Abstract
Glycosyltransferases (Gtfs) catalyze the formation of a diverse array of glycoconjugates. Small molecule inhibitors to manipulate Gtf activity in cells have long been sought as tools to understand Gtf function. Success has been limited due to challenges in designing inhibitors that mimic the negatively-charged diphosphate substrates. Here we report the mechanism of action of a small molecule that inhibits O-GlcNAc transferase (OGT), an essential human enzyme that modulates cell signaling pathways by catalyzing a unique intracellular post translational modification, β-O-GlcNAcylation. The molecule contains a five heteroatom dicarbamate core that functions as a neutral diphosphate mimic. One dicarbamate carbonyl reacts with an essential active site lysine that anchors the diphosphate of the nucleotide-sugar substrate. The lysine adduct reacts again with a nearby cysteine to crosslink the OGT active site. While this unprecedented mechanism reflects the unique architecture of the OGT active site, related dicarbamate scaffolds may inhibit other enzymes that bind diphosphate containing substrates.
A diverse range of glycoconjugates exists in nature1. These glycoconjugates play fundamental roles in cell structure, signaling processes, and cell-cell recognition, but their molecular mechanisms are challenging to study due to a lack of suitable chemical tools2. Notably missing are selective small molecule inhibitors for glycosyltransferases, the enzymes that assemble glycoconjugates from carbohydrate building blocks3–6. Most Gtfs transfer a sugar from an anionic leaving group – for example, a nucleotide – to an acceptor such as another sugar, a protein, or a lipid head group7. Efforts to identify Gtf inhibitors have focused primarily on the design of substrate or bisubstrate mimics8–10. A major hurdle has been finding suitable replacements for the anionic phosphates11–13. These phosphates contribute significantly to binding affinity and replacing them with neutral linkers usually results in weak inhibitors. On the other hand, retaining the phosphates typically prevents the inhibitors from getting into the cells. In a clever way around this dilemma, an approach has been developed to feed cells protected sugar analogs that are metabolized into non-hydrolyzable nucleotide-sugar donors14. This method allows polar donor analogs to be used as inhibitors in cells, but it offers limited opportunities to tune selectivity since the inhibitors produced resemble common cellular substrates. Thus, alternative approaches to identify cell permeable Gtf inhibitors are still needed.
O-linked N-acetylglucosamine (O-GlcNAc) transferase (OGT) is an essential vertebrate Gtf that β-O-GlcNAcylates a wide variety of nuclear and cytoplasmic proteins, including transcription factors, cytoskeletal proteins, metabolic enzymes, kinases, phosphatases, proteasome components, chaperones, and neural proteins15–17. OGT-mediated glycosylation is dynamic; there is a corresponding glycosidase, OGA, which removes O-GlcNAc residues from proteins18,19. The glycosylation/hydrolysis process, known as O-GlcNAc cycling, is sensitive to stress conditions and nutrient status, particularly glucose levels20. OGT glycosylates many protein side chains that can otherwise be phosphorylated, and O-GlcNAcylation is proposed to modulate kinase signaling21–23. Hyper-O-GlcNAcylation, due to chronically high glucose levels, is correlated with widespread transcriptional changes and a number of pathologies, including cancer24,25. Selective small molecule OGT inhibitors would be useful as probes of OGT cell biology and could validate OGT as a therapeutic target.
We previously reported a fluorescence-based high-throughput screen to identify glycosyltransferase (Gtf) inhibitors that compete with the nucleotide-sugar donor12,26,27. Using this assay, we had identified an OGT inhibitor containing a benzoxazolinone (BZX) core (Fig. 1a, compound 1); this compound was subsequently reported by others to inhibit OGT in cells28. We were curious to learn more about the mechanism of inhibition and to determine whether the molecule was suitable for cellular inhibition studies. Using biochemistry, mass spectrometry, and X-ray crystallography, we show here that an analog of 1, while not yet fully optimized for work in cells, irreversibly inactivates OGT via an unprecedented mechanism in which two active site nucleophiles sequentially attack the same carbonyl to form a C=O crosslink. The dicarbamate in the inhibitor binds in the same location as the substrate diphosphate and is proposed to function as a diphosphate isostere.
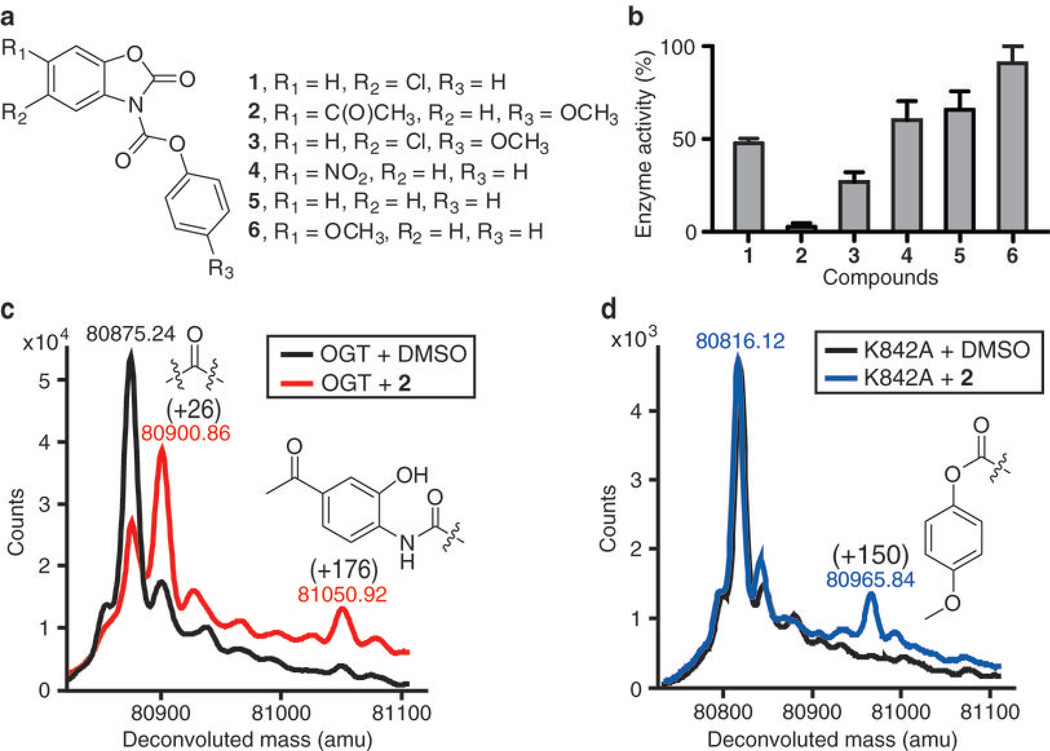
Figure 1. Inactivation of OGT by BZX compounds.
