Abstract
The transcription factor Osterix (Sp7) is essential for osteoblastogenesis and bone formation in mice. Genome wide association studies have demonstrated that Osterix is associated with bone mineral density in humans; however, the molecular significance of Osterix in human osteoblast differentiation is poorly described. In this study we have characterized the role of Osterix in human mesenchymal progenitor cell (hMSC) differentiation. We first analyzed temporal microarray data of primary hMSC treated with bone morphogenetic protein-6 (BMP6) using clustering to identify genes that are associated with Osterix expression. Osterix clusters with a set of osteoblast-associated extracellular matrix (ECM) genes, including bone sialoprotein (BSP) and a novel set of proteoglycans, osteomodulin (OMD), osteoglycin, and asporin. Maximum expression of these genes is dependent upon both the concentration and duration of BMP6 exposure. Next we overexpressed and repressed Osterix in primary hMSC using retrovirus. The enforced expression of Osterix had relatively minor effects on osteoblastic gene expression independent of exogenous BMP6. However, in the presence of BMP6, Osterix overexpression enhanced expression of the aforementioned ECM genes. Additionally, Osterix overexpression enhanced BMP6 induced osteoblast mineralization, while inhibiting hMSC proliferation. Conversely, Osterix knockdown maintained hMSC in an immature state by decreasing expression of these ECM genes and decreasing mineralization and hMSC proliferation. Overexpression of the Osterix regulated gene OMD with retrovirus promoted mineralization of hMSC. These results suggest that Osterix is necessary, but not sufficient for hMSC osteoblast differentiation. Osterix regulates the expression of a set of ECM proteins which are involved in terminal osteoblast differentiation.
Keywords: bone morphogenetic protein-6, osteoblast, human, Osterix, osteomodulin
Introduction
Bone morphogenetic proteins (BMPs) form the largest subgroup within the transforming growth factor beta (TGF-β) super family of growth and differentiation regulatory proteins. BMPs were originally identified as proteins capable of inducing ectopic cartilage and bone formation upon local injection in mammals [Reddi, 1992]. In cells of the osteoblast lineage, BMPs induce expression of alkaline phosphatase, type I collagen, and other noncollagenous bone proteins found in osteoid; a phenotype consistent with differentiated osteoblasts [Cheifetz et al., 1996; Cheng et al., 2003]. However, not every BMP family member is able to induce bone differentiation to the same degree. Previous studies demonstrated that the BMP family member, BMP6, is selectively induced by glucocorticoid treatment in human marrow-derived mesenchymal progenitor cells (hMSC). BMP6 mRNA levels are approximately 16- to 22-fold higher in dexamethasone treated hMSC cells, while the level of BMP2 is relatively unaffected [Diefenderfer et al., 2003; Liu et al., 2004]. Also, significantly lower quantities of BMP6 relative to BMP7 and BMP2 are required for healing of critical size ulnar bone defects in rabbits. This may be attributed to resistance of BMP6 to the BMP inhibitor, noggin. In comparison to BMP2 and 7, this property of BMP6 is supported by experiments showing that BMP6 carries a single amino acid difference, which when introduced at the corresponding positions in BMP2 and BMP7, results in their increased resistance to noggin [Song et al., 2010]. We previously reported that BMP6 was more potent and consistent than BMP2 and BMP7 in inducing osteoblast differentiation in primary hMSC [Friedman et al., 2006]. Taken together, these finding indicate that effects of BMP6 may be unique among the BMP family in mediating terminal osteoblast differentiation in human-derived cells.
Osterix (SP7) is a zinc finger transcription factor specifically expressed by osteoblasts which is required for osteoblast differentiation in mice. Osterix-deficient mice show an absence of osteoblasts and no bone is formed [Nakashima et al., 2002]. Runx2, a transcription factor implicated in the control of osteoblast differentiation, regulates Osterix transcription. Runx2 is expressed in Osterix-deficient mice, indicating that Osterix acts downstream of Runx2 [Celil et al., 2005]. Interestingly, Msx2 also regulates Osterix expression in Runx2-deficient cells [Matsubara et al., 2008]. Osterix was found to form a complex with the nuclear factor of activated T cells (NFAT), promoting osteoblastic bone formation through the activation of COLIA1 promoter activity [Koga et al., 2005]. Accordingly, constitutive activation of NFAT activates the Wnt signaling pathway, increasing bone formation and bone mass [Winslow et al., 2006]. Interestingly, a number of recent genome wide association studies (GWAS) have shown an association between the Osterix locus and bone density in adults and children [Timpson et al., 2009]; however, the mechanisms of Osterix transcriptional activity and control of osteoblast differentiation in humans are still poorly understood.
In our previous studies we have found that BMP6 treatment of hMSC results in the pronounced expression of Osterix -100s of fold higher than untreated cells - while having relatively little effect on Runx2 expression [Friedman et al., 2006]. In the present study, a microarray analysis was performed to evaluate genes that change coincident with Osterix expression. Retroviral mediated enforced overexpression and miRNA mediated Osterix knockdown were conducted in hMSC to determine the role of Osterix in human osteoblastogenesis. We show that Osterix regulates a set of ECM genes involved in terminal osteoblast differentiation, and that one of those molecules, osteomodulin, promotes increased mineralization when overexpressed in hMSC.
MATERIAL AND METHODS
Isolation and Culture of Human Mesenchymal Stem Cells
Primary hMSC were isolated from vertebral bodies obtained through the National Disease Research Interchange (NDRI, Philadelphia, PA) or extracted from mononuclear cells from Lonza (Walkersville, MD, USA), as previous reported [Friedman et al., 2006]. Briefly, bone marrow extracted from vertebral bodies was placed in RPMI 1640 medium ((Invitrogen, Grand Island, NY, USA) supplemented with 2% FBS. The bone marrow suspension was layered over histopaque 1077 (Sigma, St. Louis, MO, USA) and centrifuged at 400 × g for 30 min to obtain mononuclear cells. Approximately 2×108 low density cells were placed in assay medium with 20% defined FBS (Hyclone, Logan, Utah, USA). Assay medium is McCoy’s 5a medium (Invitrogen) supplemented to additionally contain 20 mg/ml asparagine, 10 mg/ml serine, 0.75× MEM vitamins, 0.38× MEM amino acids, 75 μm non-essential amino acids, 100 U/ml penicillin/streptomycin, 2.3 mM L-Glutamine, 1.3 mM Sodium pyruvate, 0.06% sodium bicarbonate, 50 mM BME (Invitrogen, Grand Island, NY, USA).
