk RA, McLaurin J, Fraser PE, Bowie JU, Chakrabartty A. Bmc Neurosci. 2008;9:17–27. doi: 10.1186/1471-2202-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kienlen-Campard P, Tasiaux B, Van Hees J, Li M, Huysseune S, Sato T, Fei JZ, Aimoto S, Courtoy PJ, Smith SO, Constantinescu SN, Octave JN. J Biol Chem. 2008;283:7733–7744. doi: 10.1074/jbc.M707142200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato T, Tang TC, Reubins G, Fei JZ, Fujimoto T, Kienlen-Campard P, Constantinescu SN, Octave JN, Aimoto S, Smith SO. P Natl Acad Sci USA. 2009;106:1421–1426. doi: 10.1073/pnas.0812261106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Multhaup G. Neurodegener Dis. 2006;3:270–274. doi: 10.1159/000095266. [DOI] [PubMed] [Google Scholar]
- 26.Battiste JL, Wagner G. Biochemistry-Us. 2000;39:5355–5365. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- 27.Iwahara J, Clore GM, Tang C. Curr Opin Struc Biol. 2007;17:603–616. doi: 10.1016/j.sbi.2007.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jahnke W. Chembiochem. 2002;3:167–173. doi: 10.1002/1439-7633(20020301)3:2/3<167::AID-CBIC167>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 29.Moore JM, Lepre CA, Peng JW. Chem Rev. 2004;104:3641–3675. doi: 10.1021/cr030409h. [DOI] [PubMed] [Google Scholar]
- 30.Oxenoid K, Kirn HJ, Jacob J, Sonnichsen FD, Sanders CR. J Am Chem Soc. 2004;126:5048–5049. doi: 10.1021/ja049916m. [DOI] [PubMed] [Google Scholar]
- 31.Kang CB, Tian CL, Sonnichsen FD, Smith JA, Meiler J, George AL, Vanoye CG, Kim HJ, Sanders CR. Biochemistry-Us. 2008;47:7999–8006. doi: 10.1021/bi800875q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weigelt J. J Am Chem Soc. 1998;120:10778–10779. [Google Scholar]
- 33.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J Biomol Nmr. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.