Abstract
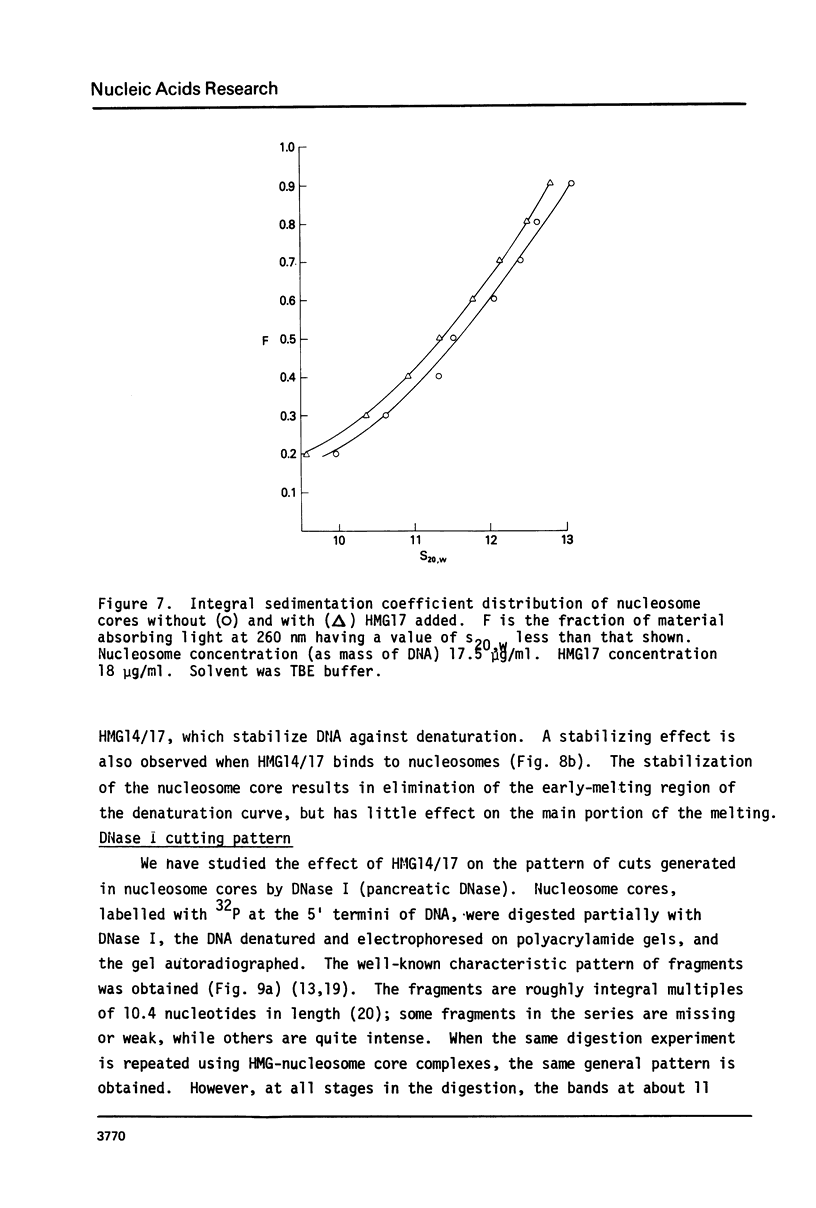
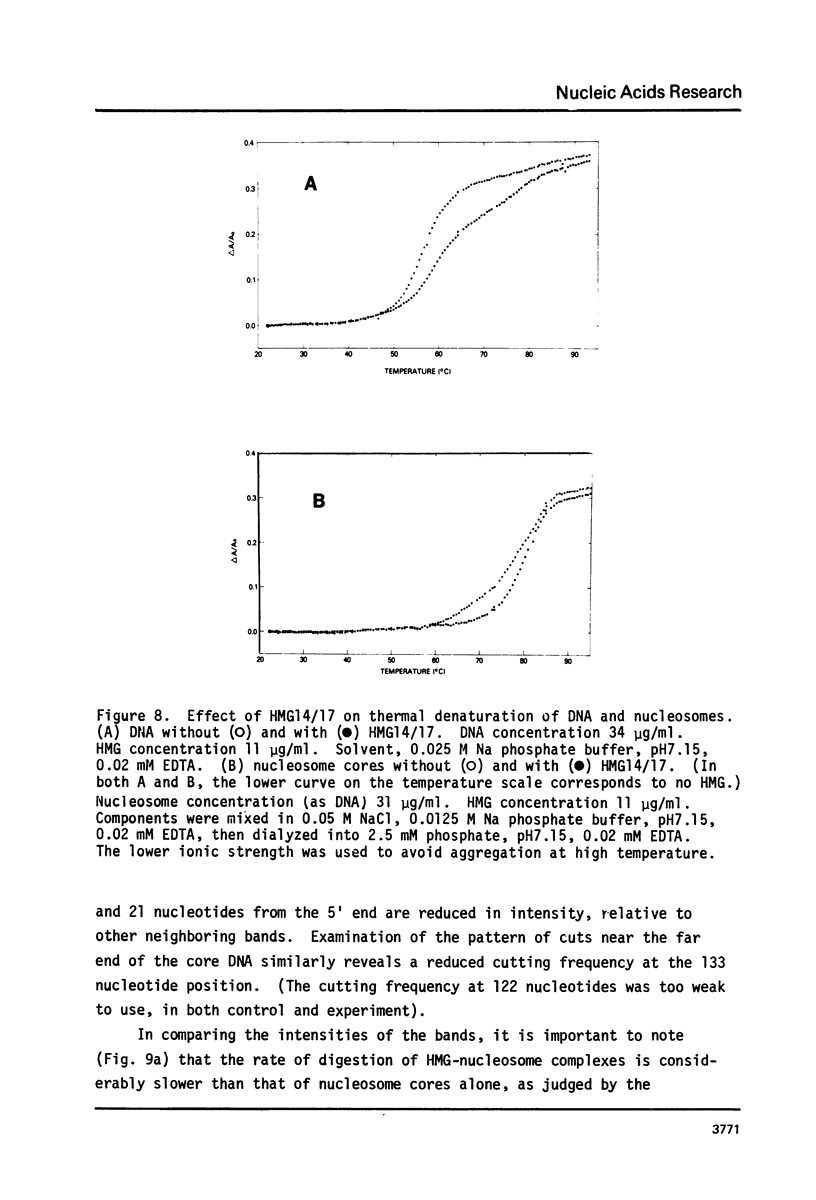
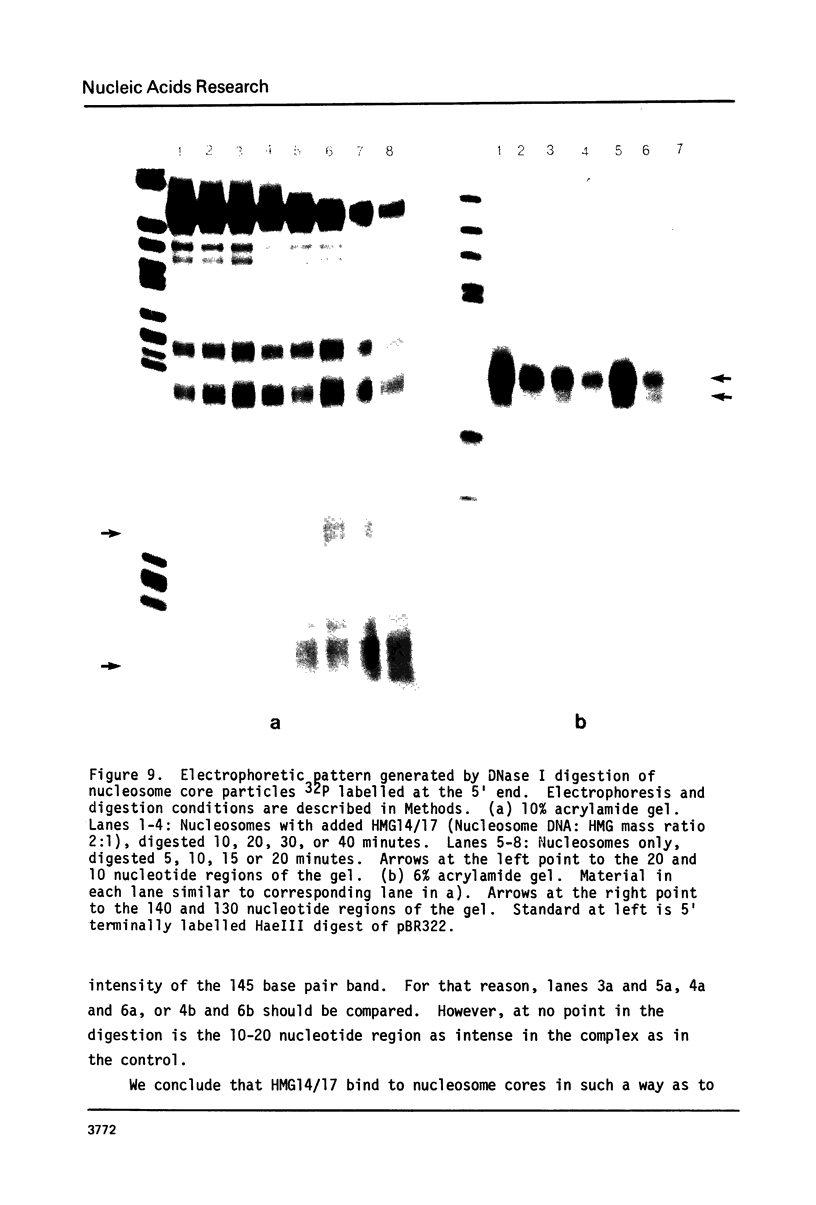
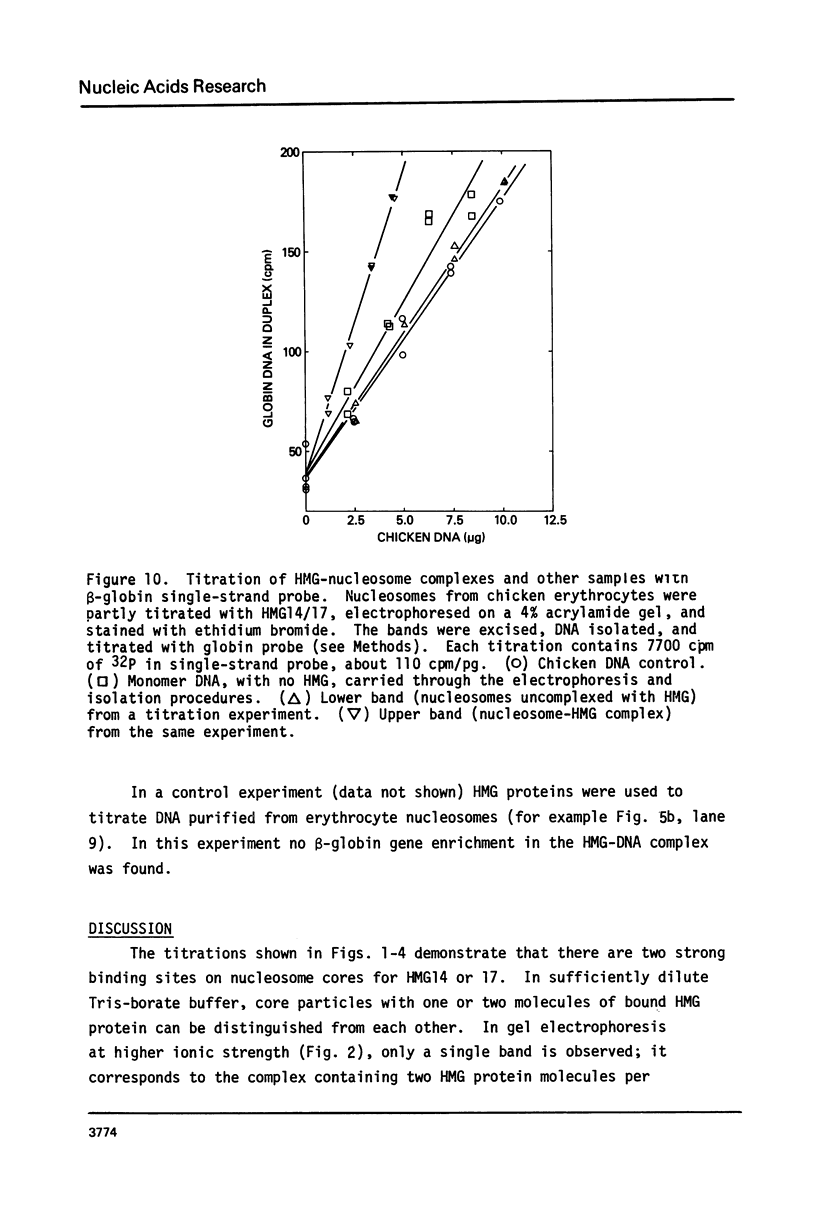
The interaction of the high mobility group proteins, HMG14 and HMG17, with nucleosome core particles has been studied. The results show that two molecules of HMG14/17 can be bound tightly but reversibly to each core particle and that their affinity for core particles is greater than their affinity for histone-free DNA of core size. Thermal denaturation and nuclease digestion studies suggest that major sites of interaction are located near the ends of the nucleosome core DNA. When nucleosome preparations from chicken erythrocyte nuclei stripped of HMG proteins are partially titrated with HMG14/17, the nucleosome-HMG complex fraction is enriched in beta-globin gene sequences.
Full text
PDF














Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abercrombie B. D., Kneale G. G., Crane-Robinson C., Bradbury E. M., Goodwin G. H., Walker J. M., Johns E. W. Studies on the conformational properties of the high-mobility-group chromosomal protein HMG 17 and its interaction with DNA. Eur J Biochem. 1978 Mar;84(1):173–177. doi: 10.1111/j.1432-1033.1978.tb12154.x. [DOI] [PubMed] [Google Scholar]
