Abstract
DNA shape variation and the associated variation in minor groove electrostatic potential are widely exploited by proteins for DNA recognition. Here we show that the hydroxyl radical cleavage pattern is a quantitative measure of DNA backbone solvent accessibility, minor groove width, and minor groove electrostatic potential, at single nucleotide resolution. We introduce maps of DNA shape and electrostatic potential as tools for understanding how proteins recognize binding sites in a genome. These maps reveal periodic structural signals in yeast and Drosophila genomic DNA sequences that are associated with positioned nucleosomes.
Specific binding of a protein to DNA is now appreciated to be influenced both by the sequence of nucleotides and by the shape of the DNA double helix.1,2 A direct connection between minor groove width and electrostatic potential recently was established, providing a physical basis for this type of shape readout. Specifically, the magnitude of the electrostatic potential in the minor groove is controlled by the width of the groove3 with narrowing of the groove associated with more negative electrostatic potential. Many proteins have been found to take advantage of this property by inserting positively charged arginine side chains into the groove where it is narrow.4
Different DNA sequences can give rise to similar DNA shapes.5 The smaller "space" of DNA structure compared to nucleotide sequence confounds typical sequence-based analyses of genomes, which may miss regions of structural similarity that are not also similar in sequence.6,7 For example, current computational strategies for finding protein binding sites in genomes, which rely on nucleotide sequence identity (or similarity),8 are not effective in identifying similarities in DNA shape. To use shape recognition to understand how a protein selects a binding site in a genome, we need a way to map DNA shape variation at both high resolution and on a large scale. Here we report that hydroxyl radical cleavage of DNA provides the information required to evaluate shape and electrostatic potential variation in a DNA molecule of any length, including the DNA of an entire genome.
We begin by comparing the experimental hydroxyl radical cleavage pattern of a DNA molecule with NMR and X-ray structures of the same DNA sequence to "calibrate" the structural information embodied in the cleavage pattern. We next construct a new type of cleavage pattern that includes information from both DNA strands, to map minor groove width and electrostatic potential. Finally, we use an experimental database of cleavage patterns as the basis of an algorithm to computationally predict the minor groove shape and electrostatic potential for entire genomes.
We first forge an explicit link between cleavage and structure through quantitative comparison of the experimental hydroxyl radical cleavage pattern and the three-dimensional structure of DNA. For this analysis we use the Drew-Dickerson dodecamer, [d(CGCGAATTCGCG)]2,9,10 undoubtedly the structurally best-characterized DNA molecule.11–15
We had previously obtained a large amount of experimental hydroxyl radical cleavage data for the Drew-Dickerson dodecamer in the context of our efforts to construct ORChID, the •OH Radical Cleavage Intensity Database.5 This database contains experimental cleavage patterns for more than 150 DNA sequences 40 base pairs in length. As a result, all 512 unique pentanucleotide sequences are represented in ORChID. Each of the 40-mers in ORChID is flanked on both sides by the Drew-Dickerson dodecamer sequence. The hydroxyl radical cleavage pattern of the dodecamer is exceptionally well determined because we have so many independent examples of the pattern.
The hydroxyl radical cleaves DNA by abstracting a hydrogen atom from a deoxyribose residue in the backbone. We showed previously that the solvent accessible surface area (SASA) of a deoxyribose hydrogen atom governs the extent of its reactivity with the hydroxyl radical.16 These experiments found that the 5', 5", and 4' hydrogen atoms, which lie on the outer edges of the DNA minor groove and are most exposed to solvent (Figure 1, panel a), react most often. We also have noted that the extent of hydroxyl radical cleavage varies at each nucleotide along a double-stranded DNA molecule,17,18 suggesting that the cleavage pattern embodies information on sequence-dependent variation in DNA shape.
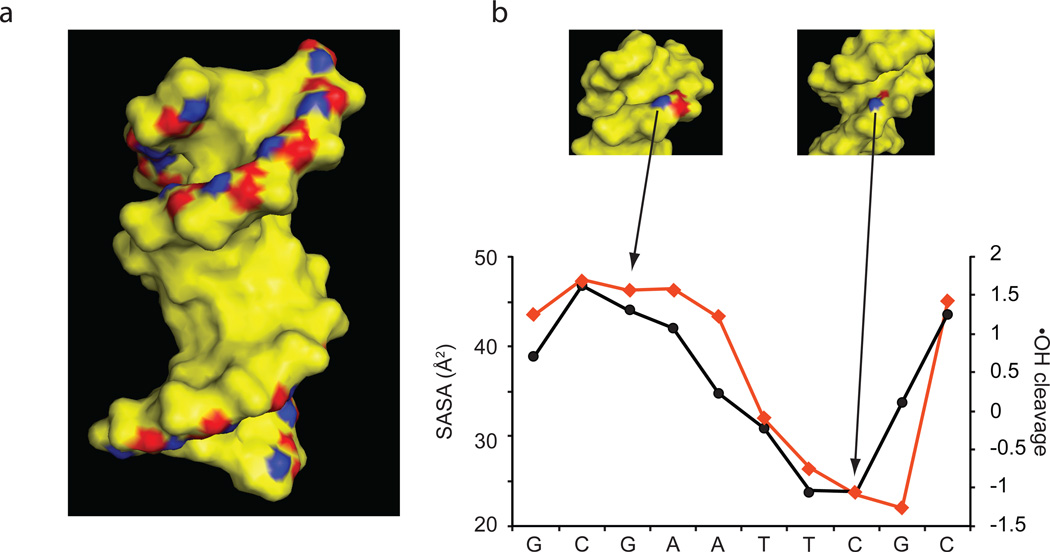
Figure 1.
