Abstract
The innate immune system, antibacterial protein Siderocalin (Scn) binds ferric carboxymycobactin (CMB) and also several catecholate siderophores. While the recognition of catecholates by Scn has been thoroughly investigated, the binding interactions of Scn with the full spectrum of CMB isoforms have not been studied. Here we show that Scn uses different binding modes for the limited subset of bound CMB isoforms, resulting in a range of binding affinities which are much weaker than other siderophore targets of Scn. Understanding the binding interaction between Scn and CMBs provides clues for the influence of Scn on mycobacterial iron acquisition.
From microbes to mammals, iron is essential for virtually all living organisms. Therefore, an organism that can efficiently acquire, utilize and store iron may out-compete other organisms for survival. (1) While iron is sequestered in the human body by intracellular ferritin and extracellular transferrin, (2) during an infection bacterial pathogens can employ siderophores (high-affinity, small-molecule chelators) with sufficient strength to remove ferric ions from the host’s iron stores. (3) Siderocalin (Scn; a.k.a. Lcn2, Ngal, and 24p3), is a mammalian anti-bacterial protein that acts as a second line of defense against iron piracy by binding siderophores and their ferric complexes, thereby intercepting the delivery of iron to the bacteria. (4) The human pathogen Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis, localizes inside host macrophages during infection (5) and taps into intracellular iron stores for survival. (6) Recent literature has shown that Scn suppresses the in vitro growth of mycobacteria, (7–9) and although this influence is attributed to Scn interfering with siderophore-mediated iron acquisition, this mechanism of action has yet to be fully elucidated.
The “stealth” siderophore phenomenon has been described in the siderophore systems of a number of human pathogens. For example, while Gram-positive pathogens Bacillus anthracis and Bacillus cereus produce both bacillibactin (BB) and petrobactin (PB), (10, 11) only PB is not sequestered by Scn, (12) and, as a result, is a virulence factor for the pathogen. Similarly, pathogenic strains of Gram-negative enteric bacteria, such as Salmonella enterica and typhimurium and Escherichia coli, produce both enterobactin (Ent) and its glycosylated derivative, salmochelin (S4), and only S4 confers virulence in the presence of the innate immune response due to its ability to evade Scn. (13) The use of a stealth siderophore under the pressure of the Scn immune response is one advantage of using a diverse, multisiderophore iron acquisition system by pathogenic bacteria.
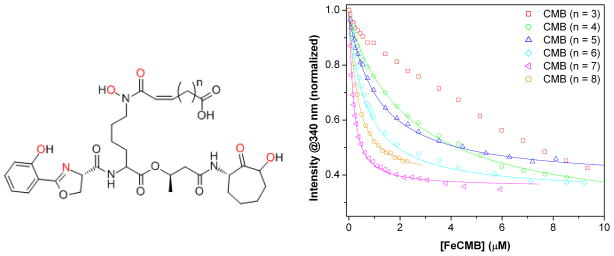
Mtb is an unusually persistent bacterium that is able to survive a number of environmental stresses including desiccation, osmotic shock and nutrient deprivation. (14) Nonetheless, survival is still dependent on the ability of the pathogen to acquire iron. (15) Carboxymycobactins (CMBs, Fig. 1) are a suite of siderophores differing in the length of the fatty acid tail (n = 3–10) produced by Mtb to sequester iron from within the host macrophage. (16) (Other siderophores produced include the structurally similar, cell-associated mycobactins and the hydroxamate-based exochelins.(17))
Figure 1.
Molecular structure of Carboxymycobactin (CMB, left, where n = 3–10 in the fatty acid tail). CMB is a hexadentate ligand that forms a mononuclear ferric complex with iron (1:1 Fe:CMB). Iron binding atoms shown in red. To the right, fluorescence binding analysis of Scn (0.1 μM) with FeIIICMB isoforms (n = 3–8) at pH 7.4. Symbols indicate fluorescence data at and lines are calculated non-linear least squares fits (DYNAFIT (25)).
