Abstract
We have developed a silicon photonic biosensing chip capable of multiplexed protein measurements in a biomolecularly complex cell culture matrix. Using this multiplexed platform combined with fast one-step sandwich immunoassays, we perform a variety of T cell cytokine secretion studies with excellent time-to-result. Using 32-element arrays of silicon photonic microring resonators, the cytokines interleukin-2 (IL-2), interleukin-4 (IL-4), interleukin-5 (IL-5), and tumor necrosis factor alpha (TNFα) were simultaneously quantified with high accuracy in serum-containing cell media. Utilizing this cytokine panel, secretion profiles were obtained for primary human Th0, Th1, and Th2 subsets differentiated from naïve CD4+ T cells, and we show the ability to discriminate between lineage commitments at early stages of culture differentiation. We also utilize this approach to probe the temporal secretion patterns of each T cell type using real-time binding analyses for direct cytokine quantitation down to ~100 pM with just a 5 min analysis.
Introduction
Cytokines constitute an important class of secreted proteins that are key regulators and effectors of the immune response.1 These small,2 low abundance3 cell-signaling proteins are secreted by both lymphocytes and epithelial cells, and they are important targets for cell-based immunological studies and in vitro diagnostics. Cytokines often have overlapping and multifaceted biological functions,4,5 motivating the development of multiplexed detection strategies. Given the importance of cytokine analysis for fundamental immunological studies and their promise as diagnostic biomarkers,6,7 it is not surprising that the development of advanced cytokine detection methods is an active area of research. Recent work in developing novel cytokine analysis platforms has involved fiber-optic microsphere arrays,8 cytometric bead assays,9 photonic crystal-enhanced fluorescent protein microarrays,10 microegraving,11 ELISpot assays,12 T cell microdevices for fluorescent cytokine detection,13 optofluidic 1-D photonic crystals,14 and microring resonators.15 Each of these new approaches features some combination of excellent sensitivity, highlevel multiplexing, fast time-to-result, real-time binding analysis, and/or demonstrated clinical or biological applicability. However, there remains a tremendous unmet need for real-time detection methods that allow extremely rapid, quantitative, and highly multiplexed cytokine analysis over a relevant dynamic range with minimal sample preparation.
Herein, we report the simultaneous detection of four cytokines secreted from primary human T cell populations using only a 5-min assay performed on an intrinsically scalable silicon photonic microring resonator analysis platform. Silicon photonic microring resonators belong to a class of high-Q optical microcavities that have recently shown promise for label-free bioanalysis.16-20 In addition to eliminating the need for fluorescent or enzymatic tags that add assay complexity and non-native perturbations to biomolecular interactions,21,22 optical ring resonators also feature real-time reaction monitoring capabilities.23 The silicon photonic microring resonator arrays utilized in this work (Figure 1a) are further distinguished by their high scalability on a small footprint, low per-device cost, and ease of fabrication by standard semiconductor processing methods.24 Resonant microcavity sensing is based on the interaction of molecules near the ring surface with the propagating evanescent field that decays with distance from the surface.25 In our platform, infrared light is inserted into linear Si waveguides that access each microring and allow efficient coupling to the ring only at wavelengths (λ) that meet the resonance condition defined as:
where m is an integer, r is the radius of the microring, and neff is the effective refractive index (RI) of the optical mode. Target binding to a receptor-modified ring surface increases neff , which leads to a corresponding shift in the resonance wavelength (Δpm). The biomolecular generality of this sensing approach makes it amenable to the detection of both nucleic acid26-28 and protein15,20,29 biomarkers.
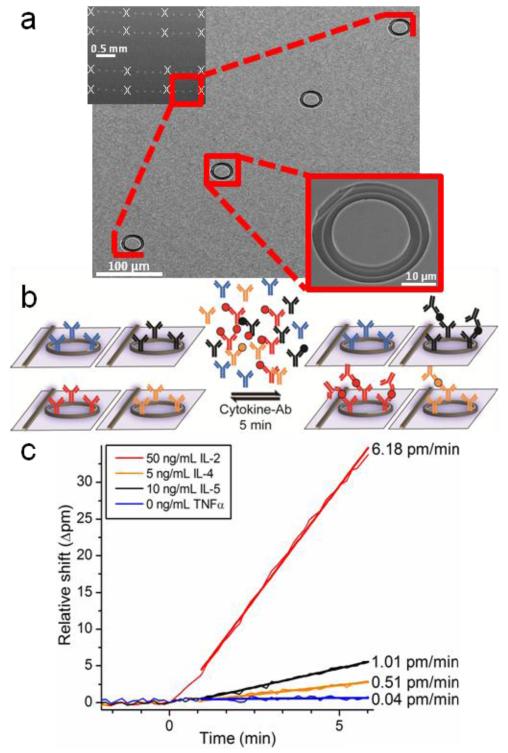
Figure 1.
Microring resonator arrays enable fast and multiplexed one-step sandwich immunoassays. (a) Scanning electron micrographs of increasing zoom depict a silicon photonic microring resonator chip with an array of 64 rings, 32 of which are monitored simultaneously. Sixteen rings are configured for use as thermal controls (indicated by white x’s) and are buried under a polymer cladding layer. Rings are arranged linearly to allow straightforward interfacing with microfluidic channels. Each 30-μm ring resonator is accessed by a separate linear waveguide. (b) Arrays of microrings can be functionalized with capture antibodies specific for various cytokine targets. Following incubation of samples with a cocktail of secondary antibodies, the onestep sandwich immunoassay is monitored in real time for each ring. Cytokines bind specifically to their capture antibody in complex with the cognate detection antibody, thus enhancing the signal. (c) Multiple cytokines at varying concentrations are simultaneously quantified based on the initial slope (Δpm/min) of the sensor response upon sample introduction. One representative ring specific for each cytokine is shown for clarity, including a TNFα negative control.
