Abstract
In vivo electroporation has become a gold standard method for DNA immunization. The method assists the DNA entry into cells, results in expression and the display of the native form of antigens to professional cells of the immune system, uses both arms of immune system, has a built-in adjuvant system, is relatively safe, and is cost-effective. However, there are challenges for achieving an optimized reproducible process for eliciting strong humoral responses and for the screening of specific immune responses, in particular, when the aim is to mount humoral responses or to generate monoclonal antibodies via hybridoma technology. Production of monoclonal antibodies demands generation of high numbers of primed B and CD4 T helper cells in lymphoid organs needed for the fusion that traditionally is achieved by a final intravenous antigen injection. The purified antigen is also needed for screening of hundreds of clones obtained upon fusion of splenocytes. Such challenges make DNA vaccination dependent on purified proteins. Here, we have optimized methods for in vivo electroporation, production, and use of cells expressing the antigen and an in-cell Western screening method. These methods resulted in (1) reproducibly mounting robust humoral responses against antigens with different cell localizations, and (2) the ability to screen for antigen eliminating a need for protein/antigen purification. This process includes optimized parameters for in vivo electroporation, the use of transfected cells for final boost, and mild fixation/permeabilization of cells for screening. Using this process, upon two vaccinations via in vivo electroporation (and final boost), monoclonal antibodies against nucleus and cytoplasmic and transmembrane proteins were achieved.
Introduction
Monoclonal antibodies (MAbs) are on the top of the list of driving forces of pharmaceutical, biotech, and academia for diagnostic and therapeutic products. Indeed, the book of business for MAbs shows billions of dollars in recent years.(1) Classical methods for generation and screening of antibodies are dependent on antigen isolation and are rather hampered by challenges in obtaining naturally/properly processed forms of protein.(2–4) Despite the advances in protein purification, it is quite common that the option of protein purification may not be preferred or affordable since (1) the native form of a protein may not be achieved when using recombinantly expressed proteins not in non-mammalian cells, and (2) refolding may not be correct in the renaturing steps. Many of the increasing list of desired monoclonal antibodies need to interact with the native form of the antigen, especially in therapeutic MAbs, for example, when the aim is to make neutralizing MAbs.(5,6)
It is well documented that gene delivery and inducing antibodies to conformational epitopes are achieved via gene-based vaccination for the native form of the protein.(5–8) The in vivo electroporation is known to result in a “danger signal” in the injection site, recruiting antigen presentation cells as well as a strong milieu of cytokines that elicit immune responses.(9) A final boost with either proteins or cells expressing the antigen has improved the titers dramatically.(5,10–12) Although one can circumvent the need for protein purification by using plasmids encoding for these antigens, one still needs the antigen for the screening. To be able to perform a protein-free screening, we have improved upon and optimized an in-cell Western method using cells expressing the antigens.(13,14)
Here we describe a process for vaccination and the screening for the mounted humoral immune responses in a “protein-free” manner. We describe the optimization of a non-viral gene-based vaccination method, in vivo electroporation, using Derma Vax™ electroporator from Cellectis (Glen Burnie, MD). Proteins/antigens encoded by inserted genes are selected to have different cell localizations, transmembrane, cytoplasm, or nucleus. This method was able to elicit strong humoral immune responses using plasmids encoding the antigens of interest.(9) We then optimized an in-cell Western that allowed us to screen the sera or positive clones against naturally processed antigens negating a need for purified antigen.(14) The improved methods described here use microplates containing cells that do or do not express the antigen. We have used mildly fixed and permeabilized cells expressing the antigens for screening via fluorophore-linked immunosorbent assay (FLISA) or immunofluorescence staining assay (IFA).(15,16) The method has also been optimized and validated so that the plated cells can be mildly-fixed, permeabilized, blocked, and stored for up to 1 month at 4°C. Ready-plated cells will be assayed in a high throughput screening (HTS) and semi-quantitative manner by an infrared colorimetric plate reader for approximately 1 h in 600 wells. Easy access to mammalian vectors expressing most antigens may make such cutting-edge screening methods universal as they will save time and resources.
Materials and Methods
Mice
Female/male BALB/c mice (Charles River Laboratories, Wilmington, MA), 4–6 weeks old, were used in these studies. The mice were bred in specific pathogen-free facilities at the University of Miami, Miller School of Medicine. All animal procedures were approved by the University of Miami's Institutional Animal Care and Use Committee.
In vivo electroporation immunization
Sera from naïve mice were collected prior to immunization and in some cases were pooled. Mice were anesthetized by injecting 100 μL of xylazine/ketamine in phosphate-buffered saline (PBS) (Phoenix Pharmaceuticals, Burlingame, CA) at concentrations of 15%, 7%, and 78%, respectively. DNA-plasmid encoding the gene of interest was prepared at concentration of ∼1–2 μg/μL in PBS. All plasmids were prepared using the endotoxin-free GenElute kit (Sigma St. Louis, MO) or PureLink kit (Invitrogen, San Diego, CA). Plasmids used for gene-based immunizations were with CMV promoter and included HL1-pcDNA3 (provided by Dr. V. Lemmon, University of Miami); pcDNA3-vGPCR (G protein-coupled receptor) and pcDNA3 empty (provided by Dr. E. Mesri, University of Miami); pEGFP-C2-uPAR (provided by Dr. J. Reiser, University of Miami); pCMV6-XL5-PYCARD V1 (OriGene Technologies, Rockville, MD); OVA-pcDNA3 (provided by Dr. V. Perez, University of Miami); pmax-GFP (Amaxa, Gaithersburg, MD), or as otherwise mentioned. Anesthetized mice received intradermal injection of 20–30 μL of DNA-plasmid in both left and right flank regions. The area of injection was then electroporated with each mouse being immunized with ∼20–40 μg of DNA. For in vivo electroporation, Derma Vax (Cellectis Bioscience, Glen Burnie, MD) was used. Needle rows of an IDE-4-4-2 electrode (4 mm gap) were inserted spanning the injection site.(9,17) Following insertion of the needles, a PulseAgile electroporation protocol consisting of 10 rectangular wave pulses (one pulse, 450 V, 50 μS duration, 0.2 mS pulse interval plus one pulse, 450 V, 50 μS duration, 50 mS pulse interval plus eight pulses, 110 V, 10 mS duration, and 20 mS pulse interval) was followed. The operated mice were warmed using a heating pad until they recovered from the anesthetic. The primary immunization was followed by a boost 2 weeks later. Sera from immunized mice were obtained 2 weeks after the boost and tested for the presence of antibodies. For the generation of monoclonal antibodies, the mouse with the highest titer was chosen for fusion. The fusion was performed 3–5 days after a final intravenous injection of 50,000 transfected Cos-7 cells.