(a) Chemical structure of BZX compounds 1–6. (b) Histogram showing OGT inactivation for BZX compounds after a five-minute preincubation with a three-fold excess of each compound. Following dilution of the preincubation mixture (see methods), enzyme activity was tested as described27 and normalized to DMSO control (data represent mean values ± s.e.m., n=3). (c) Intact protein MS overlay of OGT treated with 2 (1:1 ratio) and DMSO control shows two covalent modifications (+26 Da and +176 Da) in the treated protein. A possible structure for each modification is shown. (d) Intact protein MS of 2-treated K842A (1:1 ratio) shows that mutation of K842 to alanine abolished the +26 and +176 modifications, but a new +150 mass peak appeared and a possible structure corresponding to this adduct is shown.
RESULTS
BZX compounds irreversibly inactivate OGT
Compound 1 was identified in a high-throughput screen of 65,000 compounds (Fig. 1a)27. It contains a dicarbamate moiety that connects two aromatic groups. This compound features a conjugated chemical structure and both carbamate carbonyls are potentially reactive29. Indeed, preincubation of OGT with 1 caused an irreversible loss of enzyme activity, consistent with a covalent reaction (Fig. 1b). Therefore, a small panel of related compounds was made from commercially available building blocks in order to identify structural features that might improve potency (Fig. 1a). Time-dependent inactivation studies were performed in which OGT was preincubated with a three-fold excess of compound for 5 min, and the mixture was then diluted 100-fold into buffer containing UDP-14C-GlcNAc and the well characterized peptide substrate, CKII30. Enzyme activity was measured using a previously developed peptide capture assay27. Several inhibitors showed an irreversible reduction in enzymatic activity (Fig. 1b), with the largest effect observed for compound 2, which contains a ketone on the BZX core and a p-methoxy group on the phenyl ring.
An inactivation time course showed that 2 fully inactivated OGT within 5 minutes at a 1:1 ratio of inhibitor:enzyme (Supplementary Fig. 1). The kobs/[I] value for inactivation of purified OGT was found to be 45 min−1 mM−1 (Supplementary Fig. 2). In contrast, closely related analog 6 exhibited no inhibition, implying that enzyme inactivation by BZX compounds is not due to non-specific effects such as aggregation. The presence of UDP-GlcNAc in the preincubation mixture reduced the rate of inactivation, suggesting that binding of the substrate competes with inhibitor binding (Supplementary Fig. 3).
We compared the effects of compounds 1 and 2 on O-GlcNAcylation levels in cells and cell lysates supplemented with OGT and UDP-GlcNAc. Western analysis showed inhibition of O-GlcNAcylation by 2, but not 1, under both sets of conditions (Supplementary Fig. 4). These results are consistent with studies on purified enzyme showing better inhibitory activity for 2 compared to 1. In addition, we tested compounds 1 and 2 on two other Gtfs, ppGalNAcT231 and MurG12. Neither 1 nor 2 inactivated ppGalNAcT2 up to high micromolar concentrations (Supplementary Fig. 5), but a three-fold excess of 1 inactivated MurG by 80% while 2 inactivated it by 40% (Supplementary Fig. 6). Since altering the substituents on the BZX scaffold can improve its inhibitory properties, we studied the mode of inhibition of 2 to enable future efforts to design better OGT inhibitors.
MS analysis shows crosslinks to lysine and cysteine
To identify possible covalent adducts, we analyzed intact protein mass spectra of an OGT construct after incubation with 2 at a 1:1 ratio. The construct has the same catalytic activity towards peptide substrates as the full length enzyme, but its N-terminal domain is truncated32, which is advantageous for MS analysis. Following incubation, the enzyme mixture was analyzed using LC-ESI-Q-TOF-MS. Two covalent adducts were detected (Fig. 1c). The major adduct had a mass increase of +26 Da, consistent with the incorporation of C=O and the loss of two protons. The minor adduct had a mass increase of +176 Da, and a possible structure for this adduct is shown (Fig. 1c). The major modification can only arise by sequential attack of two active site nucleophiles on the same carbonyl of 2 (Supplementary Fig. 7).