Notably, we recently demonstrated the ability to detect the cytokine IL-2 at concentrations as low as 6 pM using a 45-min two-step sandwich immunoassay.15 Though this study demonstrated good sensitivity and precision for secreted IL-2 analysis in Jurkat T cell cultures, the (1) incorporation of multiplexed cytokine measurements and (2) significant reduction of the assay time-to-result remained critically important. Multiplexed measurements are of particular significance in cytokine analysis because of their complex interplay and overlapping roles that involve multiple signaling pathways.1,5 Higher throughput assays are also clearly needed to more practically perform detailed immunological studies that require measuring multiple cytokines from within many samples in a reasonable time period. To accomplish these goals, we designed a rapid kinetic-based one-step sandwich immunoassay that combines multiplexed primary cytokine binding with secondary antibody amplification in a single step (Figure 1b). Other manifestations of one-step sandwich immunoassays have been demonstrated to increase the speed of protein detecting assays, albeit in single-parameter assay formats, using surface plasmon resonance,30 interferometry,31 and radioimmunoassays.32
In this article, we report unprecedentedly fast, multiplexed cytokine immunoassays based on 5-min initial slope analysis (Figure 1c) using a silicon photonic microring resonator array platform. We demonstrate simultaneous quantitation of the cytokines IL-2, IL-4, IL-5, and TNFα from unknown solutions in serumcontaining cell media with high accuracy (10% average error). By monitoring real-time binding of cytokines pre-associated with a secondary antibody and subtracting the non-specific binding response on in situ control sensors, we overcome the challenges associated with detecting low-molecular weight proteins in a biomolecularly complex sample matrix. We further apply the onestep sandwich immunoassay to cytokine secretion profiling of four T cell types: primary human Th0, Th1, and Th2 subsets, as well as Jurkat T lymphocytes. The high throughput provided by the one-step sandwich assay allows for the redundant, multiplexed analysis of 10s of samples within a short timeframe. Even at short differentiation periods, commitment to particular T cell lineages is observed based upon unique cytokine signatures, and multiplexed temporal secretion profiles are also obtained. The ability to monitor multiple cytokines simultaneously with real-time binding analysis enables primary T cell immunological studies with unprecedented temporal resolution.
Results and Discussion
One-step sandwich immunoassays for simultaneous quantitation of 4 cytokines
The ring resonator platform, as shown in Figure 1a, contains a usable array of thirty-two 30-μm diameter rings (including on-chip thermal controls) for multiplexed cytokine detection. The ring resonator sensor signal (shift in resonance wavelength) is sensitive to changes in the local RI near the sensor surface, with the 1/e decay length of the evanescent field confined to 63 nm due to the high RI contrast between the waveguide and aqueous surroundings.25 This length scale is well-suited to sandwich immunoassays since antibodies have dimensions of 15 × 7 × 3.5 nm,33 while cytokines are roughly one tenth the size of antibodies.
For the experiments described herein, the rings are first covalently modified with four anti-cytokine capture antibodies that are directed by microfluidics to groups of 4-5 rings each (Figure S1). A few rings are not modified with capture antibodies and serve as controls for non-specific binding and bulk RI changes in subsequent experiments. Antibody immobilization is monitored in real time to ensure an optimal density of 3-6 ng mm−2 (Figure S2). Biomolecular binding of the target cytokine to capture antibodies on the ring surface increases neff and results in a shift of the resonance to longer wavelengths, which is monitored in real time for each ring as a relative shift (Δpm). The initial slope of this response as a function of time (Δpm/min) after sample introduction is analyte concentration dependent, as described by Fick’s first law under masstransport limiting conditions.20,34 For the one-step sandwich assays utilized in these studies, the sample was pre-incubated with all relevant detection antibodies so that cytokines were detected as larger cytokineantibody conjugates (Figure 1b). Since antibodies have a molecular weight of ~150 kDa (compared to ~5-40 kDa for cytokines),2 the signal associated with binding of the cytokine-antibody conjugate is roughly an order of magnitude larger than the cytokine alone. Multiplexed cytokine standards prepared in cell media showed concentration-dependent responses in initial slope that were fit to a linear regression from just 5 min of real-time binding data (Figure 1c).
Well-blocked chips allow highly selective binding of cytokine-antibody conjugates only to the appropriate rings. Each chip was tested for cross-reactivity by serially flowing each of the cytokines across all the rings. Figure 2 shows that no appreciable cross-reactivity exists between rings functionalized to detect specific cytokines, even at a relatively high concentration (50 ng mL−1). This is especially important since the absence of cross-reactivity is necessary for simultaneous calibration and detection of multiple cytokines from the same sample. One-step sandwich assay negative control experiments performed in cell media spiked with 1 μg mL−1 of each detection antibody (but without cytokines) indicated no non-specific binding (Figure S3). After determining that the rings are highly specific for the appropriate cytokine, sensors were calibrated by performing one-step sandwich assays on a series of multiplexed cytokine calibration standard cocktails prepared in cell media (Table S1a). Each assay involves a 6-7-min incubation with the standard followed by a low-pH capture antibody regeneration and return to cell media running buffer. The low-pH rinse disrupts protein-protein interactions to remove all bound cytokines and detection antibodies, restoring the covalently bound capture antibodies to their original state. Capture antibodies used herein were selected for their high affinity and stability, and they can be regenerated up to 30 times without substantial loss in binding activity. This allows for the consecutive analysis of many samples using a single chip. By plotting the control ring-corrected initial slopes as a function of cytokine concentration (Figure S4), a linear calibration plot was obtained simultaneously for each of the 4 cytokines assayed (IL-2, IL-4, IL-5, and TNFα). One-step sandwich immunoassays were also shown to have a linear dynamic range spanning more than 2 logs, with limits of detection in serum-containing cell media ranging from 67-119 pM for each cytokine (Table S2). Although rapid, the one-step sandwich limits of detection are inferior by roughly 0.5 orders of magnitude compared to 45-min two-step sandwich assays.