Quantitative correlation of hydroxyl radical cleavage with DNA structure. (a) H4' (blue) and H5', H5" (red) are the deoxyribose hydrogen atoms most often abstracted by the hydroxyl radical.16 (b) The extent of hydroxyl radical cleavage (black circles) at each of the 10 interior nucleotides of the Drew-Dickerson dodecamer is compared to the sum of the solvent accessible surface area (SASA) of the 4', 5', and 5" hydrogen atoms of that nucleotide (red diamonds). (These data points represent the sum of the SASAs of the blue and red deoxyribose hydrogen atoms that are shown in (a)). Two backbone deoxyriboses in different structural environments are highlighted in the insets (top). A wide minor groove results in high SASA and hydroxyl radical cleavage (left); a narrow groove is associated with low SASA and cleavage. The Pearson correlation for comparison of the hydroxyl radical cleavage pattern and SASA in Figure 1, panel b is 0.865 (p-value = 0.00123).
In Figure 1, panel b we compare the extent of hydroxyl radical cleavage for each nucleotide of the Drew-Dickerson dodecamer, with the sum of the solvent accessible surface areas of the 5', 5", and 4' hydrogen atoms of that nucleotide as determined from X-ray structures. Where the minor groove is wide, and deoxyribose backbone hydrogens are exposed, cleavage is high (Figure 1, panel b, left inset); where the groove is narrow, and backbone hydrogens are diminished in exposure, cleavage is low (Figure 1, panel b, right inset). This plot demonstrates that hydroxyl radical cleavage accurately reports the sequence-dependent variation in the shape of the DNA backbone. We call this type of hydroxyl radical cleavage pattern ORChID1, to indicate that it represents the •OH radical cleavage pattern of one strand of the DNA duplex.
We next sought to develop a method to map the shape of the DNA minor groove, since minor groove width and electrostatic potential are important recognition elements for protein binding.3,4,19 But while the ORChID1 pattern and SASA are properties associated with individual nucleotides in one of the strands of the double helix (Figure 1), the minor groove width depends on both DNA strands. To construct a metric that incorporates hydroxyl radical cleavage information from both strands, we first determined the extent of cleavage for a given nucleotide, and then averaged this value with the extent of cleavage for the residue on the opposite strand that is closest in space across the minor groove. In B-form DNA these two positions are staggered by three nucleotides in the 3' direction.7 The phosphate groups of the same two nucleotides are used to define minor groove width.20 We call this new type of hydroxyl radical cleavage pattern ORChID2, to denote that it incorporates cleavage information from both strands of the DNA duplex. This approach enables a direct comparison between hydroxyl radical cleavage and minor groove width, with both structural parameters treated as double-strand properties.
To test the correspondence of the ORChID2 pattern with minor groove width we again took advantage of the structurally well-characterized Drew-Dickerson dodecamer. We made two comparisons, first with NMR structures of the dodecamer, and then with X-ray structures. While NMR structures of nucleic acids are usually not as high in resolution as X-ray structures, they are determined in solution, and so are free of crystal packing effects. For our analysis we used an NMR structure of the Drew-Dickerson dodecamer15 that was determined using dipolar coupling and chemical shift anisotropy data, and is thus of very high quality. We observe an excellent correlation of the experimental ORChID2 pattern with the width of the minor groove derived from the NMR structure (Figure 2, panel a).