Optimization of protein-free high throughput screening using in-cell Western
We used an in-cell Western assay with fixed, permeabilized, and transfected cells displaying the antigen of interest. Titer studies were performed 2 weeks post-boost using sera from immunized mice to test for the presence of specific antibodies against the antigen, which was encoded by the DNA. Cos-7 or HEK 293 cells were transfected with 20 μg of DNA-plasmid of interest using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) and seeded in 96-well plates. The same method was used for in-cell Western and IFA for fixation and permeabilization.(15,16,18) Twenty-four hours later, the transfected cells were fixed with 4% paraformaldehyde and blocked/permeabilized using 10% goat serum with 0.01% Triton-X in PBS. However, to perform IFA on vGPCR transfected HEK 293 cells, 0.2% Triton-X in PBS was used for 20 min at 4C°.(18) Fixed/permeabilized plates were washed thrice with PBS and a final wash with distilled water, air dried, and sealed with sealing tapes for 96-well plates (Thermo Scientific, Rockford, IL) and protected from light. Such plated fixed/permeabilized transfected cells could be stored at 4°C for at least 4 weeks.
The fixed cells were now subjected to sera from immunized mice or supernatants of the clones for 90 min at room temperature. One hundred μL of solution containing samples were added. For optimization studies human neural cell adhesion molecule L1 (HL1) encoding plasmids were used for the immunizations in which we have previously used an anti-HL1 MAb.(16) In these studies, mouse ascitic fluid prepared from hybridoma of anti-HL1 MAb was used as a positive control. Sera from immunized and naïve mice were used always at a starting dilution of 1:25 in PBS, unless otherwise mentioned. When supernatants from the hybridoma cell line were tested they were used neat as starting dilution. Two types of detection antibody were used. The first method was FLISA using the Odyssey Infrared Imaging System (LI-COR, Biosciences, Lincoln, NE). FLISA was performed by the addition of goat anti-mouse IRDye 800 (LI-COR Biosciences) as the secondary. The plate was read in channel 8 of an Odyssey Infrared Imager. Positive response was quantitated in terms of integrated intensity while a visual qualitative readout was based on observation of bright green circular spots. Averages of the integrated intensity values of groups of mice are shown for the 1:100 unless otherwise mentioned. The second method of detection was an IFA, which was performed by the addition of Alexa Fluor 546 goat anti-mouse IgG (Invitrogen) as the secondary antibody. IFA utilized a fluorescent microscope to view cell morphology. IFA would allow examination of the antigen cell localization in the in-cell Western assay.
Ninety-six well plates were plated with 20,000 Cos-7 cells expressing gene of interest, fixed/permeabilized, and used for HTS. One hundred μL of neat supernatant from harvested clones were added to each well. Goat anti-mouse IRDye 800 (LI-COR Biosciences) was used as the secondary antibody. Six plates were read simultaneously using Odyssey Infrared Imager (LI-COR Biosciences). The positive wells corresponding to its respective hybridoma clone were then selected for further screening and expansion. Fixed/permeabilized plates were stored at 4°C for a period of up to 4 weeks. Positive and negative sera were used to determine effects of storage for Cos-7 cells transfected with different antigens including viral G protein-coupled receptor (vGPCR), human cell adhesion molecule L1, and urokinase receptor (uPAR).
Fusion and clone harvest
The mouse with the highest titer was given a final boost. A fusion kit (Stemcell Technologies, Vancouver, Canada) was used. Spleen cells were mixed with SP2/0 myeloma cells and polyethylene glycol (PEG) mediated fusion was performed. In some cases an improved SP2/IL-6 expressing myeloma was used (ATCC CRL-2016).(19) The pellet was gently re-suspended in 1 mL of PEG (Stemcell Technologies) followed by drop-wise addition of 4 mL of medium B (Stemcell Technologies). Ten mL of medium B were added and the cells were incubated in 37°C for 15 min. After washing, the cells were resuspended in medium C (Stemcell Technologies) and incubated (at 37°C, 5% CO2) overnight. The cells were resuspended in medium D with HAT and methylcellulose (Stemcell Technologies), and the resulting cell suspension was distributed equally in ten 100-mm Petri dishes. The Petri dishes were incubated (at 37°C, 5% CO2) for 14 days, at the end of which only hybridoma clones survived. Clones were picked and transferred to six 96-well plates (one clone per well). Each well contained 250 μL of pre-warmed Medium E (Stemcell Technologies).(20) The 96-well plates were incubated (at 37°C, 5% CO2) for 4–5 days, after which the supernatant from each well were ready to be screened.
Statistical analysis
The Odyssey Infrared Imager provides the option of quantitating the output in terms of integrated intensity. A paired Student's T test was used to compare the integrated intensity values of mouse sera in experimental wells (Cos-7 expressing antigen of interest) vs. control wells, including Cos-7 expressing irrelevant antigen. Differences were considered statistically significant at p values less than 0.01.
Results
We have employed gene-based vaccine delivery technology via in vivo electroporation. This method is known for its ability to deliver genes in vivo for the expression of the protein in the host and for the induction of a transient ”danger signal” in the electroporated area that in turn results in the expression of a milieu of cytokines inviting the involvement of innate immune responses.(21)
Mice were immunized with plasmid-DNA expressing an antigen of interest using local electroporation. Prior to immunization sera from naïve mice were pooled for control purposes. The mice were boosted 2 weeks after the first immunization. Sera from mice were collected for titer study 12–14 days post-boost.