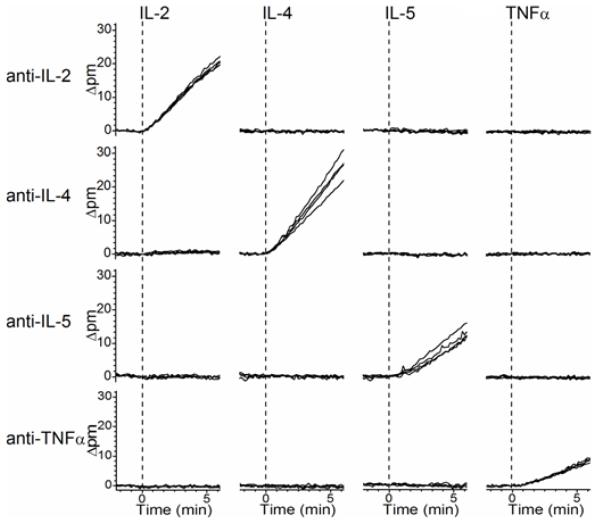
Figure 2.
Cross-reactivity diagram for multiplexed onestep sandwich immunoassay. Each of the 4 cytokines (IL-2, IL-4, IL-5, and TNFα) is introduced in series. All cross-reactivity data is collected from a single sensor chip, with a low-pH regeneration between each cytokine assay. Rows contain responses from a group of 4 rings functionalized with the same cytokine capture antibody, and each column represents a different targeted cytokine. A 50 ng mL−1 solution of a single cytokine prepared in serum-containing cell media in introduced at t = 0 min in each case, as indicated by the dashed lines. Only on-diagonal shifts in resonant wavelength are observed, confirming that there each detection event is orthogonal to other off-target cytokines.
In order to demonstrate the highly quantitative multiplexed sensing capabilities of the platform, onestep sandwich assays were performed on 3 blinded, multiplexed cytokine unknowns prepared in cell media. Based on the calibration curves shown in Figure 3a, the unknowns were quantitated via inverse regression. Figure 3b shows the strong agreement between the asprepared values on the left and the experimentally determined concentrations on the right for each of the 3 blinded unknowns. The one-step sandwich successfully quantitated each unknown (average error = 10% ; average absolute error = 1 ng mL−1), allowing easy differentiation between the unknowns based on their cytokine “fingerprint.” This demonstrates the simultaneous and highly quantitative analysis of 4 cytokines on a single chip.
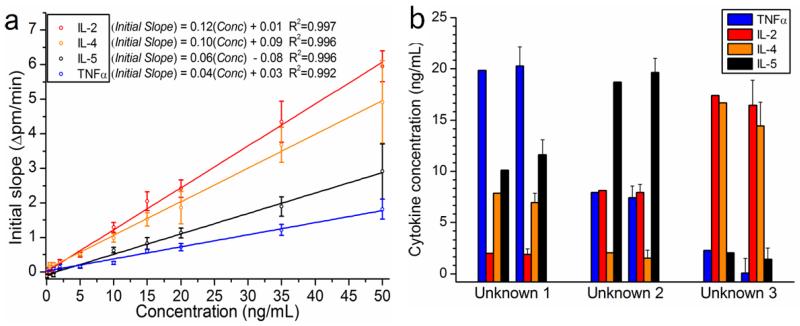
Figure 3.
Simultaneous quantitation of 4 cytokines from 3 blinded unknowns prepared in serum-containing cell media. (a) The average control ring-corrected initial slope is plotted as a function of cytokine concentration for each cytokine. All multiplexed standards were prepared in cell culture media containing 10% serum, and all calibration data was obtained on a single chip that was also used for determination of unknowns. The calibration curves used 10 standards between 0 and 50 ng mL−1. Each calibration curve is fit well with a linear regression (all R2≥0.992), and the displayed equations are used to quantitate solutions with unknown cytokine concentrations via inverse regression. (b) Cytokine cocktails of unknown concentration were prepared in RPMI 1640 + 10% FBS. After incubating with all 4 corresponding detection antibodies, the unknowns were quantitated by measuring the initial slope of the binding interaction in a one-step sandwich immunoassay. For each blinded unknown, the as-prepared values (left) show strong agreement with the values determined by the ring resonator array (right). Error bars represent the total propagated standard error that includes: (1) the standard deviation of n=4-5 rings used to analyze each cytokine and (2) the regression error of the calibration curve.
Multiplexed Jurkat cytokine secretion analysis and method validation
In previous work, temporal Jurkat T cell IL-2 secretion was quantitated by a two-step sandwich immunoassay that required ~45 min.15 Due to the longer assay time, it was difficult to achieve high temporal resolution; the Jurkat culture was thus sampled every 8 h (t = 0, 8, 16, and 24 h post-stimulation). One-step sandwich immunoassays based on a 5-min initial slope analysis enable higher throughput sampling. After removing cells by centrifugation, a cocktail containing each of the 4 appropriate detection antibodies (anti-IL-2, anti-IL-4, anti-IL-5, anti-IL-1β) was added to cell culture media aliquots taken at t = 0, 2, 4, 6, 8, 10, 15, and 24 h from both PMA/PHA-stimulated and unstimulated cultures. After calibrating the chip, Jurkat temporal secretion aliquots were assayed in series by the one-step sandwich assay. Consistent with previous work,15 the temporal secretion profile displays exclusive IL-2 secretion that increases over time, and is only observed from stimulated cells (Figure S5). Additional independent reports confirm that PMA/PHA stimulation is effective for stimulating IL-2 secretion35 and also that IL-2 transcripts and protein are absent without stimulation.36 Furthermore, the greater temporal resolution clarifies the biological nuances of the secretion profile. It was observed that appreciable IL-2 secretion was only evident after 4 h of stimulation; no IL-2 was observed at t = 2 h, signifying a delay between mitogenic stimulation and protein expression/secretion. Additionally, the IL-2 concentration leveled off and then decreased slightly after 24 h, suggesting IL-2 degradation or uptake by the delayed expression of IL-2 receptor at the cell surface.37,38 Improved temporal resolution made possible by faster assay time can have important implications for observing details about the cytokine secretion process. This multiplexed one-step sandwich assay performed on Jurkat T cells was successfully validated by conducting 4 commercial ELISAs in parallel (Figure S6).