In 2005, the crystal structure of Scn bound to FeIIICMB provided the first indication that the Scn immune response may impede the pathogenicity of Mtb by interfering with its iron acquisition. (18) Although Scn was in the crystallization solution with a mixture of ferric CMBs, only the n = 6 isoform was bound within the protein calyx in the crystal structure. This observation raised several new questions about the influence of Scn on iron acquisition in Mtb. Does Scn also bind the ferric complexes of the other CMB isoforms? Does Scn selectively bind the n = 6 isoform over the others? What is the binding affinity of Scn for the CMBs? How does the Scn-FeIIICMB interaction compare to the Scn binding interactions with other siderophores? And ultimately, how does the presence of Scn in the host versus the secretion of a suite of siderophores by the pathogen affect the balance of iron during an Mtb infection? To begin to answer these questions about the functional significance of the Mtb suite of siderophores, a mixture of CMBs was isolated from the non-pathogenic bacterium M. smegmatis (19) and the individual ferric complex isoforms (n = 3–8) were separated by HPLC (Supplementary Figure 1). CMBs of longer tail length were also detectable in the mixture, although in trace amounts. The separated compounds were characterized by LCMS and UV-visible spectroscopy (λmax = 450 nm, ε = 3,800 M−1cm−1; based on the reported A1% i.e., the absorbance of a 1% solution of CMBs) (19–21) to ensure that each isoform had been completely separated from the others in the suite. Subsequently, the interaction of each isoform with Scn was analyzed through fluorescence binding assays and protein crystallography.
The fluorescence methods used in Scn binding studies for all previous siderophore systems were employed for the ferric CMBs. (4, 12, 22) The range of binding constants determined for the suite of siderophores spans over two orders of magnitude in affinity. The strongest binding ferric CMB complex is the n = 7 FeIIICMB (Kd = 128 nM), and the affinity for the other isoforms decreases in both directions of the series such that the dissociation constants for n are in the order 7 < 8 < 6 < 5 < 4 < 3 (Figure 1 and Table 1). In general, Scn binds FeIIICMB with lower affinity (nearly 1000-fold weaker for even the tightest binder) than previously described ferric siderophore substrates BB (12) and Ent. (4)
Table 1.
Dissociation constants determined by fluorescence for Scn binding to CMB isoforms (n = 3–8)a
| FeIIICMB (by n) | Kd (μM) | Est. Std. Dev. |
|---|---|---|
| 3 | > 9 | -- |
| 4 | 2.36 | ±9.0×10−3 |
| 5 | 1.10 | ±2.6×10−2 |
| 6 | 0.654 | ±2.2×10−2 |
| 7 | 0.128 | ±1.0×10−3 |
| 8 | 0.280 | n.d.b |
Uncertainties were determined from the standard deviation of three independent titrations.
Minimal sample size allowed for only one binding experiment.
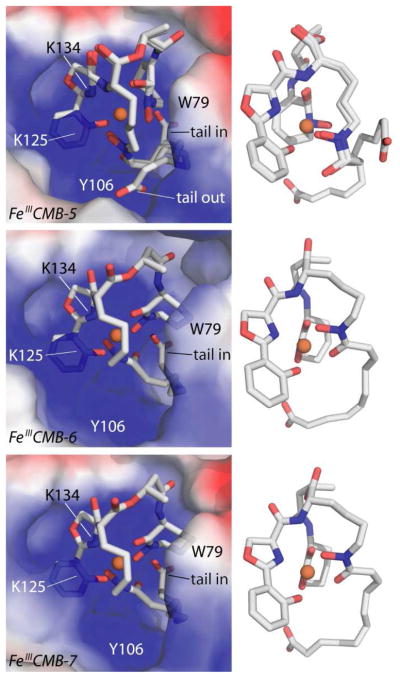
Next, the individual isoforms were cocrystallized with Scn in order to correlate the variable dissociation constants with the molecular interactions within the protein calyx. While crystals were obtained for all isolable isoforms (n = 3–8, Supplementary Figure 2), the electron density was only interpretable for CMBs with tail lengths n = 5 (FeIIICMB-5), n = 6 (FeIIICMB-6, the original structure (18)), and n = 7 (FeIIICMB-7). The structures of these three isoforms are shown in Figure 2. Crystallographic analysis of the FeIIICMBs bound to wild type human Scn show that the change in the tail length of CMB does not affect the backbone conformation of Scn locally or globally (superpositions rmds of FeIIICMB-5 onto the original FeIIICMB-6 structure (PDB accession code 1X89): 0.12Å molecule A, 0.17Å molecule B, and 0.12Å molecule C; superpositions rmds of FeIIICMB-5 onto the original FeIIICMB-6 structure: 0.19 Å molecule A, 0.30 Å molecule B, and 0.20 Å molecule C; calculated on all common Cα). There were no significant changes between the original FeIIICMB n = 6 structure and the n = 5 and n = 7 structures with the exception of the position of the bound siderophore within the calyx. Three pockets in the Scn calyx are walled off from each other by side-chains with pocket #1 defined by the side-chains of Lys125 and Lys134, pocket #2 defined by Lys134 and Trp79/Arg81 and pocket #3 defined by Trp79/Arg81 and Lys125. Previous studies have shown that the cationic calyx residues (Lys125, Lys134, Arg81) play an important role in the recognition of siderophores through electrostatic interactions. (4, 22) Similar to the original structure, the hydroxyphenyl oxazoline (HPO) group is bound in pocket #1 between lysines Lys125 and Lys134, and the seven-membered cyclic hydroxamate in pocket #2 between Lys134 and Arg81. The fatty acid tail branches from the hydroxamate bound in pocket #3 and reaches down into a sub-pocket located below pocket #2.
Figure 2.
Structures of Scn bound to FeIIICMB. The carboxylate tail is found bound in the lower portion of pocket #2 in both the n = 6 (FeIIICMB-6, middle) and n = 7 (FeIIICMB-7, bottom) structures. In contrast, the tail adapts both a “tail in” and “tail out” conformation in the case of n = 5 (FeIIICMB-5, top). To accommodate the “tail in” binding conformation, the hydroxamate in pocket #3 shifts 2.0 Å down into the calyx, disrupting ideal iron coordination.
The fatty acid tail of FeIIICMB-5 is found in two different conformations: “tail in” where the tail of CMB binds deep into pocket #2, and “tail out” where the tail of CMB moves out the backside of pocket #3 and away from the calyx. Conversely, the structure of FeIIICMB-7 has the same siderophore binding mode as the previously published FeIIICMB-6 in which the tail is tucked under the complex and the carboxylate is buried deep in pocket #2. While the electron density is difficult to interpret in the FeIIICMB n = 4 structure, it is apparent that the tail is not seen buried deep in pocket #2, likely contributing to the overall disorder of the siderophore in these data sets. There was no density for the siderophore found in the FeIIICMB n = 3 structure.
There is a correlation between the affinities determined by fluorescence and the binding modes determined by crystallography. The two binding modes of the CMBs, “tail in” and “tail out”, show that the tucked tail creates important interaction in the recognition of FeIIICMB. It has been previously observed that the positioning of the tail in the pocket mirrors the crystal structure of Scn bound to an opportunistic ligand, n-capric acid. (18) In this structure, the carboxylate of the fatty acid was bound in pocket #2 with the alkyl tail trailing into the calyx. (23) Without this interaction, the mechanism of recognition of the FeIIICMB substrate is severely hindered. We speculate that the smaller tail lengths are not long enough to tuck under the complex and bury the carboxylate, and as a result are bound with lower affinity by Scn.
The molecular structure of CMB is vastly different from catecholate siderophores (BB, Ent, parabactin etc.), yet it is still bound by Scn, emphasizing the effectiveness of the generalized recognition strategy of Scn based on hybrid electrostatic interactions (cation-π with aromatic metal-binding units and coulombic with anionic complexes). (4, 22) CMB has only a single aromatic unit, the HPO, to participate in a cation-π binding interaction within the Scn calyx, compared to the tris-catecholate Scn substrates Ent and BB. The position of the HPO unit in the CMB-Scn cocrystal structures in between the Lys125 and Lys134 in pocket #1 is conserved across all the structures, and the phenol ring of the HPO superimposes with the catecholamide of Ent from the Scn cocrystal (Supplementary Figure 4). The electrostatic potential map of the HPO binding unit (Figure 3), along with the calculated cation-binding energies (BEapoHPO = 22.51 kcal/mol, BEM-HPO = 36.68 kcal/mol), demonstrate an enhanced ability to participate in a cation-π interaction compared to the 2,3-catecholamide. (23)
Figure 3.