Plasmids containing transgenes inserted under the control of a eukaryotic promoter resulted in protein (antigen) expression in mammalian cells in vitro or in vivo. An important consideration when optimizing the efficacy of DNA vaccines is the appropriate choice of plasmid vector. The plasmid DNA backbone should contain a eukaryotic promoter, a cloning site, a polyadenylation (polyA) sequence, a selectable marker, and a bacterial origin of replication. We have used plasmids with a viral-derived promoter (e.g., cytomegalovirus [CMV]). The cloning site after the promoter enables the insertion of intended genes of antigens. The polyA sequence results in the stabilization of mRNA transcripts. A selectable marker such as an ampicillin resistance gene is normally used to prevent contamination when the plasmid is amplified. To ensure a high plasmid yield, such plasmids contain an origin of replication, such as Escherichia coli ColE1.
Protein expression in host using in vivo electroporation
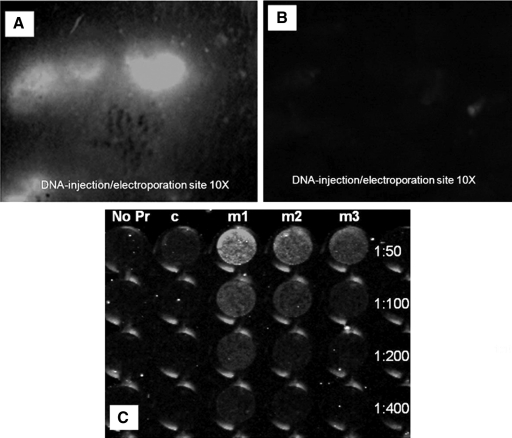
Mice were immunized via in vivo electroporation with DNA-plasmid pmax-GFP (Amaxa, Gaithersburg, MD) encoding for green fluorescence protein (GFP). Anesthetized mice were administered with 20 μg GFP plasmid intradermally after which the injection site was electroporated. Twenty-four hours later the electroporated site and its neighboring area were viewed using a stereo-fluorescence microscope. The results showed GFP expression in the electroporated region of the skin surface (Fig. 1A,B). However there was little to no expression of GFP in the area immediately outside of the electroporated region (Fig. 1C). This confirmed that the GFP plasmid was taken up by the cells after electroporation and was used by cell machinery to translate the protein resulting in the expression of the protein. The same mice showed detectable titers of anti-GFP antibodies, on day 22 after a single in vivo electroporation at the site of an intradermal injection with 20 μg GFP plasmid when followed by electroporation (Fig. 1D).
FIG. 1.
(A) Evidence of in vivo protein expression by in vivo electroporation. In vivo transduction of mouse skin 24 h post-intradermal delivery of GFP DNA followed by electroporation using Derma Vax. Stereo fluorescence microscope imaging on live mice shows electroporated area of skin expressing GFP at objective 10×. (B) Area of control skin injected by GFP plasmid immediately outside of the electroporated region. (C) FLISA image performed on pmax-GFP transfected Cos-7 cells with sera of immunized mice. No Pr, no primary antibodies added. Pooled sera from mice collected from mice immunized with DNA alone. m1-m3, sera from 3 different mice post-immunization. p < 0.01, when titers of electroporated mice are compared with those of controls.
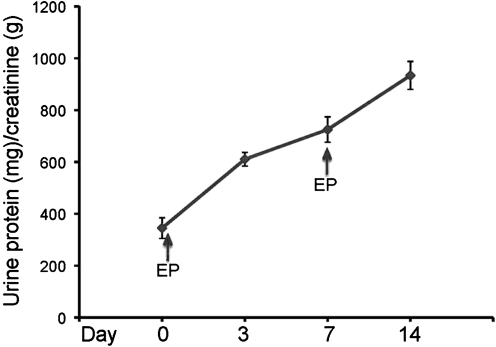
To further demonstrate the in vivo expression of delivered genes, we employed a novel strategy using plasmid encoding for uPAR. This receptor normally has low tissue expression, but under stress, injury, and inflammation, uPAR expression increases.(22) Its expression in kidneys dictates progressive glomerular disease.(22) Two in vivo electroporations, on days 0 and 7, each after 40 μg uPAR plasmid was intradermally injected, were sufficient to result in proteinuria as measured in the urine specimens from treated mice (Fig. 2) and not in control mice. Albuminuria levels (μg/dL) normalized by creatininuria levels (mg/dL) in the urine of male mice at the age of 5–9 months are shown in Figure 2. In brief, under anesthesia, mice were injected with uPAR plasmid (40 (g in PBS) intradermally and bilaterally at the hind leg, followed by in vivo electroporation with Derma Vax DNA delivery system (Cellectis Bioresearch). Blood and urine were collected before and after each gene delivery for analysis. The method may therefore be used as a transient transgene expression of uPAR resulting in an insult to kidney.
FIG. 2.
uPAR in vivo gene delivery via electroporation. Long-term overexpression of mouse uPAR was achieved in wild-type C57BL/6 mice by in vivo gene delivery accompanied with electroporation. Electroporation was performed after plasmid harboring uPAR was injected intradermally in the flank of the mice. Two plasmid injections on days 0 and 7 followed by in vivo electroporation were performed. In vivo mouse uPAR expression is indirectly shown by the persistent proteinuria in wild-type C57BL/6 mice. Albuminuria levels (μg/dL) normalized by creatininuria levels (mg/dL) were tested in male mice. In brief, under anesthesia, mouse uPAR plasmid (40 μg in PBS) was injected intradermally and bilaterally at the hind leg of the mice, followed by in vivo electroporation with Derma Vax DNA delivery system.