BMP6 Treatment
Human mesenchymal stem cells (hMSCs) at passage 4 were plated in 12-well dishes (1×10 4 Per well) and cultured for 1 day in assay medium with 20% defined FBS (Hyclone, Logan, Utah, USA). The next day, the medium was removed and the cells were cultured in osteogenic medium, which is serum-free assay medium additionally containing 1% ITS (BD Bioscience, Bedford, MA, USA), ascorbic acid (25 mg/ml) and β-glycerol phosphate (5 mM). hMSC were treated with BMP6 (R&D systems, Minneapolis, MN, USA) and cultured for 0, 8, 24, and 96 hr. Then BMP6 was removed and washed out. Cells were harvested at the indicated time points.
Microarray Analysis
Clustering of gene expression data from an existing published data base [Luo et al., 2011] was performed using the model based clustering algorithm MCLUST to cluster genes with common expression profiles across all time points and treatment conditions. From these clustering results we could identify patterns of genes that are repressed and activated by BMP6 treatment and we identified a set of genes that clustered with Osterix in a temporally dependent manner. Expression of several genes that showed changes with microarray were further validated using quantitative RT-PCR. These genes included Osterix, KROX20, HEY1, HEY2, DLX5, Osteomodulin (OMD), bone sialoprotein (BSP), and osteocalcin (OCN) from at least 3 unique donors.
Retroviral Overexpression
Full-length human cDNA sequence for Osterix (MHS1010-98684613, OpenBiosystems, Huntsville, AL, USA) was subcloned into the murine myeloproliferative sarcoma virus-based (MPSV) retroviral vector (PRLP2) vector. Stable retroviral producer lines were established using a trans-infection method as follows: Phoenix E cells were plated at a density of 2 ×104 cells/well of a 6-well plate and cultured overnight. On the following day, the culture media were aspirated and exchanged with fresh media (Dulbecco’s modified Eagle’s medium, high glucose, 10% fetal bovine serum, penicillin/streptomycin, L-glutamine). The phoenix E cells were transfected with 1 μg of either control retroviral empty vector PRLP2 or the PRLP2-Osterix with 2 μl Lipofectamine 2000 (Invitrogen, Grand Island, NY, USA). The media were changed 8–10 h later and replaced with fresh medium. On the following day, the media were changed and the cells were placed at 37 °C, 5% CO2 overnight to harvest viral supernatant on day 2 post-transfection. Fresh media were added every 24 hrs until 4 days post-transfection. On day 1 of transfection, the ecotropic retroviral packaging line, PG13, was placed in a 6-well dish (2 ×104cells/well) and cultured overnight. On day 2, retroviral supernatant was harvested from phoenix cells, filtered (0.45 μm), and supplemented with polybrene (8μg/ml). Approximately 2 ml of retroviral supernatant was added to the PG13 cells on day 2 and again on days 3 and 4. Stable producer lines were selected in puromycin when applicable (2μg/ml). PG13 cells derived from this protocol were considered stable retroviral producer cell lines with an approximate viral titer of 1 × 107 colony-forming units/ml. hMSC (passage 4) were plated in 6-well plates (2×104 cells/well) and cultured overnight at 37°C, 5% CO2. On the following day, retroviral supernatant was harvested from PG13 ecotropic retroviral producers, filtered (0.45 μm), and supplemented with polybrene (8μg/ml). Alternatively, filtered retroviral supernatant was frozen, thawed, and supplemented with polybrene. The hMSC were centrifuged for 90 minutes at 1200 × g with 3 ml of retroviral supernatant, and then the supernatant replaced with fresh medium immediately. The procedure of transduction was repeated the following day. On day 3, the transduced hMSC were trypsinized and plated to 12-well plates at a concentration of 1×104cells per well and cultured for 1 day in assay medium with 20% defined FBS (Hyclone, Logan, Utah, USA). On day 4, the medium was removed and the cells were cultured in osteogenic medium (descried above), before hMSC were treated with BMP6 for 8, 24, or 96 hours at 0, 2.5 and 10nM.
Retroviral Transduction for Knockdown Studies
The microRNAi based knockdown cassettes were designed using the Invitrogen miRNAi designer tool (K4939-00, Invitrogen, Grand Island, NY, USA). miRNAi against Osterix (Target sequence: AGAGGTTCACTCGTTCGGATG) was designed and cloned into the pCDNA6.2 GW/miR vector. After sequencing, the knockdown cassette was subcloned into the PRLP2 retroviral vector. The production of retroviral supernatant and transduction were performed as described previously.
Cell Proliferation Assay
To investigate the effect of BMP6 on cell proliferation, passage 4 hMSC were plated at a concentration of 2×104 cells per well in 6-well plates and incubated in a 37 °C 5% CO2 incubator. The following day, hMSC were cultured with 20% FBS in assay medium containing BMP6 (0, 2.5, 10nM). The fresh medium was changed every 3 days. The cells were counted with a hemocytometer (Electron Microscopy Sciences, Hatfield, PA, USA) at 6, 12, 18 days.
To study the effects of Osterix on cell proliferation, the transduced cells (forced overexpression, knock down, and control) from 2 donors were plated at a concentration of 2×104 cells per well in 6-well plates and cultured in assay medium with 20% serum. The fresh medium was changed every 3 days. The cells were counted with a hemocytometer at 6, 12, and 18 days.
Alizarin Red S staining
Cells were harvested between days 12 and 16 (unless otherwise indicated). The cells were washed in PBS and fixed in 50% ethanol for 5 min. The ethanol solution was removed and an Alizarin Red S solution (1%) was added for 5 min. Each well was rinsed three times with PBS.
Quantitative RT-PCR
1×104 cells were placed into each well of a 12-well plate and cytokine induction studies were performed (as described above). Total RNA was isolated at various time points using the RNeasy kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. RNA was dissolved in DEPC ddH2O and stored at −80°C. All primers were designed using the Primer3 program (Whitehead Institute, Cambridge, MA, USA). cDNAs were synthesized using Superscript III reverse transcriptase (Invitrogen).