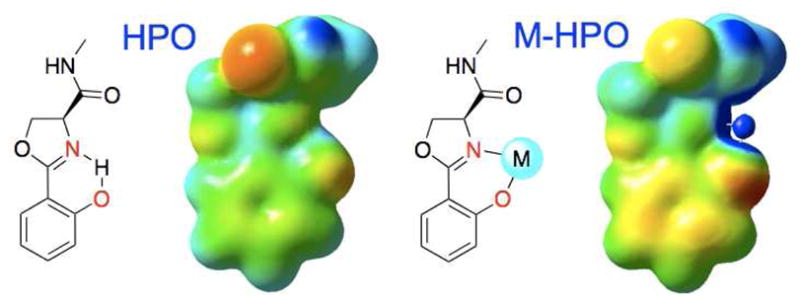
Electrostatic potentials for HPO and M-HPO units (with the input structures) were calculated at the RHF/6-311G** level of theory with iso-values of −0.2 (red) to 0.2 (blue). Cation-π interaction energies were calculated at the MP2/6-311G** level of theory and BSSE was corrected for with the counterpoise method.
Several tris-hydroxamate siderophores have been demonstrated to not bind to Scn. (18) The lack of recognition is attributed to (a) the absence of aromatic binding units, which negates the cation-π binding interaction, (b) the formation of neutral ferric complexes, which negates the coulombic binding interaction, and (c) the lack of shape complementarity with Scn. In spite of the neutral, bishydroxamate iron coordination of CMB, some isoforms are still sequestered by Scn. The HPO unit enables Scn to recognize the primary coordination sphere of the complex through enhanced cation-π interactions. Furthermore, the fatty acid tail is the source of the overall monoanionic complex charge. However, CMB complexes that cannot adopt the “tail-in” conformation in the Scn calyx eliminate the coulombic binding interaction. Therefore, while the HPO unit is critical to the Scn recognition of CMB, the length of the fatty acid tail ultimately determines whether a CMB complex is intercepted by Scn.
In addition to Scn dissociation constant, another important factor in siderophore sequestration is the rate at which the siderophore dissociates from Scn. In other words, the half-life of the protein complex ultimately determines the effectiveness of the sequestration. While the siderophore off-rate was not determined in this study, the variance binding mode among the CMBs with a corresponding change in dissociation constants suggest that varying tail lengths in the suite of CMBs is the means of immunoevasion. Moreover, the relatively weak binding affinities indicate that the function of these siderophores is more complex than simply acting as stealth siderophores. Although the dissociation constant of the Mtb receptor for FeIIICMB has not been reported, the weak affinity of Scn for the FeIIICMBs may indicate that Scn cannot interrupt iron transport in Mtb in the same way as the Ent and BB bacterial systems. This is corroborated by Halaas et al. who showed that inhibiting the synthetic pathway of the CMBs with p-aminosalycilate was more effective at growth inhibition of mycobacteria in vitro than Scn. (9)
Since Mtb is primarily an intracellular bacterium, the host-pathogen interactions are more pronounced. As a result, Mtb has developed more complex survival strategies than the typical extracellular bacteria. One example of this is the mechanism by which Mtb evades cell death by preventing fusion with late endocytic organelles in the macrophage. (14, 17) Similarly, in order to overcome host defenses, the siderophore production of Mtb must be tailored specifically to acquire iron from within the host cell. Recent in vivo studies have shown that Scn is most effective against free-living M. avium in the bloodstream during the early phase of infection, where a substantial fraction of the bacteria are extracellular, but is less effective at suppressing the growth of intracellular mycobacteria. (9) This is attributed to the fact that within macrophages, Scn is eventually trafficked to endolysosomes whereas the pathogen colocalizes with intracellular transferrin in separate compartments. While the intracellular iron acquisition of Mtb is not yet well understood, the atypical structures of CMB and mycobactin, including the cyclic hydroxamate, the HPO and the side-chains, are a testament to the strides the bacterium takes to produce a siderophore uniquely qualified for this task. One example of the structure-function correlation in these siderophores is the wide range of lipophilicities of CMBs and mycobactins, which enable the siderophores to navigate throughout the cell in search of iron stores. (24) While CMB is captured by Scn in some cases, the ultimate survival of the pathogen implies that the Scn immune response does not effectively inhibit iron acquisition by mycobacteria.