Optimization of in-cell Western method using proteins with various localizations
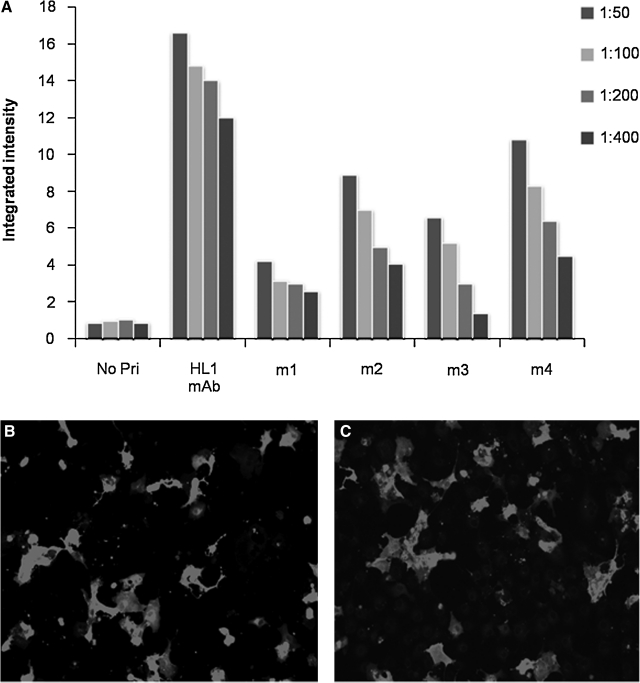
To optimize the in-cell Western method as a screening method for immunizations, mice were immunized with human L1 encoding plasmid (HL1-pcDNA3) and titers were compared to our described anti-HL1 monoclonal antibody.(16) HL1 is a previously described transmembrane protein and a neuronal cell adhesion molecule.(16) An example of transmembrane proteins was selected as such proteins show common purification challenges. Fixed/permeabilized Cos-7 cells expressing HL1-pcDNA3 were plated in 96-well plates. The best fixation and permeabilization condition was achieved using 4% paraformaldehyde and 0.01% Triton-X respectively (data not shown). The plates were then air dried and stored at 4°C. Titer studies were carried out at 1 week, 2 weeks, and 4 weeks post-storage. The resultant titer studies showed no significant difference in integrated intensity values or staining patterns (data not shown). Figure 3A shows the integrated intensity of the infrared signals of in-cell Western images using these 96-well plates. Sera from three of four immunized mice demonstrated significant anti-HL1 titers. The positive control was mouse ascitic fluid containing known anti-HL1 antibodies. The positive output confirms the efficacy of the in-cell Western method. The positive control was compared to sera from immunized mice. One of the immunized mice (m4) with the highest anti-HL1 titer in FLISA showed a similar binding pattern to that of the positive control (Fig. 3A). This is important as the positive anti-HL1 clone was prepared from hybridoma cultured in ascites that has concentrations much higher than that normally found in the sera of immunized mice.(23) As a negative control routinely, cells expressing an irrelevant protein were used to assess the level of antibodies against the plasmid backbone. In this case, mitogen-activated protein kinase (MKK7) transfected Cos-7 was used. Cos-7 cells were transfected with plasmid MKK7-pcDNA3, then plated in 96-well plates and fixed/permeabilized. MKK7(24) is a cytoplasmic protein and was chosen primarily because it has the same plasmid backbone (pcDNA3) as HL1. The reasoning behind using a different antigen encoded in the same backbone was to account for the degree of immunization efficacy against the backbone itself. The MKK7 transfected Cos-7 cells resulted in a very low background (integrated intensity <0.25 ± 0.13), thus confirming the effectiveness of in vivo electroporation. To further validate the immunization method, IFA staining was performed using secondary antibody Alexa Fluor 546. Figure 3C shows the staining pattern of positive control (ascitic fluid with known anti-HL1 antibody), and Figure 3D shows a very similar staining pattern using sera from m4 positive mouse. Both staining patterns show the antibody binding to HL1 protein that is scattered throughout the cell membrane.
FIG. 3.
Optimization of screening using known anti-HL1 MAb as positive control. (A) The integrated intensity values obtained from FLISA (in-cell Western) on Cos-7 cells transfected with HL1-pcDNA (the antigen). Negative transfected cells, Mkk7-pcDNA as an irrelevant antigen, with integrated intensity values of <0.25 ± 0.13 (not shown) where both plasmids had the same backbone, pcDNA.3. No Pr, no primary antibodies added, only secondary antibodies added; HL1 MAb, positive control, ascitic fluid containing known anti-HL1 antibody; m1-m4, sera from 4 different mice post-immunization. The p values when comparing Mkk7 and HL1 transfected Cos-7 cells was <0.01. (B) IFA staining of HL1-pcDNA3 transfected Cos-7 cells with ascitic fluid containing known anti-HL1 MAb. (C) IFA staining of HL1-pcDNA3 transfected COS-7 cells with the serum of mouse (m4).
Monoclonal antibody generation against novel genes using semi high throughput screen
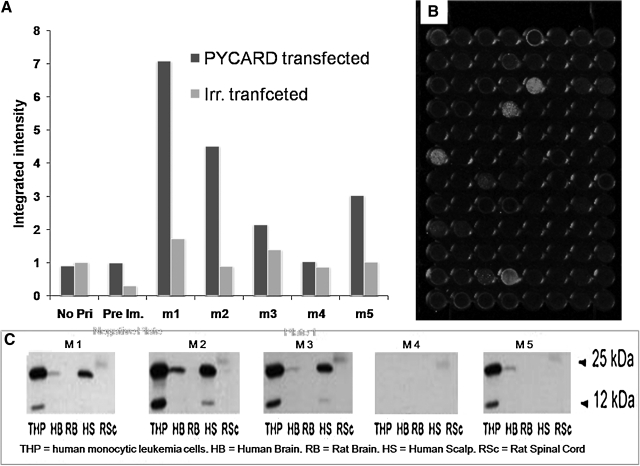
The next example is the protein encoded by PYCARD variant 1, apoptosis-associated speck-like protein with CARD domain also called ASC. This PYCARD protein appears to have cytoplasmic localization and serves as an adaptor protein in the inflammasomes.(25) Mice received two immunizations with 40 μg of mammalian plasmid encoding PYCARD, 2 weeks apart. An in-cell Western using transfected cells expressing the antigens was used to determine the antibody titers. Figure 4A shows the humoral response of PYCARD v1 (OriGene Technologies, Rockville, MD) plasmid immunized mice reacting with Cos-7 cells expressing the protein versus control Cos-7 cells. While the control serum shows weak response to both plates, four of the mice show a strong immune response against PYCARD v1 but are weak against a different antigen with the same backbone (MYC-pCMV, kindly provided by Dr. Vance Lemmon, University of Miami). The sera from immunized mice were also tested in Western blots against various cell and tissue samples expressing PYCARD v1 in its native form. Figure 4C shows the Western blots in which four of the mice show binding affinity to PYCARD v1 expressions in various tissues expressing the protein and thus confirming successful vaccination. Mouse m2 was chosen for the final boost and, as was described in the Methods section, transfected cells were administered. Four days after final boost, the mouse was sacrificed and fusion was performed. Fourteen days post-fusion, individual hybridoma colonies were picked and transferred to individual wells of 96-well plate containing ∼250 μL of Medium E. Five days later the supernatant of the harvested clones was enriched and ready for semi-HTS. Six 96-well plates (screening plates) with Cos-7 cells expressing PYCARD v1 were prepared, fixed, and permeabilized. Supernatant from the harvested clones was added to the screening plates. FLISA was performed for six plates and they were read at the same time using the Odyssey Infrared Imager. The wells in one of the six plates expressing positive signal are shown in the Figure 4B. Positive wells were correlated to its source hybridoma well, which were then chosen for further expansion. The expanded positive hybridoma cell lines (15 clones) from the primary screening were further examined in Western blot analysis. Western blot was performed for positive clones similar to the Figure 4C (not shown), and four clones that reacted with human tissues/cell lines(26,27) were selected. In vivo binding and biological activity of the selected anti-PYCARD clones are promising features and will be reported on in the future upon further confirmation (data not shown).