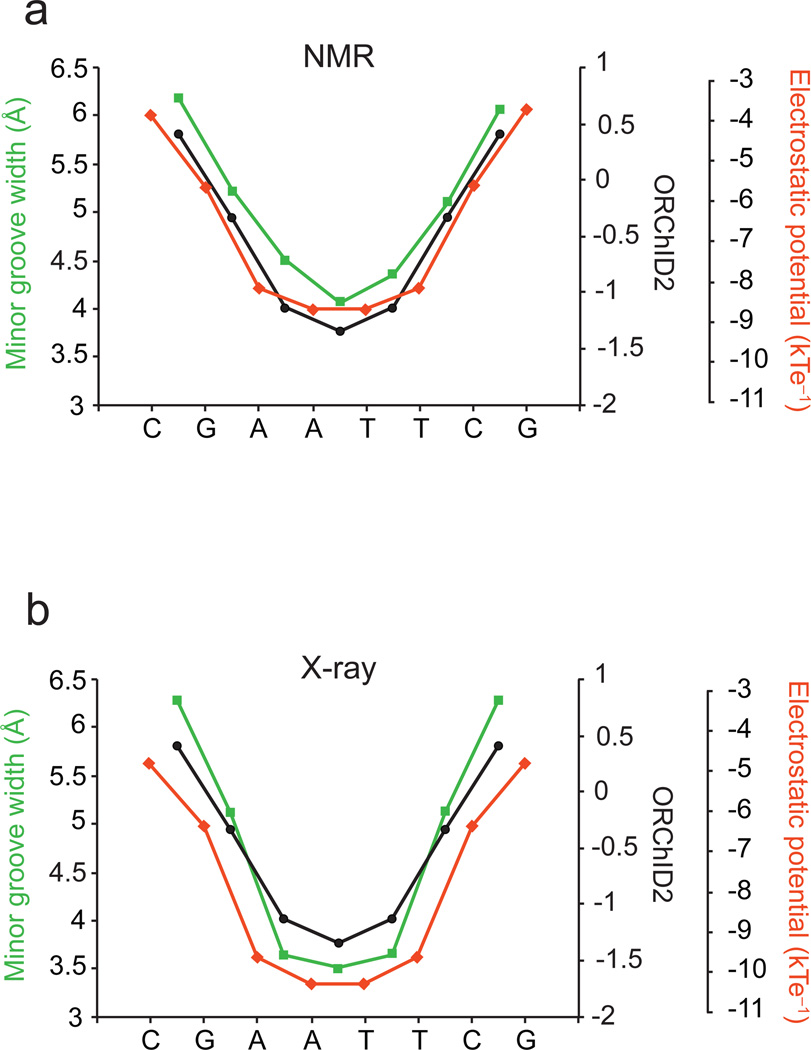
Figure 2.
(a) Quantitative correlation of the experimental ORChID2 cleavage pattern (black circles), with electrostatic potential (red diamonds) and minor groove width (green squares) determined from a set of five NMR structures of the Drew-Dickerson dodecamer. The Pearson correlation for comparison of the ORChID2 pattern with minor groove width (7 nucleotide positions) is 0.9917 (p-value = 1.21 × 10−5); for comparison of ORChID2 with electrostatic potential (7 positions), 0.868 (p-value = 0.01). (b) Quantitative correlation of the experimental ORChID2 cleavage pattern (black circles), with electrostatic potential (red diamonds) and minor groove width (green squares) determined from eight X-ray structures of the Drew-Dickerson dodecamer. Minor groove width and electrostatic potential are symmetrized to reflect the symmetry of the Drew-Dickerson sequence. The Pearson correlation for comparison of the ORChID2 pattern with minor groove width (7 positions) is 0.997 (p-value = 7.27 × 10−7); for comparison of ORChID2 with electrostatic potential (7 positions), 0.881 (p-value = 8.67 × 10−3).
Like the NMR experiment, the hydroxyl radical cleavage experiment is performed in solution, where the three-dimensional structure of the Drew-Dickerson dodecamer has the same symmetry as its palindromic nucleotide sequence.15 The ORChID2 pattern therefore exhibits the symmetry that is inherent in the palindromic sequence and structure of the dodecamer. But it is well known that crystal packing effects lead to asymmetry in the crystal structure of the dodecamer.21 To compare the ORChID2 pattern to the minor groove width determined from various X-ray structures,10,12,22–27 we symmetrized the groove width28 based on the inherent symmetry of the Drew-Dickerson dodecamer sequence. Symmetrization is a standard approach to separate crystal packing from sequence-dependent effects on DNA structure.28 We find an excellent correlation between the experimental ORChID2 pattern and the symmetrized minor groove width derived from the X-ray structures (Figure 2, panel b). We also compared the ORChID2 pattern with the minor groove width derived from all-atom Monte Carlo simulations of the Drew-Dickerson dodecamer,3,28 and observe an excellent correlation (Supplementary Figure 1).
To test the generality of the correspondence of cleavage pattern with structure, we searched the ORChID database for sequences that also have X-ray structures. We found the 9-mer sequence GATATCGCG, which is contained in the dodecamer [d(CGCGATATCGCG)]2 for which the X-ray structure has been determined.29 Despite the more limited experimental cleavage data available from ORChID for this sequence compared to the Drew-Dickerson dodecamer, we find an excellent correlation between the ORChID2 pattern and the X-ray-derived minor groove width (Supplementary Figure 2).
To extend the comparison of ORChID2 to more nucleotide sequences, for each tetranucleotide in the Protein Data Bank (PDB) we plotted the minor groove width4 versus the experimental ORChID2 value derived from the ORChID database (Supplementary Figure 3). For DNA in protein-DNA complexes we find a very good correlation of average ORChID2 value with minor groove width (Pearson correlation = 0.653, p-value < 1 × 10−16). For free DNA molecules the correlation is similar (Pearson correlation = 0.638, p-value = 4.06 × 10−8), despite the fewer number of free DNA structures in the PDB.
Since it has been shown that electrostatic potential depends on minor groove width,3 the results depicted in Figure 2 and Supplementary Figures 1 and 2 suggest that the ORChID2 pattern also embodies information on the local variation of minor groove electrostatic potential. To test this idea we solved the non-linear Poisson-Boltzmann equation to calculate the electrostatic potential at points in the center of the DNA minor groove.4,30 We symmetrized the electrostatic potential pattern that was calculated from X-ray-derived structures to remove crystal-packing effects. We did not symmetrize the electrostatic potential pattern derived from the NMR-based structures. As we anticipated, the ORChID2 pattern and electrostatic potential are highly correlated, both for NMR- and X-ray derived structures (Figure 2). We find a similarly high degree of correspondence between the ORChID2 pattern and electrostatic potential for the sequence GATATCGCG (Supplementary Figure 2).