Multiplexed analysis of primary T cell subsets based on cytokine secretion signatures
After confirming and validating the utility of one-step sandwich assays for T cell secretion analysis, a study involving multiple cytokines and multiple cell lines was undertaken. Primary naïve CD4+ T cells, which can be differentiated into various T helper subsets with unique cytokine signatures, provide an interesting application for multiplexed analysis. Naïve CD4+ T cells, which have not yet encountered an antigen, are produced in the thymus and are known to differentiate into various effector subsets including Th1, Th2, Th17, and iTregs.39 Th1 and Th2 subsets have been well-studied, and the required cytokine stimuli and genetic mechanisms for differentiation have been described.40,41 Th0 cells arise after initial activation of naïve T cells but prior to final differentiation to an effector subset. The cytokine differentiation cues, relevant pathways, and cytokine secretion signatures for Th0, Th1, and Th2 cell differentiation paths are summarized in Figure S7. In brief, activation of naïve T cells with IL-2, anti-CD3, and anti-CD28 results in Th0 cells that produce a large array of cytokines. Th0 cells differentiate to Th1 upon exposure to IL-12, while Th0 cells differentiate to Th2 upon exposure to IL-4.39,40 Th1 cells are known to produce IFNγ, IL-2, and TNFα; Th2 cells produce IL-4, IL-5, IL-13,42 and to a lesser extent, TNFα and IL-2.39 By guiding these differentiation processes in vitro starting with primary naïve CD4+ T cells isolated from healthy donors, directed differentiation toward Th0, Th1, and Th2 lineages allowed for a comparison of the various T helper cytokine secretion profiles.
Naïve CD4+ T cells were isolated via magnetic bead negative selection, then cultured, activated, and differentiated for 7 days in vitro as described previously.43,44 Based on flow cytometry experiments performed by Cousins et al, a considerable portion of cells remain undifferentiated after 7 days (especially in the Th2 subset).44 However, 7 days proved to be sufficient for observing significant differences in cytokine secretion among the subsets using this multiplexed measurement approach. At the end of 7 days, cells were washed to remove any exogenous cytokines and then stimulated. PMA and ionomycin were used as co-stimulators that act on the T-cell receptor as in vivo stimulation analogues.41,44 After 24 h of stimulation, cell aggregation and blasting were observed as evidence of stimulation (Figure S8a). Stimulated and unstimulated cell culture aliquots were obtained for each subset (Th0, Th1, Th2). Cells were removed by centrifugation, and one-step sandwich assays on a calibrated ring resonator chip (Table S1b; Figure S4b) allowed for multiplexed cytokine quantitation. In this case, ~1% DMSO present in the cell aliquots due to protease inhibitors and PMA/ionomycin addition resulted in a ~125-pm bulk RI shift. Slopes for one-step sandwich assays were fit following this large bulk shift (Figure S9). All T cell subset cytokine secretion experiments were performed three times using different healthy donors, giving consistent results.
The multiplexed T cell differentiation studies are summarized in Figure 4. For each T cell subset, stimulated cells secrete significantly more cytokines than unstimulated controls. Th0 and Th1 stimulated cells show much higher (p<0.01) levels of IL-2 and TNFα compared to the corresponding controls. Th2 stimulated cells display higher (p<0.05) levels of IL-2, IL-4, IL-5, and TNFα compared to the unstimulated Th2 controls. The stimulated secretion levels of the signature Th2 cytokines (IL-4 and IL-5), which were significantly higher in Th2 culture than Th1 culture (Figure 4, p<0.01), agree well with an earlier report by Yano et al based on the same 7-day activation/differentiation and 24-h PMA/ionomycin stimulation.43 Specifically, one-step sandwich immunoassays quantitated Th2 secretion concentrations for IL-4 (3.1 ± 1.6 ng mL−1) and IL-5 (2.9 ± 2.1 ng mL−1) that agree well with this previous study, which used a commercially-available cytometric bead array kit.
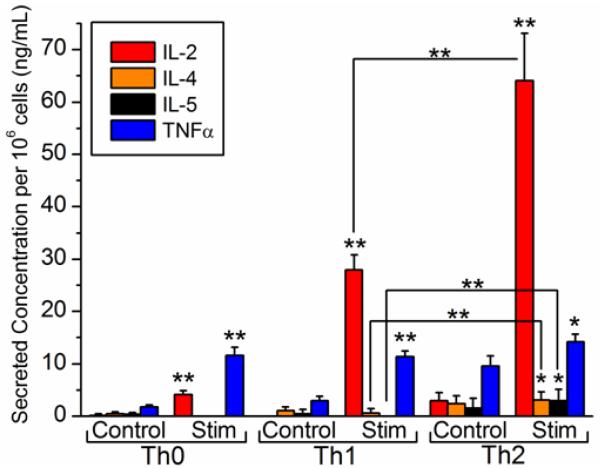
Figure 4.
Cytokine secretion profiling for differentiated primary T cell subsets. Cytokine secretion levels for three 24-h differentiated T cell subset cultures were determined by one-step sandwich immunoassay and normalized to the number of cells per unit volume in each culture. For each T cell subset, control cultures on the left are compared to PMA/ionomycin-stimulated (Stim) cultures on the right. Results of one-tailed paired difference t-tests comparing cytokine secretion levels both (1) within a T cell subset (between control and stimulated cells, as indicated above the stimulated bars) and (2) between T cell subsets (Th1 vs. Th2, as indicated above brackets joining pairs of bars) are shown. * indicates significance at the 95% confidence level, and ** indicates significance at the 99% confidence level. Error bars represent the total propagated standard error that includes: (1) the standard deviation of n=4-6 rings used to analyze each cytokine and (2) the regression error of the calibration curve.