FIG. 4.
Generation of anti-PCARDv1 MAbs. (A) The FLISA (in-cell Western) results of PCARDv-1 on transfected Cos-7 cells with sera of mice immunized with plasmid harboring PCARDv1 and control cells (Irr. transfected), cells transfected with irrelevant plasmid Mns1 (meiosis-specific nuclear structural encoding pcDNA plasmid). No Pri, no primary antibodies added, only secondary antibodies added; Pre Im, the pooled sera of mice collected prior immunizations; m1–m5, sera from 5 different immunized mice. The p values of titers of m1, m2, m3, and m5 when compared to cells transfected with irrelevant plasmid were <0.01. (B) FLISA image of transfected Cos-7 cells expressing PCARDv1 plated in one representative 96-well plate showing positive clones. Undiluted supernatants from harvested hybridoma cell lines were used to screen for clones producing anti-PCARDv-1 antibodies. (C) Western blot image of anti-PCARDv1 antibody activity against various cell and tissue samples expressing PCARDv1. Fifteen positive clones resulted in this screen.
Induction of humoral response using plasmids encoding for transmembrane and perinuclear and cytoplasmic antigens
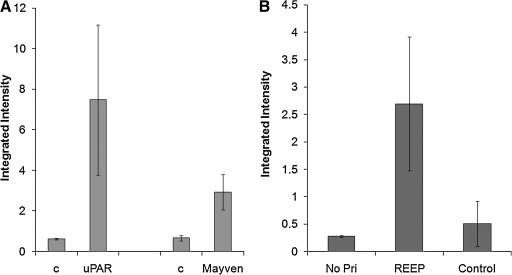
Figure 5 shows the titer results of mice vaccinated against various DNA-plasmids. Figure 5A and B shows the humoral responses of mice vaccinated against plasmid encoding murine uPAR (muPAR), Mayven, and REEP1.(28) Mayven (also called kelch like protein 2 [KLHL2]) has a cytoplasmic localization and is an actin-associated protein.(28) uPAR and murine uPAR are glycolipid anchored cell surface glycoproteins that play a role in tissue remodeling processes and cancer invasion.(29) REEP1 is a transmembrane protein residing in mitochondria and the endoplasmic reticulum.(30,31) The integrated intensities of mouse titers against REEP1 were compared side by side with antibody response against cells transfected with an irrelevant antigen at the same dilution. The binding activities of sera from immunized mice with such control transfected cells were significantly lower than those immunized mice. However, such backgrounds vary, which may be due to the different backbones. Mice immunized with uPAR and Mayven showed higher integrated intensity values than controls (pooled pre-immune sera), as are shown in Figure 5A. Similarly, all five mice had a statistically significant immune response against REEP1. As shown in Figure 5B, the integrated intensity values of titers from mice immunized with REEP1 are significantly higher than those of negative control, cells transfected with an irrelevant plasmid.
FIG. 5.
Mounting antibody responses against cytosolic, transmembrane, and receptor target antigens. (A) Units of integrated intensity of FLISA on uPAR transfected 293T cells and Mayven transfected Cos-7 with sera of mice immunized. FLISA (in-cell Western) was performed and the units of integrated intensity at channel 8 of an Odyssey Infrared Imager were graphed. (B) Serum titers (1:100 dilution) are shown as integrated intensity of titers of sera from mice immunized with plasmid REEP1 obtained in a FLISA (in-cell Western) assay compared with cells transfected with an irrelevant plasmid (control). Averages and standard deviations for five (uPAR and REEP) or three (Mayven) immunized mice of each group are shown. No Pr, no primary antibodies added, only secondary antibodies added. c: Pooled sera from mice collected prior to immunization.