Quantitative RT-PCR was performed using an ABI 7600 Fast Thermocycler with SYBR Green (BioWhittaker Molecular Application, Rockland, ME, USA). All reaction products were normalized to the expression level of mRNA. PCR primer sets used were as follows: Osterix forward, 5′-TGC TTG AGG AGG AAG TTC AC -3′; reverse, 5′-AGG TCA CTG CCC ACA GAG TA -3′; Osteomodulin (OMD) forward, 5′-TCC AAG AAA TTT GGA ACA CC -3′; reverse, 5′-TGA CCA TTA GTG CTT CGT TG -3′; Bone sialoprotein (BSP) forward, 5′-ACA ACA CTG GGC TAT GGA GA -3′; reverse, 5′-CCT TGT TCG TTT TCA TCC AC -3′; Id1 forward, 5′-CTC TAC GAC ATG AAC GGC TGT-3′; reverse, 5′-TGC TCA CCT TGC GGT TCT G-3′; β-actin forward, 5′-AGA CCT GTA CGC CAA CAC AG -3′; reverse, 5′-CGA TCC ACA CGG AGT ACT TG -3′. To ensure primer specificity, melt curves were performed after 45 cycles of PCR. Fold differences in gene expression were calculated using the ΔΔCt method with normalization to β-actin.
Western Blot
Whole cell lysates were prepared from hMSC in 100 μl lysis buffer per well (M-PER Mammalian protein Extraction reagent (Thermo Scientific, Rockford, IL, http://www.piercenet.com); 1× complete protease inhibitor (Roche Applied Science Roche Diagnostics GmbH, Mannheim, Germany, https://www.roche-applied-science.com)). The lysates were centrifuged for 10 minutes at 16,000 × g at 4°C. Protein samples were electrophoresed on a SDS polyacrylamide gel (NuPAGE 4–12% gradient gel; Invitrogen) and were transferred onto a polyvinylidene difluoride membrane (Millipore, Billerica, MA, USA). Membranes were washed with TBS/0.1% Tween 20, and then blocked with 1% BSA (Fisher Scientific, Fair Lawn, NJ, USA) in TBS/0.1% Tween 20 (Amersham Biosciences, Piscataway, NJ, USA) for 30 minutes at room temperature. Primary antibodies including rabbit polyclonal anti-P-Smad 1/5/8 (1:1000; Cell Signaling Technology, Beverly, MA, USA); rabbit polyclonal anti-SP7 (1:1000; Abcam, Cambridge, U.K.); mouse monoclonal anti-HEY1 (1:1000; Novus Biologicals, Littleton, CO, USA); mouse monoclonal anti-BSP(1:1000; CHEMICON International, Billerica, MA, USA); or mouse monoclonal anti-β-tubulin (1:10,000, SIGMA ALDRICH, St. Louis, MO, USA) were added to the membranes overnight at 4 °C. Blots were washed 3 times with TBS/0.1% Tween 20 and followed by horseradish peroxidase-conjugated anti-rabbit or anti-mouse secondary antibodies (Bio-Rad, Hercules, CA, USA) incubated for 1 hour at room temperature. After washing, the membrane was detected with ECL Detection Reagents (Thermo Scientific, Rockford, IL, USA) and exposed onto x-ray film (Denville Scientific INC, Metuchen, NJ, USA).
Statistical Analysis
Student’s t-tests were used to determine whether treated groups, within donor, were significantly different from controls at P<0.05.
RESULTS
Osterix Clusters with Extracellular Matrix (ECM) Genes during Osteoblast Differentiation Previously we showed that 12 hours of BMP6 exposure induces gene expression changes, including upregulation of the transcription factor Osterix (SP7), consistent with osteoblast differentiation. However, while short-term exposure to BMP6 was sufficient to induce expression of some osteoblastic associated genes (including Dlxs and Ids), short periods of treatment were not sufficient for maximal Osterix expression and osteoblast differentiation as determined by mineral incorporation. A maximal mineralization response was observed with 96 hours of BMP6 treatment [Friedman et al., 2006].
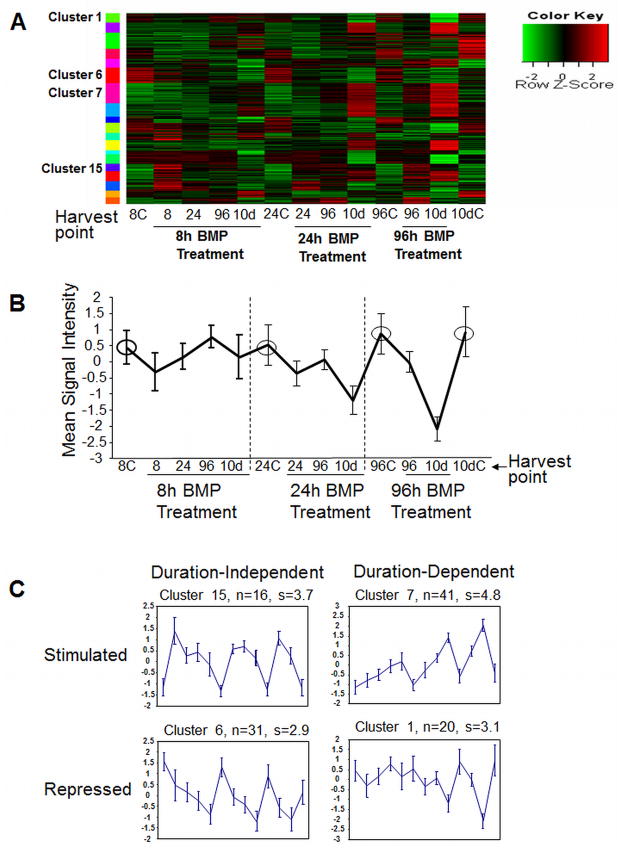
To better understand BMP6 regulation of hMSC osteoblast differentiation and to determine genes with patterns of expression similar to Osterix, we analyzed microarray data of cells treated with 8, 24, or 96 hours of BMP6 using cluster analysis. The top 392 differentially expressed genes were sorted into clusters based on similarities in the pattern of gene expression (no change, up regulation, down regulation) at thirteen discrete time points following control or BMP6 treatment. Genes were sorted into 19 clusters based on BMP treatment duration, harvest time point, and gene expression level and pattern (Figure 1 and Supplementary Figure 1). Changes in gene expression in various clusters are represented in the heat map (Figure 1A), while normalized expression patterns are represented graphically in Figures 1B and 1C and Supplementary Figure 1. Individual genes that are present in each cluster are described in Supplementary Tables 1–5. The clusters fit several different patterns: (i) Clusters 2,5,7,8,12,16,19 are BMP6-induced, but sustained expression was dependent upon treatment duration (Induced/Dependent) (Figure 1C; Supplementary Figure 1; Supplementary Table 1), (ii) Clusters 13 and 15 are BMP6-induced but sustained expression was independent of treatment duration (Induced/Independent) (Figure 1C; Supplementary Figure 2; Supplementary Table 2), (iii) Clusters 1,4, 10, 14, 18 are BMP6-repressed, but sustained repression was dependent upon longer treatment (Repressed/Dependent) (Figure 1C; Supplementary Figure 2; Supplementary Table 3), (iv) Clusters 3, 6, and 9 are BMP6-repressed but sustained repression was independent of treatment length (Repressed/Independent) (Supplementary Table 4), and (v) Clusters 11 and 17 are BMP6-induced highest at 8 hours and expression is not maintained, regardless of treatment duration (Early) (Supplementary Figure 2 and Supplementary Table 5).