The results shown in Figure 2 and Supplementary Figure 2 establish that the experimentally determined ORChID2 pattern represents a quantitative map of minor groove width and electrostatic potential. To use ORChID2 to map structural and electrostatic variation in genome-scale DNA molecules, we have developed a method to computationally predict the ORChID2 pattern. We have shown previously that a prediction tool based on experimental ORChID1 patterns, which considers the properties of a single DNA strand, can be used to predict the ORChID1 pattern for any DNA sequence, of any length, with high accuracy.5 We developed a related algorithm that computationally predicts the ORChID2 pattern for any DNA sequence of interest. The algorithm is very efficient, so predictions can be made for genome-length DNA sequences. We have deposited a dataset consisting of the ORChID2 pattern for the human genome in the UC Santa Cruz genome browser. Because of its high correlation with minor groove electrostatic potential (Figure 2), the ORChID2 pattern represents the structure-dependent variation of electrostatic potential in the DNA minor groove throughout the human genome, at single base-pair resolution. This tool allows the role of DNA shape in protein-DNA recognition to be evaluated at the whole-genome scale. We note that our approach is applicable to any genome for which sequence information is available.
To demonstrate the application of ORChID2 to genome-scale recognition of DNA shape, we have analyzed sets of nucleosome-bound sequences that were identified in the yeast31 and Drosophila32 genomes. Appreciation of the involvement of chromatin structure in gene regulation has focused widespread attention on the underlying basis for nucleosome positioning.33 Previous work has most often attempted to find nucleotide sequence motifs that are associated with positioned nucleosomes,4,31,34 with limited success. But while sequence motifs have been identified that lead to bends or kinks that facilitate nucleosome binding,4 similar structural patterns can result from different sequence motifs.5 Because the hydroxyl radical cleavage pattern has been shown to be capable of uncovering structural similarity among sets of diverse nucleotide sequences,6 we used the ORChID2 pattern to reveal structural motifs in genomic DNA sequences that form nucleosomes.
When DNA wraps around the histone octamer, the minor groove faces the histone core every 10 base pairs, a strikingly periodic structural feature. X-ray structures of nucleosome core particles reveal a corresponding periodic variation in the width of the minor groove of nucleosome-bound DNA (Supplementary Figure 4). We calculated the ORChID2 patterns for 23,076 DNA sequences from yeast that were found experimentally to be occupied by nucleosomes,31 and averaged these patterns. The resulting composite ORChID2 pattern has a clear 10 bp periodicity (Figure 3, panel a). We find a very good correlation between minor groove width and the ORChID2 value for each nucleotide in the nucleosome-binding sequences (Figure 3, panel c). Minima in the ORChID2 pattern occur at nucleotide positions at which the minor groove is most narrow in the nucleosome structure (Supplementary Figure 5).
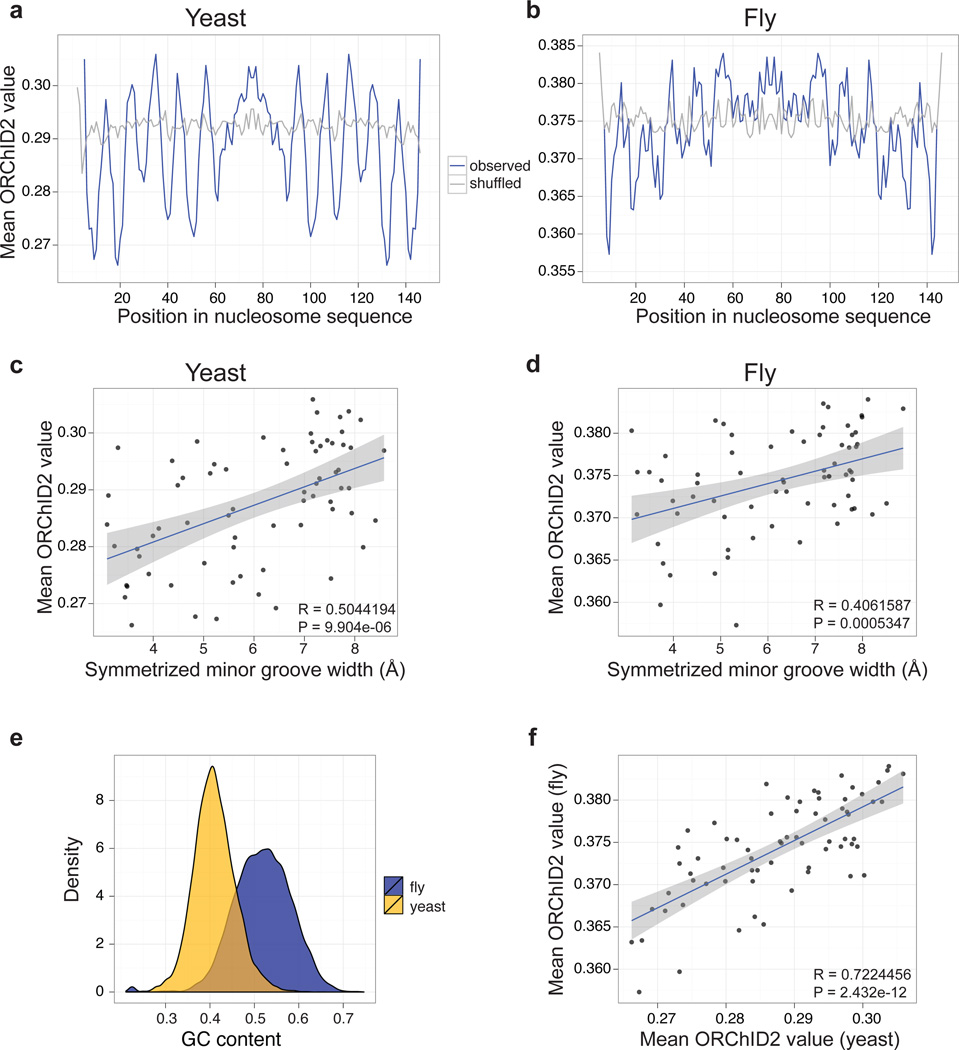
Figure 3.