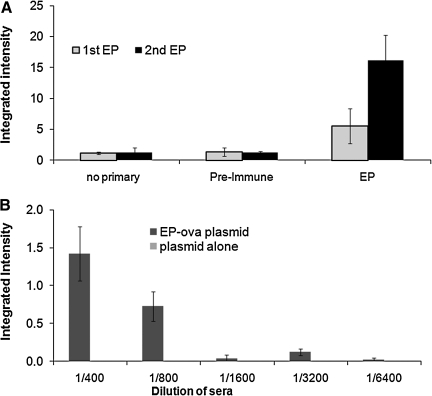
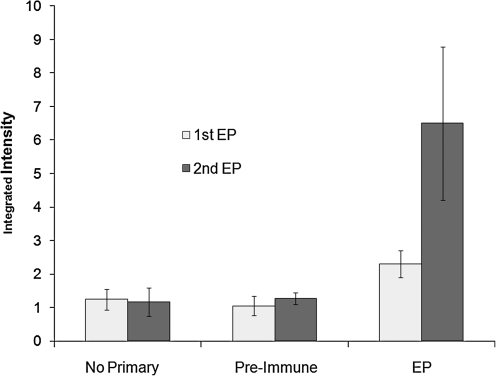
The humoral immune response resulting from a single or booster immunization are shown in Figure 6A. Mice vaccinated with plasmid OVA-pcDNA3 (encoding for ovalbumin) followed by electroporation results in significant anti-OVA titers that is noticeably enhanced upon booster immunization/electroporation (data for the serum dilution of 1:100 are shown). The increase in integrated intensity values for each mouse confirms the increase in antibody production against OVA-pcDNA3 post-boost. Figure 6B shows the integrated intensity values of mice upon a sole immunization followed by electroporation and compared with vaccination with DNA alone. In this experiment similar amounts of DNA harboring OVA gene alone were injected as a control, which resulted in no detectable titers. Similarly, Figure 7 shows that booster immunization was needed to achieve significant titers for this antigen; in this case the plasmids encoding for membrane proteins such as viral G protein-coupled receptor (vGPCR) was used. Indeed, augmenting humoral responses against vGPCR as a membrane protein is important and has been challenging.(32) As shown in Figure 7, when cells transfected with vGPCR were used, pooled pre-immune sera show higher background compared to a control plasmid construct used. It was concluded that it is helpful to use pre-immune serum from individual mice. However, all three immunized mice showed detectable titers upon a single immunization with vGPCR plasmid followed by in vivo electroporation. The titers were significantly enhanced after a boost (a second DNA immunization followed by electroporation) 2 weeks after the first immunization (Fig. 7). Moreover, two vaccinations followed by electroporation using plasmid encoding for viral interferon (IFN) regulatory factor 1 (vIRF) that is most frequently localized in the nucleus(33) resulted in positive (p value < 0.017) titers of 7.3 ± 1.9 (integrated intensity) of anti-vIRF antibodies versus 4.8 ± 0.27 in mice immunized with DNA alone.
FIG. 6.
The minimum immunization requirements. (A) Units of integrated intensity of FLISA on OVA-pcDNA3 transfected Cos-7 cells with sera of mice immunized (via in vivo electroporation) with plasmid OVA-pcDNA3 upon first immunization. (B) The integrated intensity titers after two immunizations (a boost) compared with integrated intensity titers of sera from mice immunized with DNA alone. Averages and standard deviations for three immunized mice of each group are shown. Average integrated intensity values of the 1:100 sera dilution are shown. No Primary, no primary antibodies added, only secondary antibodies added; Pre-immune, pooled sera from mice collected prior to immunization; EP, immunization via in vivo electroporation. When compared to controls, the p values of m1 and m2 titers were <0.01.
FIG. 7.
Mounting antibody responses against vGPCR, which is a membrane protein, via in vivo electroporation. Low antibody responses against vGPCR was achieved by a single DNA immunization followed by in vivo electroporation that was augmented upon a boost. The plasmid encoding for vGPCR (20 μg in PBS) was injected intradermally followed by in vivo electroporation on the injection site. Average integrated intensity values of the 1:100 sera dilution are shown. Titers were assessed as discussed in Figure 6. Integrated intensity values for five immunized mice of each group are shown as averages and standard deviations. EP, DNA immunization followed by in vivo electroporation.
Alternative screening methods or further improvement of the screening method described here is needed to more effectively screen for antibodies reacting with conformational epitopes.
Discussion
Gene-based immunizations using non-viral methods have long been used for the induction of immune responses. DNA immunization shows promise in mounting immune responses against targets where protein-based or peptide-based immunizations become challenging and costly.(34) Like the cell membrane, nucleic acids are negatively charged and have poor cellular uptake per se. In vivo electroporation not only facilitates the DNA entry into cells, it also has a built-in adjuvant activity. The plasmids have their own immune-enhancing sequences such as CpG motifs that act as an adjuvant. The method provides an alternative in that the plasmids encoding for the antigen will be delivered to the host and the naturally processed antigens will be expressed using the host cell machinery. DNA vaccines can activate arms of the immune system that protein vaccines cannot. Indeed, they may work in both therapeutic and preventative vaccine. Already a few DNA-based veterinary vaccines have been marketed including Apex-IHN made by Novartis (Aqua Health) and a DNA vaccine for infectious hematopoietic necrosis virus (IHNV) in farm-raised Atlantic salmon, approved by Health Canada.(35)
DNA vaccines take advantage of a low cost, safety, and relatively simplicity. Plasmids encoding the majority of proteins are obtainable or may be purchased with low cost. They can be easily propagated with low cost. The method also offers the flexibility of using tailored portions of a gene, such as mutated and/or hot spots, that include more T or B cell epitopes including CD4 T helper epitopes. However, one would still need purified antigens (proteins) for assessment of the humoral responses in the sera of vaccinated animals for screening for antibody titers, especially when making monoclonal antibodies. Here we show the feasibility of using non-viral gene-based vaccinations to mount humoral immune responses in mice. In addition, we show the screening of the sera of vaccinated mice or supernatants of the hybridoma clones using an in-cell Western followed by FLISA or IFA. FLISA is used predominantly for semi HT screening. However in some cases inserted genes in plasmids may be toxic to cells or may induce apoptosis leading to background and hindrance of the qualitative binding assessment due to non-specific antibody attachment. IFA was alternatively used for further qualitative assessment of immunocytostaining, verifying known localization of the antigen of interest.
In summary, we have optimized the “protein free” screening method in which we plate transfected Cos-7 cells in 96-well plates and compare it to parallel plates of Cos-7 cells transfected with irrelevant antigen-encoded plasmids with same backbone. Via the optimized method (EP and in-cell Western) described here, (1) we show in vivo heterologous expression, (2) we were able to induce humoral response against each of the targets for at least a subset of immunized mice (localized in different subcompartments), and 3) we described MAbs for one target. We have optimized a process including in vivo electroporation, and in-cell Western using cells expressing the antigen to measure such humoral responses. A series of factors dictating the outcome include the levels of expression of the plasmid vector, the plasmid size, the localization of the protein, and the degree of tolerance. The method, in particular, is ideal for the generation of antibodies that interact with native form of the protein or mild fixation. For example, in some cases the reactive sera or clones were obtained in FLISA or IFA where a 2–4% paraformaldehyde was used for fixation; however, they did not react in Western blot analyses. We observed that levels of background staining vary for various plasmids encoding for different proteins. Some genes initiate expression of other downstream genes; the plasmid also may result in some non-specific immune responses against itself. Therefore the screening must be carefully planned by using multiple controls to discriminate the right clones. Additional methods of screening such as Western blot or functional assays for the selected clones are helpful to choose the highest value clones.