Figure 1. BMP6 regulates a two-wave transcriptional process during osteoblast differentiation.
Expression results were clustered using the model based clustering algorithm MCLUST across all time points and treatment conditions. (A) Heat map of the most significant clusters. (B) Reference Clustering Plot (in this case cluster 1). Circled points are the control conditions where the expression level is essentially constant. (C) Representative profiles of the four most common cluster patterns with BMP6 treatment. In some cases BMP6 treatment induces expression (clusters 15 and 7) or represses expression (clusters 6 and 1). For some profiles, removal of BMP6 treatment results in expression reverting back to the control levels (e.g. clusters 15 and 1) (treatment duration dependent), while in other cases a transient exposure of BMP6 is sufficient to activate a pathway that remains changed even after short-term BMP6 exposure and BMP6 removal (e.g. clusters 7 and 6) (duration independent). Refer to Supplementary Figure 1 to view all 19 clusters and Supplementary Tables 1–5 for specific genes located in these clusters. Differentially expressed genes in clustered groups were validated using quantitative real-time PCR in 2–3 independent donor hMSC lines.
The transcription factors DLX2, 3, and 5; Id1, 3, and 4; and SMAD5 and 6 clustered together and were elevated with only 8 hours of BMP treatment and remained elevated for 10 days without any additional exogenous BMP (Induced/Duration Independent) (Supplementary Table 2). While 8 hours of treatment was sufficient to induce the expression of the duration dependent genes, 96 hours of treatment was required for high, sustained expression of Osterix (represented in cluster 7) and co-clustering genes throughout 10 days (Induced/Duration Dependent) (Supplementary Table 1). The ECM genes osteopontin, bone sialoprotein (IBSP), aggrecan, syndecan 1, osteomodulin (OMD), and glycoprotein M6B are located in cluster 7 with Osterix while the ECM genes osteoglycin (OGN) and asporin (ASPN) are located in the very similar cluster 8, showing expression patterns associated with terminal osteoblast function (Supplementary Table 2).
Genes that are repressed and treatment-duration dependent included a number of extracellular proteins that may modify TGF-beta, Wnt, and IGF signaling, including WISP1 and WISP2, follistatin (FST), IGFBP6 and 3, and SFRP4 (Supplementary Table 3). Genes that are repressed and treatment-duration independent included, IGF1 and IGFBP2, as well as inflammation and immune-associated genes and genes associated with proteolysis (Supplementary Table 4). Finally, there were a small number of genes expressed early with short-term treatment, but down-regulated later including AMIGO2 and SCUBE3 (Supplementary Table 5). Results for specific genes in these five primary clusters were validated by real-time qPCR in 2–3 additional independent human donors MSC isolates (data not shown).
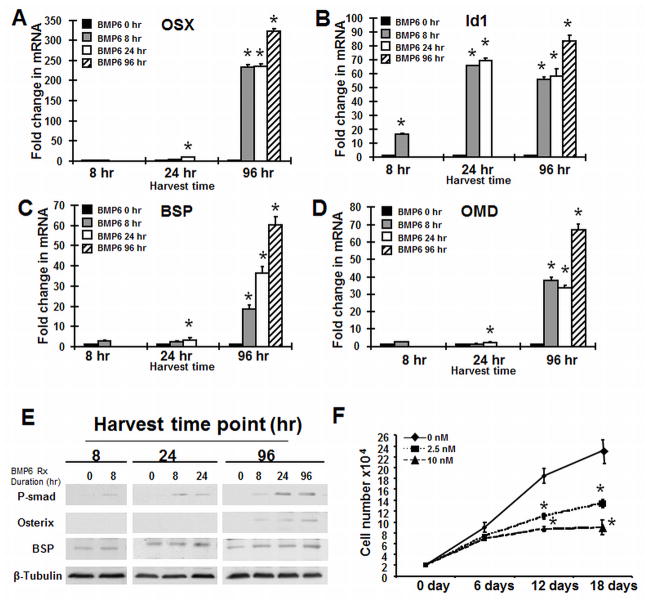
To further investigate genes associated with Osterix expression and the duration-dependent cluster we used quantitative RT-PCR analysis to evaluate hMSC treated with BMP6 or control vehicle for 8, 24, or 96 hours and harvested for RNA at 96 hours. These investigations were performed under defined serum-free conditions. Under these conditions, hMSC show little to no proliferation, while cells grown in 20% serum double approximately every 6 days (results not shown). Consistent with our microarray results, Osterix expression is dependent on length of BMP6 treatment (Fig. 2A). Id1 on the other hand, a transcription factor present in the Duration Independent cluster, shows a 20-fold increase with only 8-hours of treatment and the high-expression is maintained for 96 hours with 8 hours treatment (Fig. 2B). The mRNA level of osteoblast related ECM proteins, BSP (Fig. 2C), OPN (data not shown), as well as the proteoglycans OMD (Fig. 2D), ASPN (data not shown), and OGN (data not shown), that were also present in the Duration-dependent cluster with Osterix showed a similar increase in a time-dependent manner.
Figure 2. BMP6 upregulates Osterix and other osteoblast genes dependent upon treatment duration.
hMSC were differentiated into osteoblasts in serum-free osteogenic medium and in the presence or absence of BMP6 (20 nM) for 8, 24 and 96 hours, respectively. Real-time PCR analysis of the relative expression of the transcription factors (A) Osterix and (B) Id1 and the extracellular matrix proteins (C) bone sialoprotien (BSP) and (D) osteomodulin (OMD). The data represent the mean values ±SD of triplicate experiments normalized to the housekeeping gene β-actin. (F): BMP6 treatment decreases hMSC proliferation. hMSCs (2×104 per well in 6-well plates) were cultured in the assay medium with 20% FBS in the presence of BMP6 at 0, 2.5 and 10 nM. Cumulative cell number was counted at 6, 12 and 18 days. Statistical differences (*) of p < 0.05 between the BMP6 treated and untreated cell lines were determined using t-test.