METHODS
General
The FeIIICMBs were isolated from M. smegmatis based on published procedures. (19, 21) M. smegmatis strain mc2 155 was obtained from Prof. Hiroshi Nikaido (University of California, Berkeley). Affinities of Scn for the CMBs were determined by a fluorescence-based binding assay as previously described. (4) Electrostatic calculations (23, 26) were performed according to published procedures. Computational studies were conducted at the Molecular Graphics and Computation Facility, College of Chemistry, University of California, Berkeley.
HPLC Conditions
Separation of the ferric CMBs was achieved on an Agilent Eclipse XDB-C18 (reverse phase) 5μM, semi-preparative column with a gradient solvent system of water:methanol (50:50 @ 0 min to 30:70 at 30min) with a flow rate of 1.25 mL/min and a detection wavelength of 450 nm. The 30:70 system was maintained for an additional 50 min while the FeIIICMB fractions eluted. The fractions were analyzed by LCMS to identify the isoform (Supplementary Figure S1), condensed, and a second HPLC run was performed with the same water:methanol solvent system to further purify the complexes (with UV/VIS detection at 450, 310, and 254 nm). The fractions were dried and pure ferric CMBs were stored under vacuum in a dark cabinet until it was used in binding studies (usually within 3 days).
LCMS Characterization
Ferric CMB-containing HPLC fractions were analyzed by LCMS on an Agilent Eclipse XDB-C18 (reverse phase) 5 μM analytical column (λ = 450 nm, flow rate = 0.5 mL/min) with the following gradient: 0 min – 50:50 (H2O:MeOH); 10 min – 30:70 (H2O:MeOH); 30 min – 30:70 (H2O:MeOH); 33 min – 50:50 (H2O:MeOH).
Fluorescence Quenching Binding Assay
Fluorescence quenching of recombinant Scn was measured on a Cary Eclipse fluorescence spectrophotometer. A 5 nm slit band-pass for excitation and a 10 nm slit band-pass for emission were used with a high-voltage detector. An excitation wavelength of λexc = 281 nm was used, and emission was collected at λem = 340 nm. Measurements were made at a protein concentration of 100 nM in buffered aqueous solutions, plus 32 μg/mL ubiquitin (Sigma) and 5% DMSO. Fluorescence values were corrected for dilution upon addition of substrate. Fluorescence data were analyzed by nonlinear regression analysis of fluorescence response versus substrate concentration using a one-site binding model as implemented in DYNAFIT.25 All standard binding experiments were done at pH 7.4 using TBS aqueous buffer. Control experiments were performed to ensure the stability of the protein at experimental conditions, including dilution and the addition of ubiquitin and DMSO.
Computational Methods
To determine the quadrupole moments, Θzz, the aromatic structures were geometry optimized and characterized via frequency calculations at the RHF/6-311G** level of theory in the Gaussian 03 package. (26) To determine the aromatic-cation interaction energies the aromatic structures, the sodium cation, and the aromatic-cation complexes were characterized via a frequency calculation at the MP2/6-311++G** level of theory and the aromatic-cation interaction energies corrected for basis set superposition error (BSSE) with the counterpoise method in the Gaussian 03 package. (26) In the aromatic-cation calculations the sodium ion was fixed at a distance of 2.47 Å above the centroid of the aromatic unit. To determine the orbitals involved in the ESIPT and the triplet energy for the HPO unit, time-dependent density functional theory calculations were performed using a B3LYP/6-311++G(d,p) basis set in Gaussian 03 package. (26) Geometry optimizations were performed on the input structures without symmetry constraints.