To identify the side chains that react with 2, we analyzed tryptic digests of OGT following treatment with inhibitor. We observed a triply charged 34 residue peptide (S823QYGLPEDAIV-YCNFNQLYKIDPSTLQMWANILK856) in digests of incubated protein, which was not seen in the control sample. Based on the b and y ions generated from CID fragmentation (Supplementary Fig. 8), the +26 and +176 modifications were both assigned to the same active site residue, K842 (underlined), which plays an essential role in catalysis (see below). K842 is a trypsin cleavage site and its modification blocks proteolysis, explaining the appearance of a new, longer peptide fragment in the treated protein. When incubated with OGT at a 1:1 ratio, compound 1 also modifies K842, but unlike 2 it labels some surface cysteine residues, consistent with its weaker OGT inhibition (Supplementary Fig. 9).
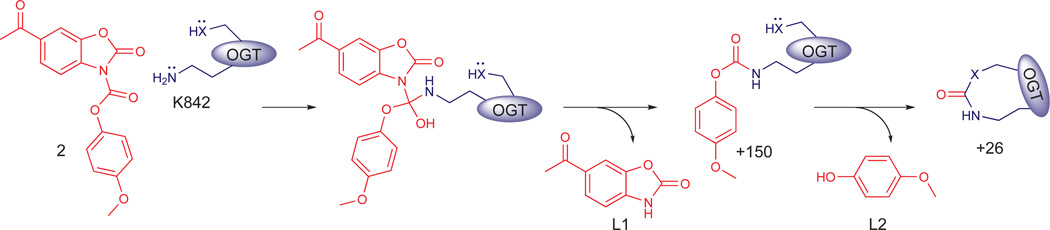
We confirmed the covalent modification site of 2-treated OGT by MS analysis of a K842A mutant. Neither the +26 nor the +176 modifications were present in the alanine mutant (Fig. 1d); however, we observed a new +150 Da mass peak, which mapped to C835 in the OGT active site (Supplementary Fig. 10). Based on the identification of K842 as the primary active site nucleophile, combined with the finding that +150 adducts are the major modifications in its absence, we propose a double displacement mechanism in which the acyclic carbonyl is the primary site of attack and the BZX moiety is the preferred leaving group (Fig. 2). Consistent with the proposed mechanism, the leaving groups generated upon incubation of 2 with OGT are 4-methoxyphenol and 6-acetyl-2-benzoxazolinone in a near 1:1 ratio (Supplementary Fig. 11).
Figure 2. Proposed double displacement mechanism for the reaction of 2 with OGT.
Mutation of K842 to alanine abolished the formation of the +26 modification and resulted in a +150 modification on a distant cysteine in the active site. A simplified double displacement mechanism to form the crosslink with K842 as the primary nucleophile first attack on the acyclic carbonyl is proposed. We cannot exclude attack of the thiol first. A more complex scheme involving several possible reaction pathways and different intermediates that produce a C=O crosslink is in Supplementary Figure 7.
Another active site nucleophile in addition to K842 must react with 2 in order to form the C=O crosslink. We speculated that mutation of the crosslinking partner would result in accumulation of a +150 adduct on K842, corresponding to a single displacement on the acyclic carbonyl. To identify the crosslinking partner, we individually mutated all residues containing a nucleophilic side chain within 10 Å of K842 in the active site of OGT, which included Y841, C911, T914, C917, T921, and T922. We also mutated C835 since it is labeled in the K842A mutant, although it is more than 10 Å away from K842. From analysis of the intact protein MS for each mutant after incubation with 2 at a 1:1 ratio, three different outcomes were observed: 1) the mutation had no effect compared with WT enzyme, e.g., Y841A and C835A (Supplementary Fig. 12a); 2) the mutation appeared to shift the ratio of the +26 and +176 modifications, e.g., T921A and T922A (Supplementary Fig. 12b); or 3) the mutation altered the product ratio and produced a new +150 mass peak (Supplementary Fig. 12c). Since only C917A led to significant accumulation of a +150 adduct on K842 (Supplementary Fig. 13), we concluded that this side chain is the major crosslinking partner.
A crystal structure confirms the crosslink
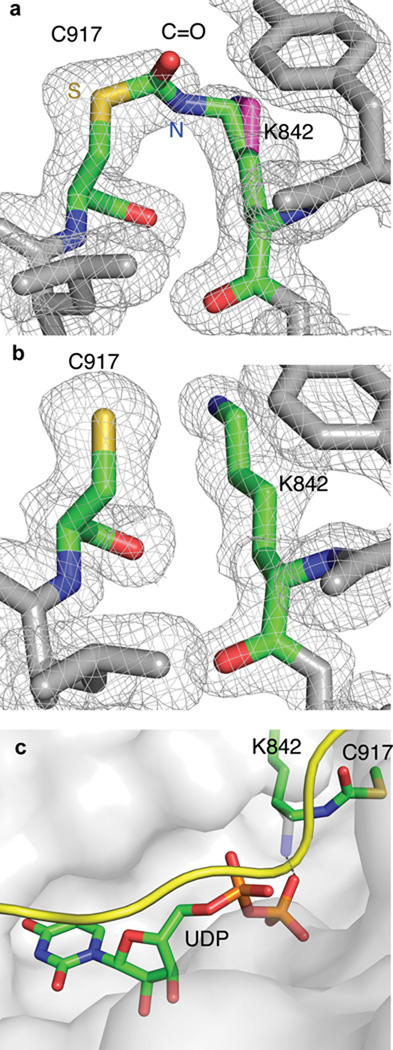
The enzyme was incubated with two equivalents of 2 and then crystallized in the presence of UDP and CKII under the same conditions as reported previously for the OGT-UDP-CKII complex32. Trapezoidal crystals diffracting to 1.88 Å were obtained (Supplementary Table 1). The inhibitor-treated and untreated OGT-UDP-CKII structures are similar, but in the inhibitor-treated structure the 2Fo-Fc omit map shows new electron density connecting K842 and C917 (Fig. 3a and 3b). The C=O and lysine side chain occupancy in the crosslinked structure refined to approximately 50%; the remaining occupancy corresponds to the unlinked lysine. The crystal structure shows that the side chain C-N bond of K842 rotates 120° towards the C917 thiol, which moves it away from the β-phosphate with which it normally interacts (Fig. 3c). The new density and rotation of the amine side chain to enable crosslink formation with C917 confirm the proposed double displacement mode of inhibition.