To verify these changes at the protein level we performed western blot analysis. BMP6 treatment induces R-smad1/5/8 phosphorylation and increases the expression of Osterix and BSP at the protein level in a duration dependent manner (Fig. 2E).
BMP6 Inhibits hMSC Proliferation
BMP6 in serum-free culture conditions, as used in the previous experiment, had no effect on cell number (data not shown); however, we evaluated the effect of BMP6 on cell proliferation by adding BMP6 ligand (0, 2.5, 10 nM) to hMSC cells that were first cultured for 24 hours in 20% growth medium. Cell number was counted at days 6, 12 and 18. BMP6 treated cells displayed 18% less growth at day 6, 40% less growth at day 12 and 42% less growth at day 18 than untreated cells at the same time points (Fig. 2F). Similarly, in the presence of 10 nM BMP6, the growth inhibition was more significant, 23% less at day 6, 53% less at day 12 and 61% less at day 18 (Fig. 2F). Taken together, BMP6 treatment, under serum conditions, decreased cell number in a dose-dependent manner, consistent with cells undergoing differentiation [Drozdoff et al., 1994; Kersten et al., 2005].
Enforced Expression of Osterix Does Not Induce Human Osteoblast Differentiation
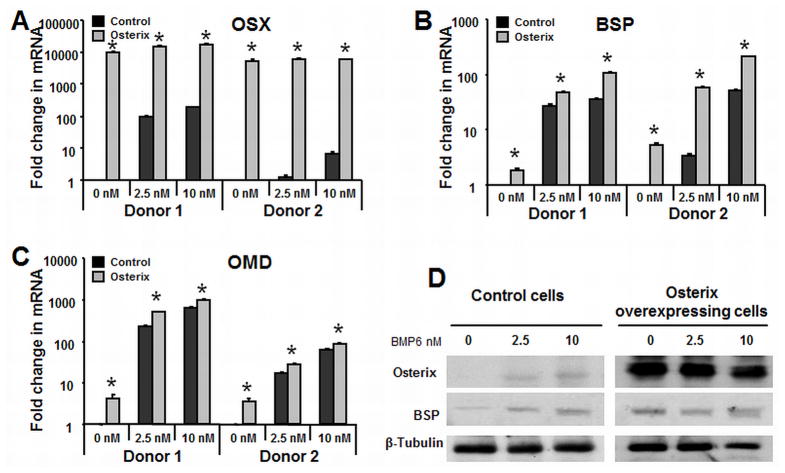
To determine the role that Osterix plays in regulating co-clustered ECM genes, retroviral transduction was used to enforce expression of Osterix. Following plating and 48 hours of transduction, cells were treated with BMP6 (0, 2.5, 10 nM) for 4 days and RNA harvested for qRT-PCR analysis. Osterix gene expression in the overexpressing cells was increased over 10,000-fold relative to control cells (Fig. 3A), which was consistent with significantly increased protein levels detected using Western blot (Fig. 3C). Despite the substantial increase in Osterix expression, there were relatively minor changes in the expression of genes that clustered with Osterix, including OMD (Fig 3B), BSP (Fig. 3C), ASPN (data not shown), and OPN (data not shown), relative to control transfected cells. As well, Osterix overexpression is not sufficient for inducing in vitro mineralization (Fig. 5A). However, when Osterix overexpressing cells were treated with a suboptimal dose of BMP6 (2.5 nM), osteogenic gene expression (Figure 3B and 3C) and mineralization was increased (Fig. 5A). This mineralization was further increased in Osterix overexpressing cells treated with a higher dose of BMP6 (10 nM). Collectively, these studies show that Osterix alone is not sufficient for driving osteoblast differentiation, but promotes terminal differentiation in conjunction with BMP-signaling.
Figure 3. Enforced expression of Osterix increases the expression of osteoblast related extracellular matrix protein genes.
hMSC were transfected with either PRLP2 control or PRLP2-Osterix retroviral supernatant and treated with BMP6 for four days. Quantitative real-time PCR was used to demonstrate the effects of Osterix expression on other duration-dependent genes in the absence of BMP2 and with suboptimal 2.5 nM and 10 nM BMP6. (A) Osterix (B) BSP (C) osteomodulin (D): Western blot was performed to demonstrate that Osterix increased BSP protein level in the osteogenic medium with BMP6 for 4 days at 0, 2.5 and 10 nM. The qPCR data represent the mean values ±SD of triplicate experiments normalized to the housekeeping gene β-actin. Statistical differences (*) of p < .05 between the Osterix-overexpressed and control PRLP2 cell lines were determined using a t-test.
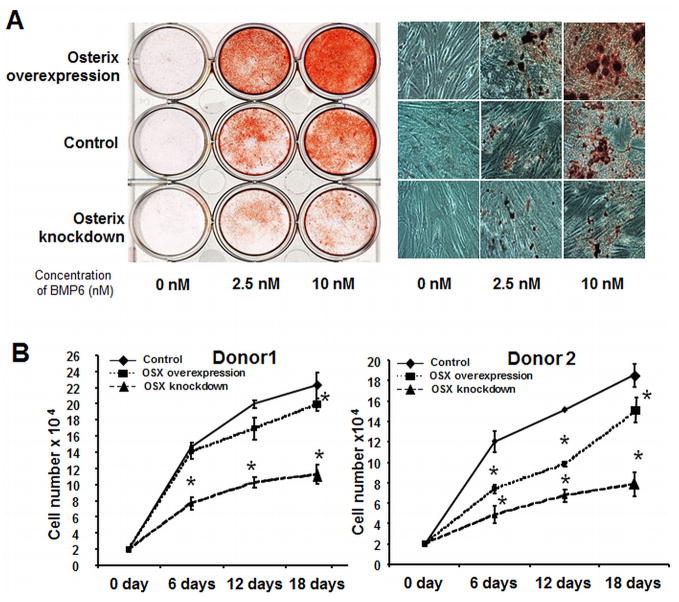
Figure 5. Osterix regulates hMSC osteoblastogenesis and proliferation.