In vivo electroporation clearly could elicit antibody responses against antigens when plasmids encoding antigens express in various cell compartments, including membrane protein or those in cytoplasm. Tuning factors, such as efficacy of the plasmid expression, plasmid/gene toxicity, and cell localization of the encoded antigen, can further improve the outcome of the gene-based vaccine efficacy.
Acknowledgments
We would like to thank Dr. Norma Kenyon and the Wallace H. Coulter Center for Translational Research for supporting this work, Cellectics Bioresearch for providing the Derma Vax, and Dr. Eckhard R. Podack for critical review of the manuscript. This study was also supported in part by the NINDS (R01NS054132 to SZ) and 5RO1HD057632 to VPL. VPL holds the Walter G. Ross Distinguished Chair in Developmental Science.
Author Disclosure Statement
The authors have no financial interests to disclose.
References
- 1.Reichert J. Pavolu A. Monoclonal antibodies market. Nat Rev Drug Discov. 2004;3:383–384. doi: 10.1038/nrd1386. [DOI] [PubMed] [Google Scholar]
- 2.Walls D. McGrath R. Loughran ST. A digest of protein purification. [review]. Methods Mol Biol. 2011;681:3–23. doi: 10.1007/978-1-60761-913-0_1. [DOI] [PubMed] [Google Scholar]
- 3.Burgess RR. Refolding solubilized inclusion body proteins. In: Burgess RR, editor; Deutscher MP, editor. Guide to Protein Purification. 2nd. Academic Press; San Diego: 2009. p. 463. [DOI] [PubMed] [Google Scholar]
- 4.Lin S-H. Guidotti G. Purification of membrane proteins. In: Burgess RR, editor; Deutscher MP, editor. Guide to Protein Purification. 2nd. Academic Press; San Diego: 2009. p. 463. [Google Scholar]
- 5.Wei CJ. Boyington JC. McTamney PM. Kong WP. Pearce MB. Xu L. Andersen H. Rao S. Tumpey TM. Yang ZY. Nabel GJ. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science. 5995;329:1060–1064. doi: 10.1126/science.1192517. [DOI] [PubMed] [Google Scholar]
- 6.Andre F. Mir L. DNA electrotransfer: its principles and an updated review of its therapeutic applications [review] Gene Ther. 2004:11. doi: 10.1038/sj.gt.3302367. [DOI] [PubMed] [Google Scholar]
- 7.Cantelli CP. Teixeira M. da Gloria Martins. Santos EA. Silva HC da. Mouta S. da Silva E. Pimenta MM. Vianna CO. de Souza NP. Batoreu NM. Galler R. de Moraes MT. Generation of monoclonal antibodies against human recombinant interferon beta using genetic immunization with simultaneous expression of IgM and IgG isotypes. Hybridoma. 2009;28:211–214. doi: 10.1089/hyb.2008.0094. [DOI] [PubMed] [Google Scholar]
- 8.Chua KY. Ramos JD. Cheong N. Production of monoclonal antibody by DNA immunization with electroporation. Methods Mol Biol. 2008;423:509–520. doi: 10.1007/978-1-59745-194-9_40. [DOI] [PubMed] [Google Scholar]
- 9.Ahmad S. Casey G. Sweeney P. Tangney M. O'Sullivan G. Optimised electroporation mediated DNA vaccination for treatment of prostate cancer. Genet Vacc Ther. 2010;8:1. doi: 10.1186/1479-0556-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong W. Hood C. Yang Z. Wei C-J. Xu L. García-Sastre A. Tumpey TM. Nabel GJ. Protective immunity to lethal challenge of the 1918 pandemic influenza virus by vaccination. Proc Natl Acad Sci. 2006;103:15987–15991. doi: 10.1073/pnas.0607564103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu TH. Yazdanbakhsh K. Oyen R. Smart E. Reid EM. Production and characterization of anti-kell monoclonal antibodies using transfected cells as the immunogen. Br J Haematol. 1999;106:817–823. doi: 10.1046/j.1365-2141.1999.01599.x. [DOI] [PubMed] [Google Scholar]
- 12.Buchan S. Gronevik E. Mathiesen I. King CA. Stevenson FK. Rice J. Electroporation as a “prime/boost” strategy for naked DNA vaccination against a tumor antigen. J Immunol. 6292;174:6292–6298. doi: 10.4049/jimmunol.174.10.6292. [DOI] [PubMed] [Google Scholar]
- 13.Bates MK. Zhang G. Sebestyen MG. Neal ZC. Wolff JA. Herweijer H. Genetic immunization for antibody generation in research animals by intravenous delivery of plasmid DNA. Biotechniques. 2006;40:199–208. doi: 10.2144/000112088. [DOI] [PubMed] [Google Scholar]
- 14.Moeller S. Jung C. Adriouch S. Dubberke G. Seyfried F. Seman M. Haag F. Koch-Nolte F. Monitoring the expression of purinoceptors and nucleotide-metabolizing ecto-enzymes with antibodies directed against proteins in native conformation. Purinergic Signal. 2007;3:359–366. doi: 10.1007/s11302-007-9084-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selkirk JV. Nottebaum LM. Ford IC. Santos M. Malany S. Foster AC. Lechner SM. A novel cell-based assay for G-protein-coupled receptor-mediated cyclic adenosine monophosphate response element binding protein phosphorylation. J Biomol Screen. 2006;11:351–358. doi: 10.1177/1087057106286608. [DOI] [PubMed] [Google Scholar]
- 16.Hlavin ML. Lemmon V. Molecular structure and functional testing of human L1CAM: an interspecies comparison. Genomics. 1991;11:416–423. doi: 10.1016/0888-7543(91)90150-d. [DOI] [PubMed] [Google Scholar]