- Albright S. C., Wiseman J. M., Lange R. A., Garrard W. T. Subunit structures of different electrophoretic forms of nucleosomes. J Biol Chem. 1980 Apr 25;255(8):3673–3684. [PubMed] [Google Scholar]
- Goodwin G. H., Nicolas R. H., Johns E. W. An improved large scale fractionation of high mobility group non-histone chromatin proteins. Biochim Biophys Acta. 1975 Oct 20;405(2):280–291. doi: 10.1016/0005-2795(75)90094-x. [DOI] [PubMed] [Google Scholar]
- Goodwin G. H., Woodhead L., Johns E. W. The presence of high mobility group non-histone chromatin proteins in isolated nucleosomes. FEBS Lett. 1977 Jan 15;73(1):85–88. [PubMed] [Google Scholar]
- Jackson J. B., Pollock J. M., Jr, Rill R. L. Chromatin fractionation procedure that yields nucleosomes containing near-stoichiometric amounts of high mobility group nonhistone chromosomal proteins. Biochemistry. 1979 Aug 21;18(17):3739–3748. doi: 10.1021/bi00584a015. [DOI] [PubMed] [Google Scholar]
- Javaherian K., Sadeghi M., Liu L. F. Nonhistone proteins HMG1 and HMG2 unwind DNA double helix. Nucleic Acids Res. 1979 Aug 10;6(11):3569–3580. doi: 10.1093/nar/6.11.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy W B., Wong N. C., Dixon G. H. Selective association of the trout-specific H6 protein with chromatin regions susceptible to DNase I and DNase II: possible location of HMG-T in the spacer region between core nucleosomes. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2810–2814. doi: 10.1073/pnas.74.7.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutter L. C. Kinetic analysis of deoxyribonuclease I cleavages in the nucleosome core: evidence for a DNA superhelix. J Mol Biol. 1978 Sep 15;124(2):391–420. doi: 10.1016/0022-2836(78)90306-6. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Reaction of nucleosome DNA with dimethyl sulfate. Proc Natl Acad Sci U S A. 1979 May;76(5):2133–2137. doi: 10.1073/pnas.76.5.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunell A., Kornberg R. D., Lutter L., Klug A., Levitt M., Crick F. H. Periodicity of deoxyribonuclease I digestion of chromatin. Science. 1979 May 25;204(4395):855–858. doi: 10.1126/science.441739. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Mechanism of a reversible, thermally induced conformational change in chromatin core particles. J Biol Chem. 1979 Oct 25;254(20):10123–10127. [PubMed] [Google Scholar]