There were several other notable findings from the cytokine secretion studies performed on Th0, Th1, and Th2 subsets. First, high TNFα levels (>10 ng mL−1) are consistently observed for each subset. Undifferentiated Th0 cells are capable of producing a wide array of cytokines including TNFα, and Th1 and Th2 cells have been known to secrete TNFα to a lesser extent.39 In each case, however, PMA/ionomycin-stimulated levels of TNFα were significantly higher than unstimulated controls, suggesting a T-cell receptor mediated activation of TNFα secretion. A second notable finding is that high IL-2 secretion levels were observed for Th2 cells. Since IL-2 is not a signature Th2 cytokine, these high levels are likely attributable to the large proportion of undifferentiated (Th0) IL-2-producing cells that remain after 7 days. Cousins et al found that ~50% of cells in Th2-differentiating conditions remained as IL-2-secreting cells even after 28 days of differentiation.44 Furthermore, lower levels of IL-2 for Th1 cells could suggest that Th1 cells differentiate faster from the IL-2-producing Th0 cells, which is supported by a previous study using intracellular staining of monensin-treated T cells and subsequent FACS analysis performed under similar in vitro differentiation conditions.44
Surprisingly, significant protease activity was observed in all cell samples, requiring the addition of protease inhibitors. Protease inhibitors were added after cell aliquot collection because they are known to affect lymphocyte stimulation and alter cytokine secretion.45,46 Without the addition of protease inhibitors prior to performing assays, no net binding was observed for any cell subsets (Figure S10). Beyond possible proteolytic degradation of secreted cytokines, proteases act to digest blocking proteins and capture antibodies on the ring surface, resulting in a progressive, negative net shift in resonance wavelength that masks cytokine binding. This interesting observation, which has profound consequences for immunoassays in general, was made possible by the real-time analysis capability afforded by the silicon photonic microring resonator platform.
Temporal secretion profiling of multiple T cell types
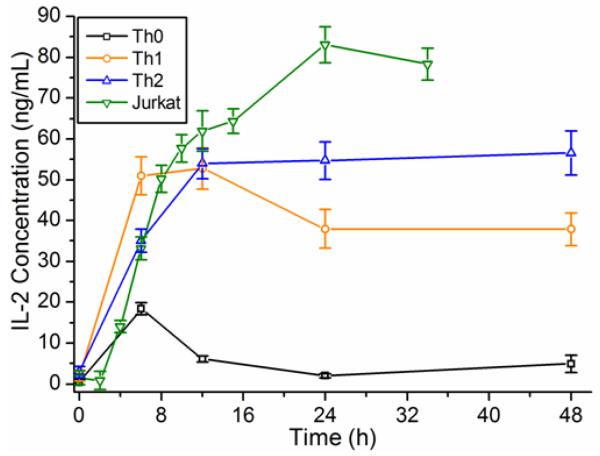
In addition to showing the utility of ring resonators for comparing T cell cytokine expression levels among subsets, multiplexed temporal cytokine secretion studies were also conducted. We monitored accumulation of secreted cytokines by sampling differentiated T cell cultures at various time points after stimulation. Micrographs of cells at various time points show increasing cellular aggregation (Figure S8b), consistent with mitogenic stimulation.37,47 Temporal secretion profiles were generated for each T cell population by again conducting one-step sandwich immunoassays. The IL-2 secretion profiles for Th0, Th1, Th2, and Jurkat T cells are shown in Figure 5. In each case, IL-2 levels increase sharply for ~12 h prior to leveling off or decreasing. It is hypothesized that after 12 h of stimulation, further IL-2 secretion is balanced by IL-2 proteolytic degradation and uptake by IL-2 receptors, which are expressed transiently following stimulation and only after encountering IL-2.37,38 It is also of note that the absolute concentration (Figure 5 concentrations normalized to 106 cells/mL) of IL-2 in Th0, Th1, and Th2 cultures consistently reaches a maximum of 50-60 ng mL−1 prior to leveling off. This effect seems to be more pronounced for Th0 cells as their accompanying IL-2 levels decrease more abruptly from the peak concentration at t = 6 h. IL-4, IL-5, and TNFα temporal cytokine secretion profiles of Th0, Th1, and Th2 are shown in Figure S11. As expected, TNFα levels are observed to be higher for Th0 and Th1 cells, reaching a peak of >50 ng mL−1 after 48 h. IL-4 levels are highest for Th2 cells, but the difference becomes less pronounced after 48 h, possibly owing to the significant protease activity. Th2 IL-5 levels peak at roughly 7 ng mL−1 after 12 h, similar to levels reported previously.44
Figure 5.
Temporal IL-2 secretion profiles for primary and Jurkat T cells. Aliquots taken from T cell cultures at several time points following stimulation were analyzed by one-step sandwich immunoassays. IL-2 temporal secretion levels for stimulated Th0, Th1, Th2 and Jurkat T cell lines are shown. All concentrations are normalized to the number of cells per unit volume in each culture (per 106 cells mL−1). Error bars represent the total propagated standard error that includes: (1) the standard deviation of n=3-5 rings used to quantitate IL-2 and (2) the regression error of the calibration curve.
Conclusions
Rapid one-step sandwich immunoassays performed on silicon photonic microring resonator arrays were demonstrated to be biomolecularly specific, highly quantitative, and thus highly amenable to detailed multiplexed cytokine analysis of T cell secretion. The ring resonator assays successfully distinguished Th0, Th1, and Th2 subsets based on their secretion profiles. Through the assay development process, the importance of observable real-time binding was evident; after unsuccessful assays without inhibitors, the negativesloping response only observable by real-time binding analysis provided evidence of protease activity. Furthermore, as shown previously,48 the ring resonator platform offers an excellent tool for screening antibodies in order to optimize sandwich pairs for the best combination of durability, cross-reactivity, and target binding kinetics. In sum, real-time multiplexed reaction monitoring with microring resonators is a powerful platform for immunoassay development and execution.