- 17.Roos AK. Eriksson F. Timmons JA. Gerhardt J. Nyman U. Gudmundsdotter L. Brave A. Wahren B. Pisa P. Skin electroporation: effects on transgene expression, DNA persistence and local tissue environment. PLoS One. 7226:4. doi: 10.1371/journal.pone.0007226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng H. Sun Z. Farzan MR. Feng P. Sulfotyrosines of the Kaposi's sarcoma-associated herpesvirus G protein-coupled receptor promote tumorigenesis through autocrine activation. J Virol. 3351;84:3351–3361. doi: 10.1128/JVI.01939-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris FJ. Hawley GR. Hawley ST. Crawford-Sharpe CG. Increased frequency of both total and specific monoclonal antibody producing hybridomas using a fusion partner that constitutively expresses recombinant Il-6. J Immunol Methods. 1992;148:199–207. doi: 10.1016/0022-1759(92)90173-q. [DOI] [PubMed] [Google Scholar]
- 20.Chua KY. Ramos JD. Cheong N. Production of monoclonal antibody by DNA immunization with electroporation. Methods Mol Biol. 2008;423:509–520. doi: 10.1007/978-1-59745-194-9_40. [DOI] [PubMed] [Google Scholar]
- 21.Chiarella P. Massi E. De Robertis M. Sibilio A. Parrella P. Fazio VM. Signori E. Electroporation of skeletal muscle induces danger signal release and antigen-presenting cell recruitment independently of DNA vaccine administration. Expert Opin Biol Therapy. 1645;8:1645–1657. doi: 10.1517/14712598.8.11.1645. [DOI] [PubMed] [Google Scholar]
- 22.Wei C. Moller CC. Altintas AA. Li L. Schwarz K. Zacchigna S. Xie L. Henger A. Schmid H. Rastaldi MP. Cowan P. Kretzler M. Parrilla K. Bendayan M. Gupta V. Nikolic B. Kalluri R. Carmeliet P. Mundel P. Reiser J. Modification of kidney barrier function by the urokinase receptor. Nat Med. 2008;14:55–63. doi: 10.1038/nm1696. [DOI] [PubMed] [Google Scholar]
- 23.Hendriksen CF. de Leeuw W. Production of monoclonal antibodies by the ascites method in laboratory animals [review] Res Immunol. 1998;149:535–542. [PubMed] [Google Scholar]
- 24.Holloway JN. Murthy S. El-Ashry D. A cytoplasmic substrate of mitogen-activated protein kinase is responsible for estrogen receptor-alpha down-regulation in breast cancer cells: the role of nuclear factor-kappaB. Mol Endocrinol. 2004;18:1396–1410. doi: 10.1210/me.2004-0048. [DOI] [PubMed] [Google Scholar]
- 25.Cheng J. Waite AL. Tkaczyk ER. Ke K. Richards N. Hunt AJ. Gumucio DL. Kinetic properties of ASC protein aggregation in epithelial cells. J Cell Physiol. 2010;222:738–747. doi: 10.1002/jcp.22005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin Y. Yan Y. Jiang X. Mai J. Chen NC. Wang H. Yang XF. Inflammasomes are differentially expressed in cardiovascular and other tissues. Intl J Immunopathol Pharmacol. 2009;22:311–322. doi: 10.1177/039463200902200208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tekippe EM. Allen IC. Hulseberg PD. Sullivan JT. McCann JR. Sandor M. Braunstein M. Ting JP. Granuloma formation and host defense in chronic Mycobacterium tuberculosis infection requires PYCARD/ASC but not NLRP3 or caspase-1. PLoS One. 2010:5. doi: 10.1371/journal.pone.0012320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montague P. Kennedy PG. Barnett SC. Subcellular localization of Mayven following expression of wild type and mutant EGFP tagged cDNAs. BMC Neurosci. 2010:11. doi: 10.1186/1471-2202-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei C. Moller CC. Altintas MM. Li J. Schwarz K. Zacchigna S. Xie L. Henger A. Schmid H. Rastaldi MP. Cowan P. Kretzler M. Parrilla R. Bendayan M. Gupta V. Nikolic B. Kalluri R. Carmeliet P. Mundel P. Reiser J. Modification of kidney barrier function by the urokinase receptor. Nat Med. 2008;14:55–63. doi: 10.1038/nm1696. [DOI] [PubMed] [Google Scholar]
- 30.Zuchner S. Wang G. Tran-Viet KN. Nance MA. Gaskell PC. Vance JM. Ashley-Koch AE. Pericak-Vance MA. Mutations in the novel mitochondrial protein REEP1 cause hereditary spastic paraplegia type 31. Am J Hum Genetics. 2006;79:365–369. doi: 10.1086/505361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park SH. Zhu PP. Parker RL. Blackstone C. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J Clin Invest. 1097;120:1097–1110. doi: 10.1172/JCI40979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masuko T. Ohno Y. Masuko K. Yagi H. Uejima S. Takechi M. Hashimoto Y. Towards therapeutic antibodies to membrane oncoproteins by a robust strategy using rats immunized with transfectants expressing target molecules fused to green fluorescent protein [review] Cancer Sci. 2011;102:25–35. doi: 10.1111/j.1349-7006.2010.01741.x. [DOI] [PubMed] [Google Scholar]
- 33.Pozharskaya VP. Weakland LL. Zimring JC. Krug LT. Unger ER. Neisch A. Joshi H. Inoue N. Offermann MK. Short duration of elevated vIRF-1 expression during lytic replication of human herpesvirus 8 limits its ability to block antiviral responses induced by alpha interferon in BCBL-1 cells. J Virol. 2004;78:6621–6635. doi: 10.1128/JVI.78.12.6621-6635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howarth M. Elliott T. The processing of antigens delivered as DNA vaccines. Immunol Rev. 2004;199:27–39. doi: 10.1111/j.0105-2896.2004.00141.x. [DOI] [PubMed] [Google Scholar]
- 35.Cai Y. Rodriguez R. Hebel H. DNA vaccine manufacture: scale and quality [review] Expert Rev Vacc. 1277;8:1277–1291. doi: 10.1586/erv.09.84. [DOI] [PubMed] [Google Scholar]