(A) Left panel: Osterix-overexpressing and knockdown hMSC were treated with BMP6 for 96 hours at 0, 2.5 and 10 nM, and stained with Alizarin red S to evaluate calcium phosphate mineral, compare to the control cells. Right panel: Alizarin red S stained wells visualized using bright-field microscopy. (B): Transduced hMSC from donor 1 and donor 2 were cultured in assay medium with 20% FBS (growth conditions). Cumulative cell number was counted at 6, 12 and 18 days. Statistical differences (*) of p< .05 between the Osterix overexpression or knockdown and control PRLP2 cell lines were determined using t-test.
Osterix is Required for BMP6 Induced hMSC Osteoblast Differentiation
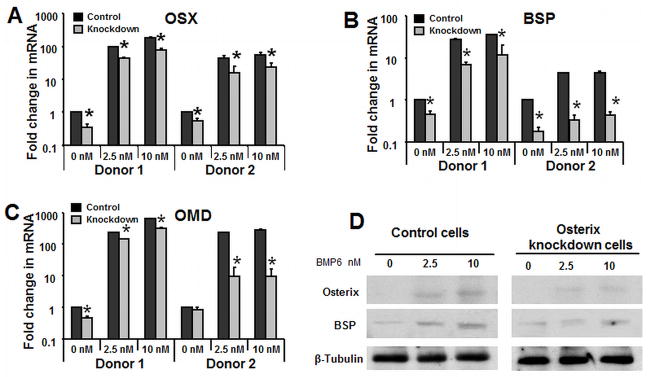
To determine whether Osterix expression was required for BMP6-induced hMSC osteoblast differentiation and mineralization, we used retrovirus-based microRNAi to knock down expression of Osterix. Knockdown cells were infected with Osterix miRNAi retrovirus, and control cells were infected with an empty-vector (PRLP2) retrovirus. By qPCR, Osterix expression level in knockdown cells was reduced approximately 65% (donor 1) or 50% (donor 2) relative to the control cells, and was also decreased with BMP6 exposure (Fig. 4B). OMD (Fig. 4B), BSP (Fig. 4C), ASPN (results not shown), and OPN (results not shown) were decreased with Osterix knockdown. Compared to the vector transfected control cells, the expression level of OMD was reduced about 40% (donor 1) or 90% (donor 2) with 2.5 nM BMP6 and about 50% (donor 1) or 90% (donor 2) with 10nM BMP6 (Fig. 4B). Furthermore, BSP was reduced 75% (donor 1) or 90% (donor 2) with 2.5 nM BMP6 treatment and 67% (donor 1) or 90% (donor 2) with 10 nM BMP6 treatment (Fig 4C). Osterix and BSP protein level was also examined by western blot and demonstrated similar alterations (Fig. 4C). To examine the effect of Osterix knockdown on osteoblast maturation, we performed a mineralization assay. The Alizarin red S staining results show that Osterix knockdown inhibits the effect of BMP6 on mineralization (Fig. 5A).
Figure 4. Osterix is required for BMP6 induced hMSC osteogenic differentiation.
hMSC were transfected with mir-Osterix retroviral supernatant treated hMSC in the osteogenic medium with BMP6 for 4 days at 0, 2.5 and 10 nM Real-time quantitative PCR analysis was performed to demonstrate that knockdown of (A) Osterix on osteoblast associated genes (B) OMD and (C) BSP relative to the vector controls. (D) Western blot was performed to demonstrate that knockdown of Osterix decreased BSP protein level. The qPCR data represent the mean values ±SD of triplicate experiments normalized to the housekeeping gene β-actin. Statistical differences (*) of p < .05 between the Osterix knockdown and control PRLP2 cell lines were determined using t-test.
Both Osterix Overexpression and Knockdown Reduce hMSC Proliferation
Triplicate studies were performed to determine the effect of Osterix on the proliferation of hMSC from 2 donors. Following transduction, the retroviral constructs (PRLP2, PRLP2-Osterix, and PRLP2-mirOsterix) transduced cells were plated to the six-well plates at 2×104/well with 20% serum. The cell numbers were examined at day 6, 12 and 18. Donor 1 cells overexpressing Osterix, showed 4% less proliferation at day 6, 15% less at day 12 and 11% less at day 18 as compared to vector control transfected cells at the same time points (Fig. 5B). For donor 2 cells, our study showed 39% less proliferation day 6, 35% less at day 12 and 20% less at day 18 with Osterix overexpression as compared to vector control transfected cells (Fig. 5B). We further examined whether cell proliferation was affected by Osterix knockdown. Surprisingly, as shown in Fig 4B, there was significant decrease in the proliferation of Osterix knockdown cells from both of donors (Fig. 5B). Considered together these results suggest that Osterix directly or indirectly regulates hMSC proliferation and that the expression of Osterix above or below a certain threshold may decrease cell proliferation.
Enforced Expression of OMD, a Downstream Gene of Osterix, Enhances BMP6 Induced Osteoblastogenesis
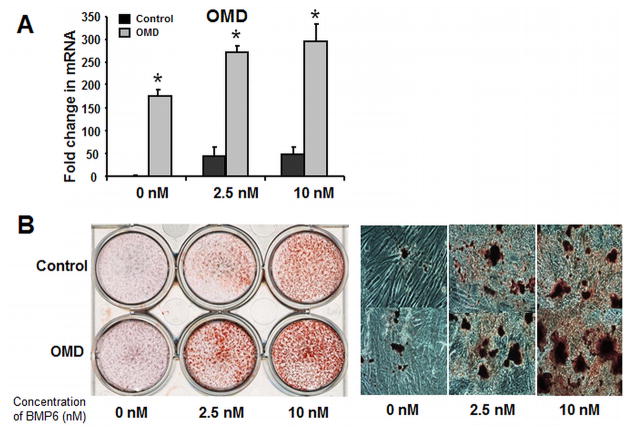
It is intriguing that a set of ECM proteins co-clusters with Osterix and that Osterix regulates their expression. OMD is a large proteoglycan that has previously been shown to be produced by osteoblasts and we postulated that it may play a role in Osterix-regulated terminal human osteoblast maturation. Retroviral transduction was performed to enforce OMD expression in hMSC (Fig. 6A). Basal OMD expression was upregluated over 150-fold, and with BMP6 treatment levels were 5–6-fold higher than in untreated cells. OMD overexpressing hMSCs show higher mineralization compared to the vector control cells (Fig. 6B), while having no significant effect on expression of other ECM genes (results not shown).