The one-step sandwich assay described herein is well-validated on account of repeating the Jurkat IL-2 analysis performed previously,15 running multiple ELISAs in parallel, and the good agreement with related literature of primary cell cytokine secretion using cytometric bead assays.43,44 It is notable that the previously used two-step IL-2 sandwich assay, which permitted ~1 assay h−1, can be replaced with a one-step sandwich assay that yields 4-5 assays h−1 (including full surface regeneration). The improved assay speed is enabled by the use of initial slope analysis instead of pseudothermodynamic net shift analysis required for two-step sandwich assays. Batch chip processing will further streamline these assays by removing the requirement for individual chip calibration.49
By combining one-step sandwich immunoassays with initial slope-based quantitation, multiplexed cytokine assays for complex immunological studies are possible without long sample incubation times. Premixing of samples with secondary antibodies essentially transforms small cytokines into larger protein complexes whose binding generates a more easily quantifiable signal in only 5 min. With the one-step cytokine immunoassay, cytokine detection limits mirror those for larger protein biomarkers detected on the microring platform without secondary amplification.20,29 Though ELISAs can feature superior sensitivity with much longer assay times, they are limited in quantitative multiplexing capabilities and cumbersome to perform in parallel. The assay developed herein provides: (1) sufficient sensitivity (~100 pM) to simultaneously detect cytokines at relevant levels, (2) rapid time-to-result (5 min), (3) ample dynamic range (2+ logs) to avoid serial dilutions, and (4) the ability to quantitate multiple cytokines from a single sample aliquot. For the first time with silicon photonic sensors, multiplexed protein measurements have been performed in primary biological samples to provide fundamental insights into immunological function. With higher-level multiplexing readily achievable, microring resonators represent an attractive technology for combining highly quantitative detection with the more qualitative protein fingerprinting capabilities of conventional antibody microarrays.
The demonstration of one-step assays also opens the possibility of performing real-time cell secretion measurements on single cells. Previous assays performed on cells in microwells have quantified cytokine levels at a defined endpoint by fluorescence staining,13,50,51 but these methods provide limited information on temporal secretion profiles. Though real-time detection techniques have predominantly been used for analyses of binding kinetics, dynamic cell secretion assays offer another outlet for leveraging real-time monitoring and multiparameter measurement capabilities. The small footprint and high scalability of microring resonators positions the technology as a promising platform for such extensions, and more generally for a number of multiplexed in vitro diagnostic applications.
Materials and Methods
Chip functionalization and covalent antibody conjugation to microrings
Ring resonator optical scanning instrumentation, software and chips were obtained from Genalyte, Inc, and have been described previously.20,52 Microfluidics and protocols for silanization and subsequent covalent multiplexed capture antibody immobilization are described in detail in the Supporting Information.
Multiplexed one-step sandwich assay protocol
Multiplexed cytokine calibration standard cocktails were prepared by dilution in RPMI 1640 + 10% fetal bovine serum (FBS) cell media from recombinant human cytokines [IL-2 (14-8029); IL-4 (34-8049); IL-5 (14-8059); TNFα (14-8329), all from eBioscience)]. Each cytokine standard cocktail contained variable and randomized concentrations of each cytokine (Table S1) and 1 μg mL−1 of each cytokine detection antibody [anti-IL-2 clone B33-2 (555040, BD Biosciences); anti-IL-4 clone MP4-25D2 (13-7048, eBioscience); anti-IL-5 clone JES1-5A10 (16-7059, eBioscience); anti-TNFα clone MAb11 (14-7349, eBioscience)]. In Jurkat analyses, IL-1β (14-8018, eBioscience) was also assayed [anti-IL-1β clones CRM56 (14-7018) and CRM57 (13-7016) from eBioscience].
Samples were incubated with detection anti-bodies (≥20-min) prior to performing the assays. Blinded unknown samples were prepared independently by a researcher who was not connected with these studies. For consistency in cell culture studies, premade standards were exposed to the same storage conditions (overnight at −20 °C) as cell culture aliquots. After soaking the chip overnight in StartingBlock PBS buffer (Fisher Scientific) at 4°C and prior to running the first assay, the chip was exposed to 10 mM glycine pH 2.2 + 150 mM NaCl for 2 min to remove excess blocking proteins by disrupting non-covalent protein-protein interactions. For each one-step sandwich immunoassay, RPMI 1640 + 10% FBS was used as running buffer. This insured RI matching between standards, samples, and running buffer as well as continuous re-blocking of the surface after each regeneration. All one-step sandwich assays were monitored in real time and involved a 6-7-min exposure to standard or sample followed by a 1-min low-pH regeneration of the capture antibody surface. The surface was re-blocked and equilibrated with cell media for 7 min prior to injecting the next standard or sample. Between 15 and 30 one-step multiplexed sandwich assays were performed in series on a single chip to collect all calibration and sample data for a given experiment. All experiments were carried out at a 30 μL min−1 flow rate controlled by an 11 Plus syringe pump (Harvard Apparatus) in withdraw mode. See the Supporting Information for more detail about initial slope-based quantitation.
Isolation of naïve CD4+ T cells from whole blood
Venous blood (15-20 mL) was collected from healthy volunteers into heparin-coated vials. Approval for the use of human volunteers in this study was obtained from the University of Illinois at Urbana-Champaign Institutional Review Board, and informed consent was documented for each blood donor. Primary blood mononuclear cells (PBMCs) were isolated from whole blood by density gradient centrifugation as described in the Supporting Information. This protocol yielded ~20 × 106 PBMCs from 18 mL whole blood, which were resuspended in 2% FBS in sterile PBS at ~50 ×106 cells mL−1 for naïve CD4+ T cell enrichment by negative selection.
Negative selection was performed using magnetic bead separation as described in the Supporting Information. In brief, a cocktail of antibodies against cell surface markers indicative of memory T cells [CD45RO+ ] and other cell types was used to remove all cells except naïve (CD4+CD45RA+) T cells. 2-3 × 106 CD4+CD45RA+ T cells that remained after magnetic negative selection were resuspended in T cell media.
T cell activation and differentiation
Anti-CD3/anti-CD28/IL-2-activated T cells were differentiated to Th0, Th1, and Th2 subsets in 24-well plates. Cytokine cues specific for each subset (Figure S7) were added to direct differentiation, with all culture conditions described in the Supporting Information.