Protein Crystallographic Studies
Human Scn was isomorphously co-crystallized with FeIIICMB with fatty acid tail lengths n = 3, 4, 5, 6, 7 and 8 using the previously reported crystallization conditions. (18) Crystals were cryoprotected by supplementing the mother liquor with 15% glycerol before flash freezing them in liquid nitrogen at −170°C. All data sets were collected at the Advanced Light Source in Berkeley, CA and processed with the HKL2000 software package. (27) Reflections chosen for calculating Rfree were matched to those used in the original wild type human structure (PDB code 1L6M). Phases were calculated using the original Scn structure, modeling was performed with Coot, and the structure was refined using Refmac5. (28–30) Relevant data collection and refinement statistics are shown in Table 1. Coordinates for both structures have been deposited with the PDB (accession codes 3PEC.pdb and 3PED.pdb).
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (Grant R01AI11744 and Grant R01AI59432). The authors thank the Advanced Light Source beamline 5.0.1 for crystallographic data collection, H. Nikaido for the M. smegmatis strain, C. Chang for use of his LCMS, and R. Abergel for helpful discussions. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinberg ED. Iron loading and disease surveillance. Emerg Infect Dis. 1999;5:346–352. doi: 10.3201/eid0503.990305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raymond KN, Dertz EA. Biochemical and Physical Properties of Siderophores. In: Crosa JH, Mey AR, Payne SM, editors. Iron transport in bacteria. ASM Press; Washington, D. C: 2004. pp. 3–17. [Google Scholar]
- 4.Goetz DH, Holmes MA, Borregaard N, Bluhm ME, Raymond KN, Strong RK. The Neutrophil Lipocalin NGAL Is a Bacteriostatic Agent that Interferes with Siderophore-Mediated Iron Acquisition. Mol Cell. 2002;10:1033–1043. doi: 10.1016/s1097-2765(02)00708-6. [DOI] [PubMed] [Google Scholar]
- 5.Kauffmann SHE. New issues in tuberculosis. Ann Rheum Dis. 2004;63:1150–1156. doi: 10.1136/ard.2004.028258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olakanmi O, Schlesinger LS, Britigan BE. Hereditary hemochromatosis results in decreased iron acquisition and growth by Mycobacterium tuberculosis within human macrophages. J Leuk Biol. 2007;81:195–204. doi: 10.1189/jlb.0606405. [DOI] [PubMed] [Google Scholar]
- 7.Saiga H, Nishimura J, Kuwata H, Okuyama M, Matsumoto S, Sato S, Matsumoto M, Akira S, Yoshikai Y, Honda K, Yamamoto M, Takeda K. Lipocalin 2-dependent inhibition of mycobacterial growth in alveolar epithelium. J Immunol. 2008;181:8521–8527. doi: 10.4049/jimmunol.181.12.8521. [DOI] [PubMed] [Google Scholar]
- 8.Martineau AR, Newton SM, Wilkinson KA, Kampmann B, Hall BM, Nawroly N, Packe GE, Davidson RN, Groffiths CJ, Wilkinson RJ. Neutrophil-mediated innate immune resistance to mycobacteria. J Clin Inv. 2007;117:1988–1994. doi: 10.1172/JCI31097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halaas Ø, Steigedal M, Haug M, Awuh J, Ryan L, Brech A, Sato S, Husebye H, Cangelosi GA, Akira S, Strong RK, Espevik T, Flo TH. Intracellular Mycobacterium avium Intersect Transferrin in the Rab11+ Recycling Endocytic Pathway and Avoid Lipocalin 2 Trafficking the Lysosomal Pathway. J Infect Dis. 2010;201:783–792. doi: 10.1086/650493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson MK, Abergel RJ, Raymond KN, Arceneau JEL, Byers BR. Siderophores of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis. Biochem Biophys Res Comm. 2006;348:320–325. doi: 10.1016/j.bbrc.2006.07.055. [DOI] [PubMed] [Google Scholar]
- 11.Koppisch AT, Browder CC, Moe AM, Shelley JT, Kinkel BA, Hersman LE, Iyer S, Ruggiero CE. Petrobactin is the Primary Siderophore Synthesized by Bacillus anthracis Str. Sterne under Conditions of Iron Starvation. BioMetals. 2005;18:577–585. doi: 10.1007/s10534-005-1782-6. [DOI] [PubMed] [Google Scholar]