Figure 6. Enforced expression of osteomodulin promotes BMP6 induced hMSC osteoblastogenesis.
(A): Real-time quantitative PCR analysis to determine OMD expression level in the PRLP2-OMD retroviral supernatant treated hMSC in osteogenic medium with BMP6 for 4 days at 0, 2.5 and 10 nM. (B): Left panel: OMD-overexpressing hMSC were treated with BMP6 for 96 hours at 0, 2.5 and 10 nM, and stained with Alizarin Red S to evaluate calcium phosphate mineral, compared to the control cells. Right panel: OMD-overexpressing hMSC were stained with Alizarin Red S to evaluate mineralization and visualized using bright-field microscopy at 14th day. The Q-PCR data represent the mean values ±SD of triplicate experiments normalized to the housekeeping gene β-actin. Statistical differences (*) of p < .05 between the Osterix knockdown and control PRLP2 cell lines were determined using t test.
DISCUSSION
The primary objective of this investigation was to explore transcriptional events associated with BMP6 regulation of hMSC osteoblast differentiation and to specifically determine the role of Osterix. Clustering analysis demonstrates two-waves of transcriptional activity that coincide with human osteoblastogenesis. Genes that are expressed at a high level with only 8 hours of BMP treatment (Duration-independent), such as Dlx2, 3, and 5 and Id1, 3, and 4 are in the first wave of expression, while Osterix clusters with a set of ECM genes, such as osteomodulin and bone sialoprotein, that are in a second wave (Duration-independent).
As a highly expressed transcription factor induced by BMP6 in hMSC, Osterix clearly plays an important role in the process of terminal hMSC osteoblastogenesis. Intriguingly, recent genome-wide association studies indicate that genetic variants in the chromosomal region of Osterix are associated with osteoporosis and bone mineral density (BMD) in children and adults [Timpson et al., 2009]. In our study, Osterix enhances BMP6-induced hMSC osteoblast differentiation while regulating the expression of co-clustering ECM genes. miRNA induced knockdown of Osterix results in a reduction in BMP6 induced hMSC mineralization that is associated with a decrease in osteoblastic ECM gene expression.
Although the precise mechanisms of Osterix regulation of osteoblastogenesis are yet to be determined, BMP signaling is required for Osterix to have a significant impact on osteoblast maturation. Our functional studies show Osterix alone had a relatively little effect on differentiation unless BMP6 signaling was active. This result is similar to a previous study reporting that Osterix induced osteogenic gene expression but not differentiation in human fetal cells [Kurata et al., 2007]. In contrast, overexpression of Osterix alone is sufficient to induce osteogenic differentiation in vitro in murine embryonic stem cells and primary murine bone marrow MSCs, but not in murine fibroblasts [Tu et al., 2006; Kim et al., 2006]. In its role in regulating terminal osteoblast differentiation Osterix likely requires co-acting transcription factors or secondary post-translation modifications for it to be fully effective. A recent study by Ortuno et al (2010) has shown that Osterix is phosphorylated by p38 in response to BMP2 and that this phosphorylation is required for maximal induction of expression of BSP and fibromodulin [Ortuno et al., 2010]. Additionally, Osterix has been shown to function cooperatively with NFAT, and has been shown to promote the activation of Col1a1 promoter [Koga et al., 2005].
Osterix was found to up-regulate a set of ECM protein genes, including but not limited to BSP, OMD, OGN, and ASPN. The regulation of BSP by Osterix is relatively well-described, at least in murine cell lines [Ortuno et al., 2010], but Osterix has not been shown to regulate these proteoglycans that are clustered on chromosome 9. While the role of BSP in terminal human osteoblast differentiation is described, the other genes are not as well characterized. Recent work has shown that ASPN binds to collagen and promotes mineralization in a MG63 cell line [Kalamajski et al., 2009] and overexpression of osteomodulin (also known as osteoadherin in mice) in MC3T3E1 cells enhances osteoblast differentiation [Rehn et al., 2008]. We extended the examination of the importance of genes regulated by Osterix by overexpressing OMD in hMSC. In our study, Alizarin red S staining data showed that OMD enhances BMP6 induced hMSC mineralization, indicating a possible role for OMD in regulating mineralization in human primary cells.
It is intriguing that both enforced overexpression and knockdown of Osterix decreased the growth of hMSC. The negative effect of Osterix on proliferation is perhaps consistent with decreased proliferation of cells as they progress through osteoblast differentiation. A previous study has shown that the inhibition of Wnt/β-catenin signaling by Osterix constitutes a possible mechanism for the inhibition of osteoblast proliferation [Zhang et al., 2008]. In contrast, other studies have shown that the enforced expression of Osterix promotes the proliferation of murine bone marrow stromal cells and NIH3T3 cells [Tu et al., 2006; Kim et al., 2006]. This may reflect a difference between cells obtained from mice and humans, or could reflect differences in stage of differentiation. The finding that knockdown of Osterix decreased growth of hMSC is supported by a recent study demonstrating that there is an almost complete absence of bone marrow cells and excessive mineralized cartilage in the femurs of postnatal Osterix inactivated mouse [Zhou et al. 2010]. These findings suggest that Osterix plays multiple functions in MSC proliferation at different stages of differentiation.
We conclude that Osterix regulates a set of ECM proteins that are involved with terminal osteoblast differentiation and/or matrix mineralization. We hypothesize that Osterix acts as a key mediator of terminal human osteoblast differentiation and that a threshold of Osterix expression, both quantity and duration, must be achieved for osteoblast maturation.
Supplementary Material
Acknowledgments
The funding for this work was provided by R01-DE017471 from the NIH to KDH and PW.