ORChID2 nucleosome patterns in yeast and fly. Mean ORChID2 values at each nucleotide position of 23,076 yeast (a) and 25,654 fly (b) nucleosome sequences (blue lines) are compared with ORChID2 values for shuffled versions of the same sequences (gray). Minor groove width measurements from nucleosome X-ray structures 1KX5 and 2PYO (Supplementary Figure 4) were aligned by dyad center to the ORChID2 patterns from panels a and b, respectively, and plotted for yeast (c) and fly (d). There is a significant correlation of the ORChID2 value with minor groove width for both yeast and fly nucleosome-bound sequences (inset, panels c and d). (e) Yeast and fly nucleosome sequences have significantly different G/C content distributions (P < 2.2 × 10−16; Wilcoxon rank sum test), but their overall ORChID2 patterns are highly correlated (f). All correlation plots (panels c, d, f) are based on one-half of the nucleosome dyad and are center-aligned by the dyad axis. Gray shading indicates the standard error around the best-fit line.
We performed a similar analysis for a dataset consisting of 25,654 sequences bound by nucleosomes in Drosophila.32 We found a very similar periodic ORChID2 pattern (Figure 3, panel b) and correlation of ORChID2 values with minor groove width (Figure 3, panel d). Despite the very different G/C contents of the Drosophila melanogaster and Saccharomyces cerevisiae nucleosome-bound sequences (Figure 3, panel e), the two ORChID2 patterns are highly similar to each other (Figure 3, panel f).
What is especially noteworthy about the distinctive ORChID2 patterns of nucleosome-binding sequences is that they reveal periodic structural features that are present in naked genomic DNA sequences that correspond to the more extreme structural deformations that DNA adopts when wrapped around the histone octamer.
Although the periodic ORChID2 nucleosome pattern (Figure 3, panels a and b) is not correlated with the pattern for shuffled sequences (Supplementary Figure 6), the pattern itself is weak. The range in ORChID2 values for the consensus nucleosome pattern is ~0.03, while the range for the Drew-Dickerson pattern is ~3. The weakness of the pattern likely is the consequence of averaging many thousands of ORChID2 patterns to give the consensus nucleosome-associated ORChID2 pattern. To investigate how ORChID2 patterns of individual nucleosome binding sites correspond to the consensus pattern, we scanned the consensus pattern across all 23,076 yeast nucleosome-bound sequences, and calculated the Pearson correlation for each sequence. As a control we scanned the consensus pattern across shuffled versions of the same sequences. We performed the same analysis for the set of 25,654 Drosophila sequences, and plotted the distribution of correlation scores for each (Supplementary Figure 7). Distributions of correlation scores for both real and shuffled sequences are shifted to the right of zero, suggesting that most nucleotide sequences have a positive correlation with the weak consensus pattern. However, the real genomic sequences are shifted significantly more to the right (p-value < 2.2 × 10−16) and have a longer right tail compared to the shuffled sequences, showing that they are more similar to the consensus. We speculate that genomic sequences on the far right of the distribution, for which the ORChID2 pattern most closely resembles the consensus periodic pattern, form stable and well-phased nucleosomes and therefore might serve as nucleation sites for nucleosomal arrays.
While this paper was being revised, a method was published for computational prediction of minor groove electrostatic potential, using uranyl photocleavage of DNA as input data.35 Uranyl yields an essentially mirror-image cleavage pattern compared to hydroxyl radical. Uranyl, a positively charged ion, binds directly to the negatively charged phosphates in the DNA backbone, and cleaves most in regions where the minor groove is narrow.36 In contrast, the hydroxyl radical cleaves least where the minor groove is narrow (Figure 1, panel a). We used the authors' web server to predict the electrostatic potential of the Drew-Dickerson dodecamer based on uranyl cleavage data. We found a Pearson correlation of 0.79 (p-value = 6.11 × 10−3, 10 nucleotide positions), when we compared the uranyl-based prediction with a Poisson-Boltzmann calculation of the electrostatic potential from eight X-ray structures of the dodecamer, a result comparable to ours using ORChID2 (Figure 2).