Figure 3. Active site comparison of crosslinked and uncrosslinked OGT.
(a) 2Fo-Fc omit map, contoured at 1 σ, of OGT-2-UDP-CKII (map calculated to 1.88 Å) shows the C=O crosslink bridging K842 and C917 together on Chain C. The reactive nitrogen and sulfur from OGT are indicated in blue (N) and yellow (S), respectively. An unbiased map was calculated with Buster48 from a model lacking both the crosslink and the reacting cysteine and lysine. The non-crosslinked lysine is shown in magenta, modeled in as an alternate conformation since 50% of protein is crosslinked, based on the occupancy refinement. The alternate conformation is similar to the confirmation of the lysine in the native structure. (b) 2Fo-Fc omit map, contoured at 1 σ, of OGT-UDP-CKII (PDB 3PE4, 1.95 Å)32 does not show density connecting K842 and C917. (c) The crosslinked active site. The OGT-2-UDP-CKII structure is shown here as a surface representation with the critical lysine crosslink highlighted (green). The CKII peptide (yellow) and UDP (green) are bound in the same conformations as in the absence of inhibitor. The uncrosslinked K842 (grey) has been overlaid to show where it normally is positioned to contact the β-phosphate. K842 must rotate 120° away from the phosphate in order to form the crosslink. The inferred H-bond between uncrosslinked K842 side chain and β-phosphate of UDP is shown with a dashed line.
The dicarbamate is a predicted neutral diphosphate mimic
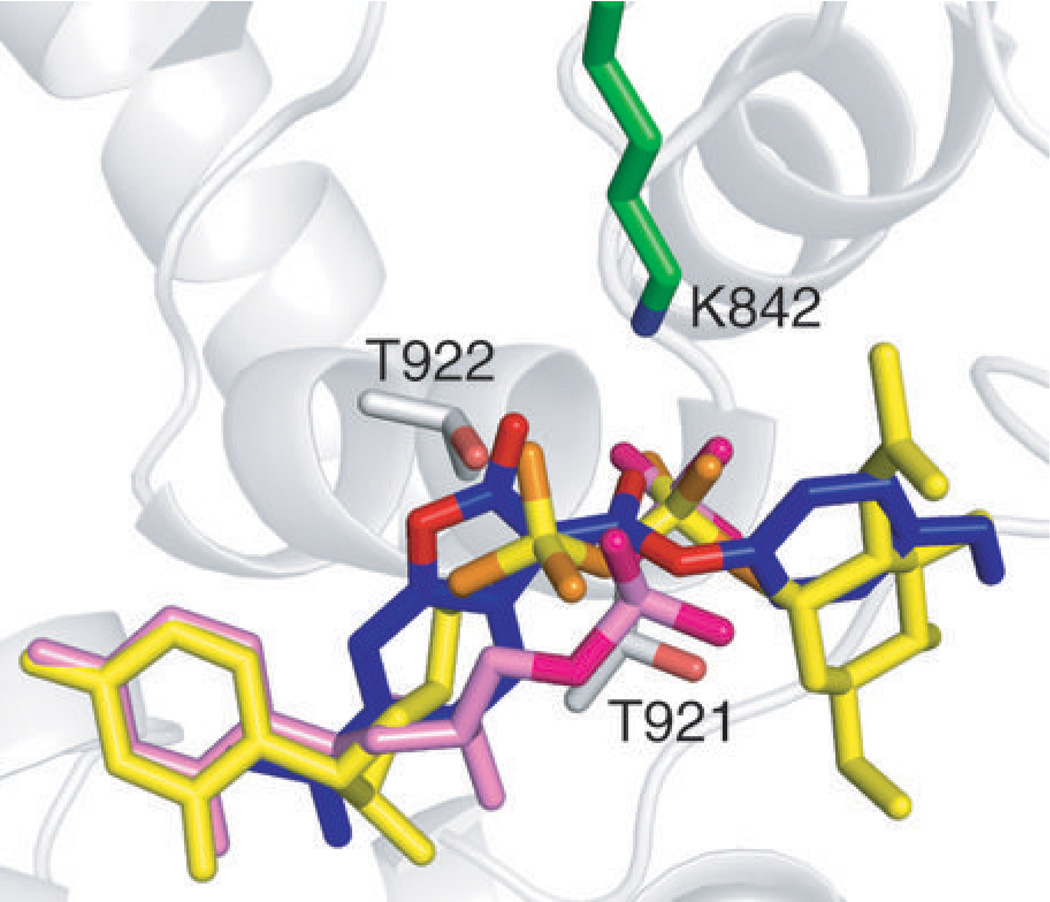
We used Glide computational modeling software33–35 to dock the pre-reactive 2 to OGT in order to obtain more insight into how the molecule may bind. Docking was carried out using protein coordinates from the OGT-UDP-CKII structure (PDB 3PE4) after removal of ligands. Computational experiments were conducted independently with 50 different starting poses of the inhibitor. The pose of the top-scoring ligand from each experiment was compared to the position of the nucleotide sugar. Despite some differences in the details, the dicarbamate of the inhibitor aligned with the substrate diphosphate in all fifty cases, with the side chain amine of lysine 842 positioned between the two carbonyls (Supplementary Fig. 14). A representative pose highlights the overlap between the dicarbamate and diphosphate in the binding site (Fig. 4). The docking models are consistent with reaction on K842 as well as MS data showing that mutation of residues flanking the diphosphate binding site perturb the major and minor product ratios, likely reflecting changes in inhibitor binding (Supplementary Fig. 12b). Based on the combined results, we conclude that the dicarbamate element functions as a neutral diphosphate mimic.