References
- Celil AB, Hollinger JO, Campbell PG. Osx transcriptional regulation is mediated by additional pathways to BMP2/Smad signaling. J Cell Biochem. 2005;95:518–528. doi: 10.1002/jcb.20429. [DOI] [PubMed] [Google Scholar]
- Cheifetz S, Li IW, McCulloch CA, Sampath K, Sodek J. Influence of osteogenic protein-1 (OP-1; BMP-7) and transforming growth factor-beta 1 on bone formation in vitro. Connect Tissue Res. 1996;35:71–78. doi: 10.3109/03008209609029176. [DOI] [PubMed] [Google Scholar]
- Cheng H, Jiang W, Phillips FM, Haydon RC, Peng Y, Zhou L, Luu HH, An N, Breyer B, Vanichalarn P, Szatkowski JP, Park JY, He TC. Osteogenic activity of the fourteen types of human bone morphogenetic proteins (BMPs) J Bone Joint Surg Am. 2003;85:1544–1552. doi: 10.2106/00004623-200308000-00017. [DOI] [PubMed] [Google Scholar]
- Diefenderfer DL, Osyczka AM, Garino JP, Leboy PS. Regulation of BMP-induced transcription in cultured human bone marrow stromal cells. J Bone Joint Surg Am. 2003;85:19–28. doi: 10.2106/00004623-200300003-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdoff V, Wall NA, Pledger WJ. Expression and growth inhibitory effect of decapentaplegic Vg-related protein 6: evidence for a regulatory role in keratinocyte differentiation. Proc Natl Acad Sci U S A. 1994;91:5528–5532. doi: 10.1073/pnas.91.12.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman MS, Long MW, Hankenson KD. Osteogenic differentiation of human mesenchymal stem cells is regulated by bone morphogenetic protein-6. J Cell Biochem. 2006;98:538–554. doi: 10.1002/jcb.20719. [DOI] [PubMed] [Google Scholar]
- Kalamajski S, Aspberg A, Lindblom K, Heinegard D, Oldberg A. Asporin competes with decorin for collagen binding, binds calcium and promotes osteoblast collagen mineralization. Biochem J. 2009;423:53–59. doi: 10.1042/BJ20090542. [DOI] [PubMed] [Google Scholar]
- Kersten C, Sivertsen EA, Hystad ME, Forfang L, Smeland EB, Myklebust JH. BMP-6 inhibits growth of mature human B cells; induction of Smad phosphorylation and upregulation of Id1. BMC Immunol. 2005;6:9. doi: 10.1186/1471-2172-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Kim HN, Park EK, Lee BH, Ryoo HM, Kim SY, Kim IS, Stein JL, Lian JB, Stein GS, van Wijnen AJ, Choi JY. The bone-related Zn finger transcription factor Osterix promotes proliferation of mesenchymal cells. Gene. 2006;366:145–151. doi: 10.1016/j.gene.2005.08.021. [DOI] [PubMed] [Google Scholar]
- Koga T, Matsui Y, Asagiri M, Kodama T, de Crombrugghe B, Nakashima K, Takayanagi H. NFAT and Osterixcooperatively regulate bone formation. Nat Med. 2005;11:880–885. doi: 10.1038/nm1270. [DOI] [PubMed] [Google Scholar]
- Kurata H, Guillot PV, Chan J, Fisk NM. Osterix induces osteogenic gene expression but not differentiation in primary human fetal mesenchymal stem cells. Tissue Eng. 2007;13:1513–1523. doi: 10.1089/ten.2006.0374. [DOI] [PubMed] [Google Scholar]
- Liu Y, Titus L, Barghouthi M, Viggeswarapu M, Hair G, Boden SD. Glucocorticoid regulation of human BMP-6 transcription. Bone. 2004;35:673–681. doi: 10.1016/j.bone.2004.04.017. [DOI] [PubMed] [Google Scholar]
- Luo W, Friedman MS, Hankenson KD, Woolf PJ. Time series gene expression profiling and temporal regulatory pathway analysis of BMP6 induced osteoblast differentiation and mineralization. BMC Syst Biol. 2011;5:82. doi: 10.1186/1752-0509-5-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara T, Kida K, Yamaguchi A, Hata K, Ichida F, Meguro H, Aburatani H, Nishimura R, Yoneda T. BMP2 regulates Osterix through Msx2 and Runx2 during osteoblast differentiation. J Biol Chem. 2008;283:29119–29125. doi: 10.1074/jbc.M801774200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, Crombrugghe BD. The novel zinc finger-containing transcription factor Osterixis required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- Ortuno MJ, Ruiz-Gaspa S, Rodriguez-Carballo E, Susperregui AR, Bartrons R, Rosa JL, Ventura F. p38 regulates expression of osteoblast-specific genes by phosphorylation of Osterix. J Biol Chem. 2010;285:31985–31994. doi: 10.1074/jbc.M110.123612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddi AH. Regulation of cartilage and bone differentiation by bone morphogenetic proteins. Curr Opin Cell Biol. 1992;4:850–855. doi: 10.1016/0955-0674(92)90110-x. [DOI] [PubMed] [Google Scholar]
- Rehn AP, Cerny R, Sugars RV, Kaukua N, Wendel M. Osteoadherin is Upregulated by Mature Osteoblasts and Enhances Their In Vitro Differentiation and Mineralization. Calcif Tissue Int. 2008;82:454–464. doi: 10.1007/s00223-008-9138-1. [DOI] [PubMed] [Google Scholar]
- Song K, Krause C, Shi S, Patterson M, Sutto R, Grgurevic L, Vukicevic S, van Dinther M, Falb D, Dijke PD, Alaoui-Ismaili MH. Identification of a key residue mediating bone morphogenetic protein (BMP)-6 resistance to noggin inhibition allows for engineered BMPs with superior-agonist activity. J Biol Chem. 2010;285:12169–12180. doi: 10.1074/jbc.M109.087197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpson NJ, Tobias JH, Richards JB, Soranzo N, Duncan EL, Sims AM. Common variants in the region around Osterix are associated with bone mineral density and growth in childhood. Hum Mol Genet. 2009;18:1510–1517. doi: 10.1093/hmg/ddp052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Q, Valverde P, Chen J. Osterix enhances proliferation and osteogenic potential of bone marrow stromal cells. Biochem Biophys Res Commun. 2006;341:1257–1265. doi: 10.1016/j.bbrc.2006.01.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winslow MM, Pan M, Starbuck M, Gallo EM, Deng L, Karsenty G, Crabtree GR. Calcineurin/NFAT signaling in steoblasts regulates bone mass. Dev Cell. 2006;10:771–782. doi: 10.1016/j.devcel.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Zhang C, Cho K, Huang Y, Lyons JP, Zhou X, Sinha K, McCrea PD, de Crombrugghe B. Inhibition of Wnt signaling by the osteoblast-specific transcription factor Osterix. Proc Natl Acad Sci U S A. 2008;105:6936–6941. doi: 10.1073/pnas.0710831105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Zhang Z, Feng JQ, Dusevich VM, Sinha K, Zhang H, Darnay BG, de Crombrugghe B. Multiple functions of Osterix are required for bone growth and homeostasis in postnatal mice. Proc Natl Acad Sci U S A. 2010;107:12919–12924. doi: 10.1073/pnas.0912855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.