Representing genome sequences as strings of letters obscures the structural biology of DNA that is the true basis for protein recognition. We have shown here that a chemical probe (the hydroxyl radical) can be used to link high-resolution three-dimensional structure with DNA shape and electrostatic potential variation at the scale of an entire genome. Our results open the way to applying the powerful idea of shape-directed DNA recognition4 to the analysis of genomes.
METHODS
Experimental hydroxyl radical cleavage data
We extracted from the ORChID database (http://dna.bu.edu/orchid) 112 copies of the hydroxyl radical cleavage pattern of the Drew-Dickerson dodecamer sequence that had been experimentally determined in diverse sequence environments. We have shown previously that the experimental hydroxyl radical cleavage data for this sequence are highly reproducible, with a standard deviation of less than 13%.5
X-ray structures
We used eight different X-ray crystal structures of the Drew-Dickerson dodecamer for structural analysis and comparison with ORChID1 and ORChID2 patterns (see Supporting Information). We chose these eight structures because they have no chemical modifications or unpaired bases. We calculated solvent accessible surface areas, minor groove widths, and electrostatic potentials (see below) for each of the eight structures, and then averaged these values for comparison with ORChID2.
NMR structures
We used a set of five NMR structures of the Drew-Dickerson dodecamer (PDB ID 1NAJ) for structural analysis and comparison with the ORChID2 pattern (see Supporting Information). We calculated minor groove widths and electrostatic potentials (see below) for each of the five structures that were submitted as a single PDB entry, and then averaged these values for comparison with ORChID2.
Calculation of solvent accessible surface area
We used the Lee and Richards algorithm,37 as implemented in Surfv,38 with a probe size of 1.4 Å, to calculate the solvent-accessible surface areas of the deoxyribose hydrogens from the X-ray coordinates of the Drew-Dickerson dodecamer. Hydrogens were added to the structure as described.4 We obtained the best correlation between solvent accessible surface area (SASA) and cleavage pattern when we considered the sum of the SASA of the 4', 5', and 5" hydrogen atoms, which are the sugar hydrogens located towards the edge of the minor groove (Figure 1, panel a).
Calculation of minor groove width
We used the program CURVES20 to calculate the width of the minor groove at each nucleotide of the Drew-Dickerson dodecamer and [d(CGCGATATCGCG)]2. We symmetrized the minor groove width pattern that was derived from X-ray structures to reflect the palindromic nature of the Drew-Dickerson sequence (see text, and below).
Calculation of the electrostatic potential
We calculated the electrostatic potential in the minor groove4,30 for the Drew-Dickerson dodecamer and for [d(CGCGATATCGCG)]2. We symmetrized the X-ray-derived datasets to reflect the palindromic nature of the sequences (see text, and below). The electrostatic potential was calculated with DelPhi39,40 using the non-linear Poisson-Boltzmann equation solved at the physiologic salt concentration of 0.145 M. The electrostatic potential was calculated in five focusing steps, and plotted along the minor groove at reference points placed approximately in the plane of a base pair at the geometric midpoints between the two nearest sugar 4' oxygen atoms on opposite strands.4
Symmetrization of minor groove width and electrostatic potential
Because the Drew-Dickerson sequence, d(CGCGAATTCGCG), and the sequence d(CGCGATATCGCG) are palindromic, we symmetrized the electrostatic potential and minor groove width datasets that were determined from X-ray structures to reflect this symmetry, by averaging the minor groove width or electrostatic potential values for sequence positions that are symmetry-related. Such symmetrization is a standard approach to separate crystal packing from sequence-dependent effects on DNA structure.28
Computational prediction of the ORChID2 pattern
Hydroxyl radical cleavage intensity predictions were performed using an in-house sliding tetramer window algorithm.5 This algorithm draws data from ORChID,5 which contains more than 150 experimentally determined hydroxyl radical cleavage patterns of 40-mer DNA sequences. ORChID1 predictions were performed on the plus strand of the DNA sequence. We have shown previously that these predictions are fairly accurate, with a Pearson correlation of 0.88 between the predicted and experimentally determined cleavage intensities.5
To produce ORChID2 patterns, two ORChID1 predictions were performed, one for the plus strand sequence and the other for the minus strand sequence. We then averaged the predicted cleavage intensities for nucleotides in close proximity across the minor groove to produce the ORChID2 value for that pair of nucleotides. These nucleotides are shifted relative to each other by three nucleotides in the 3' direction, a structural characteristic of B-form DNA.7 We assigned the ORChID2 value to the base step between the pair of nucleotides used in the calculation.
For example, to calculate the ORChID2 value for the base step ApA on the top strand of the Drew-Dickerson DNA molecule:
-
5'-CpGpCpGpApApTpTpCpGpCpG-3'
3'-GpCpGpCpTpTpApApCpGpCpG-5'
we average ORChID1 values for T7 on the top strand and C4 on the bottom strand (boldface).