- 12.Abergel RJ, Wilson MK, Arceneaux JEL, Hoette TM, Strong RK, Byers BR, Raymond KN. The Anthrax Pathogen Evades the Mammalian Immune System Through Stealth Siderophore Production. Proc Natl Acad Sci USA. 2006;103:18499–18503. doi: 10.1073/pnas.0607055103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischbach MA, Lin H, Zhou L, Abergel RJ, Liu DR, Raymond KN, Wanner BL, Strong RK, Walsh CT, Aderem A, Smith KD. The pathogen-associated iroA gene cluster mediates bacterial evasion of lipocalin 2. Proc Natl Acad Sci USA. 2006;103:16502–16507. doi: 10.1073/pnas.0604636103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomez JE, McKinney JD. M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis. 2004;84:29–44. doi: 10.1016/j.tube.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez GM, Smith I. Identification of an ABC Transporter Required for Iron Acquisition and Virulence in Mycobacterium tuberculosis. J Bacteriol. 2006;188:424–430. doi: 10.1128/JB.188.2.424-430.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Voss JJ, Rutter K, Schroeder BG, Su H, Zhu YQ, Barry CE. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc Natl Acad Sci USA. 2000;97:1252–1257. doi: 10.1073/pnas.97.3.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ratledge C. Iron, mycobacteria and tuberculosis. Tuberculosis. 2004;84:110–130. doi: 10.1016/j.tube.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 18.Holmes MA, Paulsene W, Xu J, Ratledge C, Strong RK. Siderocalin (Lcn 2) Also Binds Carboxymycobactins, Potentially Defending against Mycobacterial Infections through Iron Sequestration. Structure. 2005;13:29–41. doi: 10.1016/j.str.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 19.Ratledge C, Ewing M. The occurrence of carboxymycobactin, the siderophore of pathogenic mycobacteria, as a second extracellular siderophore in Mycobacterium smegmatis. Microbiol. 1996;142:2207–2212. doi: 10.1099/13500872-142-8-2207. [DOI] [PubMed] [Google Scholar]
- 20.Adilakshmi T, Ayling PD, Ratledge C. Mutational Analysis of a Role for Salicylic Acid in Iron Metabolism of Mycobacterium smegmatis. J Bacteriol. 2000;182:264–271. doi: 10.1128/jb.182.2.264-271.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lane SJ, Marshall PS, Upton RJ, Ratledge C. Isolation and characterization of carboxymycobactins as the second extracellular siderophores in Mycobacterium smegmatis. BioMetals. 1998;11:13–20. [Google Scholar]
- 22.Hoette TM, Abergel RJ, Xu J, Strong RK, Raymond KN. The Role of Electrostatics in Siderophore Recognition by the Immunoprotein Siderocalin. J Am Chem Soc. 2008;130:17584–17592. doi: 10.1021/ja8074665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goetz DH, Willie ST, Armen RS, Bratt T, Borregaard N, Strong RK. Ligand Preference Inferred from the Structure of Neutrophil Gelatinase Associated Lipocalin. Biochemistry. 2000;39:1935–1941. doi: 10.1021/bi992215v. [DOI] [PubMed] [Google Scholar]
- 24.Luo MK, Fadeev EA, Groves JT. Mycobactin-mediated Iron Acquisition within Macrophages. Nat Chem Biol. 2005;1:149–153. doi: 10.1038/nchembio717. [DOI] [PubMed] [Google Scholar]
- 25.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal Biochemistry. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 26.Frisch MJT, et al. Gaussian 03, Revision C.02. Gaussian, Inc; Wallingford, CT: 2004. [Google Scholar]
- 27.Otwinowski Z, Minor W. Methods in Enzymology. In: Carter CW Jr, Sweet RM, editors. Macromolecular Crystallography. part A. Vol. 276. Academic Press; New York: 1997. pp. 307–326. [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Murshudov GN, Vagin AA, Dodson EJ. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 30.Collaborative Computational Project, Number 4. The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.