- Simpson R. T., Shindo H. Conformation of DNA in chromatin core particles containing poly(dAdT)-poly(dAdT) studied by 31 P NMR spectroscopy. Nucleic Acids Res. 1979 Sep 25;7(2):481–492. doi: 10.1093/nar/7.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978 Dec 12;17(25):5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Whitlock J. P. Mapping DNAase l-susceptible sites in nucleosomes labeled at the 5' ends. Cell. 1976 Oct;9(2):347–353. doi: 10.1016/0092-8674(76)90124-0. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Felsenfeld G. Pancreatic DNAase cleavage sites in nuclei. Cell. 1977 Mar;10(3):537–547. doi: 10.1016/0092-8674(77)90040-x. [DOI] [PubMed] [Google Scholar]
- Stalder J., Groudine M., Dodgson J. B., Engel J. D., Weintraub H. Hb switching in chickens. Cell. 1980 Apr;19(4):973–980. doi: 10.1016/0092-8674(80)90088-4. [DOI] [PubMed] [Google Scholar]
- Sterner R., Boffa L. C., Vidali G. Comparative structural analysis of high mobility group proteins from a variety of sources. Evidence for a high mobility group protein unique to avian erythrocyte nuclei. J Biol Chem. 1978 Jun 10;253(11):3830–3836. [PubMed] [Google Scholar]