We have deposited ORChID1 and ORChID2 values for the hg19 version of the human genome in the UCSC genome browser (http://genome-preview.ucsc.edu/cgi-bin/hgTrackUi?hgsid=2561111&g=wgEncodeBuOrchid). These datasets are freely available to the community.
ORChID2 analysis of nucleosome-binding sequences
ORChID2 analysis of yeast DNA sequences that form nucleosomes was based on 23,076 sequences of length 146–148 bp that we obtained from a dataset of yeast in vivo nucleosome-bound sequences.31 We obtained a similar set of 25,654 sequences that were bound to nucleosomes in Drosophila.32 We aligned the sequences about their centers, using both the sequence and its reverse complement, calculated the ORChID2 pattern for each sequence, and averaged these patterns. Because even-length sequences of length 146 and 148 cannot be perfectly aligned to the center, the ORChID2 pattern for each such sequence was calculated twice, once with the center base step shifted left in the alignment, and again with the center base step shifted right. The ORChID2 pattern for each shifted alignment was multiplied by 0.5, and then included in the set of ORChID2 patterns that was averaged.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by National Institutes of Health grants R01 HG003541 (T.D.T), R01 GM30518 (B.H.), U54 CA121852 (R.S.M., B.H., and T.D.T.), and USC start-up funds (R.R.). The Tullius laboratory is a member of the ENCODE Consortium.
Footnotes
ASSOCIATED CONTENT
Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS. Origins of specificity in protein-DNA recognition. Annu. Rev. Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stella S, Cascio D, Johnson RC. The shape of the DNA minor groove directs binding by the DNA-bending protein Fis. Genes Devel. 2010;24:814–826. doi: 10.1101/gad.1900610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joshi R, Passner JM, Rohs R, Jain R, Sosinsky A, Crickmore MA, Jacob V, Aggarwal AK, Honig B, Mann RS. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell. 2007;131:530–543. doi: 10.1016/j.cell.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenbaum JA, Pang B, Tullius TD. Construction of a genome-scale structural map at single-nucleotide resolution. Genome Res. 2007;17:947–953. doi: 10.1101/gr.6073107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parker SCJ, Hansen L, Abaan HO, Tullius TD, Margulies EH. Local DNA topography correlates with functional noncoding regions of the human genome. Science. 2009;324:389–392. doi: 10.1126/science.1169050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parker SCJ, Tullius TD. DNA shape, genetic codes, and evolution. Curr. Opin. Struct. Biol. 2011;21:342–347. doi: 10.1016/j.sbi.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hannenhalli S. Eukaryotic transcription factor binding sites--modeling and integrative search methods. Bioinformatics. 2008;24:1325–1331. doi: 10.1093/bioinformatics/btn198. [DOI] [PubMed] [Google Scholar]
- 9.Wing R, Drew H, Takano T, Broka C, Tanaka S, Itakura K, Dickerson RE. Crystal structure analysis of a complete turn of B-DNA. Nature. 1980;287:755–758. doi: 10.1038/287755a0. [DOI] [PubMed] [Google Scholar]
- 10.Drew H, Wing R, Takano T, Broka C, Tanaka S, Itakura K, Dickerson R. Structure of a B-DNA dodecamer: conformation and dynamics. Proc. Natl. Acad. Sci. U.S.A. 1981;78:2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Subirana JA. Structure of d(CGCGAATTCGCG) in the presence of Ca(2+) ions. J. Biol. Chem. 1999;274:24749–24752. doi: 10.1074/jbc.274.35.24749. [DOI] [PubMed] [Google Scholar]
- 12.Sines CC, McFail-Islom L, Howerton SB, VanDerveer D, Williams LD. Cations mediate B-DNA conformational heterogeneity. J. Am. Chem. Soc. 2000;122:11048–11056. [Google Scholar]
- 13.Tereshko V, Minasov G, Egli M. The Dickerson-Drew B-DNA dodecamer revisited at atomic resolution. J. Am. Chem. Soc. 1999;212:470–471. [Google Scholar]
- 14.Tjandra N, Tate S, Ono A, Kainosho M, Bax A. The NMR structure of a DNA dodecamer in an aqueous dilute liquid crystalline phase. J. Am. Chem. Soc. 2000;122:6190–6200. [Google Scholar]
- 15.Wu Z, Delaglio F, Tjandra N, Zhurkin VB, Bax A. Overall structure and sugar dynamics of a DNA dodecamer from homo- and heteronuclear dipolar couplings and 31P chemical shift anisotropy. J. Biomol. NMR. 2003;26:297–315. doi: 10.1023/a:1024047103398. [DOI] [PubMed] [Google Scholar]
- 16.Balasubramanian B, Pogozelski WK, Tullius TD. DNA strand breaking by the hydroxyl radical is governed by the accessible surface areas of the hydrogen atoms of the DNA backbone. Proc. Natl. Acad. Sci. U.S.A. 1998;95:9738–9743. doi: 10.1073/pnas.95.17.9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tullius TD, Dombroski BA. Iron(II) EDTA used to measure the helical twist along any DNA molecule. Science. 1985;230:679–681. doi: 10.1126/science.2996145. [DOI] [PubMed] [Google Scholar]