Figure 4. The dicarbamate docks into the same site as the diphosphate.
Overlay of UDP (PDB 3PE4, pink) in the OGT active site with top-scoring poses of docked inhibitor 2 (blue) and UDP-GlcNAc (yellow) shows that the dicarbamate of 2 binds in the same location as the diphosphate. Oxygen atoms of the dicarbamate and the diphosphate are highlighted with different colors. The side chain amine of K842 (green) is located between the dicarbamate carbonyls, which are sandwiched by two neighboring threonines (T921 and T922) in the diphosphate binding site.
DISCUSSION
Small molecule inhibitors of OGT have been sought for many years as cellular probes and different approaches to identify such inhibitors have been explored14,36,37. OGT is a challenging target because it is a nucleotide-sugar glycosyltransferase and the donor sugar substrate contains a diphosphate leaving group. The proposed transition state for these types of enzymes is dissociative and the oxonium ion-like portion resembles the transition state of glycosidases38,39. A key difference in glycosidase and Gtf transition states, however, is that in the latter a negatively charged diphosphate leaving group rather than a carboxylate side chain helps stabilize the oxonium character7. Accordingly, although there are many good inhibitors of glycosidases, including OGA19,40–42, they typically do not inhibit Gtfs effectively. Efforts focusing on substrate-based inhibitors of Gtfs have been unsuccessful because identifying isosteres that mimic the negatively charged diphosphate is a major hurdle. To discover new types of inhibitors, we developed a high-throughput glycosyl donor displacement screen for OGT in which compound 1 was identified27.
A small panel of related inhibitors having different substituents from 1 was prepared and inhibition was compared under a standard set of conditions (Fig. 1). Compound 2 was the best compound in preliminary studies, inactivating OGT within 5 min at a 1:1 ratio. None of the other compounds reached full inhibition at this low ratio, and all of the others, including 1, showed some labeling of surface cysteine side chains (Supplementary Fig. 9). Therefore, compound 2 can fully inactivate OGT in vitro by modifying the active site without reacting non-specifically with other residues. Since 2 contains the most electron-donating para substituent of the compounds tested, we speculate that decreased reactivity of the acyclic carbonyl is an important factor in its improved active site selectivity. This is consistent with the observation that 2 is less labile to hydrolysis than 1 at neutral pH.
In studies to probe the potential of this family of inhibitors further, compounds 1 and 2 were tested for inactivation of two other Gtfs, ppGalNAcT2 (Supplementary Fig. 5) and MurG (Supplementary Fig. 6). The former was chosen as a representative member of the mucin family of O-glycosyltransferases, which transfers N-acetylgalactosamine (GalNAc) to Ser/Thr residues of proteins and thus resembles OGT in O-glycosylating proteins31; the latter is an essential bacterial GlcNAc transferase involved in bacterial cell wall biosynthesis, which has a very similar three dimensional structure to OGT12. Neither compound inhibited ppGalNAcT2, but the two compounds showed differential inhibition of OGT and MurG respectively, which demonstrates that altering substituents on the BZX scaffold can increase specificity towards OGT. Further manipulation of substituents on the dicarbamate scaffold may improve inhibition properties, and we undertook mechanistic studies to provide a basis for rational approaches to improve inhibitors for use in cells.
Using a combination of MS analysis, mutational studies, and X-ray crystallography, we deduced that 2 reacts via a double displacement mechanism on the same dicarbamate carbonyl to form a C=O crosslink between two active site nucleophiles, K842 and C917. Inhibitors that react sequentially with two nucleophiles in the active site of an enzyme are very rare and there are no other examples involving displacement on the same carbon atom43,44. Manipulation of carbonyl reactivity and leaving group properties can potentially be exploited in designing inhibitors.
In addition to revealing an unusual reaction mechanism, our studies suggest that the five-heteroatom core of the dicarbamate class of inhibitors functions as a diphosphate isostere. This proposal is based on several lines of evidence. First, 2 reacts with the essential catalytic lysine residue that stabilizes the glycosyl donor diphosphate45, indicating that it binds in the same location. Second, the proposed binding site is consistent with protection from inactivation by UDP-GlcNAc. Third, mutations in residues that flank the diphosphate binding site perturb the ratios of the major and minor products observed upon reaction of 2, consistent with its binding at the same site. Finally, computational studies show that the dicarbamate docks into the diphosphate binding site, and superpositions of the nucleotide and inhibitor show that the heteroatoms are aligned. We point out that the five-heteroatom core of the dicarbamate scaffold has very limited rotational mobility, and the accessible conformations have an architecture similar to the OGT-bound conformation of the UDP-GlcNAc diphosphate.
Dicarbamate scaffolds related to those reported here may serve as starting points for cellular probes for other enzymes that utilize diphosphate-containing substrates and have suitable active site architecture. The dicarbamate motif has a notable advantage over a diphosphate because it is uncharged and hence more likely to cross cell membranes. Indeed, we have shown that 2 inhibits global O-GlcNAcylation in cells, consistent with cell permeability. The concentration of 2 required for inhibition of O-GlcNAcylation in cells is relatively high, possibly due to decomposition and side reactions. The mechanistic understanding provided by the studies reported here will enable rational approaches to tune reactivity and binding in order to obtain analogs with better cellular efficacy.