On day 3, activated cells were expanded under the same conditions but in the absence of anti-CD3 and anti-CD28. Cells were split 1:4 with fresh cell media, and differentiation reagents were added again to the appropriate wells at the initial concentrations listed in the Supporting Information. On day 7, the (partially) differentiated cells were removed from the wells, washed twice in cell media to remove any secreted or exogenous cytokines, and the cells were resuspended in 1-1.5 mL cell media. The Th0, Th1, and Th2 cell cultures were counted and divided among 2-4 wells of a fresh 24-well plate for either (1) a comparison of stimulated and control (unstimulated) cells at a fixed time point following stimulation or (2) a temporal secretion study. To wells designated as stimulated cells, 10 ng mL−1 phorbol-12-myristate acetate (PMA, P 1585, Sigma-Aldrich) and 1 μg mL−1 ionomycin (I9657, Sigma-Aldrich), both in dimethylsulfoxide (DMSO), were added prior to placing the cells back in the incubator. Cell culture aliquots were harvested at defined time points after stimulation. The method for sampling cell culture aliquots is described in the Supporting Information. Jurkats were cultured, stimulated with PMA and phytohemagglutinin (PHA), and sampled as described previously.15
Supplementary Material
ACKNOWLEDGMENT
This work was funded by the NIH Director’s New Innovator Award Program, part of the NIH Roadmap for Medical Research (grant number 1-DP2-OD002190-01), and by the Camille and Henry Dreyfus Foundation. M.S.L. is supported by a National Science Foundation Graduate Research Fellowship and a Robert C. and Carolyn J. Springborn Fellowship. We acknowledge J.M. Banks for preparing cytokine unknown solutions, J-Y. Byeon for obtaining the SEM image, and Genalyte, Inc. for technical support. R.C.B. is a research fellow of the Alfred P. Sloan Foundation.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Experimental details, custom microfluidic design, capture antibody functionalization data, one-step sandwich negative control experiment, calibration plots with fittings, multiplexed Jurkat T cell cytokine secretion profiles with ELISA validation, T cell differentiation schematic, visual evidence of T cell activation by optical microscopy, real-time primary T cell secretion plots, evidence of protease activity, calibration standard details, and sensitivity metrics. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Bezbradica JS, Medzhitov R. Nat. Immunol. 2009;10:333. doi: 10.1038/ni.1713. [DOI] [PubMed] [Google Scholar]
- (2).Haddad JJ. Biochem. Biophys. Res. Commun. 2002;297:700. doi: 10.1016/s0006-291x(02)02287-8. [DOI] [PubMed] [Google Scholar]
- (3).Anderson NL, Anderson NG. Mol. Cell Proteomics. 2002;1:845. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- (4).Young HA. In: Methods in Molecular Biology: Inflammation and Cancer. Kozlov SV, editor. Vol. 511. Humana Press; New York: 2009. p. 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Seder RA, Darrah PA, Roederer M. Nat. Rev. Immunol. 2008;8:247. doi: 10.1038/nri2274. [DOI] [PubMed] [Google Scholar]
- (6).Hanrahan EO, Lin HY, Kim ES, Yan S, Du DZ, McKee KS, Tran HT, Lee JJ, Ryan AJ, Langmuir P, Johnson BE, Heymach JV. J. Clin. Oncol. 2010;28:193. doi: 10.1200/JCO.2009.22.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Gorelik E, Landsittel DP, Marrangoni AM, Modugno F, Velikokhatnaya L, Winans MT, Bigbee WL, Herberman RB, Lokshin AE. Cancer Epidemiol. Biomarkers Prev. 2005;14:981. doi: 10.1158/1055-9965.EPI-04-0404. [DOI] [PubMed] [Google Scholar]
- (8).Blicharz TM, Siqueira WL, Helmerhorst EJ, Oppenheim FG, Wexler PJ, Little FF, Walt DR. Anal. Chem. 2009;81:2106. doi: 10.1021/ac802181j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Prabhakar U, Eirikis E, Reddy M, Silvestro E, Spitz S, Pendley C, Davis HM, Miller BE. J. Immunol. Methods. 2004;291:27. doi: 10.1016/j.jim.2004.04.018. [DOI] [PubMed] [Google Scholar]
- (10).Mathias PC, Ganesh N, Cunningham BT. Anal. Chem. 2008;80:9013. doi: 10.1021/ac801377k. [DOI] [PubMed] [Google Scholar]
- (11).Bradshaw EM, Kent SC, Tripuraneni V, Orban T, Ploegh HL, Hafler DA, Love JC. Clin. Immunol. 2008;129:10. doi: 10.1016/j.clim.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Streeck H, Frahm N, Walker BD. Nat. Protoc. 2009;4:461. doi: 10.1038/nprot.2009.7. [DOI] [PubMed] [Google Scholar]
- (13).Zhu H, Stybayeva G, Macal M, Ramanculov E, George MD, Dandekar S, Revzin A. Lab Chip. 2008;8:2197. doi: 10.1039/b810244a. [DOI] [PubMed] [Google Scholar]
- (14).Mandal S, Goddard JM, Erickson D. Lab Chip. 2009;9:2924. doi: 10.1039/b907826f. [DOI] [PubMed] [Google Scholar]
- (15).Luchansky MS, Bailey RC. Anal. Chem. 2010;82:1975. doi: 10.1021/ac902725q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Arnold S, Khoshsima M, Teraoka I, Holler S, Vollmer F. Opt. Lett. 2003;28:272. doi: 10.1364/ol.28.000272. [DOI] [PubMed] [Google Scholar]