- 18.Price MA, Tullius TD. Using hydroxyl radical to probe DNA structure. Methods Enzymol. 1992;212:194–219. doi: 10.1016/0076-6879(92)12013-g. [DOI] [PubMed] [Google Scholar]
- 19.Churchill ME, Travers AA. Protein motifs that recognize structural features of DNA. Trends Biochem. Sci. 1991;16:92–97. doi: 10.1016/0968-0004(91)90040-3. [DOI] [PubMed] [Google Scholar]
- 20.Lavery R, Sklenar H. Defining the structure of irregular nucleic acids: conventions and principles. J. Biomol. Struct. Dyn. 1989;6:655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- 21.Johansson E, Parkinson G, Neidle S. A new crystal form for the dodecamer C-G-C-G-A-A-T-T-C-G-C-G: symmetry effects on sequence-dependent DNA structure. J. Mol. Biol. 2000;300:551–561. doi: 10.1006/jmbi.2000.3907. [DOI] [PubMed] [Google Scholar]
- 22.Chiu TK, Kaczor-Grzeskowiak M, Dickerson RE. Absence of minor groove monovalent cations in the crosslinked dodecamer C-G-C-G-A-A-T-T-C-G-C-G. J. Mol. Biol. 1999;292:589–608. doi: 10.1006/jmbi.1999.3075. [DOI] [PubMed] [Google Scholar]
- 23.Drew HR, Samson S, Dickerson RE. Structure of a B-DNA dodecamer at 16 K. Proc. Nat. Acad. Sci. U.S.A. 1982;79:4040–4044. doi: 10.1073/pnas.79.13.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howerton SB, Sines CC, VanDerveer D, Williams LD. Locating monovalent cations in the grooves of B-DNA. Biochemistry. 2001;40:10023–10031. doi: 10.1021/bi010391+. [DOI] [PubMed] [Google Scholar]
- 25.Shui X, McFail-Isom L, Hu GG, Williams LD. The B-DNA dodecamer at high resolution reveals a spine of water on sodium. Biochemistry. 1998;37:8341–8355. doi: 10.1021/bi973073c. [DOI] [PubMed] [Google Scholar]
- 26.Shui X, Sines CC, McFail-Isom L, VanDerveer D, Williams LD. Structure of the potassium form of CGCGAATTCGCG: DNA deformation by electrostatic collapse around inorganic cations. Biochemistry. 1998;37:16877–16887. doi: 10.1021/bi982063o. [DOI] [PubMed] [Google Scholar]
- 27.Woods KK, McFail-Islom L, Sines CC, Howerton SB, Stephens RK, Williams LD. Monovalent cations sequester within the A-tract minor groove of [d(CGCGAATTCGCG)]2. J. Am. Chem. Soc. 2000;122:1546–1547. [Google Scholar]
- 28.Rohs R, Sklenar H, Shakked Z. Structural and energetic origins of sequence-specific DNA bending: Monte Carlo simulations of papillomavirus E2-DNA binding sites. Structure. 2005;13:1499–1509. doi: 10.1016/j.str.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 29.Shatzky-Schwartz M, Arbuckle ND, Eisenstein M, Rabinovich D, Bareket-Samish A, Haran TE, Luisi BF, Shakked Z. X-ray and solution studies of DNA oligomers and implications for the structural basis of A-tract-dependent curvature. J. Mol. Biol. 1997;267:595–623. doi: 10.1006/jmbi.1996.0878. [DOI] [PubMed] [Google Scholar]
- 30.West SM, Rohs R, Mann RS, Honig B. Electrostatic interactions between arginines and the minor groove in the nucleosome. J. Biomol. Struct. Dyn. 2010;27:861–866. doi: 10.1080/07391102.2010.10508587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput. Biol. 2008;4:e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, Gilmour DS, Albert I, Pugh BF. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Field Y, Fondufe-Mittendorf Y, Moore IK, Mieczkowski P, Kaplan N, Lubling Y, Lieb JD, Widom J, Segal E. Gene expression divergence in yeast is coupled to evolution of DNA-encoded nucleosome organization. Nat. Genet. 2009;41:438–445. doi: 10.1038/ng.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Segal E, Fondufe-Mittendorf Y, Chen L, Thåström A, Field Y, Moore IK, Wang J-PZ, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindemose S, Nielsen PE, Hansen M, Møllegaard NE. A DNA minor groove electronegative potential genome map based on photo-chemical probing. Nucleic Acids Res. 2011;39:6269–6276. doi: 10.1093/nar/gkr204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Møllegaard NE, Nielsen PE. Increased temperature and 2-methyl-2,4-pentanediol change the DNA structure of both curved and uncurved adenine/thymine-rich sequences. Biochemistry. 2003;42:8587–8593. doi: 10.1021/bi0343267. [DOI] [PubMed] [Google Scholar]
- 37.Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 38.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 39.Rocchia W, Sridharan S, Nicholls A, Alexov E, Chiabrera A, Honig B. Rapid grid-based construction of the molecular surface and the use of induced surface charge to calculate reaction field energies: applications to the molecular systems and geometric objects. J. Computational Chem. 2002;23:128–137. doi: 10.1002/jcc.1161. [DOI] [PubMed] [Google Scholar]
- 40.Honig B, Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.