METHODS
Protein purification
The expression and purification of the hOGT4.5 construct (spanning residues 313–1031 based on the numbering of the full length human protein) were performed following previously described protocols32. Mutations were introduced using the Stratagene QuikChange II XL Site-Directed Mutagenesis Kit and DNA from plasmid of hOGT4.5 as template in combination with the mutagenic primers listed in Supplementary Table 2 (nucleotide substitutions are underlined). The mutants were purified similarly as WT protein.
Time-dependent inactivation of OGT with BZX compounds
BZX compounds were incubated with OGT (5 µM) in reaction buffer (125 mM NaCl, 1 mM EDTA, 20 mM potassium phosphate, pH 7.4, and 500 µM tris(hydroxypropyl)phosphine (THP)) at room temperature for varying lengths of time, then diluted 100-fold, mixed with substrates (6 µM UDP-14C-GlcNAc and 500 µM CKII peptide (KKKYPGGSTPVSSANMM)) and allowed to react for another 2 h. Enzyme activity was tested with the peptide capture assay as previously reported27. The final volume of each reaction was 20 µl and the experiments were conducted in triplicate. The activities of OGT mutants were also tested in triplicate as previously reported27.
Intact protein mass spectrometry
Each of the BZX compounds (10 µM) was incubated with OGT (10 µM) at room temperature for 5 min in the reaction buffer containing PBS (pH 7.5) and 100 µM THP, in a final volume of 20 µl. After incubation, 50 µl of PBS (pH 7.5) was added to each reaction, and the total 70 µl of mixture was passed through a PD SpinTrap G-25 column (GE Healthcare, pre-equilibrated with PBS, pH 7.5) to remove the non-enzymatic fraction. Eluted sample (40 µl each) was injected in an Agilent 6520 Q-TOF LC-MS. The mobile phase flow rate was set at 0.4 ml/min through a C4 column (Grace, Vydac 214MS C4 5u, 100 × 2.1 mm) which was initially equilibrated with solvent A (0.1% aqueous formic acid). The proteins were eluted using a linear gradient of 0 to 95% solvent B (90% acetonitrile and 0.1% aqueous formic acid) over 25 min. Mass spectrometric analyses were carried out in positive ion mode with an ESI source. The MS spectra were deconvoluted using Agilent MassHunter BioConfirm B.02.00 software with the maximum entropy algorithm.
Trypsin digestion and peptide mass spectrometry
Reaction mixture containing OGT (15 µM), 2 (200 µM), NH4HCO3 (25 mM, pH 7.8), and THP (100 µM) in a final volume of 23 µl was incubated for 30 min at room temperature. To avoid complications with over alkylation, cysteines were not “capped” and the formation of disulfides was prevented by the presence of THP. Trypsin was added to a final OGT:trypsin concentration ratio of 6:1 (w/w) and the reaction was incubated for 4 h at 37 °C. The peptides were separated using an Agilent 6520 LC/QTOF system equipped with a Phenomenex Gemini-NX C18 column (5 µm, 110 Å, 50 × 2.00 mm, pre-equilibrated with 0.1% aqueous formic acid) at a flow rate of 0.4 ml/min with a linear gradient of 0 to 60% buffer B (90% acetonitrile and 0.1% aqueous formic acid) over 50 min. Mass spectra were acquired from m/z 300–3200 for 0.25 s, followed by MS/MS scans from m/z 50–3200 of the 8 most intense species from the preceding MS scan for 0.25 s each in positive ion mode with collision energy fixed at 35 V. All spectra were processed with Mascot Distiller (Matrix Sciences) to generate the peak lists. Database searches were performed with Mascot Server 2.3 (Matrix Sciences). Error tolerant search was performed following the standard search to maximize the matches. The significance threshold was set p < 0.05, and the false discovery rate based on a decoy database was below 2%.
Crystallization and crystal structure determination
A full description is in Supplementary Methods. Briefly, inhibitor crystals were grown with the hanging drop method with hOGT4.5, seeded from native OGT-UDP-CKII crystals. The space group of OGT-2-UDP-CKII complex was I121, the same as the original OGT-UDP-CKII complex (PDB 3PE4). The data set was processed to a resolution of 1.88 Å (Supplementary Table 1). The Rfree test set array was copied and extended from the dataset for the 3PE4 coordinates. The refinement process was initiated with 3PE4 coordinates, which were adjusted by rigid body refinement, followed by several rounds of simulated annealing and atomic displacement refinement in Phenix46, and modeling in COOT47. The structure was further refined with Buster48, where the crosslink was modeled in with stereochemical restraints and occupancy refinement (See Supplementary Methods). The final Rwork/Rfree were 21.91% / 23.75%.
Molecular docking
All molecular docking was done in Schrödinger Suite 2010. The protein receptor was prepared in Protein Preparation Wizard using the hOGT4.5 crystal structure containing UDP and CKII peptide (PDB 3PE4): all water molecules and ligands were removed and hydrogen atoms were added. The docking grid was calculated in Glide v5.633,34: the rectangular docking grid (20 Å × 20 Å × 24 Å) was centered using the UDP α-phosphate. Ligands (UDP, UDP-GlcNAc, and BZX derivatives) were prepared using Ligprep 2.0 and Epik tools in Glide, at pH 7.0 ± 2.0. Ligands and hOGT4.5 were parameterized with the OPLS-2005 force field. All ligands were docked in Glide XP mode35. Molecular docking was conducted independently with 50 different starting poses of each compound. Ligand poses were analyzed in Maestro and Pymol49.