- (17).Zhu H, Dale PS, Caldwell CW, Fan X. Anal. Chem. 2009;81:9858. doi: 10.1021/ac902437g. [DOI] [PubMed] [Google Scholar]
- (18).Armani AM, Kulkarni RP, Fraser SE, Flagan RC, Vahala KJ. Science. 2007;317:783. doi: 10.1126/science.1145002. [DOI] [PubMed] [Google Scholar]
- (19).Wang S, Ramachandran A, Ja S-J. Biosens. Bioelectron. 2009;24:3061. doi: 10.1016/j.bios.2009.03.027. [DOI] [PubMed] [Google Scholar]
- (20).Washburn AL, Gunn LC, Bailey RC. Anal. Chem. 2009;81:9499. doi: 10.1021/ac902006p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Sun YS, Landry JP, Fei YY, Zhu XD, Luo JT, Wang XB, Lam KS. Langmuir. 2008;24:13399. doi: 10.1021/la802097z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Kodadek T. Chem. Biol. 2001;8:105. doi: 10.1016/s1074-5521(00)90067-x. [DOI] [PubMed] [Google Scholar]
- (23).Limpoco FT, Bailey RC. J. Am. Chem. Soc. 2011;133:14864. doi: 10.1021/ja205358g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Washburn AL, Bailey RC. Analyst. 2011;136:227. doi: 10.1039/c0an00449a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Luchansky MS, Washburn AL, Martin TA, Iqbal M, Gunn LC, Bailey RC. Biosens. Bioelectron. 2010;26:1283. doi: 10.1016/j.bios.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Qavi AJ, Bailey RC. Angew. Chem. Int. Ed. 2010;49:4608. doi: 10.1002/anie.201001712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Qavi AJ, Kindt JT, Gleeson MA, Bailey RC. Anal. Chem. 2011;83:5949. doi: 10.1021/ac201340s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Qavi AJ, Mysz TM, Bailey RC. Anal. Chem. 2011;83:6827. doi: 10.1021/ac201659p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Washburn AL, Luchansky MS, Bowman AL, Bailey RC. Anal. Chem. 2010;82:69. doi: 10.1021/ac902451b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Pieper-Fürst U, Kleuser U, Stöcklein WFM, Warsinke A, Scheller FW. Anal. Biochem. 2004;332:160. doi: 10.1016/j.ab.2004.05.047. [DOI] [PubMed] [Google Scholar]
- (31).Schneider BH, Dickinson EL, Vach MD, Hoijer JV, Howard LV. Biosens. Bioelectron. 2000;15:13. doi: 10.1016/s0956-5663(00)00056-7. [DOI] [PubMed] [Google Scholar]
- (32).Amarasiri Fernando S, Wilson GS. J. Immunol. Methods. 1992;151:47. doi: 10.1016/0022-1759(92)90104-2. [DOI] [PubMed] [Google Scholar]
- (33).Jung Y, Jeong JY, Chung BH. Analyst. 2008;133:697. doi: 10.1039/b800014j. [DOI] [PubMed] [Google Scholar]
- (34).Eddowes MJ. Biosensors. 1987;3:1. doi: 10.1016/0265-928x(87)80009-3. [DOI] [PubMed] [Google Scholar]
- (35).Weiss A, Wiskocil R, Stobo J. J. Immunol. 1984;133:123. [PubMed] [Google Scholar]
- (36).Durand D, Bush M, Morgan J, Weiss A, Crabtree G. J. Exp. Med. 1987;165:395. doi: 10.1084/jem.165.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Smith KA. Science. 1988;240:1169. doi: 10.1126/science.3131876. [DOI] [PubMed] [Google Scholar]
- (38).Greene W, Robb R, Depper J, Leonard W, Drogula C, Svetlik P, Wong-Staal F, Gallo R, Waldmann T. J. Immunol. 1984;133:1042. [PubMed] [Google Scholar]
- (39).Zhu J, Yamane H, Paul WE. Annu. Rev. Immunol. 2010;28:445. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Murphy KM, Reiner SL. Nat. Rev. Immunol. 2002;2:933. doi: 10.1038/nri954. [DOI] [PubMed] [Google Scholar]
- (41).Paul WE. Immunol. Cell Biol. 2010;88:236. doi: 10.1038/icb.2010.2. [DOI] [PubMed] [Google Scholar]
- (42).Ansel KM, Lee DU, Rao A. Nat. Immunol. 2003;4:616. doi: 10.1038/ni0703-616. [DOI] [PubMed] [Google Scholar]
- (43).Yano S, Ghosh P, Kusaba H, Buchholz M, Longo DL. J. Immunol. 2003;171:2510. doi: 10.4049/jimmunol.171.5.2510. [DOI] [PubMed] [Google Scholar]
- (44).Cousins DJ, Lee TH, Staynov DZ. J. Immunol. 2002;169:2498. doi: 10.4049/jimmunol.169.5.2498. [DOI] [PubMed] [Google Scholar]
- (45).Kelleher AD, Sewell WA, Cooper DA. Clin. Exp. Immunol. 1999;115:147. doi: 10.1046/j.1365-2249.1999.00761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Tchórzewski H, Formalczyk E, Pasnik J. Immunol. Lett. 1995;46:237. doi: 10.1016/0165-2478(95)00055-a. [DOI] [PubMed] [Google Scholar]
- (47).Nowell PC. Cancer Res. 1960;20:462. [PubMed] [Google Scholar]
- (48).Washburn AL, Gomez J, Bailey RC. Anal. Chem. 2011;83:3572. doi: 10.1021/ac200317z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Luchansky MS, Washburn AL, McClellan MS, Bailey RC. Lab Chip. 2011;11:2042. doi: 10.1039/c1lc20231f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Zhu H, Stybayeva G, Silangcruz J, Yan J, Ramanculov E, Dandekar S, George MD, Revzin A. Anal. Chem. 2009;81:8150. doi: 10.1021/ac901390j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Han Q, Bradshaw EM, Nilsson B, Hafler DA, Love JC. Lab Chip. 2010;10:1391. doi: 10.1039/b926849a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Iqbal M, Gleeson MA, Spaugh B, Tybor F, Gunn WG, Hochberg M, Baehr-Jones T, Bailey RC, Gunn LC. IEEE J. Sel. Top. Quantum Electron. 2010;16:654. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.