- Walker J. M., Goodwin G. H., Johns E. W. The primary structure of the nucleosome-associated chromosomal protein HMG 14. FEBS Lett. 1979 Apr 15;100(2):394–398. doi: 10.1016/0014-5793(79)80378-6. [DOI] [PubMed] [Google Scholar]
- Walker J. M., Hastings J. R., Johns E. W. The primary structure of a non-histone chromosomal protein. Eur J Biochem. 1977 Jun 15;76(2):461–468. doi: 10.1111/j.1432-1033.1977.tb11616.x. [DOI] [PubMed] [Google Scholar]
- Walker J. M., Stearn C., Johns E. W. The primary structure of non-histone chromosomal protein HMG17 from chicken erythrocyte nuclei. FEBS Lett. 1980 Apr 7;112(2):207–210. doi: 10.1016/0014-5793(80)80181-5. [DOI] [PubMed] [Google Scholar]
- Weisbrod S., Groudine M., Weintraub H. Interaction of HMG 14 and 17 with actively transcribed genes. Cell. 1980 Jan;19(1):289–301. doi: 10.1016/0092-8674(80)90410-9. [DOI] [PubMed] [Google Scholar]
- Weischet W. O., Tatchell K., Van Holde K. E., Klump H. Thermal denaturation of nucleosomal core particles. Nucleic Acids Res. 1978 Jan;5(1):139–160. doi: 10.1093/nar/5.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]