Supplementary Material
Acknowledgments
We thank J. Brugge (Harvard Medical School) for the MCF-10A ErbB2 cells, L. Tabak (NIH) for ppGalNAcT2 protein, T. Lupoli (Harvard University) for MurG protein, C. Walsh (Harvard Medical School), E. Jacobson and D. Ford (Harvard University) for helpful discussions, and A. Saghatelian (Harvard University) for critical reading of the manuscript. LC/MS data was acquired on an Agilent 6520 Q-TOF spectrophotometer supported by the Taplin Funds for Discovery Program (P.I., S. Walker). This work was supported in part by the NIH (GM076710), and the Harvard Biomedical Accelerator Fund.
Footnotes
Author contributions
S.W. oversaw all aspects of the experiments and manuscript preparation. J.J. carried out all experiments except those involving X-ray crystallography and molecular docking. M.B.L. obtained OGT:inhibitor crystals, acquired data, and solved the structure of the crosslinked protein. L.P. conducted molecular docking experiments. P.S. guided refinement of the X-ray crystal structure and molecular docking. J.J. and S.W. wrote the manuscript and all co-authors participated in figure preparation and editing.
Competing financial interests
The authors declare no competing financial interests.
Additional information
Supplementary information and chemical compound information is available online at http://www.nature.com/naturechemicalbiology/. The structure of the OGT-2-UDP-CKII complex has been submitted to the Protein Data Bank under the accession number 3TAX.
References
- 1.Varki A, et al. Essentials of glycobiology. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2009. [PubMed] [Google Scholar]
- 2.Kiessling LL, Splain RA. Chemical approaches to glycobiology. Annu. Rev. Biochem. 2010;79:619–653. doi: 10.1146/annurev.biochem.77.070606.100917. [DOI] [PubMed] [Google Scholar]
- 3.Wagner GK, Pesnot T. Glycosyltransferases and their assays. Chembiochem. 2010;11:1939–1949. doi: 10.1002/cbic.201000201. [DOI] [PubMed] [Google Scholar]
- 4.Brown JR, Crawford BE, Esko JD. Glycan antagonists and inhibitors: a fount for drug discovery. Crit. Rev. Biochem. Mol. Biol. 2007;42:481–515. doi: 10.1080/10409230701751611. [DOI] [PubMed] [Google Scholar]
- 5.Pesnot T, Jorgensen R, Palcic MM, Wagner GK. Structural and mechanistic basis for a new mode of glycosyltransferase inhibition. Nat. Chem. Biol. 2010;6:321–323. doi: 10.1038/nchembio.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frantom PA, Coward JK, Blanchard JS. UDP-(5F)-GlcNAc acts as a slow-binding inhibitor of MshA, a retaining glycosyltransferase. J. Am. Chem. Soc. 2010;132:6626–6627. doi: 10.1021/ja101231a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 8.Trunkfield AE, Gurcha SS, Besra GS, Bugg TD. Inhibition of Escherichia coli glycosyltransferase MurG and Mycobacterium tuberculosis Gal transferase by uridine-linked transition state mimics. Bioorg. Med. Chem. 2010;18:2651–2663. doi: 10.1016/j.bmc.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Izumi M, Yuasa H, Hashimoto H. Bisubstrate analogues as glycosyltransferase inhibitors. Curr. Top. Med. Chem. 2009;9:87–105. doi: 10.2174/156802609787354351. [DOI] [PubMed] [Google Scholar]
- 10.Skropeta D, Schworer R, Haag T, Schmidt RR. Asymmetric synthesis and affinity of potent sialyltransferase inhibitors based on transition-state analogues. Glycoconj. J. 2004;21:205–219. doi: 10.1023/B:GLYC.0000045093.96413.62. [DOI] [PubMed] [Google Scholar]
- 11.Wang R, et al. A search for pyrophosphate mimics for the development of substrates and inhibitors of glycosyltransferases. Bioorg. Med. Chem. 1997;5:661–672. doi: 10.1016/s0968-0896(97)00005-9. [DOI] [PubMed] [Google Scholar]
- 12.Helm JS, Hu Y, Chen L, Gross B, Walker S. Identification of active-site inhibitors of MurG using a generalizable, high-throughput glycosyltransferase screen. J. Am. Chem. Soc. 2003;125:11168–11169. doi: 10.1021/ja036494s. [DOI] [PubMed] [Google Scholar]
- 13.Hang HC, et al. Small molecule inhibitors of mucin-type O-linked glycosylation from a uridine-based library. Chem. Biol. 2004;11:337–345. doi: 10.1016/j.chembiol.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 14.Gloster TM, et al. Hijacking a biosynthetic pathway yields a glycosyltransferase inhibitor within cells. Nat. Chem. Biol. 2011;7:174–181. doi: 10.1038/nchembio.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 16.Love DC, Hanover JA. The hexosamine signaling pathway: deciphering the "O-GlcNAc code". Sci. STKE. 2005;2005:re13. doi: 10.1126/stke.3122005re13. [DOI] [PubMed] [Google Scholar]
- 17.Boyce M, et al. Metabolic cross-talk allows labeling of O-linked {beta}-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway. Proc. Natl. Acad. Sci. USA. 2011;108:3141–3146. doi: 10.1073/pnas.1010045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong DL, Hart GW. Purification and characterization of an O-GlcNAc selective N-acetyl-beta-D-glucosaminidase from rat spleen cytosol. J. Biol. Chem. 1994;269:19321–19330. [PubMed] [Google Scholar]
- 19.Macauley MS, Vocadlo DJ. Increasing O-GlcNAc levels: An overview of small-molecule inhibitors of O-GlcNAcase. Biochim. Biophys. Acta. 2010;1800:107–121. doi: 10.1016/j.bbagen.2009.07.028. [DOI] [PubMed] [Google Scholar]
- 20.Hanover JA, Krause MW, Love DC. The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim. Biophys. Acta. 2010;1800:80–95. doi: 10.1016/j.bbagen.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeidan Q, Hart GW. The intersections between O-GlcNAcylation and phosphorylation: implications for multiple signaling pathways. J. Cell Sci. 2010;123:13–22. doi: 10.1242/jcs.053678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 2010 doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu P, Shimoji S, Hart GW. Site-specific interplay between O-GlcNAcylation and phosphorylation in cellular regulation. FEBS Lett. 2010;584:2526–2538. doi: 10.1016/j.febslet.2010.04.044. [DOI] [PubMed] [Google Scholar]
- 24.Slawson C, Copeland RJ, Hart GW. O-GlcNAc signaling: a metabolic link between diabetes and cancer? Trends Biochem. Sci. 2010;35:547–555. doi: 10.1016/j.tibs.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang X, et al. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature. 2008;451:964–969. doi: 10.1038/nature06668. [DOI] [PubMed] [Google Scholar]
- 26.Hu Y, et al. Identification of selective inhibitors for the glycosyltransferase MurG via high-throughput screening. Chem. Biol. 2004;11:703–711. doi: 10.1016/j.chembiol.2004.02.024. [DOI] [PubMed] [Google Scholar]
- 27.Gross BJ, Kraybill BC, Walker S. Discovery of O-GlcNAc transferase inhibitors. J. Am. Chem. Soc. 2005;127:14588–14589. doi: 10.1021/ja0555217. [DOI] [PubMed] [Google Scholar]
- 28.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alexander JP, Cravatt BF. Mechanism of carbamate inactivation of FAAH: implications for the design of covalent inhibitors and in vivo functional probes for enzymes. Chem. Biol. 2005;12:1179–1187. doi: 10.1016/j.chembiol.2005.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kreppel LK, Hart GW. Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. J. Biol. Chem. 1999;274:32015–32022. doi: 10.1074/jbc.274.45.32015. [DOI] [PubMed] [Google Scholar]
- 31.Ten Hagen KG, et al. Cloning and expression of a novel, tissue specifically expressed member of the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase family. J. Biol. Chem. 1998;273:27749–27754. doi: 10.1074/jbc.273.42.27749. [DOI] [PubMed] [Google Scholar]
- 32.Lazarus MB, Nam Y, Jiang J, Sliz P, Walker S. Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature. 2011;469:564–567. doi: 10.1038/nature09638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friesner RA, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 34.Halgren TA, et al. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 35.Friesner RA, et al. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 36.Konrad RJ, et al. Alloxan is an inhibitor of the enzyme O-linked N-acetylglucosamine transferase. Biochem. Biophys. Res. Commun. 2002;293:207–212. doi: 10.1016/S0006-291X(02)00200-0. [DOI] [PubMed] [Google Scholar]
- 37.Hajduch J, et al. A convenient synthesis of the C-1-phosphonate analogue of UDP-GlcNAc and its evaluation as an inhibitor of O-linked GlcNAc transferase (OGT) Carbohydr. Res. 2008;343:189–195. doi: 10.1016/j.carres.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vocadlo DJ, Davies GJ. Mechanistic insights into glycosidase chemistry. Curr. Opin. Chem. Biol. 2008;12:539–555. doi: 10.1016/j.cbpa.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 39.Gloster TM, Davies GJ. Glycosidase inhibition: assessing mimicry of the transition state. Org. Biomol. Chem. 2010;8:305–320. doi: 10.1039/b915870g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rempel BP, Withers SG. Covalent inhibitors of glycosidases and their applications in biochemistry and biology. Glycobiology. 2008;18:570–586. doi: 10.1093/glycob/cwn041. [DOI] [PubMed] [Google Scholar]
- 41.Dorfmueller HC, et al. Cell-penetrant, nanomolar O-GlcNAcase inhibitors selective against lysosomal hexosaminidases. Chem. Biol. 2010;17:1250–1255. doi: 10.1016/j.chembiol.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim EJ, Perreira M, Thomas CJ, Hanover JA. An O-GlcNAcase-specific inhibitor and substrate engineered by the extension of the N-acetyl moiety. J. Am. Chem. Soc. 2006;128:4234–4235. doi: 10.1021/ja0582915. [DOI] [PubMed] [Google Scholar]
- 43.Vijayalakshmi J, Meyer EF, Jr., Kam CM, Powers JC. Structural study of porcine pancreatic elastase complexed with 7-amino-3-(2-bromoethoxy)-4-chloroisocoumarin as a nonreactivatable doubly covalent enzyme-inhibitor complex. Biochemistry. 1991;30:2175–2183. doi: 10.1021/bi00222a022. [DOI] [PubMed] [Google Scholar]
- 44.Drawz SM, Bonomo RA. Three decades of beta-lactamase inhibitors. Clin. Microbiol. Rev. 2010;23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martinez-Fleites C, et al. Structure of an O-GlcNAc transferase homolog provides insight into intracellular glycosylation. Nat. Struct. Mol. Biol. 2008;15:764–765. doi: 10.1038/nsmb.1443. [DOI] [PubMed] [Google Scholar]
- 46.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bricogne G, et al. BUSTER version 2.9. Cambridge, United Kingdom: Global Phasing Ltd.; 2010. [Google Scholar]
- 49.DeLano WL. The Pymol molecular graphics system. San Carlos, CA, USA: Delano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.